Abstract
Protein phosphorylation is a major post-translational modification in plants crucial for the regulation of diverse cellular functions. In the early stages of this field, efforts focused on the qualitative detection, identification, and cataloging of in vivo protein phosphorylation sites. Recently these studies have advanced into utilizing quantitative mass spectrometric measurements, capable of dynamically monitoring changes in phosphorylation levels in response to genetic and environmental alterations. This review will highlight current untargeted and targeted mass spectral technologies used for quantitative phosphoproteome measurements in plants, and provide a discussion of these phosphorylation changes in relation to important biological events.
Introduction
The protein kinase- catalyzed covalent addition of a phosphate group to the three amino acids serine, threonine and tyrosine, and subsequent removal by protein phosphatases, is one of the most important and diverse signaling mechanisms in plants and animals. The sequential phosphorylation of proteins in a signaling pathway provides the basis for complex signaling networks and regulatory processes within plants, including hormone perception and transduction [1], and environmental stress perception and response [2]. The Arabidsopsis and other sequenced plant genomes have more than 1000 predicted protein kinases [3], almost twice as many as the kinase genes predicted in humans [4], suggesting a complex phosphorylation network in plant signaling.
In this review we would like to focus on a small number of emerging questions in the area of plant phosphoproteomics. First, we will suggest criteria by which protein phosphorylation events are tied to a biological event or phenotypic response. Second, we will provide the reader with a brief discussion of the current mass spectrometry (MS) based technologies used in phosphoproteomics. Finally, since a timely and excellent review of plant phosphoproteomic papers was provided recently in this journal by Shulze ([6], 2010), we will focus on reviewing only those quantitative phosphoproteomic reports that have come out in the past year subsequent to that critique, with the goal of suggesting avenues for future work.
Connecting Protein Phosphorylation to Biological Importance
With at least 30,000 proteins encoded by plant genomes, and an estimation that as many as 30% of all proteins in such multicellular organisms can be phosphorylated at any time [5], the number of amino acid residues that are present as phosphorylated moieties may be as high as one or two million. However, it is likely that out of all of these potentially phosphorylated protein sites, only a small minority may be biologically important. Promiscuous phosphorylation is a well-known phenomenon that occurs during in vitro assays since the concentration and location of reactants can be artificially high, thereby resulting in non-natural reactivity and phosphorylation patterns unreflective of true in planta chemistry. Promiscuity caused by circumstantial proximity in vivo may also occur and thus, considerable experimentation may be required before it can be concluded that a site under study is relevant to a particular biological process.
In the early stages of the phosphoproteomic field in plants, much of the effort focused on overcoming technological hurdles, accumulating databases, and cataloging the detection of phosphorylated peptides.[7-9] These studies are now being supplanted with quantitative studies aimed at determining which of the hundreds of thousands of phosphoresidues are changing in vivo to provide a primary screen for identifying those playing a role in regulating plant processes. While the simple identification of phosphorylated peptides in biological samples is useful, we are now tackling the more difficult job of measuring the dynamic changes of these phosphopeptides in response to genetic and environmental perturbations.
One question that arises in phosphoproteomics is, if a phosphosite is shown to change in vivo in response to stimuli, then does it mean that this site is biologically important? The answer to this is no. Since there may be large-scale changes in pleiotropic cell states (e.g. cytoplasmic pH changes) that cause changes in protein kinase or phosphatase activity, it is likely that a large number of phosphoproteins show dynamic phosphorylation status while only a few such changes are necessary and/or sufficient for regulating a plant process. To conclusively show that a phosphorylated residue is directly involved, one must perform forward or reverse genetics experiments where the serine, threonine, or tyrosine residue is mutated to a non-phosphorylatable amino acid such as alanine (A), which is neutral and thus mimics a constitutively unmodified residue. Conversely, a mutation could be made to a negatively charged aspartyl residue, such as aspartate (D) mimicking a constitutively phosphorylated amino acid. If a phenotypic change is observed following the site-directed mutation, and if the A and D mutations show opposite effects correlating with what is expected to happen with and without phosphorylation, it can provide a compelling case that this phosphorylation site is important and relevant to what is going on in vivo.[10] If a phenotypic change is observed following the site-directed mutation, it can provide a compelling case that this phosphorylation site is important. While genetic redundancies and compensation can sometimes mask these effects, without this genetic underpinning it is reasonable to argue that one cannot be certain the phosphorylation changes observed are in fact biologically important, or directly linked to the phenotypic or biochemical changes observed. Of course, compelling cases for biological significance of phosphorylation site importance can sometimes be made through kinase assays, which investigate the effect of a specifically activated kinase in vitro without associated genetic experiments. However, biological relevance arguments are made stronger with independent genetic studies that support, and complement, the biochemical investigations that are performed in vitro and in vivo.
