Abstract
We provide an overview of lipid-dependent polytopic membrane protein folding and topogenesis. Lipid dependence of this process was determined by employing Escherichia coli cells in which specific lipids can be eliminated, substituted, tightly titrated or controlled temporally during membrane protein synthesis and assembly. The secondary transport protein lactose permease (LacY) was used to establish general principles underlying the molecular basis of lipid-dependent effects on protein domain folding, protein transmembrane domain (TM) orientation, and function. These principles were then extended to several other secondary transport proteins of E. coli. The methods used to follow proper conformational organization of protein domains and the topological organization of protein TMs in whole cells and membranes are described. The proper folding of an extramembrane domain of LacY that is crucial for energy dependent uphill transport function depends on specific lipids acting as non-protein molecular chaperones. Correct TM topogenesis is dependent on charge interactions between the cytoplasmic surface of membrane proteins and a proper balance of the membrane surface net charge defined by the lipid head groups. Short-range interactions between the nascent protein chain and the translocon are necessary but not sufficient for establishment of final topology. After release from the translocon short-range interactions between lipid head groups and the nascent protein chain, partitioning of protein hydrophobic domains into the membrane bilayer, and long–range interactions within the protein thermodynamically drive final membrane protein organization. Given the diversity of membrane lipid compositions throughout nature, it is tempting to speculate that during the course of evolution the physical and chemical properties of proteins and lipids have co-evolved in the context of the lipid environment of membrane systems in which both are mutually depend on each other for functional organization of proteins.
Keywords: phosphatidylethanolamine, lactose permease, protein topology, lipochaperone, positive-inside rule
1. Introduction
The Singer-Nicolson [1] fluid mosaic concept of biological membranes envisioned individual protein units moving through a sea of lipids that form a bilayer composed of a hydrophobic core bounded on each side by the hydrophilic domains of polar lipids. The main role of lipids was to provide a hydrophobic sink for membrane barrier function and residency for the hydrophobic portion of protein domains that are inserted into or traverse the membrane. Membrane proteins are now well established to be composed of multiple transmembrane domains (TM1 s) alternately oriented in opposite directions with respect to the plane of the bilayer and connected by hydrophilic domains on alternating sides of the membrane. Since many membrane proteins can be purified in a functional and structurally compact form using detergents in place of natural lipids, less attention has been paid to the native lipid environment in defining the structure and function of membrane proteins. Taking into account the different hydrophobic and hydrophilic domains of natural lipids, the diversity within the lipidome probably exceeds that of the proteome [2]. When lipids are added back to purified membrane proteins, they are usually single or simple mixtures of synthetic lipids that do not reflect the diversity of lipids found in biological membranes However, increasing evidence [3] indicates that individual native lipids and lipid composition of biological membranes play a more specific role in membrane structure and function than originally envisioned by Singer and Nicolson. Specific lipids have also been found associated with the surface and even integrated into the structure of purified membrane proteins [4]. Addressing this problem only through purification using detergents followed by reconstitution with even native lipids still requires in vivo evidence for a specific function of lipids, which has not been done extensively.
There are major obstacles to defining specific roles for lipids in vivo. Lipids have no catalytic activity so their effects are generally determined secondary to effects on biological processes usually reconstituted in vitro. In vivo importance of proteins has generally been established through gene mutation. Genes do not encode lipids so that changes in lipid composition must be done by mutations in the enzymes that define their biosynthesis. Mutations early in the pathway eliminate minor lipids leading to the major endpoint lipid and mutations late the pathway result in accumulation of minor intermediates. Changes in membrane lipid composition often affect multiple processes, especially in eukaryotic cells containing several membranes containing the same lipids. Large changes in lipid composition can result in compromising membrane barrier function resulting in cell death before affecting a specific biological process. In spite of these limitations it has been possible to define specific roles for lipids by establishing complimentary effects of lipids in vivo by genetic manipulation and in vitro through reconstitution.
This review will summarize the use of a set of mutants in Escherichia coli in which membrane lipid composition can be systematically altered while maintaining cell viability. Examples will be reviewed in which changes in lipid environment affects membrane protein structure and function in vivo with in vitro verification of a specific effect.
2. Systematic alteration in membrane lipid composition
2.1. Genetic manipulation of E. coli phospholipid metabolism
The cell envelope of E. coli is composed of an outer membrane that, due to the presence of porins, is a barrier to molecules > 600 Da [5]. The outer membrane is made up of an inner leaflet of glycerol-based phospholipids (hereafter referred to as phospholipids) and an outer leaflet made up of lipid A based-lipids [6], which are the membrane-anchoring component of lipopolysaccharides. The inner membrane bilayer is composed solely of phospholipids, represents the regulated barrier to solutes and is the site of the majority of membrane-associated cell processes [7]. The major phospholipids of E. coli (see Fig. 1 and 2) are zwitterionic (no net charge) phosphatidylethanolamine (PE, 70–75%) and the two anionic phospholipids phosphatidylglycerol (PG, 20–25%) and cardiolipin (CL, 5–10%). The remaining phospholipids are less than 5% of the total phospholipid. The inner leaflet of the outer membrane contains about 90% PE, but the distribution of PE, PG and CL in the two leaflets of the inner membrane is not known.
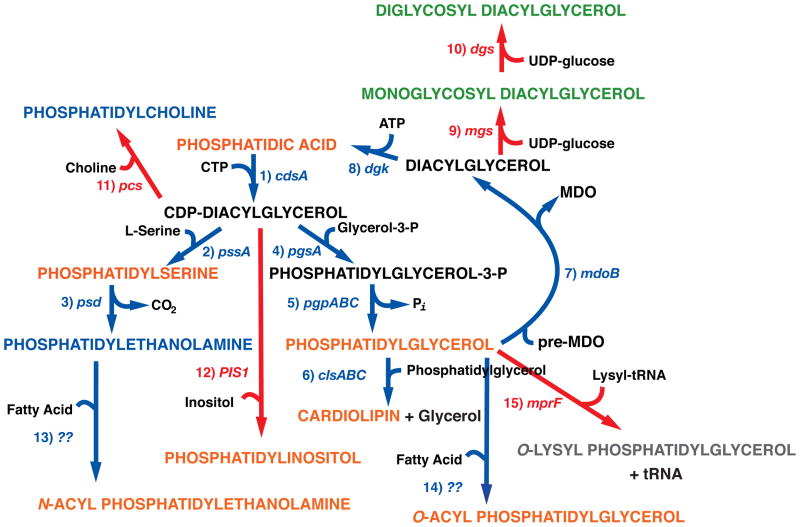
Fig. 1.
Native and foreign lipid biosynthesis in E. coli. Pathways native to E. coli are noted with blue arrows, and pathways resulting from foreign genes introduced into E. coli are noted with red arrows. Lipids emphasized in the text are color coded as zwitterionic (blue), neutral (green), anionic (orange) or cationic (gray). The genes encoding the following enzymes and associated with each biosynthetic step are listed next to the arrows: (1) CDP-diacylglycerol synthase; (2) PS synthase; (3) PS decarboxylase; (4) PG-P synthase; (5) PG-P phosphatases [8]; (6) CL synthases; (7) PG:MDO (membrane derived oligosaccharide) sn-glycerol-1-P transferase; (8) diacylglycerol kinase; (9) GlcDAG synthase (Acholeplasma laidlawii); (10) GlcGlcDAG synthase (A. laidlawii); (11) PC synthase (Legionella pneumophila); (12) PI synthase (Saccharomyces cerevisiae): (13) N-acyl PE synthase; (14) O-acyl PG synthase; O-lys PG synthase (Staphylococcus aureus) [9,10].
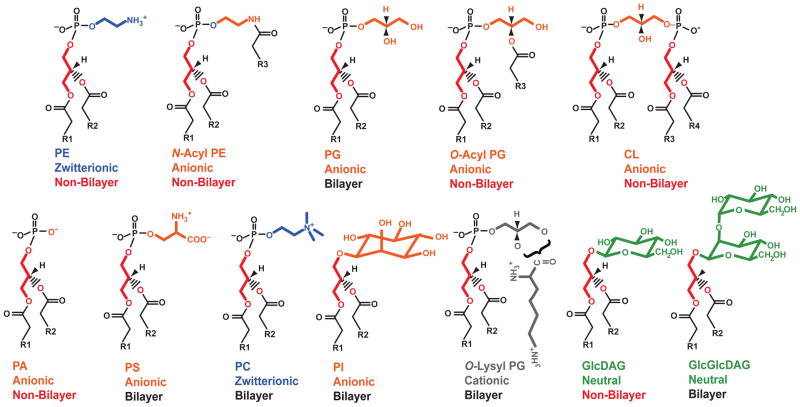
Fig. 2.
Lipid structures and properties. Stick figure representation of lipid structures with the glycerol backbone colored red and different fatty acids denoted by ‘R’. Color-coding of head groups follows the pattern in Fig. 1.
All genes responsible for the synthesis of the major lipids (Fig. 1) from the common precursor phosphatidic acid (PA) have been identified and cloned [7], and mutants exist in each locus. Null mutations in pssA [11] or pgsA [12] or clsABC (see [13] and personal communication B. Tan and C.R.H. Raetz, Duke University) genes render cells devoid of phosphatidylserine (PS) and PE or PG-phosphate and PG and CL, or CL, respectively. Surprisingly each of these null mutants is viable under selective growth conditions. Not surprising is that null mutations prior to the formation of CDP-diacylglycerol are not viable. Genetic manipulation of lipid synthesis (see Table 1) in viable strains results in dramatic alterations of membrane phospholipid composition [7] that makes these mutants useful reagents to study the role of membrane lipid composition in the structure and function of native and foreign proteins in vivo [14].
