Summary
Vaccines represent a potent tool to prevent or contain infectious diseases with high morbidity or mortality. However, despite their widespread use, we still have a limited understanding of the mechanisms underlying the effective elicitation of protective immune responses by vaccines. Recent research suggests that this represents the cooperative action of the innate and adaptive immune systems. Immunity is made of a multifaceted set of integrated responses involving a dynamic interaction of thousands of molecules, whose list is constantly updated to fill the several empty spaces of this puzzle. The recent development of new technologies and computational tools permits the comprehensive and quantitative analysis of the interactions between all of the components of immunity over time. Here, we review the role of the innate immunity in the host response to vaccine antigens and the potential of systems biology in providing relevant and novel insights in the mechanisms of action of vaccines to improve their design and effectiveness.
Keywords: innate immunity, PRRs, PAMPs, TLRs, APCs, adaptive immunity
Vaccine development
An effective vaccine needs to mimic as close as possible the ‘real’ biological entity from which it is derived (i.e. pathogen, cancer cell) in order to be recognized by the host immune system as real ‘danger’. This will eventually initiate the cascade of molecular and cellular events inducing several levels of cross-talk between the innate and adaptive immune systems for an effective immune response and immunological memory. However, most successful vaccines have been empirically derived, and the immunological mechanisms underlying the effective induction of long-term protective immunity remain largely unknown (1, 2).
Most of the current successful vaccines are based on live attenuated or inactivated pathogen ‘particles’ carrying their own unique and specific antigens. The live attenuated vaccines are characterized by a limited viral replication in the host upon injection and carry the native pathogen-associated molecular signals (PAMS) (i.e. viral genetic material), which trigger the activation of the innate immune system binding to the pathogen recognition receptors (PRRs). Live attenuated vaccines replicate and reach host immune sites where they are taken up by dendritic cells (DCs) or other antigen-presenting cells (APCs), which migrate to lymphoid organs for presenting the antigens to T and B lymphocytes. They elicit immune responses similar to those from natural infections and often are effective after a single administration (3). However, such vaccines may cause mild-to-severe adverse effects in patients, often as consequence of the limited replication in the host.
On the contrary, the inactivated vaccines do not replicate and are safer than live attenuated vaccines; however, they are generally less effective, requiring multiple administrations to boost the immune response antibody titer over time. The inactivated vaccines are made as whole cell or as subunit vaccines (i.e. individual viral proteins). In this framework, recent advances in genomics and proteomics have provided essential tools to develop alternative non-replicating vaccine strategy, including recombinant proteins, synthetic peptides, DNA, and particulate structures (i.e. virus-like particles).
Inactivated as well as non-replicating vaccines activate innate responses only at their site of injection, and intradermal skin immunization seems to induce a more effective protective immune responses (4–6), considering the high number of DCs in the skin dermis (7). In contrast to safety advantages, the major drawback of vaccines based on selected antigens is the less effective processing and presentation to the immune system. Therefore, most formulations of non-living vaccines must include an adjuvant as ‘danger’ signal to trigger a sufficient activation of the innate system and, downstream, of the adaptive immune response (8).
Innate immunity and vaccine recognition
Research over the past decade has revealed a fundamental role for the innate immune system in sensing microbes or viruses and regulating the strength and quality of the adaptive immune responses to that microbe (9, 10). The innate immune system consists of several interacting cell types, including DCs, macrophages, epithelial cells, endothelial cells, natural killer (NK) cells, NK T cells, basophils, and mast cells, which are involved in ‘sensing’ microbes or viruses and initiating immunity against them. DCs plays a key role in directly sensing the presence of pathogens and orchestrating the interactions between the other innate immune cell types and facilitating the elicitation of anti-viral defenses, such secretion of type I interferons (IFNs) and defensins (11, 12). In addition to their roles in sensing pathogens and orchestrating innate immune defenses, DCs also play a critical role in translating innate immunity into adaptive immunity (13, 14). Understanding the impact of innate immunity on the regulation of adaptive immunity, and harnessing such knowledge to induce optimal immunity to human immunodeficiency virus (HIV), was recognized as an area of the highest importance.
The innate immune system is able to sense microbial or viral stimuli by the expression of so-called PRRs, which are expressed constitutively in the host on cells of the innate immune system, such as DCs (9, 10, 14–16), activating specific signaling pathways to drive biological and immunological responses. Among the PRRs, a key role is played by Toll-like receptors (TLRs), which are widely expressed on innate immune cells (including DCs, macrophages, mast cells, neutrophils), endothelial cells, and fibroblasts (9, 10, 16–20).
TLRs are a family of 12 type I integral membrane glycoproteins with extracellular domains containing varying numbers of leucine-rich-repeat motifs and a cytoplasmic signaling domain homologous to that of the interleukin 1 receptor (IL-1R), termed the Toll/IL-1R homology (TIR) domain (21). TLRs 1, 2, 4, 5, and 6 are expressed on the cell surface, whereas TLR3, 7, 8, and 9 are found almost exclusively in intracellular compartments such as endosomes (9, 10, 14–16).
TLRs recognize structural components shared by many bacteria, viruses, and fungi (17). Examples of such components include lipopolysaccharides (LPSs) (recognized by TLR4) (22, 23), lipopeptides (by cooperation of TLR2 with TLR1 or TLR6) (24–27), viral single- or double-stranded RNA (by TLR7 with TLR8 and by TLR3, respectively) (28–31), bacterial or viral DNA containing CpG motifs (by TLR9) (32, 33), and flagellin (by TLR5) (34).
In addition to TLRs, other families of PRRs such as the C-type lectins and nuclear-binding oligomerization domain (NOD) proteins are involved in sensing microbial stimuli and modulating immune responses (15, 35–37). The C-type lectins such as DC-specific inter-cellular adhesion molecule-3 (ICAM-3) grabbing non-integrin (DC-SIGN) and DC-associated C-type lectin-1 (Dectin-1) recognize molecules of pathogens such as HIV, hepatitis C virus (HCV), Helicobacter pylori, and Mycobacterium tuberculosis (38). NOD proteins recognize components of intracellular bacteria (36). Furthermore, intracellular RNA helicases RIG-I and melanoma differentiation-associated gene 5 (also called helicard) can sense dsRNA (39–41).
The interaction between PRRs and components of microbes or viruses triggers a downstream signaling cascade leading to several cellular processes, including production of proinflammatory cytokines and chemokines (42). Signaling intermediates of TLR activation include myeloid differentiation factor-88 (MyD88), TIR-associated-protein (TIRAP), also known as MAL, Toll receptor-associated activator of interferon, Toll receptor-associated molecule, IL-1 receptor-associated kinases (IRAK), and tumor necrosis factor (TNF) receptor-associated factor 6 (15, 42). The endpoint of this signaling cascade is the activation of transcription factors [IFN regulatory factor (IRF)3, IRF7, AP-1, NF-κB] inducing the activation of inflammatory cytokine genes, such as TNF-β, IL-6, IL-1β, and IL-12, as well as the upregulation of costimulatory molecules such as CD80, CD86, CD40 on DCs.
Many of the best empirically derived vaccines and adjuvants mediate their efficacy by activating specific innate immune receptors. For example, the highly effective yellow fever vaccine-17D, one of the most successful vaccines that has been administered to over half a billion people globally, signals via at least four different TLRs as well as RIG-I like receptors to elicit a broad spectrum of T-cell responses (43, 44). This suggests that the immune response generated by a live attenuated vaccine can be effectively mimicked by adjuvants composed of the appropriate TLR and/or non-TLR ligands. Consistent with this, it was recently shown that the superior immunogenicity of the inactivated whole virus H5N1 influenza vaccine is primarily controlled by TLR signaling (45), and the Haemophilus influenzae type b-outer membrane protein complex glycoconjugate vaccine induces cytokine production by engaging human TLR2 and requires the presence of TLR2 for optimal immunogenicity (46). In addition, the Bacillus Calmette–Guerin (BCG) has been shown to engage TLR2 and TLR4 (47, 48), although the consequence of this engagement for adaptive immunity is not known. Furthermore, recent work suggests that some adjuvants can induce robust adaptive immunity in a TLR-independent manner, perhaps through other receptors in the innate immune system (49). For example, it was recently demonstrated that alum, the only adjuvant that was licensed for several decades, signals through the NALP3 inflammasome (50–52), as DCs or macrophages stimulated in vitro with alum plus LPS induce IL-1β and IL-18 in a manner dependent on caspase-1 and NALP3 (50–52). However, whether NALP3 is required for the adjuvanticity of alum remains controversial, with some studies demonstrating abrogation of antibody responses in Nalp3−/− mice and other studies showing partial or no effects.
Exploiting vaccine recognition by PRRs
For most currently licensed vaccines, the degree of engagement of TLRs has not been studied, with a few exceptions. In particular, the live attenuated yellow fever vaccine has been demonstrated to activate multiple DC subsets via TLRs 2, 7, 8, and 9 (43). For many other licensed vaccines, the engagement of TLRs has not been documented, but it is possible to guess based on studies performed on the original virus from which the vaccine is derived. Single-stranded RNA of live attenuated (cold adapted) influenza vaccines, indeed, are likely to activate TLR3 and TLR7 during intracellular replication, leading to the upregulation of inflammatory cytokines (53, 54). Similarly, bacterial and viral DNA containing unmethylated CpG motifs activate TLR9 (55, 56).
