Abstract
Purpose of review
The use of systems biology approaches to understand and predict vaccine-induced immunity promises to revolutionize vaccinology. For centuries vaccines were developed empirically, with very little understanding of the mechanisms by which they mediate protective immunity. The so-called systems vaccinology approach employs high-throughput technologies (e.g. microarrays, RNA-seq and mass spectrometry-based proteomics and metabolomics) and computational modeling to describe the complex interactions between all the parts of immune system, with a view to elucidating new biological rules capable of predicting the behavior of the system.
Recent findings
Systems biology successfully applied to yellow-fever and influenza vaccines has led to the discovery of signatures that predict vaccine immunogenicity, and promises to advance basic immunology research by providing novel mechanistic insights about immune regulation. However a major challenge of systems vaccinology concerns the analyses and interpretation of the large and noisy data sets generated by high-throughput techniques. Overcoming these issues, we envision that systems vaccinology will have a potential impact on vaccine development, including HIV vaccines.
Summary
High-throughput technologies allow the investigation of vaccine-induced immune responses at system and molecular levels. These are currently being used to unravel new molecular insights about the immune system, and are on the verge of being integrated into clinical trials to enable rational vaccine design and development.
Keywords: blood transcriptomics, systems biology, systems vaccinology, vaccinology
INTRODUCTION
The immune system comprises an intricate network of specialized cells and organs, which participate in an immune response [1]. During an immune response, cells need to constantly and intensely intercommunicate, migrate, differentiate and die in a complex and dynamic fashion. At the same time, millions of functional molecules (i.e. genes, RNA molecules, proteins and metabolites) inside each cell or tissue will form an intricate system of pathways, deeply coordinated to produce a biological phenomenon (e.g. antibody production, cell differentiation, interferon-α release, activation of endoplasmic reticulum stress pathway, and so on). Although extremely valuable, reductionist approaches trying to study each component of the system in isolation can only provide a narrow and simplified representation of the immune system. Systems vaccinology utilizes high-throughput technologies to capture a holistic view of thousands of RNA molecules, proteins and metabolites in parallel with a view to understanding the global architecture of the biological networks that drive an immune response to vaccination [2▪,3,4].
Recently, systems vaccinology has been successfully used to identify signatures that predict the immunogenicity of vaccines and is beginning to yield mechanistic insights about immune regulation [5•,6,7▪▪]. The first examples of such studies aimed to understand and to predict the immune responses induced by the live-attenuated yellow fever virus vaccine YF-17D [6,7▪▪]. By analyzing the gene expression profiles induced in the blood of humans a few days after vaccination with YF-17D, these two independent groups identified innate gene signatures composed of type I interferon, inflammasome and complement genes [6,7▪▪]. In addition, we identified early gene expression signatures that predict the magnitude of adaptive immune outcomes to YF-17D (i.e. of CD8+ T-cell and neutralizing antibody responses) [6]. These results provided the first proof of concept demonstration that systems approaches can indeed be used to predict the immunogenicity of vaccines.
A key question was whether a similar approach could lend itself to identifying predictive signatures of immunogenicity against other vaccines and, if so, whether such signatures would be different from the signatures that predicted the immunogenicity of YF-17D. This issue was particularly relevant, given the fact that YF-17D, unlike inactivated vaccines, is a live virus that causes an acute viral infection after vaccination and, thus, comes into direct contact with blood cells. We, thus, applied the systems vaccinology approach to evaluating immune responses to an inactivated vaccine, the seasonal trivalent inactivated influenza virus (TIV) vaccine [5▪]. Unlike the situation with YF-17D, the majority of the individuals vaccinated against seasonal influenza had already been exposed to the virus, through previous infections of vaccinations. Thus, a potential further caveat was whether this approach could be used to predict the immunogenicity of recall immune responses. Initially, we compared the global expression changes that occur after vaccination with TIV and with a live-attenuated influenza virus (LAIV) vaccine and revealed that the expression of genes from inflammasome and antimicrobial pathways was similarly altered by both influenza vaccines [5▪]. Not unexpectedly, LAIV induced the expression of several interferon-related genes, similar to that observed with YF-17D [5▪]. TIV, however, induced a signature that was characteristic of plasma B cell response [5▪]. Using participants from three independent influenza seasons, we also identified sets of two to five genes that were able to accurately classify with up to 90% of the vaccinees as being high (four-fold increase in antibody titers 30 days after vaccination) or low-responders (less than four fold) to TIV vaccination [5▪].
Despite these encouraging advances, it is important to consider several challenges and potential pitfalls that need to be solved, for the successful integration of systems approaches in vaccine development. One major challenge concerns the sheer biological complexity underlying the immune system itself and how it regulates and controls the innate and adaptive immunity to vaccination. The other comes from the technical aspects of dealing with the large, noisy and multidimensional data generated by high-throughput techniques. In this review, we will describe some of these challenges and offer potential approaches to addressing them. We also discuss the emerging impact of systems vaccinology in HIV vaccine design.