Table 1.
Summary of phospholipid biosynthetic genes manipulated in E. coli
| E. coli Genes | ||
|---|---|---|
| Gene | Gene Product | Mutant phenotype |
| pssA | PS Synthase | Divalent metal ion requirement, filamentous growth, aberrant membrane protein topology and function, induction of stress response, outer membrane permeability, electron transport defects |
| pgsA | PG-P Synthase | Induction of stress responses, shorter cells, reduced rate of protein secretion, temperature sensitive, lack of mature lipoproteins |
| clsABC | CL Synthases | None known |
| psd | PS decarboxylase | Divalent metal ion requirement, filamentous growth |
| Foreign Genes | ||
|---|---|---|
| Gene | Source | Gene Product |
| pcs | Legionella pneumophila | PC Synthase |
| mgs | Acholeplasma laidlawii | GlcDAG Synthase |
| dgs | Acholeplasma laidlawii | GlcGlcDAG Synthase |
| PIS | Saccharomyces cerevisiae | PI Synthase |
| mrpF | Staphylococcus aureus | O-lys PG Synthase |
The viability and inner membrane barrier function of null mutants of the pssA gene, which completely lack all amino-containing phospholipids, is dependent on 10–50 mM concentrations of divalent cation ions Ca2+, Mg2+ or Sr2+ (Ba does not substitute) in the growth medium [11]. The remaining phospholipids are all anionic and composed of mostly PG and CL in equal amounts with about 5% PA (normally < 1%) when cells are grown in the presence of Mg2+. Growth in the presence of Ca2+ reduces in the CL level in half in favor of an increase in PG levels [15]. The divalent cation requirement appears to be outside of the inner membrane since Mg2+ concentration is 50–100 mM in the cytoplasm and Ca2+ and Sr2+ (μM range) are actively pumped out of the cytoplasm but do enter the periplasmic space between the inner and outer membranes. Although PE is not absolutely required for cell viability, the cells display multiple defects in addition to the requirement for divalent cations. Synchrony between cell growth and division is lost at the point of final constriction and separation [16]. As a result, PE-lacking cells display filamentous growth with multiple nuclei separated by cell division machinery assemblies (the divisome) and constriction occurring at the ends of filaments. Many secondary transporters are defective in energy-dependent uphill transport of substrate (coupled to downhill transport of protons) but not energy-independent downhill transport (equilibration of substrate across the membrane) [3]. The outer membrane permeability barrier to macromolecules is compromised as evidenced by leakage of periplasmic proteins to the growth medium. The cells show highly induced levels of systems that respond to membrane stress [17], and there is reduction in the rate of electron flow through succinate dehydrogenase [18]. The cells are not mobile due to failure to assembly flagella [18]. Like pssA null mutants, growth and viability of psd mutants require millimolar levels of Mg2+ in the growth medium and cells display filamentous growth but no defects in secondary transport function indicating this phenotype can be suppressed by either PS or PE [19]. PS is normally < 0.1%, but in a psdts (temperature sensitive mutant) grown at the restrictive temperature, PS nearly replaces PE (residual level about 5%). Null mutants of psd are not viable, which may be due to a downstream gene of unknown function within the same operon [20].
Introduction of a null pgsA mutation was originally thought to be lethal [21] but was later shown to only render cells temperature sensitive for growth. However, viability also depends on inactivation of several stress response systems in cells lacking PG and CL [12]. Therefore, use of a complex suppressor genetic background results in cells completely lacking of PG and CL, which now contain elevated levels of PA from < 1% to 5% with the remainder of phospholipid being PE. These cells also contain an elevated level of the minor phospholipid N-acyl PE [22]. The mutant cells display few phenotypes. They undergo normal cell division even though anionic phospholipids have been implicated as necessary for optimal cell division and protein secretion. This may be due to the inability to completely eliminate anionic lipids such as PA and CDP-diacylglycerol since they are the precursors to all phospholipids. CL, due to its non-bilayer prone property (Fig. 2), is enriched in lipid domains found at the cell poles and the mid-cell where the divisome assembles for cell division. These lipid domains have been proposed to be necessary for compartmentalization of cell functions through interaction with specific proteins [23]. Interestingly, lipids such as PA and N-acyl PE, which display the same physical properties (non-bilayer prone anionic lipids) as CL (see Fig. 2), are enriched in pgsA null mutants and localize to the cell poles and the mid-cell [22]. Cells in which PG and CL levels are drastically reduced are shorter than normal. They display reduced rates of protein secretion possibly due to a requirement of CL for optimal function of the protein translocation machine [24] and anionic lipid for membrane association of the SecA protein component of this system [25]. The molecular bases for a requirement of mutations in stress response systems and the temperature sensitivity of pgsA mutants is not understood but may be related to several cellular requirements of PG. PG is a precursor to the periplasmic membrane derived oligosaccharide [26] that is induced in cells grown in medium of low osmolarity. PG is the donor of glycerol in a posttranslational modification of the major outer membrane lipoprotein (lpp gene product), which tethers the outer membrane to the peptidoglycan of the periplasmic space. Viability of pgsA null strains is dependent on null lpp mutations to prevent accumulation of the underivatized gene product in the inner membrane [27]. Growth at higher temperatures places added stress on cells, which may not be suppressed in the mutant strains thus far used. Therefore, conditions are available to eliminate PG and CL, but these lipids are still required under normal growth conditions and for optimal cell function.
Three genes, all of which must be mutated to prevent CL synthesis, encode CL synthases (see [28] and personal communication B. Tan and C.R.H. Raetz, Duke University). Unfortunately many experiments have been done with strains in which only the first discovered clsA gene was inactivated, which complicates the determination of the importance of CL in cell processes. The triple mutant has not been extensively characterized but its lipid composition reflects the loss of CL with a rise of PG levels to compensate (unpublished results B. Tan, M. Bogdanov, W. Dowhan, and C.R.H. Raetz). CL has been implicated in non-bilayer prone anionic lipid domain formation, localization of ProP to the cell poles [29] and optimal function of the translocon responsible for membrane insertion of proteins and export from the cytoplasm [24]. The clsA gene product contributes to almost all of the CL present in rapidly growing cells while all three gene products contribute to the CL pool found in stationary cells.
The pssA [30,31] and pgsA [21] genes have been placed under regulation of promoters subject to inducer regulation. ParaB (inducible by arabinose) has been used to regulate all or none presence of PE, and Plac (inducible by isopropyl-β-D-thiogalactoside) has been used for regulated synthesis of PG and CL. Ptet (inducible by anhydrotetracycline) has also been used for regulation of PE levels. Use of Ptet results in a dose response relationship between inducer and PE at any point in the cell cycle in each cell in the culture rather than an increasing percentage of cells displaying full induction levels of PE against a background of cells lacking PE (M. Bogdanov and W. Dowhan, unpublished result). These inducible promoters have been used to study the dynamic effects of changes in lipid composition on proteins post-assembly.
2.2. Introduction of foreign lipids into E. coli
Further alterations in the physical and chemical properties of the E. coli membrane has been possible through the introduction of foreign genes that synthesize lipids not found in E. coli (Fig. 1). Thus far genes have been introduced for the synthesis of phosphatidylcholine (pcs, PC) [32], phosphatidylinositol (PIS, PI) [33], O-lysyl PG (mrpF, O-lys-PG) (P. N. Heacock, M. Bogdanov, W. Dowhan, unpublished) and [9,10], monoglucosyl diacylglycerol (mgs, GlcDAG) [34,35] and diglucosyl DAG (dgs, GlcGlcDAG) [36]. PI levels reach 10% of total phospholipids in E. coli. The other lipids have been synthesized in wild type cells as well as in PE-lacking cells (ΔpssA). In cells lacking PE, O-lys-PG or PC reach about 70% of total lipid and the two glycolipids reach 30–40% of total lipid. Using Ptet regulation the percent of each lipid can be systematically controlled uniformly in each cell (unpublished result, M. Bogdanov, P. Heacock and W. Dowhan). PC and GlcDAG restore uphill transport function to PE-lacking cells but only the latter along with GlcGlcDAG to some extent suppress filamentous growth; GlcGlcDAG does not support uphill transport function. PI does not suppress the requirement of PG and CL for cell viability. Therefore, genetic manipulation of native and foreign genes in E. coli results in viable cells with large changes in steady state and temporal membrane phospholipid composition making this collection of mutants useful as reagents to study the role of lipids in cell processes.
2.3. Physical and chemical properties of lipids
Each lipid has unique physical and chemical properties, some of which are partially overlapping. The effects of these lipids can be through specific interaction with proteins or through a more global effect on the collective properties of the lipid bilayer. Fig. 2 summarizes some of the properties of these lipids. As will be discussed latter, using different lipid mixtures both in vivo and in vitro to support native function of proteins allows conclusions as to which properties of the native lipids are important to support function. Lipids with a net negative charge are the anionic lipids PA, PG, CL, PS, PI, N-acyl PE and O-acyl PG. All of these except PS contain no free amino groups. Amine-containing lipids are PS, PC, PE, N-acyl PE and O-lys PG. However, their head groups have different properties. PC and PE are charged but have no net charge. Of this group only PC cannot form hydrogen bonds due to the quaternary amine. PS and N-acyl PE have a net negative charge while O-lys PG has a net positive charge, which is not found in any E. coli lipid.