There is little or no evidence that the immunogenicity of vaccines composed of killed pathogens or subunit elements is mediated by TLR engagement, possibly explaining the generally observed lower and short-lasting immune response. Adjuvanting formulation, in these cases, may significantly improve vaccine effectiveness. However, the number of adjuvants approved for human use is quite limited, including alum, MF59 (57), monophosphoryl lipid A (MPL) (58), AS04 (consisting of MPL adsorbed on alum) (59, 60), immunopotentiating reconstituted influenza virosomes (61, 62), and cholera toxin B subunit (63). Of these, only MPL is known to engage a TLR (TLR4), being a non-toxic derivative of the LPS of Salmonella minnesota. Consequently, many new TLR-specific vaccine adjuvants are under development and are being evaluated in preclinical and human clinical trials (64–68).
TLR signaling for potent and prolonged adaptive immune responses
Adaptive immune responses are initiated in the T-cell-rich areas of lymph nodes, where naive T cells undergo to clonal expansion and differentiation into effector cells upon activation mediated by migrating antigen-bearing DCs. In particular, antigen-specific activated CD4+ T-helper (Th) cells can be directed into a Th1, Th2, or T-regulatory (Treg) polarization upon direct contact with antigens and induction by specific cytokines (69–74). Consequently, the T-helper cells’ polarization will ultimately lead the adaptive immune system toward either a cellular T cell, sustained by CD8+ cytotoxic T lymphocytes (Th1), or a humoral antibody (Th2), or a tolerance (Treg) response.
To establish immunological memory able to promptly respond to subsequent encounters with the same antigen, a subset of polarized activated effector T cells will differentiate into long-lasting memory T cells (75), which is the ultimate goal of vaccination strategies. Although most successful vaccines in human medicine induce a long-lasting protective humoral adaptive immune response (76–78), antibodies seem to only partially contribute to protective immune response in other vaccines (79, 80). In the last years, indeed, the development of vaccines able to elicit an effective cellular adaptive immune response is considered of high priority for chronic infections, such as HCV and HIV infections (81–84) or cancer (85–87).
In this respect, the engagement of TLRs by the vaccine antigen would be extremely beneficial, resulting mainly in the production of Th-1 cytokines (88). The TLR3 ligand polyI:C, a synthetic analog of dsRNA, has been shown to have a potent adjuvanting effect in several experimental systems (89–91). TLR7/8 agonists have been shown to improve the magnitude and quality of memory T-cell responses elicited by HIV Gag protein (92). The TLR9 agonist bacterial unmethylated CpG DNA has shown a significant therapeutic potential in cancer, infectious disease, and asthma (93–97).
Besides having the capacity of inducing a Th1-type immune response, TLR ligands have been shown to amplify Th2 responses (98–101). In particular, bacterial lipoproteins exert adjuvant function in Lyme disease vaccination through TLR1 (24, 102), and flagellin has been shown to enhance immune response to influenza vaccine through TLR5 (103). Furthermore, polyI:C and CpG DNA improve the survival of activated CD4+ Th cells even in the absence of APCs (104) as well as generate fully functional CD8+ memory cells without any CD4+ T-cell help (105, 106). Furthermore, TLR7/8 agonist has been shown to enhance both effector and memory T-cell responses to HIV gag antigen (92).
Systems biology in vaccine studies
The innate immune system is at the interface between the vaccine antigen and the host’s adaptive immune response; therefore, the evaluation of the molecular effects induced by vaccines on PRRs biology is of high relevance. Studying molecular signatures that are induced rapidly after vaccination will identify causal elements of the adaptive immune response, which may be useful in ultimately predicting protective immune responses. Such prediction will enable the evaluation of the efficacy or immunogenicity of untested vaccines in the general population or the identification of unresponsive individuals to vaccination. Furthermore, the predictive signatures would uncover new correlates of protection and further decipher the biological mechanisms by which such molecular signatures modulate vaccine-induced immunity and protection.
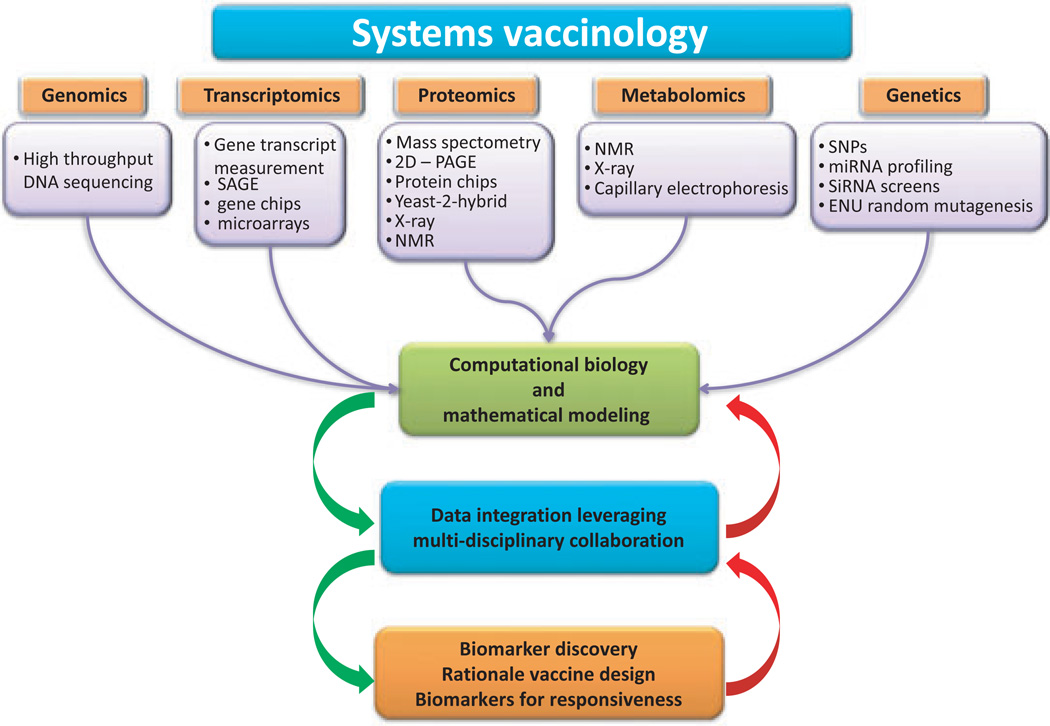
Systems biology approaches provide detailed level of investigation to better and fully analyze the network of interactions within the ménage à trois of this game (vaccine, innate, and adaptive immunity). Conversely to traditional ‘reductionist’ approach, the paradigm of systems biology is to look at a biological system as a whole, evaluating interactions among biological elements and their relationship with the surrounding environment. Systems biology has been increasingly applied to oncology (107–109), autoimmunity and infections (110, 111), and only recently to vaccinology (Fig. 1).
Fig. 1.
Systems biology approaches for vaccine studies interactions and the implications on translational research.
Transcriptomics, proteomics, and genetics
Transcriptomics applied to the immune response enables the identification of specific set of genes and pathways differentially regulated upon encounter with a foreign antigen, and several new insights into interactions between pathogens and innate immunity have been identified (112–117). However, to reduce the number of meaningless observations, results generated by transcriptomics studies need to be integrated by meta-analysis performed on multiple independent datasets, which requires access to several datasets. Several publicly available databases of immunology-related transcriptomic datasets have been created in the recent years (118–121). Furthermore, to improve integration of immunology datasets of these different databases, the Immunological Genome Project initiative has been established recently with the ambitious goal to combine immunology and computational biology laboratories in a systems-level approach (122).
The possibility of performing meta-analysis has enabled the identification of an expression signature, designated as the ‘common host response’, characterized by a cluster of 511 genes selectively and consistently induced in several cell types upon exposure to different pathogens (114). The described expression signature includes cytokine genes, IFN-stimulated genes, transcriptional factors and components of signal transduction pathways, genes that limit the pathogenetic consequences of the immune response, and, more interestingly, genes that have not previously been associated with the immune response. The identification of this shared transcriptional program to foreign antigens among different host cells, even outside of the immune system, suggests the evolution of a multi-cellular and multi-compartmental line of host defense to infection.
Proteomics gives a comprehensive picture of the immune interactome, representing the interactions involving the host and pathogen genes and gene products known to participate in the immune response. Several viral proteins, in particular of HCV, have been identified to interact with key proteins in innate immunity pathways and are involved in the viral immune evasion (123–125).
In this respect, databases on interactions between proteins of innate immunity and pathogens have been developed in the last few years, representing invaluable tools for studying the immune interactome (125–129). In particular, the Innate DB includes up to 7000 innate immunity-relevant interactions involving 2000 human and mouse genes (130).
In addition to studies on protein-to-protein interactions, the so-called ‘phosphoproteomics’ is focused on the dynamics of the innate immune response protein phosphorylation signaling cascades, to identify the complete ‘kinome’ of a cell (131, 132). A further approach is represented by large-scale RNA interference screens, where genes are progressively knocked down using appropriate inhibitory RNA molecules, which have enabled the identification of host proteins involved in HIV and West Nile virus infections (133, 134). Similarly, the profiling of microRNA (miRNA) expression can provide valuable information about the role of these posttranscriptional regulators of gene expression on the modulation of genes relevant to innate immunity and, therefore, the immune response itself, as recently shown for the leukocyte miRNA response to LPS (135–137).
