Abstract
Background
Although previous studies have shown the presence of Porphyromonas endodontalis in chronic periodontitis associated with periapical lesions, the occurrence of this pathogen in diseased periodontal sites without periapical lesions has been poorly investigated.
Objective
The aims of this study were to quantify P. endodontalis in patients with chronic periodontitis without periapical lesions, to evaluate the potential correlation of P. endodontalis with Porphyromonas gingivalis and Tannerella forsythia, and to evaluate the ability of periodontal treatment to reduce these pathogens.
Design
Patients with generalized chronic periodontitis were selected by recording clinical attachment level (CAL), probing depth (PD), and bleeding on probing (BOP). Subgingival samples from 30 diseased nonadjacent sites (CAL≥5 mm, PD between 5 and 7 mm and positive BOP) and 30 healthy nonadjacent sites (PD≤3 mm and negative BOP) were collected and subjected to microbial analysis by quantitative polymerase chain reaction (qPCR) The variables of age, PD, CAL and BOP of all individuals were analyzed using the paired t-test (GrapPad Prism5®). Data of bacteria quantification were subjected to a normality test (D'Agostino-Pearson Test). For bacterial correlation analysis, the Spearman correlation was used.
Results
Our results showed that diseased sites had significantly higher levels of P. endodontalis compared to healthy sites, similar to the results obtained for P. gingivalis and T. forsythia. The numbers of all bacterial species were reduced significantly after mechanical periodontal treatment. P. endodontalis was significantly correlated with the presence of T. forsythia and P. gingivalis in the diseased group.
Conclusion
Our results suggest that there is a high prevalence of P. endodontalis, P. gingivalis and T. forsythia in periodontitis sites and that mechanical periodontal treatment is effective at reducing the pathogens studied.
Keywords: periodontitis, polymerase chain reaction, porphyromonas endodontalis, porphyromonas gingivalis, Tannerella forsythia
Periodontitis is an inflammatory disease initiated by specific bacterial species that colonize the subgingival area between the tooth surface and the marginal gingiva. This disease causes the destruction of tooth-supporting tissues, including connective tissue and alveolar bone, and can lead to tooth loss (1).
The main periodontal pathogens of periodontal disease include Gram-positive and Gram-negative bacteria, facultative and strictly anaerobic bacteria (2). Porphyromonas gingivalis, Treponema denticola and Tannerella forsythia have been strongly associated with the chronic form of periodontitis and are thought to play important roles in pathogenesis (1, 3, 4). Although individual bacterial species and groups of bacteria have been identified as etiological factors of periodontitis, interactions between microorganisms and the host also play a key role in the etiopathogenesis of the disease (5).
Several reports have shown that the presence of other bacterial species, including Porphyromonas endodontalis, is associated with chronic periodontitis (5, 6). Further studies are required to verify the relationship of these species with the disease and their exact contribution to the etiology and progression of periodontitis. P. endodontalis is primarily found in infections that originate in the pulp, but has also been isolated from the tonsillar area, the dorsum of the tongue and the periodontal pockets of patients with periodontal and pulp lesions (6–8). Tran et al. (6) were the first to report the detection of this species in periodontal pockets, although it was present at a low concentration. P. endodontalis is an asaccharolytic, black-pigmented, Gram-negative anaerobic bacterium. Originally known as Bacteroides endodontalis, P. endodontalis is highly sensitive to oxygen and is therefore difficult to cultivate from clinical samples (9). This species, along with other black-pigmented anaerobic rods, has been implicated in the etiology of infected root canals and periodontitis (4). However, additional studies are required to determine the role of P. endodontalis in patients with chronic periodontitis but without periapical lesions, as well as the contribution of this microorganism to the pathogenesis of periodontitis (10).