Another important property of lipids is their bilayer or non-bilayer prone tendencies. Bilayer prone lipids present a cylindrical shape with the cross-sectional area of the hydrophilic head group and the hydrophobic domain being similar. Such lipids form bilayers and do not induce any curvature stress into the bilayer. In non-bilayer prone lipids the cross-sectional area of the hydrophobic domain is larger than that of the head group. In pure form, such lipids do not form bilayers but rather a series of inverted micelle structures (hydrophobic domains outside). In mixtures with bilayer prone lipids, non-bilayer prone lipids induce membrane curvature and introduce stress into the bilayer structure due to negative curvature properties. The non-bilayer prone anionic lipid property of CL has been proposed as necessary for forming CL-enriched domains at the mid-cell during cell division and at the poles of cells where membrane curvature is greatest [23,37,38]. Non-bilayer properties have been proposed as necessary for translocation of proteins across membranes, membrane fusion/fusion events, and the interface between proteins and the bulk lipid phase. The non-bilayer prone lipids are PE, N-acyl PE, O-acyl PG, GlcDAG, PA and CL; the remaining lipids are bilayer prone. The fatty acid content is also a determinant of shape with unsaturated fatty acids favoring non-bilayer prone properties. For the anionic lipids, the presence of divalent cations enhances non-bilayer properties by reducing the effective size of the head group. Except for special cases such as fusion/fission sites of membranes, any formation of a non-bilayer domain would compromise cell integrity so the major contribution of nonbilayer prone lipids is to change the lateral pressure within the bilayer and introduce membrane curvature stress.
As can be seen from the summary in Fig. 2, each lipid has it own unique physical-chemical signature. Properties are overlapping between different lipids but rarely the same for any two lipids. For instance by comparing the effects of PE, PC, GlcDAG and GlcGlcDAG, a distinction between hydrogen bonding, bilayer/non-bilayer and charge properties can be reached. Examples of the use of these lipids mutants to determine which lipid property is required to support a cellular process will be described.
3. E. coli requires PE for normal cell function
As noted above, cells lacking PE display aberrant behavior characterized by a loss of synchrony between cell growth and cell division resulting in multi-nucleated filamentous cells [16]. Cell integrity and viability is dependent on high concentrations of divalent cations in the growth medium [11]. Therefore, E. coli requires PE but conditions can be found to propagate cells lacking PE to determine other functions that require PE. PE is also required to support energy-dependent uphill transport by a number of amino acid and sugar secondary transporters such as permeases for lactose (LacY), sucrose (CscB), maltose (MalB), proline (PutP), phenylalanine (PheP), γ-aminobutyrate (GabP), lysine (LysP), and tryptophan (AroP) [3]. Energy-independent downhill transport in PE-lacking cells is not affected. Of this group of permeases LacY is the most extensively studied. Over 30 years ago it was observed that purified LacY displayed full energy-dependent uphill and energy-independent downhill transport of substrates when reconstituted into proteoliposomes made of E. coli total lipids [39,40]. However, LacY reconstituted in liposomes lacking PE or in which dioleoyl-PC replaced PE only displayed downhill transport. Monomethyl-PE was less effective than PE and dimethyl-PE was even less effective. Since psd mutants (< 5% PE with 65% PS) carry out uphill transport by LacY [19], it was concluded that an ionizable amine or hydrogen bonding capability of the head group was required for uphill transport. The other possibility was that the in vitro result did not reflect in vivo requirements for LacY. The PE-lacking E. coli mutant provided the in vivo verification of the PE requirement for LacY by mimicking the proteoliposomes results with and without PE [41]. The negative results with PC in liposomes will be discussed Section 4.3.
What is the molecular basis for the PE requirement of LacY and presumably the other secondary permeases? One possibility was an alteration or significant reduction in PE-lacking cells of the membrane potential, which drives uphill transport. However, all components of the proton electrochemical potential are normal in PE-lacking cells, which suggested a lack of coupling of substrate uptake to the potential [41]. A structural defect in LacY was the most reasonable explanation for the defect. Two methods were used to compare the structure of LacY in membranes derived from PE-containing and PE-lacking membranes. One approach was to probe the structure of an epitope of LacY whose proper folding is tightly associated with uphill transport function. The second method was to probe the topological orientation of TMs of LacY as a function of membrane lipid composition.
4. Probing epitope structure related to function
4.1. A conformation-specific antibody against LacY
Fortuitously a conformation-specific monoclonal antibody (mAb) 4B1 is available that when interacting with LacY in native membranes or reconstituted into proteoliposomes blocks uphill transport but not downhill transport [42]. In wild type cells LacY assembles into the membrane as two six-TM domain helical bundles connected by a long cytoplasmic domain (C6) as shown in Fig. 4B. The epitope recognized by 4B1 lies in periplasmic domain P7 that connects TMs VI and VII and is characterized by high Phe content. The recognition depends both on the sequence of the epitope and its proper folding into an α-helix making it a highly conformation-dependent epitope. The molecular basis for mAb 4B1 inhibition of uphill transport activity appears to be a long-range effect on the carboxyl of E325 in TM X resulting in a lowering of its abnormal pKa of < 9 [43]. This residue is part of the proton wire that couples the symport of a proton with substrate in uphill transport. The antibody does not recognize mutants of LacY defective in uphill transport, LacY assembled in cell membranes lacking PE [44], or LacY reconstituted into liposomes lacking PE [45]. This is the case for direct binding studies using isolated membranes or on Western blots after extraction and separation by SDS gel electrophoresis. LacY in or isolated from cells containing PE is recognized by mAb 4B1.
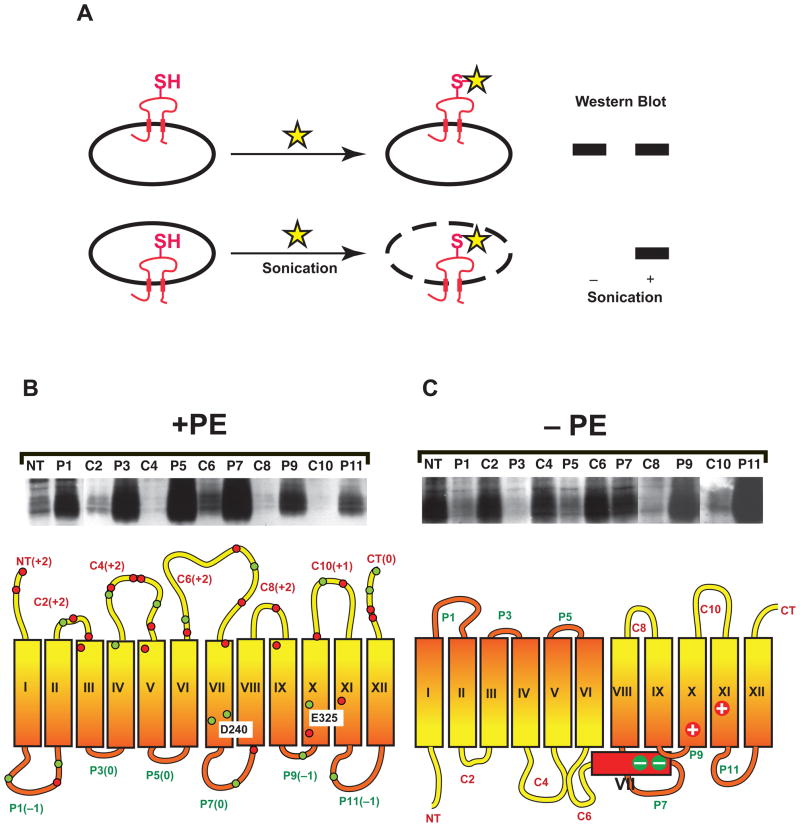
Fig. 4.
SCAM™ analysis of LacY as a function of membrane lipid composition. (A). Illustrates SCAM™ where a single cysteine replacement (SH) is exposed to either the periplasm (top) or cytoplasm (bottom). Addition of a sulfhydryl reagent (STAR) that can be detected by Western blotting to whole cells only derivatized periplasmically exposed cysteine residues with or without sonication of cells. A cytoplasmically exposed cysteine is only derivatized during sonication of cells. (B) (PE-containing cells) and (C) (PE-lacking cells) upper panels show SCAM™ Western blots after biotinylated sulfhydryl reagent treatment of whole cells expressing LacY containing single cysteine replacements in the indicated cytoplasmic (C) or periplasmic (P) domains based on orientation in PE-containing cells [30]. The lower panels illustrate the topological orientation of LacY TMs (rectangles numbered with Roman numerals) relative to the plane of the bilayer with the top of the figures facing the cytoplasm. The net charge of extra-membrane domains is shown next to the domain names. The approximate position of negatively (green) and positively (red) charged amino acids in the extramembrane domains and at selective sites within TMs is shown. The two acidic residues in TM VII are salt bridged to basic residues in TMs X and XI. D240 is the residue noted in the text that was changed to D240I, and E325 is the residue with an abnormally high pKa that is affected by binding of mAb 4B1 to domain P7. The periplasmic location of TM VII in PE-lacking cells was established in a separate set of SCAM™ experiments [31].
These results strongly suggested a structural change in the region of the P7 domain as a result of assembly of LacY in the absence of PE. In fact this structural defect in LacY is reversible by post-assembly exposure of LacY to PE [46]. LacY was synthesized in vitro in the presence of E. coli inside-out inner membrane vesicles from either wild type cells or cells lacking PE (ΔpssA). Western blotting analysis using mAb 4B1 confirmed that the P7 domain was properly folded only if wild type vesicles were used. In vitro synthesis of PE was reconstituted in the vesicles from PE-lacking cells by addition of soluble substrates and purified PS synthase. Recognition by mAb 4B1 was restored by synthesis of PE either co- or post-assembly of LacY in PE-lacking vesicles. This result suggested that PE is required in a late step along the folding pathway for LacY and could also function after final folding into a compact structure of LacY (as indicated by downhill transport function) when assembled in the absence of PE.