Variability and reproducibility issues
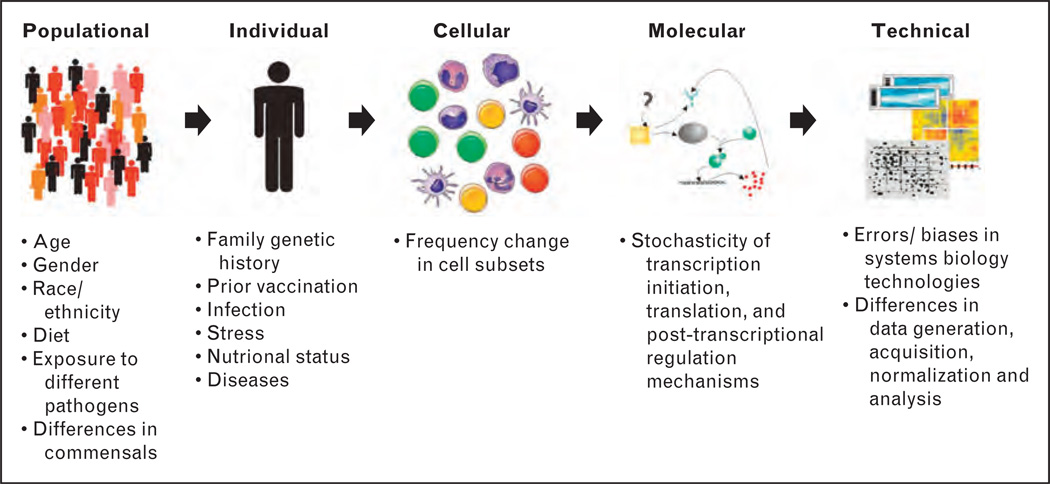
The capacity of the immune system to mount a protective response after vaccination depends on several factors. These include the immune history of an individual (e.g. prior infections or vaccinations, immune-related diseases and drugs that affect immune system), genetic diversity, as well as a plethora of other confounding variables and events (e.g. the nutritional status, physical or mental stress), which could impinge on vaccine-induced immunity [8] (Fig. 1). All this makes the immune system of each human being practically unique and, therefore, poses huge challenges to vaccine development and testing.
FIGURE 1.
Potentially confounding variables in the practice of systems vaccinology.
Additional biological variation is found at the cellular and molecular levels and it is conferred by the stochasticity of many biological processes. The stochastic component of transcription initiation, translation, and posttranscriptional regulation mechanisms gives rise to noise in the temporal amounts of proteins and RNA molecules in cells [9–11] (Fig. 1). These fluctuations (noise) may impact the dynamic behavior of gene regulatory networks and cellular fate [11,12]. Chang et al. [13] showed that stochastic gene expression may control lineage choice in mammalian progenitor cells and Tay et al. [14] demonstrated that NF-κB activation is partly governed by a stochastic process. How this stochastic component may be quantified and supplemented in deterministic models applied to systems vaccinology remains a challenge.
Dealing with the large amount of data generated from high-throughput technologies represents a critical technical issue. By measuring the outputs of entire transcriptomes, proteomes or metabolomes, there will inevitably be errors, noise and biases that, if not corrected, may affect downstream analyses [15–18] (Fig. 1). Even if different systems biology studies measure the same biological output, they will obtain variable results, which will reflect the differences in protocols, experimental design, probes used to detect the expression of genes, statistical tests, data normalization and detection methods [19–21]. This can be exemplified in the comparison of HIV host factors identified by three independent genome-wide RNAi-based screens [22–24]. In each screen, a genome-wide RNAi library was utilized to identify cellular genes that are critical for the replication of HIV-1. Although all three studies shared the same objective, they differed in the assay system used and experimental design [25]. Together, the three siRNA screens called 842 genes as diminishing HIV replication when knocked down. Of those, only three genes were called in all three screens (RELA, MED6, MED7) [26▪]. Although statistically significant, pair-wise overlaps between screens were also quite modest (ranging from 3 to 6% of the shared genes) [26▪]. Nevertheless, analyzing host factors that participate in similar cellular processes (functional analysis) yielded a greater overlap in gene ontology categories than was seen for individual genes [26▪]. Thus, the combination of human heterogeneity and the intrinsic noise and stochasticity of systems biology approaches and gene regulatory networks poses a major challenge for the field of systems vaccinology. Here, we will highlight strategies that can be applied to experimental design and data analyses of systems vaccinology studies, in order to mitigate the impact of such variables.
One critical requirement lies in the experimental design itself. In addition to an appropriate sample size [27,28], factors such as age, sex, ethnicity, prior and current disease conditions, vaccination history, and baseline levels of immune parameters should be taking into account in selecting the volunteers and in analyzing the data. Thus, studies should be performed on populations that are relatively uniform with respect to such variables. Conversely, carefully controlled comparative studies that examine immune responses, and molecular signatures induced by vaccination in populations that differ with regards to age groups (e.g. healthy young adults versus elderly versus infants), or geographical or ethnic origins, or disease states (e.g. those with autoimmune diseases or HIV) will be particularly useful in delineating the impact of such variables on the immune response.
Most importantly, additional independent trials should be included to validate predictive signatures of infection [29,30▪] or vaccination [5▪,6,7▪▪]. Ideally, robust predictors of vaccine immunogenicity or potential molecular mechanisms of vaccination revealed by one group should be able to be validated by other groups. However, statistical robustness will not always correlate with higher reproducibility. Comparing the gene lists between two groups, even if both groups have applied a rigorous statistical test in their data will usually show a relatively low overlap (5–50%) [31]. This low overlap is usually explained by variations in the protocol and experimental design (e.g. time points, whole blood versus peripheral blood mononuclear cells (PBMCs), age group, platforms used), data analysis (e.g. filtering thresholds, statistical tests, cutoffs) and the intrinsic technical noise associate with systems biology approaches. Analyzing groups of genes that have similar function (e.g. cellular pathways and gene ontology categories), have similar expression patterns or genes that form biochemical complexes may not only improve the overlap between groups, but also make the results easier to interpret [25,32,33]. An alternative way of handling heterogeneous data sets consists in combining P-values from independent studies to test whether the genes can be collectively used to reject a common null hypothesis [34–36]. This last meta-analysis approach was successfully applied in cancer field [37,38], and can offer an elegant solution to compare systems vaccinology studies generated in different laboratories.