Recent advances in diagnostic methods using molecular biology techniques that are more sensitive than bacterial culture assays have led to a better understanding of the roles that previously unidentified microbial species play in the pathogenesis of periodontitis. Most conventional microbiological tests, such as standard bacterial culture, do not distinguish between P. endodontalis and other closely related black-pigmented pathogens. However, molecular techniques, such as polymerase chain reaction (PCR), allow for the specific identification of P. endodontalis and other Gram-negative anaerobic bacteria found in the oral cavity (6, 11). Therefore, the purpose of this study was to use quantitative PCR (qPCR) to detect and quantify P. endodontalis in patients with chronic periodontitis but without infected root canals, and to investigate how these data correlate with data on pathogens that have been previously shown to be related to chronic periodontitis, i.e. P. gingivalis and T. forsythia. In addition, the levels of P. endodontalis, P. gingivalis and T. forsythia following a mechanical periodontal treatment were determined.
Material and methods
Subject population
This study was designed as a double-blind, controlled trial with a two-month duration to quantify pathogens in chronic periodontitis before and after basic periodontal treatment. The study was approved by the Ethics Committee in Human Research (Protocol # 26/08), and all study participants signed a free and informed consent form. The sample size calculation was based on previous studies using qPCR for the detection of periodontal pathogens (12, 13). For this calculation, we used the following criteria: 90% test power, 5% significance level and 1.0 mm detection difference in probing depth (PD). The standardized difference of 0.95 in study power (1−β=0.95) and a confidence level of a=0.05 necessitated a sample size of at least 17 patients that needed to receive periodontal treatment.
This study comprised 20 randomly selected patients (8 men and 12 women of 35–55 years of age) with moderate chronic periodontitis according to the general criteria established by the American Academy of Periodontology (14). Inclusion criteria were as follows: the presence of at least 20 teeth; a minimum of three non-adjacent teeth with bleeding on probing (BOP), a clinical attachment level (CAL)≥5 mm and a probing depth (PD) between 5 and 7 mm; and three healthy teeth with no BOP and a PD≤3 mm. Radiographic analyses were performed to complement the periodontal diagnosis and to exclude teeth with periapical lesions. No patients had a history of systemic disease or antibiotic therapy in the 6 months prior to the study.
Study design
We selected 30 non-adjacent posterior sites from 20 patients with a PD of 5–7 mm, positivity for BOP and a CAL≥5 mm as well as 30 healthy non-adjacent anterior sites with no clinical attachment loss or BOP and a PD≤3 mm. The selected teeth had no dysfunctions in relation to occlusion and had no prostheses.
Alginate molds of the dental arches were made to prepare acetate stents for standardizing the position of the manual probe (Williams®, São Paulo, Brazil) of sites selected for microbiological analyzes. All of the periodontal clinical measurements were performed by a single trained examiner, while another oral health professional performed the basic periodontal treatment followed by oral hygiene instruction and microbiological assessment. The clinical measurements and the sample plaque collections were performed at baseline and at 60 days after the periodontal treatment.
Oral hygiene program
Forty-five days before beginning the periodontal treatment, patients received oral hygiene instructions every week according to the individual needs of each patient. After 30 days of oral hygiene instruction, periodontal examination was performed for patients with a visible plaque index of less than 30%.
Clinical parameters
Clinical examination was performed by a trained, calibrated examiner whose intra-examiner repeatability was determined at baseline (Kappa score=0.91). The clinical parameters assessed included the Visible Plaque Index (VPI, 0–1) and Marginal Bleeding Index (MBI, 0–1) (15) determined at four points per tooth (mesial, distal, buccal and lingual), and the Probing Depth (PD), Clinical Attachment Level (CAL) and Bleeding on Probing (BOP, 0–1) (16) determined at six different sites per tooth (mesiobuccal, buccal, distobuccal, distolingual, lingual and mesiolingual) using a periodontal probe (Williams®, São Paulo, Brazil).