The study of genetic polymorphisms represents an additional level of analysis for the global evaluation of factors involved in the host response to foreign antigens. Polymorphisms can adversely affect expression of genes as well as proteins of the innate immune system and, consequently, host-pathogen interactions as well as molecular signaling (138–141). Implications of polymorphisms in TLR genes on infectious diseases progression have been reported (142–150). In particular, the role of polymorphisms in TLR9 gene and clinical course of HIV-1 infection (151) as well as susceptibility to tuberculosis and specific polymorphisms in the TLR2 gene have been described (152–154). These findings are controversial and not consistently confirmed (155–157).
Multiparametric analyses for prediction of vaccine immunogenicity
Systems level studies have been recently performed to identify gene ‘signatures’ in humans predicting immune responses to yellow fever vaccine (YF-17D) in humans (158, 159). Upon vaccination with YF-17D, a variable response over time is observed, in terms of both magnitude of the antigen-specific CD8+ T-cell responses and neutralizing antibody titers. In parallel, gene transcriptional profile in PBMCs from vaccinated individuals showed the induction of a molecular signature, including several immune genes involved in innate sensing of viruses and antiviral immunity, which lasted for more than 2 weeks post vaccination (158, 159). Among these, genes were identified encoding innate sensing receptors (i.e. TLR7, RIG-I), transcription factors that regulate the expression of type I IFNs, IRF7, and signal transducer and activator of transcription 1 (STAT1). Furthermore, genes encoding proteins in the complement pathway (i.e. C1QB) and the inflammasome were induced. In particular, a group of transcription factors were identified as key regulators of the early innate immune response to the YF-17D vaccine (158, 159), such as ETS2, whose expression is upregulated in activated and proliferating T cells (160, 161) and induces IL-12 p40 (Th1) and IL-5 (Th2) gene expression (162, 163). The enhanced transcription of downstream genes involved in the maturation and differentiation of T cells, B cells, NK cells, and macrophages was observed (159).
There was no significant correlation, however, between the induction of these genes and the magnitude of the CD8+ T-cell or neutralizing antibody response, suggesting that the observed molecular signature might be consequent to the replication of the vaccine live attenuated virus. However, two genes – solute carrier family 2, member 6 (SLC2A6), and eukaryotic translation initiation factor 2 α kinase 4 (EIF2AK4) – were found to strongly correlate with the magnitude of antigen-specific CD8+ T-cell responses and antibody titers in a second YF-17D vaccine trial (158).
EIF2AK4 regulates protein synthesis in response to environmental stresses by phosphorylating elongation initiation factor 2α (eIF2α) (164, 165). Indeed, YF-17D vaccination induced the phosphorylation of eIF2α as well as the formation of stress granules, and other genes involved in the stress response pathway correlated with the CD8+ T-cell response (158). Moreover, the TNF receptor superfamily, receptor 17 (TNFRSF17), which is a receptor for B-cell-activating factor belonging to the TNF family (BAFF), predicted the neutralizing antibody response with highly significant accuracy (158). Finally, PBMCs isolated from YF17D-vaccinated volunteers display a mixed T-helper cell phenotype, with the induction of a mixed Th1/Th2 profile (159, 166). These studies provide a global description of the innate and adaptive immune responses induced by live attenuated (YF-17D) vaccine, showing the networking of the innate immune response that is required for the induction of effective long-lasting immune protection.
Systems approaches to understand molecular basis of virus-like particles (VLP)-HIV vaccines
Such approaches have been used to identify signatures of activation of a VLP-based HIV vaccine ex vivo on monocyte-derived DCs (MDDCs) as well as on PBMCs (167–170). Studies have been performed using a baculovirus-expressed HIV-VLPs (171), which have been shown to induce HIV-1-specific CD4+ and CD8+ T-cell responses and cross-clade neutralizing antibodies at systemic and mucosal level in immunized BALB/c mice (172–174).
Baculovirus-expressed HIV-VLPs induced maturation and activation of MDDCs from HIV-1 seronegative subjects, and this effect was partially mediated by the internal TLRs, TLR3 and TLR9. The HIV-VLP-activated MDDCs produced a pattern of cytokines indicative of both Th1 and Th2 pathways and induced primary and secondary responses in autologous human CD4+ T cells in an ex vivo immunization assay (168). A specific pattern of cellular maturation and activation induced by HIV-VLPs was observed also on whole PBMCs from both HIV-1 seronegative and seropositive subjects, without further manipulation and differentiation to MDDCs (169, 170, 175). This analysis identified a number of HIV-1-seropositive subjects who showed a complete lack of maturation induced by HIV-VLPs in CD14+ cells, confirming the relevance of this multiparametric approach to identify possible non-responders (169). HIV-VLPs induced a significantly increased production of Th2 cytokines only, strongly suggesting that specific Th1 adjuvants would be required for therapeutic effectiveness in HIV-1-infected subjects (169, 176).
In parallel, the baculovirus-expressed HIV-VLPs induced specific transcriptional profiles of genes involved in the morphological and functional changes characterizing innate and early adaptive immune response. This immune signature was observed in MDDCs (167) as well as in PBMCs from HIV-1 seronegative and seropositive subjects (170, 175). As described for the yellow fever live attenuated YF-17D vaccine, HIV-VLPs induced a molecular signature including several genes involved in innate sensing of viruses and antiviral immunity. Expression of proinflammatory mediators CXC-chemokine ligand 10 and IL-1α genes were found upregulated. Similarly, several genes were identified encoding innate sensing receptors (i.e. TLR2), transcription factors that regulate the expression of type I IFNs, IRF1, and STAT2.
The gene signature predictive of both humoral and cellular adaptive immune response included several genes. The CD83 and CD28 genes indicate a strong activation of the Th2 development and B lymphocytes (177–179). TNFRSF1B and TNFRSF6B are markers for T and B-cell activation (TNFRSF1B) (180) and for blocking the pro-apoptotic activity of the FAS-ligand (TNFRSF6B) (181). The TNFSF9 is a T-cell activation marker (182, 183), and CD40 is one of the key players in activation of both humoral and cell-mediated immune responses (184, 185).
These studies provide a global description of the innate and early adaptive immune signatures induced by non-replicating VLPs (HIV-VLPs) in MDDCs as well as PBMCs. Commonalities between these signatures and those induced by the live attenuated vaccine (YF-17D) suggest the possible identification of specific shared predictive gene expression meta-signatures with a broad application in vaccinology.
Polymorphisms and response to vaccines
Studies on polymorphisms of TLRs gene have enabled to recognize the influence of single nucleotide polymorphisms (SNPs) in TLRs on immune response to vaccines (186–188). In particular, associations between SNPs in both TLRs and the downstream intracellular MyD88 and MD2 signaling molecules with antibody and cellular responses to measles vaccination have been described recently (186). A SNP in the 3′UTR of TLR3 (rs5743305 at −976 bp of TLR promoter) has been identified and the heterozygous variant AT correlates with and low humoral and cellular responses in vaccinees. Similarly, the GA variant of a non-synonymous SNP also in the TLR3 gene was associated with lower antibody production. Heterozygous variants for two non-synonymous SNPs (Gly299Asp and Ile399Thr) in the TLR4 gene, already known to be associated with septic shock after infection with Gram-negative bacteria, premature birth, myocardial infarction, and allograft rejection (189), have been identified and associated with higher IL-4 secretion to the measles vaccine strain (186).
Associations between SNPs in genes of TLRs intracellular signaling molecules and the immune response to measles vaccine have been also investigated (186). A minor allele variant for a SNP in the 3′UTR of MyD88, the intracellular adapter molecule that signals for most of the TLRs, was found to be associated with a lower antibody response to measles vaccine, while intronic SNPs in TLRs and their associated intracellular molecule genes were significantly associated with variations in cellular immune responses to measles vaccine (186).
Similar results have been reported for rubella vaccination (187). Polymorphisms in promoter and intronic regions of TLR3 and TLR4 genes have been found associated with rubella virus-specific cytokine immune responses, such as IFN-γ, IL-2, TNF-α, and granulocyte macrophage-colony stimulation factor (GM-CSF). In particular, two SNPs in the TLR3 gene appear to be significantly associated with lower rubella IFN-γ secretion in an allele dose-related manner. Interestingly, the same promoter polymorphism (rs5743305, −8441 A > T) in the TLR3 gene, associated with rubella virus-induced GM-CSF secretion, is considered a risk factor for lower antibody and low lymphoproliferative responses to measles vaccine (186). This finding strongly suggests that this SNP in the TLR3 gene may play a more general role in viral immunity and represent a checkpoint for humoral and cellular immune responses to both measles and rubella vaccines.