Correlates of immunogenicity and protection
Gene expression profiling approaches have led to the discovery of prognostic and predictive signatures of breast cancer that were later utilized in commercial multigene assays [39–41]. Despite the potential applications of biomarkers and the large number of reports on novel biomarkers generated by systems biology studies, only very few of them were translated into clinical diagnostics for patient care [42]. The considerations for a successful study include: a sample size that is adequately powered; the existence of clearly defined endpoints and the requirement of a robust and reliable assay [43]. Given the fact that microarray experiments can be reproducible and comparable [44], and that correlates of immunogenicity can be used as vaccine endpoints, the major challenge for systems vaccinology remains in obtaining a large number of vaccinees and independent trials. Toward this, our group is trying to identify and validate predictors of immunogenicity for the influenza vaccine in four trials of young adults and one trial of elderly vaccinees (Nakaya et al. unpublished observation).
Most successful vaccines seem to act through the production of antibodies that neutralize the virus or toxins or opsonize the bacteria that cause infection [45,46]. In such cases, the correlation of protection is defined by the minimum quantities of antibodies that are closely related to prevent the infection or disease. Some vaccines (e.g. diphtheria, tetanus, hepatitis A, measles, and rubella) have well defined correlates of protection [2•], suggesting that vaccine protection can be considered categorical in nature. For example, participants with antibody levels of 10 mIU/ml or higher in the serum are almost always protected against hepatitis A disease [47]. Nevertheless, correlates of protection are not always well defined. For example, a hemagglutination-inhibition antibody titer of 1/40 is often used as a correlate of protection for the influenza TIV vaccine [48]. Notwithstanding, efficacy data showed that only 70% of participants are protected at that titer, but up to 90% with higher titers [45]. Antibody titers of 200 mIU/ml or more after vaccination with measles vaccine are protective against infection and titers lower than 120 mIU/ml are not protective at all. Intermediate titers (120–200 mIU/ml) protect against clinical signs of disease but not against infection [45]. It is also important to mention that high quantity of antibodies with low avidity may not mediate protection.
Protective immunity provided by vaccines is more likely to be elicited by the combination of strong humoral and cell-mediated immune responses [49]. Although antibody titers are the predominant protective correlate, cellular immunity may also play a critical role in protection against intracellular infections (CD8+ T cells) or in helping B cell development (CD4+ T cells). This is exemplified by vaccination with influenza vaccine in the elderly population. In the elderly, strong influenza-specific T-cell responses are better (if not the only) correlates of protection than antibody titers [50].
For systems vaccinology, the immunogenicity of a vaccine is generally considered a categorical parameter. Therefore, a correlate divides vaccinees into ‘high-responders’ or ‘low-responders’ when their immune responses are above or below an absolute threshold (e.g. antibody titer or antibody fold-increase after vaccination). To identify gene signatures that predict these distinct phenotypic ‘classes’, a range of class prediction methods can be utilized, such as discriminant analysis of mixed integer programming [51], k-nearest neighbour [30▪], predictive analysis of microarray [52], classification to nearest centroids [53], Support Vector Machines [54], and Random Forest [55]. If no clear-cut correlate of protection is defined for a vaccine, prediction analysis can be performed using different thresholds. For example, in addition to the widely defined threshold for seroconversion to TIV (at least four-fold increase in the hemagglutination-inhibition antibody titers after vaccination [56]), we also identified gene signatures that accurately classify vaccinees with very high increase in antibody titers (at least eight fold) versus low-increase (two fold or less) [5•]. This approach gave us slightly different but complementary sets of predictive genes that could reveal unappreciated gene functions related to the magnitude of antibody response. A more direct way of gaining insight into the potential mechanisms underlying this variation in immunogenicity is to look for gene signatures that correlate with the magnitude of the immune response. In our influenza study, we found gene pathways associated with innate immunity, such as the natural killer cell signaling network, interferon-related genes, and network for the production of nitric oxide and reactive oxygen species in macrophages as being positively correlated to antibody response, suggesting a link between these pathways of early innate responses (day 3 and 7 postvaccination) and the later antibody adaptive response (day 28 post-vaccination) [5▪].
Profiling the blood
A dilemma commonly faced when designing blood transcriptomic studies involves the choice of sample types to be screened. Blood (e.g. whole blood or PBMCs) is a mixed population tissue, which is easily accessible and is relatively simple to store and process. Additionally, it represents a comprehensive view of the status of the immune system in health and disease [57]. Transcriptional profiling of blood tissue, however, will capture the changes of both cellular and transcript abundance, making results hard to interpret [57]. On the contrary, measuring the global expression changes that occur on cell subsets isolated from blood can provide valuable mechanistic insights of specific cells [58]. However, the high cost and great amount of work associated with isolating and screening cell subsets (that are not even unambiguously defined) make this approach unpractical to most clinical studies. For this reason, many genome-wide expression studies utilize blood to investigate the human immune system.