4.2. Eastern-Western blot
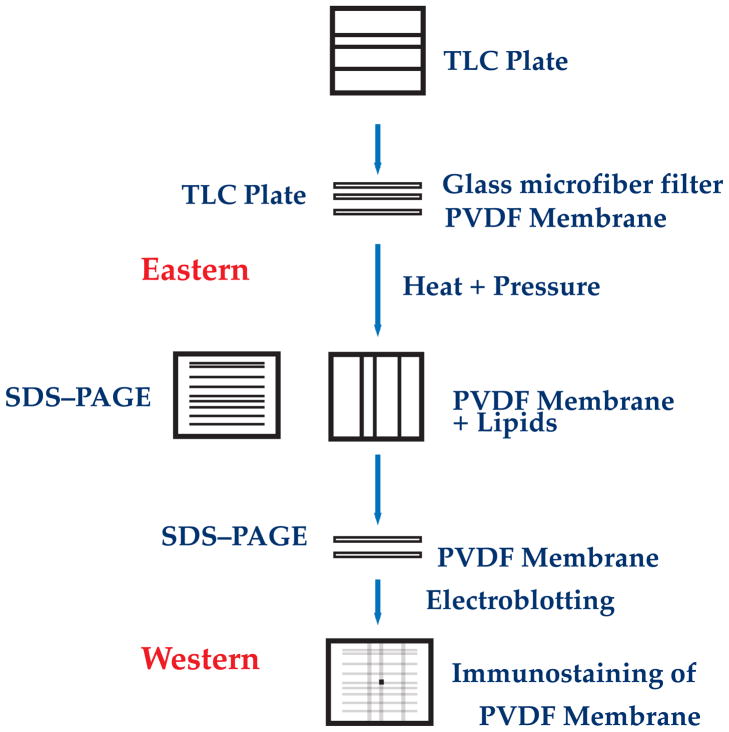
Since aberrant folding of the P7 domain can be reversed post-assembly, a method (called an Eastern-Western blot) was developed to screen for lipids that reverse misfolding [44,46,47]. Such lipids should have the properties required to support native structure and uphill transport. In this method (Fig. 3) phospholipids are transferred from a silica thin layer chromatography plate to a solid support used for Western blotting by using mild heat and pressure (Eastern blot). Lipid extracts from a cell can be applied to plates as horizontal bands and separated by solvent chromatography or individual lipids can be applied to plates as small patches. The former method is useful for screening a lipid extract of cells, and the latter method is useful for screening known lipids. A SDS extract of membranes is subjected to SDS gel electrophoresis either as a single lane or as a long horizontal band. Proteins are then electrophoretically transferred from the gel to the solid support on to which lipids have been transferred as normally done for a Western blot. If a single protein lane and patches of lipid have been used, then the position of the protein of interest (LacY) is aligned with the lipid patch on the solid support. For general screening of lipids the horizontal gel bands are turned 90° with respect to the lipid bands during transfer. In this manner the effect of all extracted lipids on all extracted proteins can be screened. During electrophoretic transfer, proteins, lipids and SDS are mixed. As the SDS is removed, the proteins fold in the presence of lipids. This method mimics on a reduced scale reconstitution of detergent extracted proteins with lipids by removal of detergent with dialysis in the presence of lipids. In general membrane proteins are not fully unfolded in SDS. Only their tertiary structure and packing of helices are disrupted. The yield of properly folded membrane protein after Eastern-Western blotting can be estimated by conformation-specific antibodies or by direct assay of enzyme activity. The Eastern-Western blot was useful as a screening technique to survey the role of specific lipids in protein folding and function of LacY [32,47].
Fig. 3.
Schematic of Eastern-Western procedure. Lipids are separated by thin layer chromatography and transferred to a solid supported (PVDF membrane) used for Western blotting (Eastern blot). An SDS extract of membranes is separated by gel electrophoresis and the proteins transferred to the above solid support (rotated 90° with respect to the direction of chromatography). The final solid support is subjected to immunodetection by specific antibody and visualized by standard Western blotting procedures.
4.3. Lipid dependence of restoration of LacY structure
Using the combined Eastern-Western procedure, the lipid properties that support P7 domain proper folding were established [44,47]. Regain of epitope conformation by LacY assembled in PE-lacking cells was attained using total E. coli phospholipids. All mixtures of PG and/or CL (natural or foreign) were not effective. Natural E. coli PE by itself was effective, as were commercial PE’s that were bilayer prone (i.e. containing saturated fatty acids). Non-bilayer prone PE’s (i.e. containing unsaturated fatty acids, plasmalogen form or lyso-PE) restored P7 conformation if mixed with PG, which is bilayer promoting. PS alone restored epitope conformation and was diastereomer-specific requiring the native stereochemistry for serine and the sn-2 position of glycerol; the addition of PG to ineffective PS diastereomers supported epitope restoration. Monomethyl- and dimethyl-PE were progressively less effective. Dioleolyl-PC and PC from several eukaryotic sources did not restore 4B1 recognition, in agreement with lack of uphill transport function in proteoliposomes containing PC in place of PE. In fact incorporating PC into the Eastern-Western blot with LacY from wild type cells prevented recognition by mAb 4B1 indicating the PC had a denaturing effect on native LacY.
These results indicate that an ionizable amine-containing bilayer prone phospholipid is required for the proper folding of domain P7. However, later results showed that E. coli lipids lacking PE and PS and containing GlcDAG but not GlcGlcDAG restored epitope conformation [34–36]. Consistent with this finding, LacY displays uphill transport in cells where PE is replaced by GlcDAG but not GlcGlcDAG. Finally, LacY is fully functional when expressed in cells where PC replaced PE [32]. Total lipid extracts from this mutant also restored P7 domain formation but E. coli derived PC required a binary mixture with PG to restore P7 conformation. Chemically synthesized PC’s containing at least one saturated fatty acid also restored the P7 domain conformation but only in the presence of PG. Therefore, an ionizable amine appears not to be required (provided PG is present) but the requirement for a bilayer prone lipid environment is required. Clearly the highly negative surface charge contributed by PG and CL is not supportive and requires some attenuation by net neutral lipids (PE, PC and GlcDAG) or the amine of PS. Why GlcGlcDAG is not effective is not clear unless the large head group prevents proper interaction with LacY. In the case where PC alone did not support P7 domain formation, the addition of lipids with hydrogen bonding capability was effective.
The lack of uphill transport function [39] and associated lack of recognition by mAb 4B1 of LacY in proteoliposomes where PE was replaced by PC [45] is an artifact of the in vitro reconstitution introduced by the use of PC species with the wrong fatty acid composition. The fact that LacY assembled in vivo in the presence of PC [32] is functional demonstrates the necessity of comparing in vitro results with in vivo results to arrive at a correct conclusion about lipid support of cell processes.
Two observations strongly suggest that PE acts as a molecular chaperone or lipochaperone at a late stage of final structural maturation at least for the folding of the P7 domain [48]. More classical protein molecular chaperones do not interact with random coils, at early steps of protein folding or with the fully folded native form of proteins but interact with late folding intermediates and assist in folding to the lowest energy minimum of the native state. In fulfilling these criteria, PE appears to act on a folding intermediate of LacY since extensive denaturation of LacY with urea-SDS prevents restoration of mAb 4B1 recognition of LacY by the Eastern-Western blotting procedure using PE. Renaturation of LacY depends on the removal of SDS in presence of different lipids rather than the simple exposure of partially denatured LacY to lipids since solubilization of PE-deficient membranes in the presence of added PE followed by Western blot analysis did not result in restoration of mAb 4B1 recognition [44].
Once final structure is attained chaperones are no longer required to maintain that structure. PE can induce the native structure of LacY even after it is folded into a compact structure sufficient to carry out downhill transport. Most important is that the conformation of the P7 domain attained in the presence of PE prior to partial denaturation by SDS and removal of PE during Western blotting is maintained as evidenced by detection by mAb 4B1. Lack of PE after Western blotting was confirmed by expression of LacY in cells where phospholipids were radiolabeled to a high specific activity [46].
The complexity of the above results illustrates the difficulty in determining the molecular basis for positive lipid protein interactions. Perturbations in the head group, the fatty acid composition, and the physical and chemical properties of lipids can have dramatic effects. However, an important point is to establish physiological significance of the in vitro observation. Thus far for LacY there is a close correlation between epitope conformation and activity as a function of lipid composition between in vivo and in vitro experiments. Given this close correlation, in vitro experiments aimed at a more detailed understanding of how lipids support the proper conformation and function of LacY will be based a solid foundation. Without the in vivo results, the effectiveness of PC with a quaternary amine in supporting epitope formation and function would not have been uncovered.
5. Investigation of membrane protein topological organization in whole cells
Lipid-dependent recognition of domain P7 by mAb 4B1 suggested a structural change in LacY in the absence of PE, which might be obscured after detergent solubilization and purification. The substituted cysteine accessibility method as applied to TMs (SCAM™) allows determination in whole cells and membranes of amino acid distribution between TMs and extra-membrane domains, the orientation of TMs with respect to the plane of the membrane bilayer, and local structural perturbations in protein domains (see [49,50] for detailed methodology and review of other methods used to determine protein topology). Beginning with a hydropathy plot of a protein to identify potential TMs, selected residues every 5 to 20 residues apart are replaced one at a time by cysteine in a protein derivative lacking native cysteines. Preferable residues to replace are serine and threonine, which are most similar to cysteine. Each single replacement derivative is checked for retention of expression and function. Derivatives are expressed in wild type cells and mutants in which lipid composition has been genetically altered. Whole cells are treated with an inner membrane impermeable sulfhydryl reagent, which can pass through the pores of the outer membrane (Fig. 4A). A parallel sample is treated during cell disruption by sonication to label solvent-exposed cysteines on both sides of inner membrane. A useful sulfhydryl reagent containing biotin is 3-(N-maleimidylpropionyl) biocytin, which can be detected on Western blots using Avidin-coupled reagents.