In addition to genes of TLRs and their intracellular signaling, the same study described associations of polymorphisms in promoter and intronic regions of vitamin A and vitamin D receptor genes and their downstream mediators of signaling with different immune response to rubella vaccination (187). The influence of SNPs in the vitamin D receptor (VDR) genes on proinflammatory immune responses to viral infection or live viral vaccination might be more general, considering that polymorphisms in the VDR gene are also associated with protection from HIV-1 infection (190). Polymorphisms in the TRIM5 gene have been associated with variations in rubella virus-specific immune responses, in accordance with recent findings on the role of the same TRIM5-gene SNPs in the immune response to retroviral (HIV-1) infection (191).
Concerning the pertussis (PT) vaccine, specific SNPs in the promoter region of the TLR4 gene as well as haplotype-tagging SNPs in genes of the TLR signaling pathway have been shown to influence the antibody response to vaccination (188, 192). SNPs in the TOLLIP gene were the most consistent in three independent analyses. The relevance of this association is due to the biological role of TOLLIP, which is a small protein that binds the activated IL-1 receptor type I (IL-1RI) complex as well as TLR2 and TLR4 complexes, coordinating optimal signaling through IL-1RI and TLR4 (193, 194). Furthermore, associations of SNPs in TIRAP and TICAM1 as well as IRAK3 and IRAK4 genes and immune response to PT vaccine have been also observed. In particular, TIRAP and TICAM1 belong to the TIR domain-containing adapters, including MyD88, that modulate TLR signaling pathways, while IRAK3 and IRAK4 are signal transduction mediators of the Toll and IL-1R families (188, 192).
These results give strong indications for the involvement of the TLR signaling pathway in the response to vaccination as well as for the cooperation of its genes in a functional interacting network. More important, they provide evidences that genetic variants are involved in the mechanisms underlying heterogeneous immune responses to vaccines and propose the possible identification of specific shared predictive polymorphisms with a broad application in vaccinology. All these studies need to be replicated in independent and larger cohorts to validate the findings, increasing the statistical association between SNPs and host immune response to vaccines.
These results indicate that a comprehensive analysis at the system-level would greatly facilitate screening for responsiveness to vaccines and an understanding of eventual failures in individuals enrolled in clinical trials. It will also guide the identification of optimal antigens and antigen formulations (i.e. adjuvanted antigens) to induce the sought cluster of genes and immune pathways leading to the required adaptive immune response.
Conclusions
In recent years, the field has seen a remarkable explosion of information about the components of innate immunity and their role in guiding and shaping the adaptive immune response. A significantly improved fund of knowledge has been accomplished regarding the PRRs, mainly the TLR family members, as well as the associated pathways and their role in the host responses upon exposure to a foreign antigen, in the form of vaccine or pathogen. In particular, many tiles have been added to the mosaic describing the molecular processing and recognition of antigens by cells of the innate immune system and how this will impact on the nature as well as the duration (immune memory) of the adaptive T and B-cell immune responses.
The development of safer but less potent recombinant vaccines, to minimize the possible biological risks linked to traditional inactivated or killed vaccines, is boosting mandatory studies aimed to improve their immunogenic and protective activity. In this regard, the use of TLR or non-TLR PRR signaling to boost vaccine efficacy is heavily explored and documented in several established vaccines. To this aim, systems biology holds considerable promise for discovery and new insights into complex networking processes such as interaction between foreign vaccine antigens, innate immunity, and the downstream adaptive immune response.
Systems biology not only has the potential to accelerate the discovery of new regulators of innate immunity but also will provide more comprehensive insights into the kinetics of regulation at the transcriptional, protein–protein interaction, and post-transcriptional levels. All this information will be of high impact on vaccine development, providing molecular prediction markers of the immunogenicity of a vaccine, uncovering new correlates of vaccine efficacy, as well as guiding the design of new vaccine antigens or formulations. Moreover, such system-level approaches could permit the identification of vaccine responders versus non-responders, allowing a better immunological coverage of the licensed vaccines.
Recent pioneering studies describe results showing that system-level analyses may provide this invaluable information. Broader application of this strategy to other vaccines will definitely allow the possible identification and validation of common markers (i.e. gene signatures, SNPs) applicable to all or clusters of vaccines. The novel and improved knowledge on molecular components of the innate immune system together with the development of more potent high throughput computational analysis will lead to the switch from the ‘empirical’ to the ‘knowledge-based’ age of the vaccinology, enabling the development of even more successful vaccines for preventive as well as therapeutic intervention strategies.
References
- 1.Plotkin SA. Vaccines: past, present and future. Nat Med. 2005;11 Suppl:S5–S11. doi: 10.1038/nm1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rappuoli R. From Pasteur to genomics: progress and challenges in infectious diseases. Nat Med. 2004;10:1177–1185. doi: 10.1038/nm1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachmann MF, Zinkernagel RM, Oxenius A. Immune responses in the absence of costimulation: viruses know the trick. J Immunol. 1998;161:5791–5794. [PubMed] [Google Scholar]
- 4.Pancharoen C, Mekmullica J, Thisyakorn U, Kasempimolporn S, Wilde H, Herzog C. Reduced-dose intradermal vaccination against hepatitis A with an aluminum-free vaccine is immunogenic and can lower costs. Clin Infect Dis. 2005;41:1537–1540. doi: 10.1086/497266. [DOI] [PubMed] [Google Scholar]
- 5.Van Damme P, Oosterhuis-Kafeja F, Van der WM, Almagor Y, Sharon O, Levin Y. Safety and efficacy of a novel microneedle device for dose sparing intradermal influenza vaccination in healthy adults. Vaccine. 2009;27:454–459. doi: 10.1016/j.vaccine.2008.10.077. [DOI] [PubMed] [Google Scholar]
- 6.Mikszta JA, et al. Microneedle-based intradermal delivery of the anthrax recombinant protective antigen vaccine. Infect Immun. 2006;74:6806–6810. doi: 10.1128/IAI.01210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 8.Pashine A, Valiante NM, Ulmer JB. Targeting the innate immune response with improved vaccine adjuvants. Nat Med. 2005;11:S63–S68. doi: 10.1038/nm1210. [DOI] [PubMed] [Google Scholar]
- 9.Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327:291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pulendran B, Tang H, Manicassamy S. Programming dendritic cells to induce T(H)2 and tolerogenic responses. Nat Immunol. 2010;11:647–655. doi: 10.1038/ni.1894. [DOI] [PubMed] [Google Scholar]
- 11.Steinman RM, Banchereau J. Taking dendritic cells into medicine. Nature. 2007;449:419–426. doi: 10.1038/nature06175. [DOI] [PubMed] [Google Scholar]
- 12.Steinman RM. Dendritic cells in vivo: a key target for a new vaccine science. Immunity. 2008;29:319–324. doi: 10.1016/j.immuni.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 13.Steinman RM. Linking innate to adaptive immunity through dendritic cells. Novartis Found Symp. 2006;279:101–109. [PubMed] [Google Scholar]
- 14.Pulendran B, Ahmed R. Translating innate immunity into immunological memory: implications for vaccine development. Cell. 2006;124:849–863. doi: 10.1016/j.cell.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 15.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 16.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 17.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 18.Germain RN. An innately interesting decade of research in immunology. Nat Med. 2004;10:1307–1320. doi: 10.1038/nm1159. [DOI] [PubMed] [Google Scholar]
- 19.Beutler B. Inferences, questions and possibilities in Toll-like receptor signalling. Nature. 2004;430:257–263. doi: 10.1038/nature02761. [DOI] [PubMed] [Google Scholar]
- 20.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 21.Bowie A, O’Neill LA. The interleukin-1 receptor/Toll-like receptor superfamily: signal generators for pro-inflammatory interleukins and microbial products. J Leukoc Biol. 2000;67:508–514. doi: 10.1002/jlb.67.4.508. [DOI] [PubMed] [Google Scholar]