Several methods have been developed to analyze blood microarray data. Deconvolution methods were applied to accurately quantify the constituents of blood samples [59] or to identify subset-specific differential expression, when cell subset frequency is known [60]. Other methods compare the blood signature to gene sets or modules of particular cell types to find significant enrichment of specific cell types [5▪,30▪,61]. Recently, a computational method was developed by Bolen et al. [62] to predict using only the transcriptional profiling data from total PBMCs the most likely cellular source for a predefined gene expression signature. These techniques will be of great value to systems vaccinology studies.
Data → knowledge → understanding
The biggest challenge in systems vaccinology is how to extract knowledge and ultimately understanding, from a sea of data. Toward this, key bioinformatics analyses (reviewed in Nakaya et al. 2011, in press) and modeling methods, such as ordinary and partial differential equations (mostly applied to metabolomics) [63,64], stochastic schemes [65,66], petri nets [67] and Boolean logic [68,69], can be potentially applied. Human knowledge and intuition, and experimental validation can also drive the meaningful interpretation of results, and the creation and validation of imaginative hypotheses [2▪]. These were applied in the studies with yellow fever [6] and influenza vaccines [5▪]. A gene named EIF2AK4 (also known as GCN2) was presented in most of the predictive signatures of the CD8+ T cell response against YF-17D [6]. GCN2 plays a key role on the formation of stress granules in response to certain types of cellular stresses (e.g. amino acid starvation) [70–72]. Consistently with the microarray results, experiments with GCN2 knockout mice vaccinated with YF-17D resulted not only with reduction of stress granules but also with impaired CD8+ T-cell responses (Khan et al., in preparation). Another example, a gene encoding the kinase CaMKIV, was found in the predictive signatures of antibody responses against TIV [5▪]. Despite the involvement of CaMKIV in several processes of the immune system, such as inflammatory responses [73,74], T-cell development [75–77] and the maintenance of hematopoietic stem cells [78], nothing was known about its possible role in B-cell responses. In our study, we observed that the expression levels of CaMKIV at day 3 post-TIV vaccination were inversely correlated with the magnitude of antibody responses at day 28 [5▪], indicating that participants with the lowest levels of CaMKIV in their blood were the ones generating the highest antibody responses. Using in-vitro experiments and mice deficient in CaMKIV, we demonstrated that TIV induces the phosphorylation of CaMKIV and that CaMKIV knockout mice had a significantly greater antibody response than that of wild-type mice [5▪]. Although these experiments suggest an unappreciated role of CaMKIV in B-cell responses, further work is needed to delineate the cellular mechanisms involved.
POTENTIAL APPLICATIONS FOR HIV VACCINE
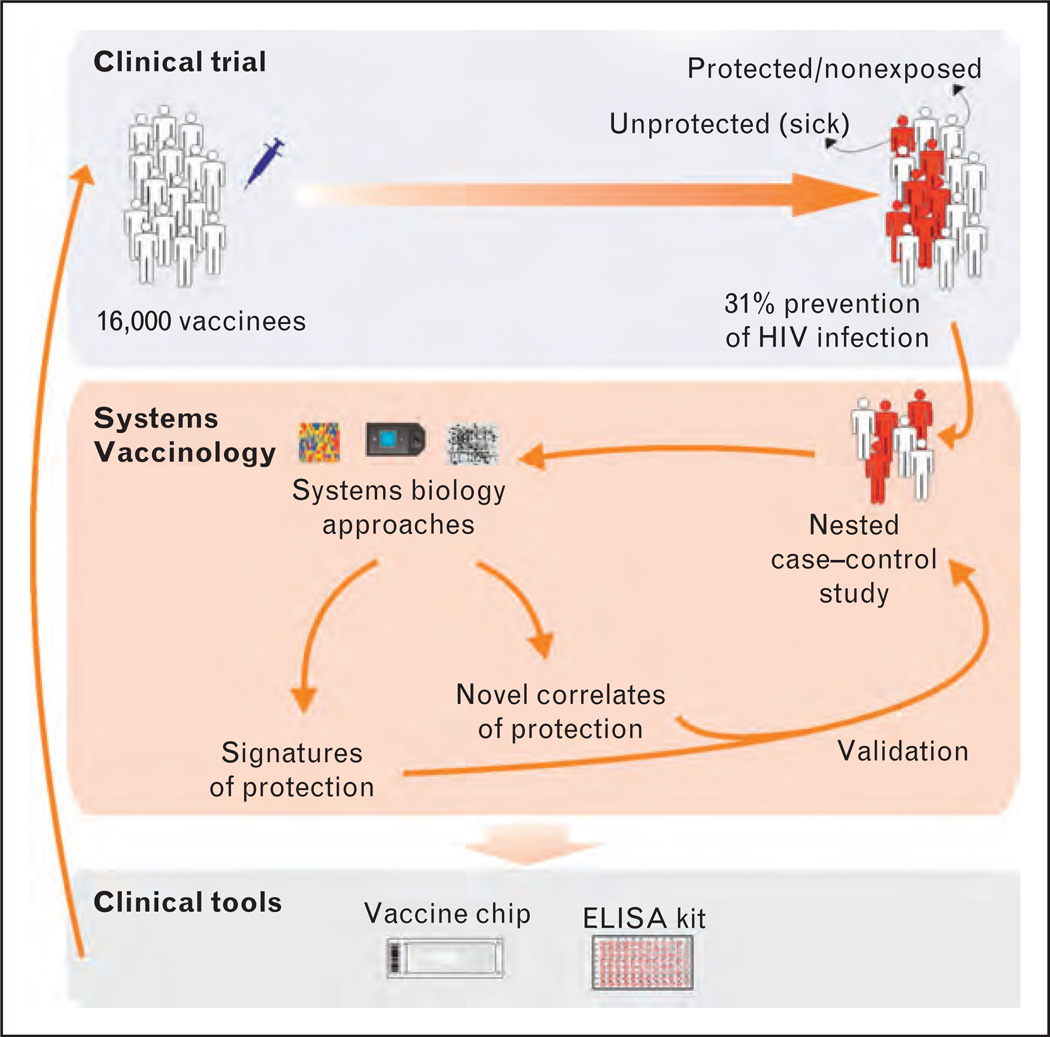
The value of systems vaccinology can be extended to vaccines that do not have clear correlates of protection or immunogenicity, such as the HIV vaccine tested in RV144 trial [79▪▪]. This trial was comprised of 16 000 heterosexuals in Thailand who received a prime–boost regime, priming with a canarypox expressing the subtype BHIV Gag, Pro and the subtype E gp120 (ALVAC-HIV) and boosting with the alum adjuvanted mix of gp120 AIDSVAX B/E [79▪▪,80]. Despite the modest level of efficacy (31% prevention of HIV infection after 3 years follow-up [79▪▪]), these results represent a major hope for the development of HIV vaccines.