Subgingival sample collection
Samples for microbiological analysis were collected seven days after the initial clinical examination and 60 days after completion of the basic periodontal treatment. After removal of the supragingival biofilm, the selected sites were isolated with cotton rolls and gently air-dried. Subgingival fluid was collected using two sterile paper points (no. 30; Dentsply, Maillefer, Petrópolis, RJ, Brazil) that were inserted in the gingival pocket up to the apical portion for 30 seconds. The paper points were immediately placed in sterile Eppendorf vials containing 500 µL of TE buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, pH 8.0) and stored at −20°C until DNA was extracted for microbial analysis by qPCR.
Basic periodontal treatment
Seven days after the microbiological sample collection, patients underwent a non-surgical periodontal treatment including scaling and root planing (SRP) followed by oral hygiene instruction; the treatment was performed under local anesthesia and with manual instruments (McCall and Gracey Curettes and Hirschfeld File Scaler – Hu Friday®). Polishing was performed immediately after each session of SRP with rubber cups and paste. After SRP, supragingival biofilm control (maintenance phase) was performed via prophylaxis and oral hygiene instruction was conducted weekly for 60 days, at which time the second collection of crevicular fluid and periodontal examination occurred.
Bacterial strains and growth conditions
The bacterial species P. endodontalis (E203), P. gingivalis (ATCC 33277) and T. forsythia (ATCC 43037) were grown anaerobically on Tryptone soy blood agar plates supplemented with hemin (5 mg/l) and menadione (1 mg/l) for 7–15 days at 37°C in 85% N2, 5% CO2 and 10% H2 in an anaerobic chamber (Plas Labs, Lansing, MI, USA). Escherichia coli strain DH5-alpha was used as the host for plasmids and was grown aerobically at 37°C in LB medium (17).
DNA extraction
The extraction of DNA from subgingival fluid samples and from bacterial reference cultures was performed using standard methods. Briefly, cells were lysed by boiling, and DNA was extracted with phenol:chloroform:iso-amyl alcohol (25:24:1). After the addition of 3 M NaCl, the DNA was precipitated with ethanol (99%) and re-suspended in TE buffer. The DNA concentration was determined by UV spectrophotometry (Biomate 3 Spectrophotometer, Thermo Electron Corporation), and the relationships between the absorbances at 260 and 280 nm were analyzed. As a quality parameter, A260/A280 values between 1.8 and 2.0 were considered appropriate.
Quantitative PCR reactions
For the quantitative analysis, plasmids containing the target genes were used as standards. PCR amplification was initially performed for the 16S rRNA of P. endodontalis, P. gingivalis and T. forsythia. The amplicons were cloned using the TOPO TA Cloning Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions and plasmids were transformed into E. coli. After growth of the transformants, the plasmids were extracted using a PureLink Quick sHRNA Miniprep kit (Invitrogen) and then sequenced. The plasmid solutions were diluted in sterile water to a concentration of 108 copies/μL and then serially diluted (final concentrations from 104 to 101 copies/µL). The dilutions were then used as template DNA in the qPCR reactions. In each reaction, data obtained from the standard curve were used to convert the CT scores (cycle threshold) obtained with patient samples into the exact numbers of DNA copies (18, 19).
The detection and quantification by qPCR was performed using universal (20) and specific primers for P. endodontalis (5′-GCT GCA GCT CAA CTG TAG TCT TG-3′, 5′-TCA GTG TCA GAC GGA GCC TAG TAC-3′-110 pb) (21), P. gingivalis (5′-ACC TTA CCC GGG ATT GAA ATG-3′, 5′-CAA CCA TGC AGC ACCT AC ATA GAA-3′- 83 pb) (22), and T. forsythia (5′-AGC GAT GGT AGC AAT ACC TGT C-3′, 5′-TTC GCC GGG TTA TCC CTC-3′- 88 pb) (22) (Applied Biosystems®). The species-specific primer sets were designed based on the variable regions of each target gene. The specificities of the primers were confirmed by multiple alignments of relevant sequences from closely related species and by a Basic Local Alignment Search Tool (BLAST) homology search (1).