- 22.Shimazu R, et al. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J Exp Med. 1999;189:1777–1782. doi: 10.1084/jem.189.11.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poltorak A, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 24.Alexopoulou L, et al. Hyporesponsiveness to vaccination with Borrelia burgdorferi OspA in humans and in TLR1- and TLR2-deficient mice. Nat Med. 2002;8:878–884. doi: 10.1038/nm732. [DOI] [PubMed] [Google Scholar]
- 25.Ozinsky A, et al. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll-like receptors. Proc Natl Acad Sci USA. 2000;97:13766–13771. doi: 10.1073/pnas.250476497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takeuchi O, et al. Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int Immunol. 2001;13:933–940. doi: 10.1093/intimm/13.7.933. [DOI] [PubMed] [Google Scholar]
- 27.Takeuchi O, et al. Cutting edge: role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J Immunol. 2002;169:10–14. doi: 10.4049/jimmunol.169.1.10. [DOI] [PubMed] [Google Scholar]
- 28.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 29.Hemmi H, et al. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol. 2002;3:196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 30.Heil F, et al. Species-specific recognition of single-stranded RNA via toll- like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 31.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 32.Ahmad-Nejad P, Hacker H, Rutz M, Bauer S, Vabulas RM, Wagner H. Bacterial CpG-DNA and lipopolysaccharides activate Toll-like receptors at distinct cellular compartments. European J Immunol. 2002;32:1958–1968. doi: 10.1002/1521-4141(200207)32:7<1958::AID-IMMU1958>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 33.Latz E, et al. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat Immunol. 2004;5:190–198. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- 34.Hayashi F, et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 35.Geijtenbeek TB, van Vliet SJ, Engering A, ‘t Hart BA, van Kooyk Y. Self- and nonself-recognition by C-type lectins on dendritic cells. Annu Rev Immunol. 2004;22:33–54. doi: 10.1146/annurev.immunol.22.012703.104558. [DOI] [PubMed] [Google Scholar]
- 36.Inohara N, Chamaillard M, McDonald C, Nunez G. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu Rev Biochem. 2005;74:355–383. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- 37.Ting JP, Duncan JA, Lei Y. How the noninflammasome NLRs function in the innate immune system. Science. 2010;327:286–290. doi: 10.1126/science.1184004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.den Dunnen J, Gringhuis SI, Geijtenbeek TB. Innate signaling by the C-type lectin DC-SIGN dictates immune responses. Cancer Immunol Immunother. 2009;58:1149–1157. doi: 10.1007/s00262-008-0615-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 40.Yoneyama M, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 41.Yoneyama M, Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol Rev. 2009;227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- 42.Takeda K, Akira S. Toll-like receptors. Curr Protoc Immunol. 2007;Chapter 14(Unit 14):12. doi: 10.1002/0471142735.im1412s77. [DOI] [PubMed] [Google Scholar]
- 43.Querec T, et al. Yellow fever vaccine YF-17D activates multiple dendritic cell subsets via TLR2, 7, 8, and 9 to stimulate polyvalent immunity. J Exp Med. 2006;203:413–424. doi: 10.1084/jem.20051720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pulendran B. Learning immunology from the yellow fever vaccine: innate immunity to systems vaccinology. Nat Rev Immunol. 2009;9:741–747. doi: 10.1038/nri2629. [DOI] [PubMed] [Google Scholar]
- 45.Geeraedts F, et al. Superior immunogenicity of inactivated whole virus H5N1 influenza vaccine is primarily controlled by Toll-like receptor signalling. PLoS Pathog. 2008;4:e1000138. doi: 10.1371/journal.ppat.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Latz E, Franko J, Golenbock DT, Schreiber JR. Haemophilus influenzae type b-outer membrane protein complex glycoconjugate vaccine induces cytokine production by engaging human toll-like receptor 2 (TLR2) and requires the presence of TLR2 for optimal immunogenicity. J Immunol. 2004;172:2431–2438. doi: 10.4049/jimmunol.172.4.2431. [DOI] [PubMed] [Google Scholar]
- 47.Tsuji S, et al. Maturation of human dendritic cells by cell wall skeleton of Mycobacterium bovis bacillus Calmette-Guerin: involvement of toll-like receptors. Infect Immun. 2000;68:6883–6890. doi: 10.1128/iai.68.12.6883-6890.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uehori J, et al. Simultaneous blocking of human Toll-like receptors 2 and 4 suppresses myeloid dendritic cell activation induced by Mycobacterium bovis bacillus Calmette-Guerin peptidoglycan. Infect Immun. 2003;71:4238–4249. doi: 10.1128/IAI.71.8.4238-4249.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mbow ML, De GE, Valiante NM, Rappuoli R. New adjuvants for human vaccines. Curr Opin Immunol. 2010;22:411–416. doi: 10.1016/j.coi.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 50.Eisenbarth SC, Colegio OR, O’Connor W, Sutterwala FS, Flavell RA. Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature. 2008;453:1122–1126. doi: 10.1038/nature06939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kool M, et al. Cutting edge: alum adjuvant stimulates inflammatory dendritic cells through activation of the NALP3 inflammasome. J Immunol. 2008;181:3755–3759. doi: 10.4049/jimmunol.181.6.3755. [DOI] [PubMed] [Google Scholar]
- 52.Li H, Willingham SB, Ting JP, Re F. Cutting edge: inflammasome activation by alum and alum’s adjuvant effect are mediated by NLRP3. J Immunol. 2008;181:17–21. doi: 10.4049/jimmunol.181.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lund JM, et al. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci USA. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Diebold SS. Recognition of viral single-stranded RNA by Toll-like receptors. Adv Drug Deliv Rev. 2008;60:813–823. doi: 10.1016/j.addr.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 55.Hemmi H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 56.Hemmi H, Kaisho T, Takeda K, Akira S. The roles of Toll-like receptor 9, MyD88, and DNA-dependent protein kinase catalytic subunit in the effects of two distinct CpG DNAs on dendritic cell subsets. J Immunol. 2003;170:3059–3064. doi: 10.4049/jimmunol.170.6.3059. [DOI] [PubMed] [Google Scholar]
- 57.Ott G, Barchfeld GL, Chernoff D, Radhakrishnan R, van HP, Van NG. MF59. Design and evaluation of a safe and potent adjuvant for human vaccines. Pharm Biotechnol. 1995;6:277–296. doi: 10.1007/978-1-4615-1823-5_10. [DOI] [PubMed] [Google Scholar]
- 58.Baldridge JR, Crane RT. Monophosphoryl lipid A (MPL) formulations for the next generation of vaccines. Methods. 1999;19:103–107. doi: 10.1006/meth.1999.0834. [DOI] [PubMed] [Google Scholar]
- 59.Tong NK, et al. Immunogenicity and safety of an adjuvanted hepatitis B vaccine in prehemodialysis and hemodialysis patients. Kidney Int. 2005;68:2298–2303. doi: 10.1111/j.1523-1755.2005.00689.x. [DOI] [PubMed] [Google Scholar]
- 60.Keam SJ, Harper DM. Human papillomavirus types 16 and 18 vaccine (recombinant, AS04 adjuvanted, adsorbed) [Cervarix] Drugs. 2008;68:359–372. doi: 10.2165/00003495-200868030-00007. [DOI] [PubMed] [Google Scholar]
- 61.Holzer BR, Hatz C, Schmidt-Sissolak D, Gluck R, Althaus B, Egger M. Immunogenicity and adverse effects of inactivated virosome versus alum-adsorbed hepatitis A vaccine: a randomized controlled trial. Vaccine. 1996;14:982–986. doi: 10.1016/0264-410x(96)00042-4. [DOI] [PubMed] [Google Scholar]
- 62.Gluck R, Moser C, Metcalfe IC. Influenza virosomes as an efficient system for adjuvanted vaccine delivery. Expert Opin Biol Ther. 2004;4:1139–1145. doi: 10.1517/14712598.4.7.1139. [DOI] [PubMed] [Google Scholar]
- 63.Ryan ET, Calderwood SB. Cholera vaccines. Clin Infect Dis. 2000;31:561–565. doi: 10.1086/313951. [DOI] [PubMed] [Google Scholar]
- 64.Lahiri A, Das P, Chakravortty D. Engagement of TLR signaling as adjuvant: towards smarter vaccine and beyond. Vaccine. 2008;26:6777–6783. doi: 10.1016/j.vaccine.2008.09.045. [DOI] [PubMed] [Google Scholar]
- 65.Kang SM, Compans RW. Host responses from innate to adaptive immunity after vaccination: molecular and cellular events. Mol Cells. 2009;27:5–14. doi: 10.1007/s10059-009-0015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reed SG, Bertholet S, Coler RN, Friede M. New horizons in adjuvants for vaccine development. Trends Immunol. 2009;30:23–32. doi: 10.1016/j.it.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 67.Kwissa M, Kasturi SP, Pulendran B. The science of adjuvants. Expert Rev Vaccines. 2007;6:673–684. doi: 10.1586/14760584.6.5.673. [DOI] [PubMed] [Google Scholar]
- 68.Fraser CK, Diener KR, Brown MP, Hayball JD. Improving vaccines by incorporating immunological coadjuvants. Expert Rev Vaccines. 2007;6:559–578. doi: 10.1586/14760584.6.4.559. [DOI] [PubMed] [Google Scholar]
- 69.McGuirk P, Mills KH. Pathogen-specific regulatory T cells provoke a shift in the Th1/Th2 paradigm in immunity to infectious diseases. Trends Immunol. 2002;23:450–455. doi: 10.1016/s1471-4906(02)02288-3. [DOI] [PubMed] [Google Scholar]