Quantitative MS Platforms for Phosphoproteomics
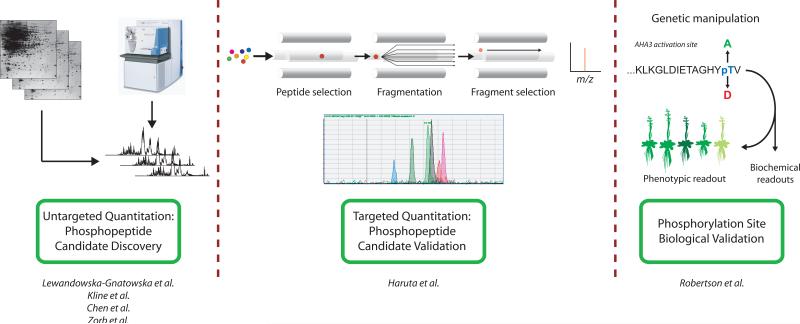
Quantitative proteomic measurements can be performed in one of two ways: via an untargeted platform, or by specifically analyzing a subset of proteins using a targeted MS approach (Figure 1). The untargeted MS platform is aimed at the global identification of in planta phosphorylation changes and frequently utilizes high-resolution mass spectrometers with the capability of performing tandem mass spectrometry (MS/MS) fragmentation experiments (see reviews [11-12] for details). Phosphopeptide quantitation in the untargeted MS platform can be achieved using a diverse panel of methods including 2D gel electrophoresis, precursor ion intensity comparison, spectral counting, metabolic labeling, or chemical isotopic tagging. However, if a biologist wishes to initiate the costly venture of genetic and biochemical validation of a small number of phosphopeptide targets from these survey experiments, further follow-up studies should be performed to confirm and validate these candidate phosphopeptide changes.
Figure 1.
Generalized quantitative phosphoproteomic workflow. Quantitative changes in phosphopeptide levels can be identified in biological samples using an untargeted mass spectrometry approach, such as done by Kline et al. [17], Zorb et al. [16], and Chen et al. [15]. Candidate phosphopeptides identified by the untargeted MS approach can then be targeted for further confirmation, analysis and quantitation using a targeted SRM-MS method, such as that shown by Haruta et al. [18]. Finally, site-specific genetic mutations and/or biochemical methods can be utilized to assess actual biological importance and impact of each phosphosite and/or phosphoprotein in the plant. [10]
Phosphopeptide validation studies are typically performed using a quantitative targeted MS platform, which utilizes the discriminating power of triple quadrupole mass spectrometers to select specific peptides in a complex sample mixture for detection and quantitation ([13], reviewed in [12]). This technology, known as selective reaction monitoring mass spectrometry (SRM-MS), facilitates the sensitive and specific quantitation of hundreds of peptides on a chromatographic time scale in a single MS run. Due to the high-throughput nature of this method, specific phosphopeptides of interest can be monitored in biological samples under many different conditions and time-points with greater ease. This allows for validation of phosphorylation changes prior to initiating slow and potentially costly biochemical and genetic mutation experiments for determination of biological importance. The move to targeted SRM-MS analyses is intensifying due to the ability to monitor target proteins of interest in highly complex samples in a more high-throughput manner, and we believe its use in the plant phosphoproteomic field will continue to grow in the near future.