The qPCR reactions were performed with the use of a Step OneTM qPCR System (Applied Biosystems®). All reactions were performed in duplicate, and average values were used to calculate the bacterial load. The total volume of each reaction was 10μL containing 5μL of SYBR Green ER qPCR SuperMix Universal (Invitrogen Tech-LineSM), 0.1μM of each primer pair (Applied Biosystems®) and 50 ng/uL template DNA. The thermocycling program included an incubation at 95°C for 10 minutes, 40 cycles of 95°C for 15 seconds and an incubation at 60°C for 1 minute. After the qPCR reaction, the dissociation curve (melting curve) was obtained using temperatures between 60°C and 95°C to determine primer specificity. Melting curve analysis revealed only one peak of amplification. All reactions were performed in 48-well MicroAmp optical plates covered with optical adhesive (Applied Biosystems). Data were analyzed by Step OneTM software (Applied Biosystems®).
Statistical analysis
The clinical and microbiological data were statistically analyzed using the software GraphPad Prism 5®. Inter- and intra-group analyses were conducted for the various periods. The variables age, PD, CAL and BOP of all individuals were analyzed using the paired t-test. Quantitative variables (PD and CAL) were subjected to the normality test (D'Agostino-Pearson Test). PD and CAL data were analyzed by the Wilcoxon test for comparisons between groups and between periods. To analyze the BOP categorical variable, the Cochran test was used. To identify statistically significant results, we used the exact McNemar test. Bacterial quantification data were subjected to a normality test (D'Agostino-Pearson Test) and showed a non-normal distribution. Thus, the Mann-Whitney test was applied. For bacterial correlation analysis, the Spearman rank correlation was used. p<0.05 was considered statistically significant.
Results
Clinical parameters
The general characteristics of the sampled population are summarized in Table 1. Since no statistically significant differences in age or number of teeth were observed in the sample set, it was considered homogeneous. The average CAL was divided into three CAL scores (1–2 mm, 3–4 mm and ≥5 mm), and the average PD was divided into three PD scores (1–3 mm; 4–5 mm; ≥6 mm). After treatment, there was a statistically significant improvement of both parameters (p<0.001).
Table 1.
Mean (± SD) clinical parameters of full mouth of subjects in the study
| N=20 subjects | BASELINE | 60 DAYS |
|---|---|---|
| Age (years) | 46±7.49 | |
| Gender (M/F) | 8/12 | |
| Average number of teeth | 23.55±2.95 | |
| % of sites with | ||
| Probing depth (1–3 mm) | 86% | 94% |
| Probing depth (4–5 mm) | 10% | 5.3% |
| Probing depth (≥6 mm) | 4% | 0.7% |
| Clinical attachment level (1–2 mm) | 60% | 78% |
| Clinical attachment level (3–4 mm) | 28.5% | 20% |
| Clinical attachment level (≥5 mm) | 11.5% | 2% |
| Bleeding on probing | 47% | 10% |
The collection sites for the microbiological analysis had a prevalence of bleeding on probing of 100% at baseline that decreased to 13.33% after treatment in the disease group (p<0.0001). In addition, PD (5.33±0.54 mm, baseline) and CAL (5.4±0.62 mm, baseline) showed statistically significant reductions (p<0.0001) after periodontal therapy in the diseased group (3.63±0.76 mm and 3.83±0.95 mm for PD and CAL, respectively). In the healthy sites, PD (1.6±0.56 mm) and CAL (1.63±0.56) showed no significant reduction (p=0.035) over the 60-day period.
Detection of microorganisms by qPCR
The standard curve for each microorganism was obtained with the use of specific primers and four serial dilutions (101–104 of the genomic DNA of P. endodontalis, P. gingivalis and T. forsythia. The reaction efficiencies for each organism were 97.1 (P. endodontalis), 96.3 (P. gingivalis) and 97.8 (T. forsythia). The correlation coefficient for the mean CT values was R2≥0.99. All 3 Sybr Green assays were highly specific and amplified only DNA extracted from periodontopathogens.