- 70.Moser M, Murphy KM. Dendritic cell regulation of TH1–TH2 development. Nat Immunol. 2000;1:199–205. doi: 10.1038/79734. [DOI] [PubMed] [Google Scholar]
- 71.O’Garra A, Vieira P. Regulatory T cells and mechanisms of immune system control. Nat Med. 2004;10:801–805. doi: 10.1038/nm0804-801. [DOI] [PubMed] [Google Scholar]
- 72.O’Garra A, Robinson D. Development and function of T helper 1 cells. Adv Immunol. 2004;83:133–162. doi: 10.1016/S0065-2776(04)83004-9. [DOI] [PubMed] [Google Scholar]
- 73.Mowen KA, Glimcher LH. Signaling pathways in Th2 development. Immunol Rev. 2004;202:203–222. doi: 10.1111/j.0105-2896.2004.00209.x. [DOI] [PubMed] [Google Scholar]
- 74.Groux H, et al. A CD4+ T-cell subset inhibits antigen-specific T-cell responses and prevents colitis. Nature. 1997;389:737–742. doi: 10.1038/39614. [DOI] [PubMed] [Google Scholar]
- 75.Kaech SM, Wherry EJ, Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nat Rev Immunol. 2002;2:251–262. doi: 10.1038/nri778. [DOI] [PubMed] [Google Scholar]
- 76.Rappuoli R. Bridging the knowledge gaps in vaccine design. Nat Biotechnol. 2007;25:1361–1366. doi: 10.1038/nbt1207-1361. [DOI] [PubMed] [Google Scholar]
- 77.Robbins JB, Schneerson R, Szu SC. Perspective: hypothesis: serum IgG antibody is sufficient to confer protection against infectious diseases by inactivating the inoculum. J Infect Dis. 1995;171:1387–1398. doi: 10.1093/infdis/171.6.1387. [DOI] [PubMed] [Google Scholar]
- 78.Robbins JB, Schneerson R, Szu SC. Hypothesis: how licensed vaccines confer protective immunity. Adv Exp Med Biol. 1996;397:169–182. doi: 10.1007/978-1-4899-1382-1_22. [DOI] [PubMed] [Google Scholar]
- 79.Kimberlin DW, Whitley RJ. Varicella-zoster vaccine for the prevention of herpes zoster. N Engl J Med. 2007;356:1338–1343. doi: 10.1056/NEJMct066061. [DOI] [PubMed] [Google Scholar]
- 80.Belshe RB, et al. Live attenuated versus inactivated influenza vaccine in infants and young children. N Engl J Med. 2007;356:685–696. doi: 10.1056/NEJMoa065368. [DOI] [PubMed] [Google Scholar]
- 81.Houghton M, Abrignani S. Prospects for a vaccine against the hepatitis C virus. Nature. 2005;436:961–966. doi: 10.1038/nature04081. [DOI] [PubMed] [Google Scholar]
- 82.Sallberg M, Frelin L, Weiland O. DNA vaccine therapy for chronic hepatitis C virus (HCV) infection: immune control of a moving target. Expert Opin Biol Ther. 2009;9:805–815. doi: 10.1517/14712590902988444. [DOI] [PubMed] [Google Scholar]
- 83.Korber BT, Letvin NL, Haynes BF. T-cell vaccine strategies for human immunodeficiency virus, the virus with a thousand faces. J Virol. 2009;83:8300–8314. doi: 10.1128/JVI.00114-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rerks-Ngarm S, et al. Vaccination with ALVAC and AIDSVAX to Prevent HIV-1 Infection in Thailand. N Engl J Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 85.Draper SJ, Heeney JL. Viruses as vaccine vectors for infectious diseases and cancer. Nat Rev Microbiol. 2010;8:62–73. doi: 10.1038/nrmicro2240. [DOI] [PubMed] [Google Scholar]
- 86.Banchereau J, Klechevsky E, Schmitt N, Morita R, Palucka K, Ueno H. Harnessing human dendritic cell subsets to design novel vaccines. Ann NY Acad Sci. 2009;1174:24–32. doi: 10.1111/j.1749-6632.2009.04999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sioud M. Does our current understanding of immune tolerance, autoimmunity, and immunosuppressive mechanisms facilitate the design of efficient cancer vaccines? Scand J Immunol. 2009;70:516–525. doi: 10.1111/j.1365-3083.2009.02326.x. [DOI] [PubMed] [Google Scholar]
- 88.Brightbill HD, et al. Host defense mechanisms triggered by microbial lipoproteins through toll-like receptors. Science. 1999;285:732–736. doi: 10.1126/science.285.5428.732. [DOI] [PubMed] [Google Scholar]
- 89.Asahi-Ozaki Y, et al. Intranasal administration of adjuvant-combined recombinant influenza virus HA vaccine protects mice from the lethal H5N1 virus infection. Microbes Infect. 2006;8:2706–2714. doi: 10.1016/j.micinf.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 90.Navabi H, et al. A clinical grade poly I:C-analogue (Ampligen) promotes optimal DC maturation and Th1-type T cell responses of healthy donors and cancer patients in vitro. Vaccine. 2009;27:107–115. doi: 10.1016/j.vaccine.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 91.Longhi MP, et al. Dendritic cells require a systemic type I interferon response to mature and induce CD4+ Th1 immunity with poly IC as adjuvant. J Exp Med. 2009;206:1589–1602. doi: 10.1084/jem.20090247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wille-Reece U, et al. Toll-like receptor agonists influence the magnitude and quality of memory T cell responses after prime-boost immunization in nonhuman primates. J Exp Med. 2006;203:1249–1258. doi: 10.1084/jem.20052433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Krieg AM. Therapeutic potential of Toll-like receptor 9 activation. Nat Rev Drug Discov. 2006;5:471–484. doi: 10.1038/nrd2059. [DOI] [PubMed] [Google Scholar]
- 94.Krieg AM. Toll-like receptor 9 (TLR9) agonists in the treatment of cancer. Oncogene. 2008;27:161–167. doi: 10.1038/sj.onc.1210911. [DOI] [PubMed] [Google Scholar]
- 95.Ma R, Du JL, Huang J, Wu CY. Additive effects of CpG ODN and R-848 as adjuvants on augmenting immune responses to HBsAg vaccination. Biochem Biophys Res Commun. 2007;361:537–542. doi: 10.1016/j.bbrc.2007.07.028. [DOI] [PubMed] [Google Scholar]
- 96.Kwissa M, et al. Adjuvanting a DNA vaccine with a TLR9 ligand plus Flt3 ligand results in enhanced cellular immunity against the simian immunodeficiency virus. J Exp Med. 2007;204:2733–2746. doi: 10.1084/jem.20071211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Speiser DE, et al. Rapid and strong human CD8+ T cell responses to vaccination with peptide, IFA, CpG oligodeoxynucleotide 7909. J Clin Invest. 2005;115:739–746. doi: 10.1172/JCI23373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Agrawal S, et al. Cutting edge: different Toll-like receptor agonists instruct dendritic cells to induce distinct Th responses via differential modulation of extracellular signal-regulated kinase-mitogen-activated protein kinase and c-Fos. J Immunol. 2003;171:4984–4989. doi: 10.4049/jimmunol.171.10.4984. [DOI] [PubMed] [Google Scholar]
- 99.Dillon S, et al. Yeast zymosan, a stimulus for TLR2 and dectin-1, induces regulatory antigen-presenting cells and immunological tolerance. J Clin Invest. 2006;116:916–928. doi: 10.1172/JCI27203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dillon S, et al. A Toll-like receptor 2 ligand stimulates Th2 responses in vivo, via induction of extracellular signal-regulated kinase mitogen-activated protein kinase and c-Fos in dendritic cells. J Immunol. 2004;172:4733–4743. doi: 10.4049/jimmunol.172.8.4733. [DOI] [PubMed] [Google Scholar]
- 101.Redecke V, et al. Cutting edge: activation of Toll-like receptor 2 induces a Th2 immune response and promotes experimental asthma. J Immunol. 2004;172:2739–2743. doi: 10.4049/jimmunol.172.5.2739. [DOI] [PubMed] [Google Scholar]
- 102.Thomas V, Fikrig E. The Lyme disease vaccine takes its toll. Vector Borne Zoonotic Dis. 2002;2:217–222. doi: 10.1089/153036602321653798. [DOI] [PubMed] [Google Scholar]
- 103.Huleatt JW, et al. Potent immunogenicity and efficacy of a universal influenza vaccine candidate comprising a recombinant fusion protein linking influenza M2e to the TLR5 ligand flagellin. Vaccine. 2008;26:201–214. doi: 10.1016/j.vaccine.2007.10.062. [DOI] [PubMed] [Google Scholar]
- 104.Gelman AE, Zhang J, Choi Y, Turka LA. Toll-like receptor ligands directly promote activated CD4+ T cell survival. J Immunol. 2004;172:6065–6073. doi: 10.4049/jimmunol.172.10.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hervas-Stubbs S, Olivier A, Boisgerault F, Thieblemont N, Leclerc C. TLR3 ligand stimulates fully functional memory CD8+ T cells in the absence of CD4+ T-cell help. Blood. 2007;109:5318–5326. doi: 10.1182/blood-2006-10-053256. [DOI] [PubMed] [Google Scholar]
- 106.Assudani D, Cho HI, DeVito N, Bradley N, Celis E. In vivo expansion, persistence, and function of peptide vaccine-induced CD8 T cells occur independently of CD4 T cells. Cancer Res. 2008;68:9892–9899. doi: 10.1158/0008-5472.CAN-08-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Potti A, et al. Genomic signatures to guide the use of chemotherapeutics. Nat Med. 2006;12:1294–1300. doi: 10.1038/nm1491. [DOI] [PubMed] [Google Scholar]
- 108.Alizadeh AA, Staudt LM. Genomic-scale gene expression profiling of normal and malignant immune cells. Curr Opin Immunol. 2000;12:219–225. doi: 10.1016/s0952-7915(99)00078-3. [DOI] [PubMed] [Google Scholar]
- 109.Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ramilo O, et al. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood. 2007;109:2066–2077. doi: 10.1182/blood-2006-02-002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chaussabel D, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ricciardi-Castagnoli P, Granucci F. Opinion: Interpretation of the complexity of innate immune responses by functional genomics. Nat Rev Immunol. 2002;2:881–889. doi: 10.1038/nri936. [DOI] [PubMed] [Google Scholar]