A case–control study of RV144 trial is being prepared in order to assess correlates of risk of HIV-1 infection in the vaccine group and to test whether and how well these identified correlates can serve as surrogate endpoints for HIV-1 infection [81]. Toward these goals, several immunogenicity assays are being tested and those that prove to be relevant will be performed on the plasma and PBMC samples from vaccine recipients of RV144 trial [81,82]. However, additional and innovative explorations should be considered. Systems biology approaches can be used to identify a signature of protection and/or novel correlates of protection based on gene expression, protein or metabolite levels or even miRNA expression (Fig. 2). The next step would be the validation of such signature or correlates in additional trials or group of participants (Fig. 2). If a signature or correlates demonstrate they are robust and reliable, the set of genes or proteins that comprise them can be represented in a vaccine chip or ELISA kit, and utilized to assess immune responses in a high throughput manner (Fig. 2). Additionally, the generation of mechanistic insights derived from the application of systems biology approaches in RV144 case–control study would help to relate the molecular effects of vaccination to HIV protection. Finally, systems biology has a potentially major impact on HIV vaccine design. Much can be learned from studies from several groups who have analyzed the gene expression profiles in the blood of nonhuman primates infected with pathogenic SIV, and have identified key differences in the innate immune response to SIV between pathogenic and natural hosts (reviewed in Steven et al. this issue of Current Opinion in HIV and AIDS). Similarly, gene expression profiling of HIV nonprogressors and HIV elite controllers can yield immunological surrogates of protection (reviewed in Zak and Aderem, this issue of Current Opinion in HIV and AIDS).
FIGURE 2.
Integrating systems biology into HIV vaccine trials.
CONCLUSION
Recent work on the application of systems biology to vaccinology [2▪,5▪,6,7▪▪] (Nakaya et al. 2011, in press) is beginning to yield novel insights into mechanisms of vaccine immunity, and demonstrates the utility of using such approaches in predicting vaccine immunogenicity and efficacy. Despite the technical and biological challenges associated to systems studies, one important aspect of systems vaccinology is that it strategically directs efforts in human immunology. The recent establishment of the NIAID Human Immunology Project Consortium has capitalized on such advances in order to create a novel public resource that characterizes diverse states of the human immune system following vaccination, or during an infection, or following treatment with an immune adjuvant that targets a known innate immune receptor(s). The consortium comprises seven important research centers and universities (Emory University, Dana-Farber Cancer Institute, Seattle Biomedical Research Institute, Stanford University, Mayo Clinic, Yale University and the Baylor Research Institute, USA) that will utilize cutting edge high throughput technologies to profile immune responses in humans, against a broad range of vaccines and infections. Such common effort is likely to catalyze the identification of signatures of vaccine immunogenicity, and facilitate the rapid integration of systems biological approaches in vaccine development.
KEY POINTS.
The modest success observed with the recent RV144 HIV vaccine trial in Thailand has re-energized the field and underscored the imperative to delineate the correlates of immune protection against HIV.
The recent application of systems biological approaches to identifying molecular signatures induced early after vaccination, which correlate with and predict the later immunogenicity of vaccines, has highlighted the potential utility of such approaches in vaccine design and testing.
Such ‘systems vaccinology’ approaches are being applied to different vaccines, and are beginning to be integrated into clinical trials of vaccines, to identify ‘biomarkers’ or ‘signatures’ of vaccine efficacy.
However, several major challenges need to be addressed. These include refining the analytical approaches to analyze and interpret the large and noisy datasets generated by high throughput technologies.
In addition to facilitating the identification of signatures of vaccine efficacy, systems vaccinology is also beginning to offer novel mechanistic insights about the networks that mediate vaccine-induced immunity.