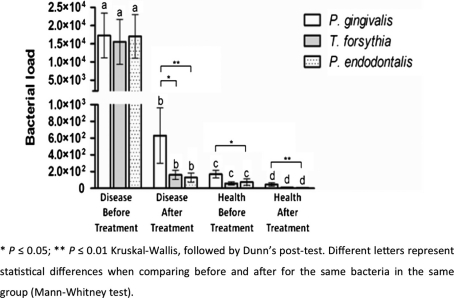
The average numbers of P. endodontalis, P. gingivalis and T. forsythia detected in the patient group were 1.7.104, 1.72.104 and 1.5.104 cells, respectively. There were no statistically significant differences among the three pathogens tested. However, after periodontal treatment, we observed significant reductions of these pathogens to 1.3. 102, 6.33.102 and 1.6.102, respectively, for P. endodontalis P. gingivalis and T. forsythia. P. gingivalis remained in higher concentrations after periodontal treatment (Fig. 1).
Fig. 1.
Quantification of P. endodontalis, P. gingivalis and T. forsythia in subgingival plaque of patients with chronic periodontitis by qPCR. Letters represented the evaluating of each bacterium separately before and after periodontal treatment. So those bacteria that had statistically significant reduction after periodontal treatment were represented with different letters.
In the healthy sites, we detected average numbers of 70, 160 and 58 cells for P. endodontalis, P. gingivalis and T. forsythia, respectively. After periodontal treatment, there were significant reductions of all bacteria tested (Fig. 1). These data suggest that even in the healthy sites, P. gingivalis is present at higher levels than T. forsythia and P. endodontalis.
Correlations in the detection of analyzed bacteria
When the correlations between the three bacterial species were assessed, we observed statistically significant correlations between P. gingivalis and T. forsythia (r=0.624, p≤0.001) and between T. forsythia and P. endodontalis (r=0.756, p≤0.01) in the diseased group (baseline). However, there was only a moderate correlation between P. gingivalis and P. endodontalis (r=0.385, p≤0.05). We found similar patterns of correlation in the diseased group after 60 days of periodontal treatment, although the correlation values were less significant. The correlations between T. forsythia and P. gingivalis (r=0.390, p≤0.05) and between T. forsythia and P. endodontalis (r=0.483, p≤0.01) were statistically significant, but the correlation was not significant between P. gingivalis and P. endodontalis. The healthy group showed no correlations between the analyzed bacteria between both periods (baseline and 60 days) possibly due to low levels of detection (data not shown).
Discussion
The primary goal of this study was to detect and quantify the pathogen P. endodontalis in patients with chronic periodontitis without periapical lesions and to determine a possible correlation with the pathogens that are already related to chronic periodontitis, i.e. P. gingivalis and T. forsythia. Microorganisms and their products are the initiators of periodontal lesions. The main pathogens associated with periodontal disease include P. gingivalis, T. forsythia and T. denticola. However, Kumar et al. (5), Tran et al. (6) and Dahlén and Leonhardt (7) suggested that other poorly characterized microorganisms (e.g. P. endodontalis) might also be associated with chronic periodontitis.
P. endodontalis was initially found in endodontic infections by Sundqvist et al. (23) and Van Winkelhoff et al. (8). This pathogen has already been identified as a member of the subgingival microbiota in humans; however, its association with periodontal disease is not well established. Few studies have reported the prevalence of this microorganism in diseased periodontal sites in patients without periapical lesions or the effect of a periodontal treatment in reducing the levels of P. endodontalis (6, 24).
Our study showed a high prevalence of P. endodontalis in addition to P. gingivalis and T. forsythia, in diseased periodontal sites when compared to healthy sites, with a statistically significant reduction after periodontal therapy. A previous study also has shown that P. endodontalis can be found in diseased sites in higher proportions than in healthy sites and in proportions similar to those of other pathogens already associated with periodontitis, such as P. gingivalis (13). On the other hand, Tran et al. (6) reported the presence of P. endodontalis in periodontal pockets but only at a low concentration.