- 113.Elkon R, Linhart C, Halperin Y, Shiloh Y, Shamir R. Functional genomic delineation of TLR-induced transcriptional networks. BMC Genomics. 2007;8:394. doi: 10.1186/1471-2164-8-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jenner RG, Young RA. Insights into host responses against pathogens from transcriptional profiling. Nat Rev Microbiol. 2005;3:281–294. doi: 10.1038/nrmicro1126. [DOI] [PubMed] [Google Scholar]
- 115.McCaffrey RL, et al. A specific gene expression program triggered by Gram-positive bacteria in the cytosol. Proc Natl Acad Sci USA. 2004;101:11386–11391. doi: 10.1073/pnas.0403215101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Leber JH, Crimmins GT, Raghavan S, Meyer-Morse NP, Cox JS, Portnoy DA. Distinct TLR- and NLR-mediated transcriptional responses to an intracellular pathogen. PLoS Pathog. 2008;4:e6. doi: 10.1371/journal.ppat.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hijikata A, et al. Construction of an open-access database that integrates cross-reference information from the transcriptome and proteome of immune cells. Bioinformatics. 2007;23:2934–2941. doi: 10.1093/bioinformatics/btm430. [DOI] [PubMed] [Google Scholar]
- 119.Abbas AR, et al. Immune response in silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes Immun. 2005;6:319–331. doi: 10.1038/sj.gene.6364173. [DOI] [PubMed] [Google Scholar]
- 120.Korb M, et al. The Innate Immune Database (IIDB) BMC Immunol. 2008;9:7. doi: 10.1186/1471-2172-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gardy JL, Lynn DJ, Brinkman FS, Hancock RE. Enabling a systems biology approach to immunology: focus on innate immunity. Trends Immunol. 2009;30:249–262. doi: 10.1016/j.it.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 122.Heng TS, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 123.Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol. 2008;8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.de Chassey B, et al. Hepatitis C virus infection protein network. Mol Syst Biol. 2008;4:230. doi: 10.1038/msb.2008.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Navratil V, et al. VirHostNet: a knowledge base for the management and the analysis of proteome-wide virus-host interaction networks. Nucleic Acids Res. 2009;37:D661–D668. doi: 10.1093/nar/gkn794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ceol A, et al. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010;38:D532–D539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chatr-aryamontri A, et al. VirusMINT: a viral protein interaction database. Nucleic Acids Res. 2009;37:D669–D673. doi: 10.1093/nar/gkn739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Driscoll T, Dyer MD, Murali TM, Sobral BW. PIG – the pathogen interaction gateway. Nucleic Acids Res. 2009;37:D647–D650. doi: 10.1093/nar/gkn799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Oda K, Kitano H. A comprehensive map of the toll-like receptor signaling network. Mol Syst Biol. 2006;2:2006. doi: 10.1038/msb4100057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lynn DJ, et al. InnateDB: facilitating systems-level analyses of the mammalian innate immune response. Mol Syst Biol. 2008;4:218. doi: 10.1038/msb.2008.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Stahl S, et al. Proteomics and pathway analysis identifies JNK signaling as critical for high linear energy transfer radiation-induced apoptosis in non-small lung cancer cells. Mol Cell Proteomics. 2009;8:1117–1129. doi: 10.1074/mcp.M800274-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bakal C, et al. Phosphorylation networks regulating JNK activity in diverse genetic backgrounds. Science. 2008;322:453–456. doi: 10.1126/science.1158739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Brass AL, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 134.Krishnan MN, et al. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhou R, Hu G, Liu J, Gong AY, Drescher KM, Chen XM. NF-kappaB p65-dependent transactivation of miRNA genes following Cryptosporidium parvum infection stimulates epithelial cell immune responses. PLoS Pathog. 2009;5:e1000681. doi: 10.1371/journal.ppat.1000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Schmidt WM, Spiel AO, Jilma B, Wolzt M, Muller M. In vivo profile of the human leukocyte microRNA response to endotoxemia. Biochem Biophys Res Commun. 2009;380:437–441. doi: 10.1016/j.bbrc.2008.12.190. [DOI] [PubMed] [Google Scholar]
- 137.Vasilescu C, et al. MicroRNA fingerprints identify miR-150 as a plasma prognostic marker in patients with sepsis. PLoS ONE. 2009;4:e7405. doi: 10.1371/journal.pone.0007405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Georgel P, Macquin C, Bahram S. The heterogeneous allelic repertoire of human toll-like receptor (TLR) genes. PLoS ONE. 2009;4:e7803. doi: 10.1371/journal.pone.0007803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bochud PY, Bochud M, Telenti A, Calandra T. Innate immunogenetics: a tool for exploring new frontiers of host defence. Lancet Infect Dis. 2007;7:531–542. doi: 10.1016/S1473-3099(07)70185-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Misch EA, Hawn TR. Toll-like receptor polymorphisms and susceptibility to human disease. Clin Sci (Lond) 2008;114:347–360. doi: 10.1042/CS20070214. [DOI] [PubMed] [Google Scholar]
- 141.Dickinson AM, Holler E. Polymorphisms of cytokine and innate immunity genes and GVHD. Best Pract Res Clin Haemato. 2008;21:149–164. doi: 10.1016/j.beha.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 142.Pine SO, McElrath MJ, Bochud PY. Polymorphisms in toll-like receptor 4 and toll-like receptor 9 influence viral load in a seroincident cohort of HIV-1-infected individuals. AIDS. 2009;23:2387–2395. doi: 10.1097/QAD.0b013e328330b489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Velez DR, et al. Variants in toll-like receptors 2 and 9 influence susceptibility to pulmonary tuberculosis in Caucasians, African-Americans, and West Africans. Hum Genet. 2010;127:65–73. doi: 10.1007/s00439-009-0741-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wurfel MM, et al. Toll-like receptor 1 polymorphisms affect innate immune responses and outcomes in sepsis. Am J Respir Crit Care Med. 2008;178:710–720. doi: 10.1164/rccm.200803-462OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Nguyen TH, et al. Toll-like receptor 4 (TLR4) and typhoid fever in Vietnam. PLoS ONE. 2009;4:e4800. doi: 10.1371/journal.pone.0004800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rezazadeh M, et al. TLR4 polymorphism in Iranian patients with brucellosis. J Infect. 2006;53:206–210. doi: 10.1016/j.jinf.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 147.Lorenz E, Mira JP, Frees KL, Schwartz DA. Relevance of mutations in the TLR4 receptor in patients with gram-negative septic shock. Arch Intern Med. 2002;162:1028–1032. doi: 10.1001/archinte.162.9.1028. [DOI] [PubMed] [Google Scholar]
- 148.Miyairi I, DeVincenzo JP. Human genetic factors and respiratory syncytial virus disease severity. Clin Microbiol Rev. 2008;21:686–703. doi: 10.1128/CMR.00017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ferwerda B, et al. TLR4 polymorphisms, infectious diseases, and evolutionary pressure during migration of modern humans. Proc Natl Acad Sci USA. 2007;104:16645–16650. doi: 10.1073/pnas.0704828104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ferwerda B, et al. Functional consequences of toll-like receptor 4 polymorphisms. Mol Med. 2008;14:346–352. doi: 10.2119/2007-00135.Ferwerda. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bochud PY, et al. Polymorphisms in Toll-like receptor 9 influence the clinical course of HIV-1 infection. AIDS. 2007;21:441–446. doi: 10.1097/QAD.0b013e328012b8ac. [DOI] [PubMed] [Google Scholar]
- 152.Yoshida A, Inagawa H, Kohchi C, Nishizawa T, Soma G. The role of toll-like receptor 2 in survival strategies of Mycobacterium tuberculosis in macrophage phagosomes. Anticancer Res. 2009;29:907–910. [PubMed] [Google Scholar]
- 153.Yim JJ, et al. The association between microsatellite polymorphisms in intron II of the human Toll-like receptor 2 gene and tuberculosis among Koreans. Genes Immun. 2006;7:150–155. doi: 10.1038/sj.gene.6364274. [DOI] [PubMed] [Google Scholar]
- 154.Ogus AC, et al. The Arg753GLn polymorphism of the human toll-like receptor 2 gene in tuberculosis disease. Eur Respir J. 2004;23:219–223. doi: 10.1183/09031936.03.00061703. [DOI] [PubMed] [Google Scholar]
- 155.Smirnova I, et al. Assay of locus-specific genetic load implicates rare Toll-like receptor 4 mutations in meningococcal susceptibility. Proc Natl Acad Sci USA. 2003;100:6075–6080. doi: 10.1073/pnas.1031605100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Allen A, et al. Variation in Toll-like receptor 4 and susceptibility to group A meningococcal meningitis in Gambian children. Pediatr Infect Dis J. 2003;22:1018–1019. doi: 10.1097/01.inf.0000095431.15606.68. [DOI] [PubMed] [Google Scholar]