Cysteines exposed to the periplasm will be derivatized in whole cells and all extra-membrane cysteines (periplasmic and cytoplasmic) will be derivatized in sonicated cells. Sulfhydryls in TMs are not derivatized due to their high pKa’s. Those in extra-membranes can be inaccessible due to local secondary structure, proximity to the membrane or local charge effects that raise the pKa of thiols. The latter effects can be overcome by carrying out the analysis at pH 9–10.5, which disrupts local secondary structure, guarantees ionization of the thiol and does not compromise cell integrity. Cysteines that lie in mini-loops that are within the membrane bilayer but do not traverse the bilayer are operationally released from the membrane and exposed by treatment with NaOH. This differentiates such domains from true TMs that remain membrane embedded under strong alkaline conditions. Generally the derivatization of a periplasmically exposed cysteine is very similar with and without sonication. There are conditions where a particular domain displays a mixed topological orientation. Pretreatment of whole cells with a hydrophilic sulfhydryl reagent transparent to subsequent detection by Avidin-linked reagents can be used to determine the extent of mixed topology. To detect the derivatized cysteine residues, membranes are solubilized, the target protein is immunoprecipitated by specific antibody, and protein subjected to SDS gel electrophoresis and Western blotting using Avidin-linked fluorescent reagents. A complete set of cysteine derivatives before and after cell disruption and as a function of membrane lipid composition is used to generate the topological organization and orientation of the protein domains with respect to the plane of the membrane. The method has the advantage of minimal perturbation of protein structure and function by the use of cysteine replacements. Since the whole protein is expressed and analyzed, long-range interactions are preserved. The derivatization step is preformed in whole living cells and sonicated cells, which minimizes perturbation of protein structure prior to analysis.
6. Lipid-dependent topological organization of membrane proteins
Fig. 4B shows the Western blot after SCAM™ of single cysteine residues in then putative periplasmic and cytoplasmic domains of LacY assembled in PE-containing and PE-lacking E. coli cells [30]. The results are consistent with topological organization in wild type cells as suggested previously by other chemical means [51] and by subsequent X-ray crystallography [52]. However in PE-lacking cells (Fig. 4C), the N-terminal six TM helical bundle including neighboring extra-membrane domains is uniformly inverted with respect to the plane of the bilayer and the last five TMs [30]. These results were further confirmed by blocking periplasmically exposed cysteine residues prior to SCAM™ and verifying membrane integrity and inaccessibility of cysteines within TMs. Orientation of LacY was also tested in right side out and inside out inner membrane vesicles isolated from cells with and without PE. The exposure of TM VII to the periplasm in PE-lacking cells was deduced by cysteine scanning along the domain and SCAM™ [31]. Derivatization of these cysteines required pH 9 reaction conditions in PE-lacking cells suggesting a close membrane association or steric hindrance from secondary structure but not a TM location; no derivatization of cysteines occurred within TM VII in PE-containing cells.
TM VII is abnormally hydrophilic (ΔG = −5.2 kcal/mol of insertion) relative to most TMs (ΔG of −12 to −25 kcal/mol) due to two aspartate residues normally salt-bridged to positive residues in TMs X and XI [52]. A D240I replacement in TM VII increases its hydrophobicity (ΔG = −17.7 kcal/mol) and prevents inversion of the N-terminal bundle in PE-lacking cells [31]. Therefore, TM VII behaves as a necessary molecular hinge between the two halves of LacY that respond differently to the lipid environment. There is a long-range thermodynamic balance between the favorable energetics of inversion of the N-terminal bundle and the unfavorable energetics of expose of TM VII (containing D240I) to the solvent. This result also establishes that final topological organization occurs during late folding events and is not determined at the time of emergence of the nascent protein chain from the ribosome-translocon complex, which is consistent with previously results using mAb 4B1. The lack of recognition by the structure-dependent mAb 4B1 is also explained by the disruption of secondary structure in the C6-TM VII-P7 region of LacY assembled in the absence of PE.
Previous results using mAb 4B1 suggested that mis-folding events during assembly of LacY in the absence of PE could be corrected by post-assembly addition of PE [46]. This also proved to be the case with topological organization. Expression of LacY was first induced from Plac in a strain in which PE levels were < 3% due to the absence of inducers for the pssA gene under regulation Ptet or ParaB [30,31]. After new synthesis of LacY was terminated by removal of its inducer, PE synthesis was induced and cells were grown until lipid composition returned to normal. SCAM™ demonstrated that previously synthesized and folded LacY (as demonstrated by downhill transport) returned to a near normal topological organization including the re-insertion of TM VII into the membrane. Only TM I remained inverted and TM II now formed a mini-loop and acted as a new molecular hinge. Recognition by mAb 4B1 and uphill transport function were restored. Demonstration that old and not new LacY was being analyzed came from lack of radiolabeled LacY synthesis after induction was stopped, no change in radiolabeled LacY content after induction was stopped, and the unique topological organization (TM I remaining inverted) of LacY after post-assembly exposure to PE.
Clearly the activation energy for TM flipping is low and must be driven thermodynamically by the change in lipid environment, which now displaces the aberrantly folded protein from its energy minimum. Since it is highly unlikely that the translocon is involved in post-assembly TM flipping [53], protein-lipid interactions are an important determinant of final membrane protein organization and provide their input after exit of protein domains from the translocon [54,55]. This conclusion was further supported by reconstitution of purified LacY into liposomes [45]. The source of LacY, either from PE-containing or PE-lacking cells, was not a factor in the structure and function of the reconstituted protein. Reconstitution into liposomes of total wild type E. coli phospholipids resulted in wild type topological organization, uphill and downhill transport and recognition by mAb 4B1. Reconstitution into liposomes made from phospholipids extracted from PE-lacking cells resulted in aberrant topological organization in the C6-TM VII-P7 region, only downhill transport and lack of recognition by mAb 4B1. Addition of dioleoyl-PC to the latter liposomes corrected topology in the central domain of LacY but did not correct other properties. Finally these results demonstrate the potential for large topological changes in membrane proteins post-assembly due to temporal local changes in lipid environment or changes in lipid environment as proteins undergo intracellular trafficking.
7. Lipid properties that influence TM orientation and function
As summarized in Fig. 2, PE has several unique properties that could be the molecular basis for its influence on LacY topological organization. PE is non-bilayer prone (when high in unsaturated fatty acids), has an ionizable amine, is capable of hydrogen bonding through its head group, and is charged but has a net charge of zero at physiological pH’s. The above reconstitution experiments with PC suggested that non-bilayer and hydrogen bonding properties were not important at least in vitro for establishing proper topological organization. Further details of the lipid requirement were established using several of the PE-lacking strains expressing lipids foreign to E. coli. LacY displays wild type topological organization in PE-lacking cells expressing PC [32], GlcDAG [35] and GlcGlcDAG [34]. These results suggest that the most important property of PE is its net neutral charge that dampens the high negative charge density of the membrane surface imparted by PG and CL. Although topology is correct in these strains, cysteine scanning coupled with SCAM™ showed a different pattern of accessibility and pH-dependence for reactivity of cysteine in the periplasmic domains compared to LacY expressed in cells containing PE. Therefore, local subtle differences exist in protein structure dependent on the lipid environment.
The lipid requirements for supporting native function of LacY are more complicated and not yet fully resolved as noted in Section 4.3. Downhill transport is not lipid or topological dependent [3,32]. Native topological organization (except in the N-terminal region of NT through TM II) is necessary but not sufficient for uphill transport. Thus far there is a very close correlation between proper folding of the P7 domain and uphill transport function [32]. Binding of mAb 4B1, which can be extrapolated to aberrant effects of lipids on P7, results in a lowering of the pKa of E325 and conformational changes in several TMs [56]. It is not clear why E. coli derived PC supports uphill transport and P7 domain conformation while PC from several other sources does not [32]. Similarly the difference between GlcDAG and GlcGlcDAG in supporting LacY function is not clear [36]. However, cysteine scanning of the periplasmic domains of LacY assembled in the presence of foreign lipids did show lipid-dependent structure differences [32,36]. The more subtle effects of lipid environment on activity and the conformation of the P7 domain will require more detailed structural analysis of LacY in different lipid environments.
8. Lipid sensitive protein topological determinants
What are the features of LacY that make its topological organization sensitive to the lipid environment? As with 85% of integral membrane proteins, the net-charge of the cytoplasmic domains of LacY follows the positive-inside rule (see Fig. 4B). According to the positive-inside rule, positive residues are cytoplasmic retention signals and negative residues are domain translocation signals [57,58]. In wild type cells positive residues are dominant over negative residues resulting in cytoplasmic retention of some net-negative domains that contain positive residues. Negative residue translocation potential is stronger closer to the membrane surface and falls off further away. Since there are more negatively charged amino acids on the cytoplasmic surface of the N-terminal six TM bundle of LacY than on the C-terminal bundle, it was proposed that membrane lipid composition may affect the relative potency of these charged topological determinants resulting in topological inversion of LacY in the absence of PE. Manipulation of the charges within normally cytoplasmic domains of LacY assembled in different lipid environments was used to address this hypothesis [3,30,31].