Acknowledgements
We are very grateful for the generous support offered by the National Institutes of Health and the Bill and Melinda Gates Foundation for their very generous support of our work.
Footnotes
Conflicts of interest
There are no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
Additional references related to this topic can also be found in the Current World Literature section in this issue (pp. 88–89).
- 1.Ricciardi-Castagnoli P, Granucci F. Opinion: interpretation of the complexity of innate immune responses by functional genomics. Nat Rev Immunol. 2002;2:881–889. doi: 10.1038/nri936. [DOI] [PubMed] [Google Scholar]
- 2. Pulendran B, Li S, Nakaya HI. Systems vaccinology. Immunity. 2010;33:516–529. doi: 10.1016/j.immuni.2010.10.006. A comprehensive review of the opportunities and challenges posed by systems vaccinology.
- 3.Pulendran B. Learning immunology from the yellow fever vaccine: innate immunity to systems vaccinology. Nat Rev Immunol. 2009;9:741–747. doi: 10.1038/nri2629. [DOI] [PubMed] [Google Scholar]
- 4.Oberg AL, Kennedy RB, Li P, et al. Systems biology approaches to new vaccine development. Curr Opin Immunol. 2011;23:436–443. doi: 10.1016/j.coi.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nakaya HI, Wrammert J, Lee EK, et al. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12:786–795. doi: 10.1038/ni.2067. This article is a successful example of systems vaccinology applied to the influenza vaccine. In addition to describing the identification of molecular signatures that predict immunogenicity, it also reveals the power of systems vaccinology in obtaining mechanistic insights.
- 6.Querec TD, Akondy RS, Lee EK, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gaucher D, Therrien R, Kettaf N, et al. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med. 2008;205:3119–3131. doi: 10.1084/jem.20082292. References [6] and [7▪▪] represent the first examples of the use of systems biological approaches to identifying molecular signatures of vaccine-induced immunity.
- 8.Pulendran B, Ahmed R. Immunological mechanisms of vaccination. Nat Immunol. 2011;131:509–517. doi: 10.1038/ni.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ribeiro AS. Stochastic and delayed stochastic models of gene expression and regulation. Math Biosci. 2010;223:1–11. doi: 10.1016/j.mbs.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 10.Eldar A, Elowitz MB. Functional roles for noise in genetic circuits. Nature. 2010;467:167–173. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balazsi G, van Oudenaarden A, Collins JJ. Cellular decision making and biological noise: from microbes to mammals. Cell. 2011;144:910–925. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mettetal JT, Muzzey D, Pedraza JM, et al. Predicting stochastic gene expression dynamics in single cells. Proc Natl Acad Sci U S A. 2006;103:7304–7309. doi: 10.1073/pnas.0509874103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang HH, Hemberg M, Barahona M, et al. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tay S, Hughey JJ, Lee TK, et al. Single-cell NF-kappaB dynamics reveal digital activation and analogue information processing. Nature. 2010;466:267–271. doi: 10.1038/nature09145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang L, Schlesinger F, Davis CA, et al. Synthetic spike-in standards for RNAseq experiments. Genome Res. 2011;21:1543–1551. doi: 10.1101/gr.121095.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balazsi G, Oltvai ZN. A pitfall in series of microarrays: the position of probes affects the cross-correlation of gene expression profiles. Methods Mol Biol. 2007;377:153–162. doi: 10.1007/978-1-59745-390-5_9. [DOI] [PubMed] [Google Scholar]
- 17.Barla A, Jurman G, Riccadonna S, et al. Machine learning methods for predictive proteomics. Brief Bioinform. 2008;9:119–128. doi: 10.1093/bib/bbn008. [DOI] [PubMed] [Google Scholar]
- 18.Chandra H, Reddy PJ, Srivastava S. Protein microarrays and novel detection platforms. Expert Rev Proteomics. 2011;8:61–79. doi: 10.1586/epr.10.99. [DOI] [PubMed] [Google Scholar]
- 19.Painter MW, Davis S, Hardy RR, et al. Transcriptomes of the B and T lineages compared by multiplatform microarray profiling. J Immunol. 2011;186:3047–3057. doi: 10.4049/jimmunol.1002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marioni JC, Mason CE, Mane SM, et al. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mane SP, Evans C, Cooper KL, et al. Transcriptome sequencing of the Microarray Quality Control (MAQC) RNA reference samples using next generation sequencing. BMC Genomics. 2009;10:264. doi: 10.1186/1471-2164-10-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konig R, Zhou Y, Elleder D, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brass AL, Dykxhoorn DM, Benita Y, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 24.Zhou H, Xu M, Huang Q, et al. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 25.Pache L, Konig R, Chanda SK. Identifying HIV-1 host cell factors by genome-scale RNAi screening. Methods. 2011;53:3–12. doi: 10.1016/j.ymeth.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 26. Bushman FD, Malani N, Fernandes J, et al. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000437. e1000437. The article is a good example of how to compare the results from different systems biology approaches sharing the same overall goal.
- 27.Plikaytis BD, Carlone GM. Statistical considerations for vaccine immunogenicity trials. Part 2: noninferiority and other statistical approaches to vaccine evaluation. Vaccine. 2005;23:1606–1614. doi: 10.1016/j.vaccine.2004.06.047. [DOI] [PubMed] [Google Scholar]
- 28.Tong T, Zhao H. Practical guidelines for assessing power and false discovery rate for a fixed sample size in microarray experiments. Stat Med. 2008;27:1960–1972. doi: 10.1002/sim.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramilo O, Allman W, Chung W, et al. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood. 2007;109:2066–2077. doi: 10.1182/blood-2006-02-002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Berry MP, Graham CM, McNab FW, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. This article nicely shows how gene module analyses can be applied to blood transcriptomics of a human important disease.