- 157.Read RC, et al. A functional polymorphism of toll-like receptor 4 is not associated with likelihood or severity of meningococcal disease. J Infect Dis. 2001;184:640–642. doi: 10.1086/322798. [DOI] [PubMed] [Google Scholar]
- 158.Querec TD, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Gaucher D, et al. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med. 2008;205:3119–3131. doi: 10.1084/jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gallant S, Gilkeson G. ETS transcription factors and regulation of immunity. Arch Immunol Ther Exp (Warsz) 2006;54:149–163. doi: 10.1007/s00005-006-0017-z. [DOI] [PubMed] [Google Scholar]
- 161.Bhat NK, et al. Reciprocal expression of human ETS1 and ETS2 genes during T-cell activation: regulatory role for the protooncogene ETS1. Proc Natl Acad Sci USA. 1990;87:3723–3727. doi: 10.1073/pnas.87.10.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sun HJ, et al. Transcription factors Ets2 and Sp1 act synergistically with histone acetyltransferase p300 in activating human interleukin-12 p40 promoter. Acta Biochim Biophys Sin (Shanghai) 2006;38:194–200. doi: 10.1111/j.1745-7270.2006.00147.x. [DOI] [PubMed] [Google Scholar]
- 163.Blumenthal SG, Aichele G, Wirth T, Czernilofsky AP, Nordheim A, Dittmer J. Regulation of the human interleukin-5 promoter by Ets transcription factors. Ets1 and Ets2, but not Elf-1, cooperate with GATA3 and HTLV-I Tax1. J Biol Chem. 1999;274:12910–12916. doi: 10.1074/jbc.274.18.12910. [DOI] [PubMed] [Google Scholar]
- 164.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 165.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 166.Santos AP, Matos DC, Bertho AL, Mendonca SC, Marcovistz R. Detection of Th1/Th2 cytokine signatures in yellow fever 17DD first-time vaccinees through ELISpot assay. Cytokine. 2008;42:152–155. doi: 10.1016/j.cyto.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 167.Aricò E, et al. Immature monocyte derived dendritic cells gene expression profile in response to Virus-Like Particles stimulation. J Transl Med. 2005;3:45. doi: 10.1186/1479-5876-3-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Buonaguro L, et al. Baculovirus-derived human immunodeficiency virus type 1 virus-like particles activate dendritic cells and induce ex vivo T-cell responses. J Virol. 2006;80:9134–9143. doi: 10.1128/JVI.00050-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Buonaguro L, Tornesello ML, Gallo RC, Marincola FM, Lewis GK, Buonaguro FM. Th2 Polarization in Peripheral Blood Mononuclear Cells from Human Immunodeficiency Virus (HIV)-Infected Subjects, as Activated by HIV Virus-Like Particles. J Virology. 2009;83:304–313. doi: 10.1128/JVI.01606-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Monaco A, et al. Molecular immune signatures of HIV-1 vaccines in human PBMCs. FEBS Lett. 2009;583:3004–3008. doi: 10.1016/j.febslet.2009.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Buonaguro L, et al. High efficient production of Pr55gag Virus-like Particles expressing multiple HIV-1 epitopes, including a gp120 protein derived from an Ugandan HIV-1 isolate of subtype A. Antiviral Res. 2001;49:35–47. doi: 10.1016/s0166-3542(00)00136-4. [DOI] [PubMed] [Google Scholar]
- 172.Buonaguro L, et al. Induction of neutralizing antibodies and CTLs in Balb/c mice immunized with Virus-like Particles presenting a gp120 molecule from a HIV-1 isolate of clade A (HIV-VLPAs) Antiviral Res. 2002;54:189–201. doi: 10.1016/s0166-3542(02)00004-9. [DOI] [PubMed] [Google Scholar]
- 173.Buonaguro L, Visciano ML, Tornesello ML, Tagliamonte M, Biryahwaho B, Buonaguro FM. Induction of systemic and mucosal cross-clade neutralizing antibodies in BALB/c mice immunized with human immunodeficiency virus type 1 clade A virus-like particles administered by different routes of inoculation. J Virology. 2005;79:7059–7067. doi: 10.1128/JVI.79.11.7059-7067.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Buonaguro L, et al. DNA-VLP prime-boost intra-nasal immunization induces cellular and humoral anti-HIV-1 systemic and mucosal immunity with cross-clade neutralizing activity. Vaccine. 2007;25:5968–5977. doi: 10.1016/j.vaccine.2007.05.052. [DOI] [PubMed] [Google Scholar]
- 175.Buonaguro L, et al. Gene expression profile of peripheral blood mononuclear cells in response to HIV-VLPs stimulation. BMC Bioinformatics. 2008;9 Suppl:S5. doi: 10.1186/1471-2105-9-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Buonaguro L, Tornesello ML, Jewis GK, Buonaguro FM. Short communication: limited induction of IL-10 in PBMCs from HIV-infected subjects treated with HIV-VLPs. AIDS Res Hum Retroviruses. 2009;25:819–822. doi: 10.1089/aid.2008.0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Magistrelli G, et al. Identification of three alternatively spliced variants of human CD28 mRNA. Biochem Biophys Res Commun. 1999;259:34–37. doi: 10.1006/bbrc.1999.0725. [DOI] [PubMed] [Google Scholar]
- 178.Andres PG, et al. Distinct regions in the CD28 cytoplasmic domain are required for T helper type 2 differentiation. Nat Immunol. 2004;5:435–442. doi: 10.1038/ni1044. [DOI] [PubMed] [Google Scholar]
- 179.Kozlow EJ, Wilson GL, Fox CH, Kehrl JH. Subtractive cDNA cloning of a novel member of the Ig gene superfamily expressed at high levels in activated B lymphocytes. Blood. 1993;81:454–461. [PubMed] [Google Scholar]
- 180.Beltinger CP, et al. Physical mapping and genomic structure of the human TNFR2 gene. Genomics. 1996;35:94–100. doi: 10.1006/geno.1996.0327. [DOI] [PubMed] [Google Scholar]
- 181.Pitti RM, et al. Genomic amplification of a decoy receptor for Fas ligand in lung and colon cancer. Nature. 1998;396:699–703. doi: 10.1038/25387. [DOI] [PubMed] [Google Scholar]
- 182.Alderson MR, et al. Molecular and biological characterization of human 4-1BB and its ligand. Eur J Immunol. 1994;24:2219–2227. doi: 10.1002/eji.1830240943. [DOI] [PubMed] [Google Scholar]
- 183.Stephan MT, et al. T cell-encoded CD80 and 4-1BBL induce auto- and transcostimulation, resulting in potent tumor rejection. Nat Med. 2007;13:1440–1449. doi: 10.1038/nm1676. [DOI] [PubMed] [Google Scholar]
- 184.Kawabe T, et al. The immune responses in CD40-deficient mice: impaired immunoglobulin class switching and germinal center formation. Immunity. 1994;1:167–178. doi: 10.1016/1074-7613(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 185.Schonbeck U, Libby P. The CD40/CD154 receptor/ligand dyad. Cell Mol Life Sci. 2001;58:4–43. doi: 10.1007/PL00000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Dhiman N, et al. Associations between SNPs in toll-like receptors and related intracellular signaling molecules and immune responses to measles vaccine: preliminary results. Vaccine. 2008;26:1731–1736. doi: 10.1016/j.vaccine.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Ovsyannikova IG, et al. Rubella vaccine-induced cellular immunity: evidence of associations with polymorphisms in the Toll-like, vitamin A and D receptors, and innate immune response genes. Hum Genet. 2010;127:207–221. doi: 10.1007/s00439-009-0763-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Banus S, et al. Toll-like receptor 4 polymorphism associated with the response to whole-cell pertussis vaccination in children from the KOALA study. Clin Vaccine Immunol. 2007;14:1377–1380. doi: 10.1128/CVI.00175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Schroder NW, Schumann RR. Single nucleotide polymorphisms of Toll-like receptors and susceptibility to infectious disease. Lancet Infect Dis. 2005;5:156–164. doi: 10.1016/S1473-3099(05)01308-3. [DOI] [PubMed] [Google Scholar]
- 190.de la Torre MS, et al. Vitamin D receptor gene haplotypes and susceptibility to HIV-1 infection in injection drug users. J Infect Dis. 2008;197:405–410. doi: 10.1086/525043. [DOI] [PubMed] [Google Scholar]
- 191.van Manen D, Rits MA, Beugeling C, van DK, Schuitemaker H, Kootstra NA. The effect of Trim5 polymorphisms on the clinical course of HIV-1 infection. PLoS Pathog. 2008;4:e18. doi: 10.1371/journal.ppat.0040018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kimman TG, et al. Association of interacting genes in the toll-like receptor signaling pathway and the antibody response to pertussis vaccination. PLoS ONE. 2008;3:e3665. doi: 10.1371/journal.pone.0003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Didierlaurent A, et al. Tollip regulates pro-inflammatory responses to interleukin-1 and lipopolysaccharide. Mol Cell Biol. 2006;26:735–742. doi: 10.1128/MCB.26.3.735-742.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Bulut Y, Faure E, Thomas L, Equils O, Arditi M. Cooperation of Toll-like receptor 2 and 6 for cellular activation by soluble tuberculosis factor and Borrelia burgdorferi outer surface protein A lipoprotein: role of Toll-interacting protein and IL-1 receptor signaling molecules in Toll-like receptor 2 signaling. J Immunol. 2001;167:987–994. doi: 10.4049/jimmunol.167.2.987. [DOI] [PubMed] [Google Scholar]