Increasing the net positive charge by one in a position independent manner in any of the cytoplasmic domains (C2, C4 or C6) of the N-terminal bundle prevented inversion of LacY in PE-lacking cells. Adding a positive charge or removing a negative charge had the same effect, but removing both a positive and negative charge made no difference. Making the N-terminal bundle more negative also resulted in inversion of the bundle in PE-containing cells. However, consistent with the dominance of positive over negative residues, the net charge of the bundle surface (all three C domains) had to be changed from +6 to −6 to induce inversion. The balance between opposing thermodynamic forces was also further confirmed. Although the D240I substitution in TM VII prevented inversion of otherwise wild type LacY in PE-lacking cells and in the −6 charged cytoplasmic derivative of LacY in PE-containing cells, the latter D240I −6 charged derivative of LacY was inverted in PE-lacking cells where the higher translocation potential of multiple negative residues overcomes the hydrophobic barrier imposed by the D240I replacement. Therefore, an important role for net-neutral lipids like PE, PC, GlcDAG and GlcGlcDAG is to dampen the translocation potential of negative residues (or strengthen the retention potential of positive residues) in cytoplasmic domains in order to maintain required topological organization when important functional negative residues are present on the cytoplasmic face. Elimination of several of these negative residues of LacY resulted in loss of function in PE-containing cells even though topology was normal.
These results form the basis for the “Charge Balance Hypothesis” [3]. Membrane proteins have co-evolved with membrane lipid composition to balance the net charge of the cytoplasmic surface of membrane proteins with the net surface charge of the membrane surface contributed by the lipid head groups. Perturbation in the charge of either results in topological and other aberrant changes in membrane protein structure and function strongly supporting protein-lipid interactions as important determinants of membrane protein structure.
9. Generality of the Charge Balance Hypothesis
Thus far only a subset of proteins, mainly secondary transporters in E. coli, has been shown to be affected by the membrane lipid composition. Given the other phenotypes of PE-lacking cells, proteins involved in other cell processes may be affected in a similar manner. However, no absolutely essential process is completely inactivated. As noted for LacY, complete transport function is not lost so that topological inversion may compromise but not completely render other processes non-functional. Three other permeases (PheP, GabP and CscB) of E. coli have been characterized with respect to their PE requirement for function and assembly. All fail to carry out uphill transport in PE-lacking cells but there are differences in effects on topology.
PheP [59] and GabP [60] topology is lipid dependent. Both show inversion in PE-lacking cells of only the N-terminal TM hairpin (TMs I and II) with TM III acting as a mini-loop and molecular hinge to the remainder of the proteins with normal topology. Regain of normal topology and function of PheP was demonstrated by post-assembly synthesis of PE. The N-terminal C2 domain of PheP is net negative and that of GabP is net neutral so these domains do not follow the positive inside rule, but the dominant positive residues must provide the cytoplasmic retention potential in PE-containing cells. However, increasing the net positive charge of the NT domain by +2 or decreasing the net negative charge of the C2 domain from −1 to +1 results in PheP of mixed topology in PE-lacking cell with 60% in favor of wild type topology [61]. Combining these two changes in the net positive charge of the hairpin bundle results in completely wild type topology in PE-lacking cells indicating adherence to the Charge Balance Hypothesis.
The situation with CscB is more complex since the protein exhibits normal topological organization in PE-containing and PE-lacking cells [61]. However, derivatives with altered charges in the cytoplasmic face follow the Charge Balance Hypothesis. CscB is similar to LacY in that the cytoplasmic face of the N-terminal six TM bundle contains a mixture of positive and negative residues, but it differs in that all of the domains have more charged residues. C2 has a net neutral charge, NT contains no acidic residues and TM VII contains only one aspartate and is highly hydrophobic (ΔG = −25.1 kcal/mol). The higher charge density of C6 in CscB (13 versus 6 for LacY) imparts higher hydrophilicity (ΔG = +98.1 kcal/mol vs. +26.5 kcal/mol for LacY). The higher hydrophobicity of TM VII might prevent its acting as a molecular hinge as the D240I substitution does for LacY. Introduction of a second aspartate residue at N243 did not induce a topological inversion of CscB in PE-lacking cells, since this replacement only lowered hydrophobicity to ΔG = −20.7 kcal/mol. However, as shown in Table 2, progressively increasing the net negative charge of the cytoplasmic domains of CscB resulted first in a mixed topological organization and then complete inversion in PE-lacking cells based on diagnostic cysteine residues in domains NT and C6. Inversion was also induced in PE-containing cells but required a more negative cytoplasmic face for initial mixed topology as well as full inversion to occur. The higher negative charge translocation potential required in both PE-containing and PE-lacking cells for CscB relative to LacY, appears to be due to the larger thermodynamic barrier imposed by above ΔG values. Thus increasing the translocation potential either by increasing the negative charge of cytoplasmic domains or eliminating PE can overcome the thermodynamic barrier to inversion of hydrophobic TM VII of either CscB or LacY. These results further support the Charge Balance Hypothesis, the dominance of positive residue retention signals over negative residue translocation signals in PE-containing cells, the dampening effect of neutral lipids on translocation potential of negative residues, and short-range lipid-protein interactions and long-range interactions within the protein as topological determinants in late folding events.
Table 2.
Dependence of CscB domain C6 and NT topology on domain charge and membrane phospholipid composition.
| C2 Chargea | C4 Chargea | C6 Chargea | +PE Topology C6/NTb | −PE Topology C6/NTb |
|---|---|---|---|---|
| +2 (2) | 0 (3) | +1 (7) | N/nd | N/nd |
| 0 (0) | 0 (3) | −2 (4) | N/nd | N/nd |
| 0 (0) | −2 (1) | −2 (4) | N/nd | M/nd |
| −2 (0) | 0 (3) | −2 (4) | N/M | M/M |
| −2 (0) | −2 (1) | −2 (4) | N/I | I/I |
| −2 (0) | −2 (1) | −5 (1) | N/nd | nd |
| −2 (0) | −2 (1) | −3 (2) | N/nd | nd |
| −2 (0) | −2 (1) | −6 (0) | I/nd | nd |
Net charge of domain with number of plus charges in parentheses
N=Normal, nd=not determined, M=mixed, I=inverted in PE-containing (+PE) or PE lacking (−PE) cells.
Table reproduced in part from [61].
The effects of lipids on transport function also appear to extend beyond these three permeases. Several other secondary transporters have also been shown to require PE for uphill transport after reconstitution into proteoliposomes, namely the multidrug transporter (LmrP) of Lactococcus lactis [62,63], the leucine permease of Pseudomonas aeruginosa [64], the branched chain amino acid transporter of Streptococcus cremoris [65], the ABC transporter HorA from Lactococcus lactis [66], and the Ca2+ ATPase of the sarcoplasmic reticulum [67]. Yeast mutants in which PE and/or PS levels have been drastically reduced show defects in the transport of tryptophan and tyrosine (Tat1p and Tat2p) [68], arginine (Can1p) [69,70], siderophore (Arn1p and Arn3p) [13], proline (Put4p) and general amino acids (Gap1p) [71]. Lipids have been implicated in the proper folding or in inducing changes in protein structure of many membrane-associated proteins in mammalian cells such as prion protein PrP, amyloid peptide implicated in Alzheimer’s disease, cystic fibrosis transmembrane conductance regulator (CFTR), and maturation of pro-insulin [4]. In these examples changes in lipid environment dramatically alter protein function and presumably structure. Ductin (component of vacuolar ATPase and connexon channel subunit) [72] and epoxide hydrolase/bile acid transporter [73] are two proteins that exhibit dual topologies upon initial assembly in the ER after which each form moves to different organelles where they stably exhibit unique but different topologies and functions. Differences in organelle lipid composition could very well stabilize different forms of these proteins. Although topological heterogeneity could be achieved in the endoplasmic reticulum by an unknown translocon-dependent insertion mechanism, it is more likely that such a mixed topology is governed by lipid-protein interactions since multiple topological forms of PheP and CscB have been generated in E. coli by manipulation of protein topological signals and membrane lipid composition [61].
10. Summary
A collection of mutants in E. coli lipid metabolism is available in which membrane lipid composition can be systematically controlled both in the steady state and temporally to observe the dynamic changes in protein topology in vivo as a function of lipid composition using different experimental strategies.
Lipids can interact with proteins as determinants of final structural organization during late folding events outside the translocon.
Lipids have molecular chaperone properties that assist in the folding of membrane proteins similar to more classical protein-based chaperones.
Membrane protein TM organization once attained is not static but can change post-assembly in response to changes in the lipid environment.
Net neutral lipids attenuate the translocation potential of negative amino acids making the retention potential of positive amino acids the dominant determinant of TM orientation thereby providing a molecular basis for the operation of the positive inside rule for domains containing a mixture of positive and negative residues.
An important physiological role for PE and other lipids with net zero charge is to allow the functional presence of negatively charged amino acids in cytoplasmic domains of membrane proteins without affecting protein topology.
Protein sequence determines protein organization but final topological organization is dependent on the lipid composition of the host organism.
During the course of evolution both membrane proteins and lipids have co-evolved in the context of the lipid environment of the host to establish a set of interdependent determinants of final protein organization.
After release from the translocon final protein folding events are governed co- and post-assembly by short-range charge interactions between the nascent protein chain and both the translocon and lipids as well as long-range interactions within the protein that thermodynamically balance the relative strength of hydrophobic forces and charge effects.
Highlights.
E. coli strains in which membrane lipid composition can be systematically altered.
Charge interactions with lipids are determinants of membrane protein topogenesis.
Late folding events outside the translocon determine membrane protein structure.
Lipids act as molecular chaperone that assist in membrane protein folding.
Membrane protein organization can change due to changes in the lipid environment.