- 31.Suarez-Farinas M, Noggle S, Heke M, et al. Comparing independent microarray studies: the case of human embryonic stem cells. BMC Genomics. 2005;6:99. doi: 10.1186/1471-2164-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee E, Chuang HY, Kim JW, et al. Inferring pathway activity toward precise disease classification. PLoS Comput Biol. 2008;4 doi: 10.1371/journal.pcbi.1000217. e1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitlock MC. Combining probability from independent tests: the weighted Z-method is superior to Fisher’s approach. J Evol Biol. 2005;18:1368–1373. doi: 10.1111/j.1420-9101.2005.00917.x. [DOI] [PubMed] [Google Scholar]
- 35.Marot G, Foulley JL, Mayer CD, Jaffrezic F. Moderated effect size and P-value combinations for microarray meta-analyses. Bioinformatics. 2009;25:2692–2699. doi: 10.1093/bioinformatics/btp444. [DOI] [PubMed] [Google Scholar]
- 36.Hong F, Breitling R. A comparison of meta-analysis methods for detecting differentially expressed genes in microarray experiments. Bioinformatics. 2008;24:374–382. doi: 10.1093/bioinformatics/btm620. [DOI] [PubMed] [Google Scholar]
- 37.Choi JK, Yu U, Kim S, Yoo OJ. Combining multiple microarray studies and modeling interstudy variation. Bioinformatics. 2003;19 Suppl 1:i84–i90. doi: 10.1093/bioinformatics/btg1010. [DOI] [PubMed] [Google Scholar]
- 38.Rhodes DR, Barrette TR, Rubin MA, et al. Meta-analysis of microarrays: interstudy validation of gene expression profiles reveals pathway dysregulation in prostate cancer. Cancer Res. 2002;62:4427–4433. [PubMed] [Google Scholar]
- 39.van ’t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 40.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 41.Sotiriou C, Wirapati P, Loi S, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Z, Chan DW. The road from discovery to clinical diagnostics: lessons learned from the first FDA-cleared in vitro diagnostic multivariate index assay of proteomic biomarkers. Cancer Epidemiol Biomarkers Prev. 2010;19:2995–2999. doi: 10.1158/1055-9965.EPI-10-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Schaeybroeck S, Allen WL, Turkington RC, Johnston PG. Implementing prognostic and predictive biomarkers in CRC clinical trials. Nat Rev Clin Oncol. 2011;8:222–232. doi: 10.1038/nrclinonc.2011.15. [DOI] [PubMed] [Google Scholar]
- 44.Shi L, Campbell G, Jones WD, et al. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat Biotechnol. 2010;28:827–838. doi: 10.1038/nbt.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plotkin SA. Vaccines: correlates of vaccine-induced immunity. Clin Infect Dis. 2008;47:401–409. doi: 10.1086/589862. [DOI] [PubMed] [Google Scholar]
- 46.Germain RN. Vaccines and the future of human immunology. Immunity. 2010;33:441–450. doi: 10.1016/j.immuni.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 47.Shouval D, Ashur Y, Adler R, et al. Single and booster dose responses to an inactivated hepatitis A virus vaccine: comparison with immune serum globulin prophylaxis. Vaccine. 1993;11 Suppl 1:S9–S14. doi: 10.1016/0264-410x(93)90151-m. [DOI] [PubMed] [Google Scholar]
- 48.Hobson D, Curry RL, Beare AS, Ward-Gardner A. The role of serum haemagglutination-inhibiting antibody in protection against challenge infection with influenza A2 and B viruses. J Hyg (Lond) 1972;70:767–777. doi: 10.1017/s0022172400022610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amanna IJ, Slifka MK. Contributions of humoral and cellular immunity to vaccine-induced protection in humans. Virology. 2011;411:206–215. doi: 10.1016/j.virol.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McElhaney JE, Xie D, Hager WD, et al. T cell responses are better correlates of vaccine protection in the elderly. J Immunol. 2006;176:6333–6339. doi: 10.4049/jimmunol.176.10.6333. [DOI] [PubMed] [Google Scholar]
- 51.Lee EK. Large-scale optimization-based classification models in medicine and biology. Ann Biomed Eng. 2007;35:1095–1109. doi: 10.1007/s10439-007-9317-7. [DOI] [PubMed] [Google Scholar]
- 52.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dabney AR. Classification of microarrays to nearest centroids. Bioinformatics. 2005;21:4148–4154. doi: 10.1093/bioinformatics/bti681. [DOI] [PubMed] [Google Scholar]
- 54.Brown MPS, Grundy WN, Lin D, et al. Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc Natl Acad Sci U S A. 2000;97:262–267. doi: 10.1073/pnas.97.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Diaz-Uriarte R, de Andres SA. Gene selection and classification of microarray data using random forest. BMC Bioinformatics. 2006;7:3. doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sullivan SJ, Jacobson R, Poland GA. Advances in the vaccination of the elderly against influenza: role of a high-dose vaccine. Expert Rev Vaccines. 2010;9:1127–1133. doi: 10.1586/erv.10.117. [DOI] [PubMed] [Google Scholar]