Similar reports have shown that putative pathogens may be present in healthy periodontal sites but at lower levels than in diseased sites. This would explain why the pathogenicity of the microbiota differs from area to area and also from one individual to another (25). Haffajee et al. (25) demonstrated that the presence of pathogenic species is essential during the development of periodontal disease, but often they are not sufficient in numbers to establish disease and the onset of periodontitis depends on the presence of other predisposing factors (25).
The similarity between P. endodontalis and P. gingivalis, one of the main bacterial species found in diseased periodontal sites, is well known. However, the microorganisms possess distinct phenotypes, and no cross-reactions between them have been detected using molecular techniques. Thus, the results obtained in this study could not be related to the detection of P. gingivalis, as specific primers for P. endodontalis were used (7).
As noted in our results, P. gingivalis showed the smallest reduction after periodontal treatment among the microorganisms studied. At the end of the 60-day period between collections, it remained present at higher concentrations than the other bacteria. Reports in the literature indicated that P. gingivalis have fimbriae, which are potent virulence factors involved in bacterial adhesion to oral tissues, allowing the microorganism to invade the periodontal tissue more easily and increasing its survival in the gingival tissue (26).
The adhesion of P. endodontalis to the epithelial cells of the gingival sulcus differs from the adhesion of P. gingivalis. P. endodontalis is less able to adhere to gingival cells compared to P. gingivalis because it does not have fimbriae (27). P. endodontalis also does not produce a trypsin-like enzyme that other pathogens use for the destruction of periodontal tissue and does not exhibit hemagglutination activity (6). However, P. endodontalis has collagenases and proteases, enzymes that can also aid in the destruction of supporting tissues (27). T. forsythia also does not have fimbriae, reducing its capacity to colonize and invade periodontal tissue. However, it expresses a protein, BspA, that has a domain similar to a domain present in the fimbriae protein of P. gingivalis and induces inflammatory cytokine production in mammalian cells. These characteristics may contribute to the greater correlation between P. endodontalis and T. forsythia when compared to the correlation between P. endodontalis and P. gingivalis (3).
The present study demonstrated significant correlations between the numbers of bacterial cells of the three pathogens tested in the diseased group. The diseased sites had increased numbers of pathogens compared to the healthy sites. Therefore, the presence and amounts of P. endodontalis, P. gingivalis and T. forsythia in the subgingival environment may influence the local development of periodontal disease. Our data support the study of Dahlén and Leonhardt (7) that proposed the addition of P. endodontalis to the list of microorganisms that are used in the microbiological diagnosis of periodontal disease.
Thus, through a sensitive and specific qPCR method that is routinely used as a diagnostic tool for periodontitis, the association of P. gingivalis and T. forsythia was confirmed; these pathogens were found in higher proportions in affected sites than in healthy sites. We also noted a higher incidence of P. endodontalis in sites with periodontal disease than in healthy sites.
Despite the short term evaluation and small number of patients in this study, our results suggest that P. endodontalis, in addition to P. gingivalis and T. forsythia, can be found at high levels in diseased periodontal sites of periodontitis patients without periapical lesions when compared to healthy sites. Furthermore, a significant reduction in the numbers of P. endodontalis, P. gingivalis, and T. forsythia after mechanical periodontal therapy was observed.
Longitudinal studies with longer evaluation periods of 6 months to 1 year after periodontal treatment and controlled studies involving higher numbers of patients should be conducted to confirm our data and to clearly assess the pathogenic potential of P. endodontalis during the course of periodontal disease.
Acknowledgements
The authors thank the financial support from FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo, grants 2009/53308-7; São Paulo, Brazil).
Conflicts of interest and funding
This manuscript does not have any kind of conflicts of interest and funding came from support by the FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo) – Grant 2009/53308-7.