Acknowledgments
The authors dedicate this review to the memory of Christian R. H. Raetz, M.D. Ph.D. who passed away in August of 2011 after a three-year battle with cancer. Chris was a friend and colleague who contributed immensely over the past 40 years to the lipid and membrane fields. The work of the authors cited in this review was supported in part by grant GM020478 from the National Institutes of General Medical Science, U.S.A. and the John Dunn Research Foundation awarded to W.D.
Footnotes
Abbreviations: TM, transmembrane domain; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; CL, cardiolipin; PA, phosphatidic acid; PS, phosphatidylserine; lys, lysyl; PC, phosphatidylcholine; GlcDAG, monoglucosyl diacylglycerol; GlcGlcDAG, diglucosyl diacylglycerol; PI, phosphatidylinositol; LacY, lactose permease; CscB, sucrose permease; PheP, phenylalanine permease; γ-aminobutyrate permease, GabP; mAb, monoclonal antibody; SCAM™, substituted cysteine accessibility method as applied to TMs;
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Singer SJ, Nicolson GL. The fluid mosaic model of the structure of cell membranes. Science. 1972;175:720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 2.Dowhan W. Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu Rev Biochem. 1997;66:199–232. doi: 10.1146/annurev.biochem.66.1.199. [DOI] [PubMed] [Google Scholar]
- 3.Dowhan W, Bogdanov M. Lipid-dependent membrane protein topogenesis. Annu Rev Biochem. 2009;78:515–540. doi: 10.1146/annurev.biochem.77.060806.091251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogdanov M, Mileykovskaya E, Dowhan W. Lipids in the assembly of membrane proteins and organization of protein supercomplexes: implications for lipid-linked disorders. Subcell Biochem. 2008;49:197–239. doi: 10.1007/978-1-4020-8831-5_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nikaido H. Outer membrane. In: Neidhardt FC, editor. E coli and S typhimurium: Cellular and Molecular Biology. 2. Amer. Soc. Microbiol; Washington, DC: 1996. pp. 29–47. [Google Scholar]
- 6.Raetz CRH, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in Gram-negative bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dowhan W. Molecular genetic approaches to defining lipid function. J Lipid Res. 2009;50(Suppl):S305–310. doi: 10.1194/jlr.R800041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu YH, Guan Z, Zhao J, Raetz CR. Three phosphatidylglycerol-phosphate phosphatases in the inner membrane of Escherichia coli. J Biol Chem. 2011;286:5506–5518. doi: 10.1074/jbc.M110.199265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ernst CM, Staubitz P, Mishra NN, Yang SJ, Hornig G, Kalbacher H, Bayer AS, Kraus D, Peschel A. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog. 2009;5:e1000660. doi: 10.1371/journal.ppat.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oku Y, Kurokawa K, Ichihashi N, Sekimizu K. Characterization of the Staphylococcus aureus mprF gene, involved in lysinylation of phosphatidylglycerol. Microbiology. 2004;150:45–51. doi: 10.1099/mic.0.26706-0. [DOI] [PubMed] [Google Scholar]
- 11.DeChavigny A, Heacock PN, Dowhan W. Phosphatidylethanolamine may not be essential for the viability of Escherichia coli. J Biol Chem. 1991;266:5323–5332. [PubMed] [Google Scholar]
- 12.Nagahama H, Sakamoto Y, Matsumoto K, Hara H. RcsA-dependent and - independent growth defects caused by the activated Rcs phosphorelay system in the Escherichia coli pgsA null mutant. J Gen Appl Microbiol. 2006;52:91–98. doi: 10.2323/jgam.52.91. [DOI] [PubMed] [Google Scholar]
- 13.Guo Y, Au WC, Shakoury-Elizeh M, Protchenko O, Basrai M, Prinz WA, Philpott CC. Phosphatidylserine is involved in the ferrichrome-induced plasma membrane trafficking of Arn1 in Saccharomyces cerevisiae. J Biol Chem. 2010;285:39564–39573. doi: 10.1074/jbc.M110.177055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bogdanov M, Xie J, Dowhan W. Lipid-protein interactions drive membrane protein topogenesis in accordance with the positive inside rule. J Biol Chem. 2009;284:9637–9641. doi: 10.1074/jbc.R800081200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Killian JA, Koorengevel MC, Bouwstra JA, Gooris G, Dowhan W, de Kruijff B. Effect of divalent cations on lipid organization of cardiolipin isolated from Escherichia coli strain AH930. Biochim Biophys Acta. 1994;1189:225–232. doi: 10.1016/0005-2736(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 16.Mileykovskaya E, Dowhan W. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J Bacteriol. 2000;182:1172–1175. doi: 10.1128/jb.182.4.1172-1175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mileykovskaya E, Dowhan W. The Cpx two-component signal transduction pathway is activated in Escherichia coli mutant strains lacking phosphatidylethanolamine. J Bacteriol. 1997;179:1029–1034. doi: 10.1128/jb.179.4.1029-1034.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mileykovskaya EI, Dowhan W. Alterations in the electron transfer chain in mutant strains of Escherichia coli lacking phosphatidylethanolamine. J Biol Chem. 1993;268:24824–24831. [PubMed] [Google Scholar]
- 19.Hawrot E, Kennedy EP. Phospholipid composition and membrane function in phosphatidylserine decarboxylase mutants of Escherichia coli. J Biol Chem. 1978;253:8213–8220. [PubMed] [Google Scholar]
- 20.Shi W, Bogdanov M, Dowhan W, Zusman DR. The pss and psd genes are required for motility and chemotaxis in Escherichia coli. J Bacteriol. 1993;175:7711–7714. doi: 10.1128/jb.175.23.7711-7714.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heacock PN, Dowhan W. Alterations of the phospholipid composition of Escherichia coli through genetic manipulation. J Biol Chem. 1989;264:14972–14977. [PubMed] [Google Scholar]
- 22.Mileykovskaya E, Ryan AC, Mo X, Lin CC, Khalaf KI, Dowhan W, Garrett TA. Phosphatidic acid and N-acyl phosphatidylethanolamine form membrane domains in Escherichia coli mutant lacking cardiolipin and phosphatidylglycerol. J Biol Chem. 2009;284:2990–3000. doi: 10.1074/jbc.M805189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mileykovskaya E, Dowhan W. Cardiolipin domains in prokaryotes and eukaryotes. Biochim Biophys Acta-Biomembranes. 2009;1778:2084–2091. doi: 10.1016/j.bbamem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gold VA, Robson A, Bao H, Romantsov T, Duong F, Collinson I. The action of cardiolipin on the bacterial translocon. Proc Nat’l Acad Sci, U S A. 2010;107:10044–10049. doi: 10.1073/pnas.0914680107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lill R, Dowhan W, Wickner W. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell. 1990;60:271–280. doi: 10.1016/0092-8674(90)90742-w. [DOI] [PubMed] [Google Scholar]
- 26.Goldberg DE, Rumley MK, Kennedy EP. Biosynthesis of membrane-derived oligosaccharides: a periplasmic phosphoglyceroltransferase. Proc Natl Acad Sci U S A. 1981;78:5513–5517. doi: 10.1073/pnas.78.9.5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki M, Hara H, Matsumoto K. Envelope disorder of Escherichia coli cells lacking phosphatidylglycerol. J Bacteriol. 2002;184:5418–5425. doi: 10.1128/JB.184.19.5418-5425.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo D, Tropp BE. A second Escherichia coli protein with CL synthase activity. Biochim Biophys Acta. 2000;1483:263–274. doi: 10.1016/s1388-1981(99)00193-6. [DOI] [PubMed] [Google Scholar]
- 29.Romantsov T, Helbig S, Culham DE, Gill C, Stalker L, Wood JM. Cardiolipin promotes polar localization of osmosensory transporter ProP in Escherichia coli. Mol Microbiol. 2007;64:1455–1465. doi: 10.1111/j.1365-2958.2007.05727.x. [DOI] [PubMed] [Google Scholar]
- 30.Bogdanov M, Heacock PN, Dowhan W. A polytopic membrane protein displays a reversible topology dependent on membrane lipid composition. EMBO J. 2002;21:2107–2116. doi: 10.1093/emboj/21.9.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bogdanov M, Xie J, Heacock P, Dowhan W. To flip or not to flip: lipid-protein charge interactions are a determinant of final membrane protein topology. J Cell Biol. 2008;182:925–935. doi: 10.1083/jcb.200803097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bogdanov M, Heacock P, Guan Z, Dowhan W. Plasticity of lipid-protein interactions in the function and topogenesis of the membrane protein lactose permease from Escherichia coli. Proc Natl Acad Sci U S A. 2010;107:15057–15062. doi: 10.1073/pnas.1006286107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia W, Dowhan W. Phosphatidylinositol cannot substitute for phosphatidylglycerol in supporting cell growth of Escherichia coli. J Bacteriol. 1995;177:2926–2928. doi: 10.1128/jb.177.10.2926-2928.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wikstrom M, Xie J, Bogdanov M, Mileykovskaya E, Heacock P, Wieslander A, Dowhan W. Monoglucosyldiacylglycerol, a foreign lipid, can substitute for phosphatidylethanolamine in essential membrane-associated functions in Escherichia coli. J Biol Chem. 2004;279:10484–10493. doi: 10.1074/jbc.M310183200. [DOI] [PubMed] [Google Scholar]