- 57.Chaussabel D, Pascual V, Banchereau J. Assessing the human immune system through blood transcriptomics. BMC Biol. 2010;8:84. doi: 10.1186/1741-7007-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grigoryev YA, Kurian SM, Avnur Z, et al. Deconvoluting posttransplant immunity: cell subset-specific mapping reveals pathways for activation and expansion of memory T, monocytes and B cells. PLoS One. 2010;5:e13358. doi: 10.1371/journal.pone.0013358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abbas AR, Wolslegel K, Seshasayee D, et al. Deconvolution of blood microarray data identifies cellular activation patterns in systemic lupus erythematosus. PLoS One. 2009;4:e6098. doi: 10.1371/journal.pone.0006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shen-Orr SS, Tibshirani R, Khatri P, et al. Cell type-specific gene expression differences in complex tissues. Nat Methods. 2010;7:287–289. doi: 10.1038/nmeth.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chaussabel D, Quinn C, Shen J, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bolen CR, Uduman M, Kleinstein SH. Cell subset prediction for blood genomic studies. BMC Bioinformatics. 2011;12:258. doi: 10.1186/1471-2105-12-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kell DB. Systems biology, metabolic modelling and metabolomics in drug discovery and development. Drug Discov Today. 2006;11:1085–1092. doi: 10.1016/j.drudis.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 64.Fell DA. Metabolic control analysis: a survey of its theoretical and experimental development. Biochem J. 1992;286(Pt 2):313–330. doi: 10.1042/bj2860313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McAdams HH, Arkin A. Stochastic mechanisms in gene expression. Proc Natl Acad Sci U S A. 1997;94:814–819. doi: 10.1073/pnas.94.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arkin A, Ross J, McAdams HH. Stochastic kinetic analysis of developmental pathway bifurcation in phage lambda-infected Escherichia coli cells. Genetics. 1998;149:1633–1648. doi: 10.1093/genetics/149.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goss PJ, Peccoud J. Quantitative modeling of stochastic systems in molecular biology by using stochastic Petri nets. Proc Natl Acad Sci U S A. 1998;95:6750–6755. doi: 10.1073/pnas.95.12.6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Watterson S, Ghazal P. Use of logic theory in understanding regulatory pathway signaling in response to infection. Future Microbiol. 2010;5:163–176. doi: 10.2217/fmb.10.8. [DOI] [PubMed] [Google Scholar]
- 69.Watterson S, Marshall S, Ghazal P. Logic models of pathway biology. Drug Discov Today. 2008;13:447–456. doi: 10.1016/j.drudis.2008.03.019. [DOI] [PubMed] [Google Scholar]
- 70.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 71.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 72.Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- 73.Illario M, Giardino-Torchia ML, Sankar U, et al. Calmodulin-dependent kinase IV links Toll-like receptor 4 signaling with survival pathway of activated dendritic cells. Blood. 2008;111:723–731. doi: 10.1182/blood-2007-05-091173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sato K, Suematsu A, Nakashima T, et al. Regulation of osteoclast differentiation and function by the CaMK-CREB pathway. Nat Med. 2006;12:1410–1416. doi: 10.1038/nm1515. [DOI] [PubMed] [Google Scholar]
- 75.Anderson KA, Means AR. Defective signaling in a subpopulation of CD4(+) T cells in the absence of Ca(2+)/calmodulin-dependent protein kinase IV. Mol Cell Biol. 2002;22:23–29. doi: 10.1128/MCB.22.1.23-29.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Krebs J, Wilson A, Kisielow P. Calmodulin-dependent protein kinase IV during T-cell development. Biochem Biophys Res Commun. 1997;241:383–389. doi: 10.1006/bbrc.1997.7823. [DOI] [PubMed] [Google Scholar]
- 77.Wang SL, Ribar TJ, Means AR. Expression of Ca(2+)/calmodulin-dependent protein kinase IV (caMKIV) messenger RNA during murine embryogenesis. Cell Growth Differ. 2001;12:351–361. [PubMed] [Google Scholar]
- 78.Kitsos CM, Sankar U, Illario M, et al. Calmodulin-dependent protein kinase IV regulates hematopoietic stem cell maintenance. J Biol Chem. 2005;280:33101–33108. doi: 10.1074/jbc.M505208200. [DOI] [PubMed] [Google Scholar]
- 79. Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. This article reports a moderate efficacy of 31% for HIV vaccine tested in RV144 trial. It provides evidence that a vaccine for HIV can be developed.
- 80.Rappuoli R, Aderem A. A 2020 vision for vaccines against, HIV, tuberculosis and malaria. Nature. 2011;473:463–469. doi: 10.1038/nature10124. [DOI] [PubMed] [Google Scholar]
- 81.Rolland M, Gilbert P. Evaluating immune correlates in HIV-1 vaccine efficacy trials: what RV144 may provide. AIDS Res Hum Retroviruses. 2011 doi: 10.1089/aid.2011.0240. [Epub ahead of print]. Available at http://www.liebertonline.com/doi/pdf/10.1089/aid.2011.0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim JH, Rerks-Ngarm S, Excler JL, Michael NL. HIV vaccines: lessons learned and the way forward. Curr Opin HIV AIDS. 2010;5:428–434. doi: 10.1097/COH.0b013e32833d17ac. [DOI] [PMC free article] [PubMed] [Google Scholar]