References
- 1.Mineoka T, Awano S, Rikimaru T, Kurata H, Yoshida A, Ansai T, et al. Site specific development of periodontal disease is associated with increased levels of Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia in subgingival plaque. J Periodontol. 2008;79:670–76. doi: 10.1902/jop.2008.070398. [DOI] [PubMed] [Google Scholar]
- 2.Jarvensivu A, Hietanen J, Rautemaa R, Sorsa T, Richardson M. Candida yeast in chronic periodontitis tissues and subgingival microbial biofilms in vivo. Oral Dis. 2004;10:106–12. doi: 10.1046/j.1354-523x.2003.00978.x. [DOI] [PubMed] [Google Scholar]
- 3.Holt SC, Ebersole JL. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: The ‘red complex’, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol. 2005;38:72–122. doi: 10.1111/j.1600-0757.2005.00113.x. 2000. [DOI] [PubMed] [Google Scholar]
- 4.Siqueira JF, Jr, Rôças IN. Diversity of endodontic microbiota revisited. J Dent Res. 2009;88:969–81. doi: 10.1177/0022034509346549. [DOI] [PubMed] [Google Scholar]
- 5.Kumar PS, Griffen AL, Barton JA, Paster BJ, Moeschberger ML, Leys EJ. New bacterial species associated with chronic periodontitis. J Dent Res. 2003;82:338–44. doi: 10.1177/154405910308200503. [DOI] [PubMed] [Google Scholar]
- 6.Tran T, Flynn MJ, Chen C, Slots J. Porphyromonas endodontalis in subgingival plaque. Clin Infect Dis. 1997;25:222–23. doi: 10.1086/516232. [DOI] [PubMed] [Google Scholar]
- 7.Dahlén G, Leonhardt A. A new checkerboard panel for testing bacterial markers in periodontal disease. Oral Microbiol Immunol. 2006;21:6–11. doi: 10.1111/j.1399-302X.2005.00243.x. [DOI] [PubMed] [Google Scholar]
- 8.Van Winkelhoff AJ, Carlee AW, de Graaff J. Bacteroides endodontalis and other black-pigmented bacteroides species in odontogenic abscesses. Infect Immun. 1985;49:494–49. doi: 10.1128/iai.49.3.494-497.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Winkelhoff AJ, Van Steenbergen TJM, de Graaff J. Oxygen tolerance of oral and non-oral black-pigmented Bacteroides species. FEMS Microbiol Lett. 1986;33:215–18. [Google Scholar]
- 10.Pereira CV, Stipp RN, Fonseca DC, Pereira LJ, Höfling JF. Detection and clonal analysis of anaerobic bacteria associated to endodontic-periodontal lesions. J Periodontol. 2011;82:1767–75. doi: 10.1902/jop.2011.110063. [DOI] [PubMed] [Google Scholar]
- 11.Ashimoto A, Chen C, Bakker I, Slots J. Polymerase chain reaction detection of 8 putative periodontal pathogens in subgingival plaque of gingivitis and advanced periodontitis lesions. Oral Microbiol Immunol. 1996;1:266–73. doi: 10.1111/j.1399-302x.1996.tb00180.x. [DOI] [PubMed] [Google Scholar]
- 12.Huse SM, Dethlefsen L, Huber JA, Mark Welch D, Relman DA, Sogin ML. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verner C, Lemaitre P, Daniel A, Giumelli B, Lakhssassi N, Sixou M. Carpegen real-time polymerase chain reaction vs. anaerobic culture for periodontal pathogen identification. Oral Microbiol Immunol. 2006;21:341–46. doi: 10.1111/j.1399-302X.2006.00297.x. [DOI] [PubMed] [Google Scholar]
- 14.Armitage GC. Development of a classification system for periodontal diseases and conditions. Ann Periodontol. 1999;4:1–6. doi: 10.1902/annals.1999.4.1.1. [DOI] [PubMed] [Google Scholar]