Recent Studies Utilizing Quantitative Phosphoproteomic Technologies
An excellent review of quantitative proteomics in plants has been published recently [11], which covers many practical aspects of quantitative proteomics such as the underlying technologies, the precision of the methods, and the software available for data analysis. Below we will review several examples of untargeted and targeted quantitative analyses of phosphoproteins performed in the past two years.[14-18] Each of these papers utilizes a different approach, and we will attempt to critique each method and thus illustrate how the technology led the investigators to their conclusions on quantitative changes in the plant phosphoproteome.
In one study, Zorb et al. ([16], 2010) used two-dimensional (2D) gel electrophoresis and fluorescent, phosphate specific dyes to measure fold-changes in maize roots following a one-hour salt treatment. Protein gel spots that showed a greater than two-fold change between control and salt treated samples were excised, and proteins were identified using high-resolution tryptic peptide mass fingerprinting. Overall, sixteen proteins were identified as being differentially phosphorylated in this study. This paper can be critiqued due to several limitations in technology. First, the gold standard for identifying and quantifying phosphopeptides is by means of tandem MS, where direct peptide sequence information can be used for a database search. Second, the identification of phosphorylation events using this technology is limited to the protein level, since individual phosphopeptides are not identified. This means specific phosphosites and localization of phosphorylation on the protein are not determined and therefore cannot be easily studied in a more comprehensive manner to assess biological significance.
Similarly, Lewandowska-Gnatowska et al. ([14], 2011) used 2D-gel technology to visualize changes in the phosphoproteome of maize leaves after short periods (10, 20, 30, 60 and 180 minute) of mechanical wounding. In this study, MS/MS fragmentation together with precursor ion exact mass were used for protein identification, resulting in the identification of 21 phosphoproteins in the samples, although no fold-change in phosphorylation levels were reported. Despite the ease of accessibility of this technology to individual laboratories, the capacity to visualize and identify differentially phosphorylated proteins is limited using a gel-based approach, as it necessitates a sufficient shift in pI and molecular weight for comparison between treatment groups. This is manifested in the low number of differentially modified proteins identified in these studies compared to other untargeted approaches such as metabolic labeling, where identification of phosphorylation change is assessed at the peptide level.
In Chen et al. ([15], 2010), the authors performed an untargeted proteomic analysis of the Arabidopsis phosphoproteome using a label-free LC-MS/MS approach on an LTQ-Orbitrap MS. The authors examined three time points (1 hour, 2 hours and 6 hours) after treatment of Arabidopsis cell cultures with a single concentration of five individual hormones (abscisic acid (ABA), auxin, gibberellin, jasmonic acid and kinetin). For quantification, precursor ions were aligned and independent component analysis (ICA) done to identify phosphorylated peptides that independently segregated between treatment and control groups. This analysis identified over 2000 phosphopeptides, which were ultimately assigned to 500 unique peptide sequences. Out of this set, 152 phosphopeptides were found to be differentially phosphorylated in at least one hormone treatment and time point. While this study does not report magnitudes of change for the phosphopeptides identified, it does use ICA to identify those that seem to be differentially regulated in response to each hormone treatment. Since only those peptides that were identified in all three replicates were considered, and there was discrimination of these phosphopeptides based on experimental treatment, this data may be considered reliable. However, it would be interesting to note the actual quantitative fold changes of these phosphopeptides in each treatment compared to the control samples in order to give an indication of the magnitude of change found in these phosphopeptides.
A separate study published by Kline et al. ([17], 2010), used 15N reciprocal metabolic labeling of Arabidopsis tissue to identify and quantifying changes in the phosphoproteome following exposure to the phytohormone ABA. In this strategy, one control Arabidopsis population was grown in natural abundance medium (14N), and the experimental Arabidopsis population (ABA treated) was grown in medium enriched in 15N heavy isotope.[19] To control for isotope effects and provide a biological replicate, a second experiment was performed where the treatment group and isotope label were reversed. Following treatment with ABA, control and treated plant tissue were combined at a 1:1 ratio and underwent protein extraction, phosphopeptide enrichment, and MS analysis. Phosphopeptides were identified on an LTQ-Orbitrap MS using both collision-induced and electron transfer dissociation fragmentation methods. In total, 17,201 phosphopeptides were identified representing 2,185 unique phosphoproteins. Of these, 50 phosphopeptides were found to have a significantly altered abundance ratio, including known ABA-responsive transcription factors and kinases. This quantification method with heavy isotope reciprocal labeling is attractive because an internal standard is present for each peptide in the sample. Furthermore, because the plant treatment groups are combined upstream of most sample handling procedures, an internal control for systematic error in sample processing is provided. Due to the internal control provided by the isotope labeling and the MS reproducibility, this method allowed for identification of statistically significant phosphorylation changes down to 10% between control and ABA treated samples. This study resulted in the identification of previously known and novel phosphopeptide potentially involved in the ABA signaling pathway. As with each of the other recent untargeted phosphoproteomic MS studies, further follow up and confirmation of phosphopeptide/protein changes can be done using a targeted SRM-MS approach. Future targeted studies would provide a validation of phosphorylation change following treatment in a larger number of biological replicates and result in a high confidence candidate list for biological validation utilizing genetic or biochemical approaches.