- 35.Xie J, Bogdanov M, Heacock P, Dowhan W. Phosphatidylethanolamine and monoglucosyldiacylglycerol are interchangeable in supporting topogenesis and function of the polytopic membrane protein lactose permease. J Biol Chem. 2006;281:19172–19178. doi: 10.1074/jbc.M602565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wikstrom M, Kelly AA, Georgiev A, Eriksson HM, Klement MR, Bogdanov M, Dowhan W, Wieslander A. Lipid-engineered Escherichia coli membranes reveal critical lipid headgroup size for protein function. J Biol Chem. 2009;284:954–965. doi: 10.1074/jbc.M804482200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang KC, Mukhopadhyay R, Wingreen NS. A curvature-mediated mechanism for localization of lipids to bacterial poles. PLoS Comput Biol. 2006;2:e151. doi: 10.1371/journal.pcbi.0020151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mukhopadhyay R, Huang KC, Wingreen NS. Lipid localization in bacterial cells through curvature-mediated microphase separation. Biophys J. 2008;95:1034–1049. doi: 10.1529/biophysj.107.126920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen CC, Wilson TH. The phospholipid requirement for activity of the lactose carrier of Escherichia coli. J Biol Chem. 1984;259:10150–10158. [PubMed] [Google Scholar]
- 40.Seto-Young D, Chen CC, Wilson TH. Effect of different phospholipids on the reconstitution of two functions of the lactose carrier of Escherichia coli. J Membr Biol. 1985;84:259–267. doi: 10.1007/BF01871389. [DOI] [PubMed] [Google Scholar]
- 41.Bogdanov M, Dowhan W. Phosphatidylethanolamine is required for in vivo function of the membrane-associated lactose permease of Escherichia coli. J Biol Chem. 1995;270:732–739. doi: 10.1074/jbc.270.2.732. [DOI] [PubMed] [Google Scholar]
- 42.Sun J, Wu J, Carrasco N, Kaback HR. Identification of the epitope for monoclonal antibody 4B1 which uncouples lactose and proton translocation in the lactose permease of Escherichia coli. Biochemistry. 1996;35:990–998. doi: 10.1021/bi952166w. [DOI] [PubMed] [Google Scholar]
- 43.Frillingos S, Kaback HR. Monoclonal antibody 4B1 alters the pKa of a carboxylic acid at position 325 (helix X) of the lactose permease of Escherichia coli. Biochemistry. 1996;35:10166–10171. doi: 10.1021/bi960995r. [DOI] [PubMed] [Google Scholar]
- 44.Bogdanov M, Sun J, Kaback HR, Dowhan W. A phospholipid acts as a chaperone in assembly of a membrane transport protein. J Biol Chem. 1996;271:11615–11618. doi: 10.1074/jbc.271.20.11615. [DOI] [PubMed] [Google Scholar]
- 45.Wang X, Bogdanov M, Dowhan W. Topology of polytopic membrane protein subdomains is dictated by membrane phospholipid composition. EMBO J. 2002;21:5673–5681. doi: 10.1093/emboj/cdf571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bogdanov M, Dowhan W. Phospholipid-assisted protein folding: phosphatidylethanolamine is required at a late step of the conformational maturation of the polytopic membrane protein lactose permease. EMBO J. 1998;17:5255–5264. doi: 10.1093/emboj/17.18.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bogdanov M, Umeda M, Dowhan W. Phospholipid-assisted refolding of an integral membrane protein. Minimum structural features for phosphatidylethanolamine to act as a molecular chaperone. J Biol Chem. 1999;274:12339–12345. doi: 10.1074/jbc.274.18.12339. [DOI] [PubMed] [Google Scholar]
- 48.Bogdanov M, Dowhan W. Lipid-assisted protein folding. J Biol Chem. 1999;274:36827–36830. doi: 10.1074/jbc.274.52.36827. [DOI] [PubMed] [Google Scholar]
- 49.Bogdanov M, Heacock PN, Dowhan W. Study of polytopic membrane protein topological organization as a function of membrane lipid composition. Methods Mol Biol. 2010;619:79–101. doi: 10.1007/978-1-60327-412-8_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bogdanov M, Zhang W, Xie J, Dowhan W. Transmembrane protein topology mapping by the substituted cysteine accessibility method (SCAM™): application to lipid-specific membrane protein topogenesis. Methods. 2005;36:148–171. doi: 10.1016/j.ymeth.2004.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaback HR, Sahin-Toth M, Weinglass AB. The kamikaze approach to membrane transport. Nat Rev Mol Cell Biol. 2001;2:610–620. doi: 10.1038/35085077. [DOI] [PubMed] [Google Scholar]
- 52.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 53.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 54.Dowhan W, Bogdanov M. Lipid-protein interactions as determinants of membrane protein structure and function. Biochem Soc Trans. 2011;39:767–774. doi: 10.1042/BST0390767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Driessen AJ, Nouwen N. Protein Translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- 56.Frillingos S, Wu J, Venkatesan P, Kaback HR. Binding of ligand or monoclonal antibody 4B1 induces discrete structural changes in the lactose permease of Escherichia coli. Biochemistry. 1997;36:6408–6414. doi: 10.1021/bi970233b. [DOI] [PubMed] [Google Scholar]
- 57.von Heijne G. Membrane-protein topology. Nat Rev Mol Cell Biol. 2006;7:909–918. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- 58.von Heijne G, Gavel Y. Topogenic signals in integral membrane proteins. Eur J Biochem. 1988;174:671–678. doi: 10.1111/j.1432-1033.1988.tb14150.x. [DOI] [PubMed] [Google Scholar]
- 59.Zhang W, Bogdanov M, Pi J, Pittard AJ, Dowhan W. Reversible topological organization within a polytopic membrane protein is governed by a change in membrane phospholipid composition. J Biol Chem. 2003;278:50128–50135. doi: 10.1074/jbc.M309840200. [DOI] [PubMed] [Google Scholar]
- 60.Zhang W, Campbell HA, King SC, Dowhan W. Phospholipids as determinants of membrane protein topology. Phosphatidylethanolamine is required for the proper topological organization of the gamma-aminobutyric acid permease (GabP) of Escherichia coli. J Biol Chem. 2005;280:26032–26038. doi: 10.1074/jbc.M504929200. [DOI] [PubMed] [Google Scholar]
- 61.Vitrac H, Bogdanov M, Heacock P, Dowhan W. Lipids and topological rules of membrane protein assembly: balance between long- and short-range lipid-protein interactions. J Biol Chem. 2011;286:15182–15194. doi: 10.1074/jbc.M110.214387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hakizimana P, Masureel M, Gbaguidi B, Ruysschaert JM, Govaerts C. Interactions between phosphatidylethanolamine headgroup and LmrP, a multidrug transporter: a conserved mechanism for proton gradient sensing? J Biol Chem. 2008;283:9369–9376. doi: 10.1074/jbc.M708427200. [DOI] [PubMed] [Google Scholar]
- 63.Gbaguidi B, Hakizimana P, Vandenbussche G, Ruysschaert JM. Conformational changes in a bacterial multidrug transporter are phosphatidylethanolamine-dependent. Cell Mol Life Sci. 2007;64:1571–1582. doi: 10.1007/s00018-007-7031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Uratani Y, Aiyama A. Effect of phospholipid composition on activity of sodium-dependent leucine transport system in Pseudomonas aeruginosa. J Biol Chem. 1986;261:5450–5454. [PubMed] [Google Scholar]
- 65.Driessen AJ, Zheng T, In’t Veld G, Op den Kamp JA, Konings WN. Lipid requirement of the branched-chain amino acid transport system of Streptococcus cremoris. Biochemistry. 1988;27:865–872. doi: 10.1021/bi00403a005. [DOI] [PubMed] [Google Scholar]
- 66.Gustot A, Smriti, Ruysschaert JM, McHaourab H, Govaerts C. Lipid composition regulates the orientation of transmembrane helices in HorA, an ABC multidrug transporter. J Biol Chem. 2010;285:14144–14151. doi: 10.1074/jbc.M109.079673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Navarro J, Toivio-Kinnucan M, Racker E. Effect of lipid composition on the calcium/adenosine 5′-triphosphate coupling ratio of the Ca2+-ATPase of sarcoplasmic reticulum. Biochemistry. 1984;23:130–135. doi: 10.1021/bi00296a021. [DOI] [PubMed] [Google Scholar]
- 68.Abe F, Iida H. Pressure-induced differential regulation of the two tryptophan permeases Tat1 and Tat2 by ubiquitin ligase Rsp5 and its binding proteins, Bul1 and Bul2. Mol Cell Biol. 2003;23:7566–7584. doi: 10.1128/MCB.23.21.7566-7584.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Opekarova M, Malinska K, Novakova L, Tanner W. Differential effect of phosphatidylethanolamine depletion on raft proteins: further evidence for diversity of rafts in Saccharomyces cerevisiae. Biochim Biophys Acta. 2005;1711:87–95. doi: 10.1016/j.bbamem.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 70.Opekarova M, Robl I, Tanner W. Phosphatidyl ethanolamine is essential for targeting the arginine transporter Can1p to the plasma membrane of yeast. Biochim Biophys Acta. 2002;1564:9–13. doi: 10.1016/s0005-2736(02)00455-8. [DOI] [PubMed] [Google Scholar]
- 71.Robl I, Grassl R, Tanner W, Miroslava, Opekarova M. Construction of phosphatidylethanolamine-less strain of Saccharomyces cerevisiae. Effect on amino acid transport. Yeast. 2001;18:251–260. doi: 10.1002/1097-0061(200102)18:3<251::AID-YEA667>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 72.Dunlop J, Jones PC, Finbow ME. Membrane insertion and assembly of ductin: a polytopic channel with dual orientations. EMBO J. 1995;14:3609–3616. doi: 10.1002/j.1460-2075.1995.tb00030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu Q, von Dippe P, Xing W, Levy D. Membrane topology and cell surface targeting of microsomal epoxide hydrolase. Evidence for multiple topological orientations. J Biol Chem. 1999;274:27898–27904. doi: 10.1074/jbc.274.39.27898. [DOI] [PubMed] [Google Scholar]