- 15.Ainamo J, Bay I. Problems and proposals for recording gingivitis and plaque. Int Dent J. 1975;25:229–35. [PubMed] [Google Scholar]
- 16.Muhlemann HR, Son S. Gingival sulcus bleeding – a leading symptom in initial gingivitis. Helv Odontolol Acta. 1971:107–13. [PubMed] [Google Scholar]
- 17.Umeda JE, Missailidis C, Longo PL, Anzai D, Wikström M, Mayer MP. Adhesion and invasion to epithelial cells by fimA genotypes of Porphyromonas gingivalis . Oral Microbiol Immunol. 2006;21:415–19. doi: 10.1111/j.1399-302X.2006.00312.x. [DOI] [PubMed] [Google Scholar]
- 18.Persson GR, Hitti J, Paul K, Hirschi R, Weibel M, Rothen M, et al. Tannerella forsythia and Pseudomonas aeruginosa in subgingival bacterial samples from parous women. J Periodontol. 2008;79:508–16. doi: 10.1902/jop.2008.070350. [DOI] [PubMed] [Google Scholar]
- 19.Teixeira SR, Mattarazo F, Feres M, Figueiredo LC, de Faveri M, Simionato MR, et al. Quantification of Porphyromonas gingivalis and fimA genotypes in smoker chronic periodontitis. J Clin Periodontol. 2009;36:482–87. doi: 10.1111/j.1600-051X.2009.01411.x. [DOI] [PubMed] [Google Scholar]
- 20.Nonnenmacher C, Dalpke A, Rochon J, Flores-de-Jacoby L, Mutters R, Heeg K. Real-time polymerase chain reaction for detection and quantification of bacteria in periodontal patients. J Periodontol. 2005;76:1542–9. doi: 10.1902/jop.2005.76.9.1542. [DOI] [PubMed] [Google Scholar]
- 21.Martin EF, Nadkarni MA, Jacques NA, Hunter N. Quantitative microbiological study of human carious dentine by culture and Real-Time PCR: association of anaerobes with histopathological changes in chronic pulpitis. J Clin Microbiol. 2002;40:1698–1704. doi: 10.1128/JCM.40.5.1698-1704.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuboniwa M, Amano A, Kimura RK, Sekine S, Kato S, Yamamoto Y, et al. Quantitative detection of periodontal pathogens using real-time polymerase chain reaction with TaqMan probes. Oral Microbiol Immunol. 2004;19:168–76. doi: 10.1111/j.0902-0055.2004.00135.x. [DOI] [PubMed] [Google Scholar]
- 23.Sundqvist GK, Eckerbom MI, Larsson AP, Sjögren UT. Capacity of anaerobic bacteria from necrotic dental pulps to induce purulent infections. Infect Immun. 1979;25:685–9. doi: 10.1128/iai.25.2.685-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hutter G, Schlagenhauf U, Valenza G, Horn M, Burgemeister S, Claus H, et al. Molecular analysis of bacteria in periodontitis: evaluation of clone libraries, novel phylotypes and putative pathogens. Microbiology. 2003;149:67–75. doi: 10.1099/mic.0.25791-0. [DOI] [PubMed] [Google Scholar]
- 25.Haffajee AD, Cugini MA, Dibart S, Smith C, Kent Jr RL, Socransky SS. The effect of SRP on the clinical and microbiological parameters of periodontal diseases. J Clin Periodontol. 1997;24:324–34. doi: 10.1111/j.1600-051x.1997.tb00765.x. [DOI] [PubMed] [Google Scholar]
- 26.Hiramine H, Watanabe K, Hamada N, Umemoto T. Porphyromonas gingivalis 67-kDa fimbriae induced cytokine production and osteoclast differentiation utilizing TLR2. FEMS Microbiol Lett. 2003;229:49–55. doi: 10.1016/S0378-1097(03)00788-2. [DOI] [PubMed] [Google Scholar]
- 27.Kon A. Pathogenic factors of Porphyromonas endodontalis . Dent J Iwate Med Univ. 2002;27:187–196. [Google Scholar]