Phosphorylation Stoichiometry
While untargeted MS work is important for identifying phosphorylation sites that may be altered during a particular perturbation, these experiments cannot determine phosphorylation stoichiometry, or the fold increases in phosphorylation state compared to the unmodified protein. In other words, what percentage of the particular protein is phosphorylated? Has the target protein gone from 0.1% phosphorylation to 0.2% phosphorylation, or from 50% phosphorylation to 100% phosphorylation? While both represent a 2-fold increase in phosphorylation, these two alternatives may have very different biological interpretations concerning the role of the site in biology. Conventional Western blotting techniques with antibodies have been used to estimate phosphorylation stoichiometry, but this method requires a protein migration shift on the gel and sensitive and specific antibodies for proteins of interest, which may not always be available. In contrast, a technique we believe will become more extensively applied in the field is the use of targeted SRM-MS to measure phosphosite stoichiometry in a high-throughput and automated fashion in biological samples. SRM-MS can quantify with great precision and sensitivity, the level of a phosphorylated and non-phosphorylated peptides to determine what percentage of molecules are present in each form. This is done using isotope labeled internal standard peptides and newly developed HPLC-Chip microfluidic chromatography systems that utilize titanium dioxide (TiO2) material for the automated online enrichment of phosphorylated peptides. This microfluidic based enrichment column consists of sandwiched C18/TiO2/C18 stationary phases, which allows both phosphorylated peptides and non-phosphorylated peptides to be trapped and eluted separately from the same initial sample injection. Quantitative stoichiometry of peptide target phosphorylation can then be assessed using this data.
Using this type of targeted MS method, Haruta et al. ([18], 2010) documented phosphorylation stoichiometry changes in plasma membrane proton pumps (AHA) proteins in wild-type and mutant Arabidopsis plants using heavy isotope labeled AHA C-terminal peptides in both phosphorylated and unmodified forms. Resulting data suggested an increase in kinase-mediated phosphorylation behaves as a posttranslational means of compensating for the loss of one proton pump, by increased phosphorylation of the other. This implies that multiple AHA proteins isoforms together perform a required function and that the effects of their single mutations may be masked, at least in part, by compensation at the phosphorylation level. As online phosphopeptide enrichment technologies become more widely available and optimized, we believe we will see this novel technology become a key method for stoichiometry determination within the plant phosphoproteomic field.
Conclusion
Out of the thousands to millions of possible phosphorylation sites in plants, untargeted MS studies are continually assembling lists of those that have been observed in planta. In order for biologists to identify the small number of these sites that are involved in a particular aspect of plant function, quantitative MS based studies must be performed. Additionally, we believe that high-throughput targeted SRM-MS methodologies will increase in use and popularity as a means to confirm quantitative changes prior to initiation of genetic based validation studies, which are necessary to confirm the biological importance of these sites.
Highlights.
Plant phosphoproteomics is transitioning from qualitative large-scale phosphoprotein identification to quantitative measurements that monitor dynamic changes in phosphorylation events.
Quantitative untargeted MS phosphoproteomic methods identify changes in protein phosphorylation in a global, undirected manner and are useful for first identification of potentially important signaling changes.
Quantitative targeted MS methods are useful for high-throughput secondary screening of potentially important protein phosphorylation events under a multitude of environmental or genetic perturbations.
To confirm the importance of a phosphosite in a biological event, genetic based studies, such as site directed phosphosite mutagenesis, should also be performed.
Acknowledgments
Funding was provided by the National Science Foundation for MRS and GBW (MCB-0929395), and the NHGRI training grant to the Genomic Sciences Training Program for KGK (5T32HG002760).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Recommended Reading
Papers of particular interest, published within the period of review, have been highlighted as;
* of special interest
** of outstanding interest
- 1.Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324(5930):1068–71. doi: 10.1126/science.1173041. PMCID: 2827199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mishra NS, Tuteja R, Tuteja N. Signaling through MAP kinase networks in plants. Arch Biochem Biophys. 2006;452(1):55–68. doi: 10.1016/j.abb.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Lukowitz W, Gillmor CS, Scheible WR. Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol. 2000;123(3):795–805. doi: 10.1104/pp.123.3.795. PMCID: 1539260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 5.Hubbard MJ, Cohen P. On target with a new mechanism for the regulation of protein phosphorylation. Trends Biochem Sci. 1993;18(5):172–7. doi: 10.1016/0968-0004(93)90109-z. [DOI] [PubMed] [Google Scholar]
- 6 *.Schulze WX. Proteomics approaches to understand protein phosphorylation in pathway modulation. Curr Opin Plant Biol. 2010;13(3):280–87. doi: 10.1016/j.pbi.2009.12.008. [Review article covering analysis of large-scale plant phosphoproteomic datasets acquired from 2007 to early 2010. Great general overview of phosphopeptide enrichment strategies, databases, and current literature within the field.] [DOI] [PubMed] [Google Scholar]
- 7.Heazlewood JL, Durek P, Hummel J, Selbig J, Weckwerth W, Walther D, et al. PhosPhAt: a database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor. Nucleic Acids Res. 2008;36:D1015–21. doi: 10.1093/nar/gkm812. PMCID: 2238998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8 *.Grimsrud PA, den Os D, Wenger CD, Swaney DL, Schwartz D, Sussman MR, et al. Large-scale phosphoprotein analysis in Medicago truncatula roots provides insight into in vivo kinase activity in legumes. Plant Physiol. 2010;152(1):19–28. doi: 10.1104/pp.109.149625. [PMCID: 2799343 Untargeted phosphoproteomic profiling of Medicago truncatula root tissue. This is the first study to utilize electron transfer dissociation for plant phosphoprotein analysis, and first reporting of phosphorylation motifs unobserved in other plant species.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9 *.Nakagami H, Sugiyama N, Mochida K, Daudi A, Yoshida Y, Toyoda T, et al. Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 2010;153(3):1161–74. doi: 10.1104/pp.110.157347. [PMCID: 2899915. Untargeted phosphoproteomic study utilizing whole cell lysates from rice cell wall, resulting in the identification of 3,393 phosphoproteins. Phosphopeptide quantitation was not performed, but a comparative analysis of site specific phosphorylation of orthologous proteins between rice, Arabidopsis, and Medicago was done.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10 **.Robertson WR, Clark K, Young JC, Sussman MR. An Arabidopsis thaliana plasma membrane proton pump is essential for pollen development. Genetics. 2004;168(3):1677–87. doi: 10.1534/genetics.104.032326. [First paper showing different phenotypic changes when a knockout mutant for the phloem specific plasma membrane proton pump (H+-ATPase) of Arabidopsis (AHA3) is complemented with a genomic clone containing the penultimate threonine is mutated to either an A or D. This threonine is one of the most readily observable phosphorylation events in the pump, and previous biochemical studies with growth regulators such as fusicoccin and with expression studies in yeast, had implicated it in regulating the catalytic properties of the pump.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11 **.Schulze WX, Usadel B. Quantitation in mass-spectrometry-based proteomics. Annu Rev Plant Biol. 2010;61:491–516. doi: 10.1146/annurev-arplant-042809-112132. [Comprehensive review of quantitative proteomics techniques with examples of application to plant biology. Includes evaluation of the accuracy, dynamic range, and proteome coverage of the various technologies.] [DOI] [PubMed] [Google Scholar]
- 12 *.Kline KG, Sussman MR. Protein quantitation using isotope-assisted mass spectrometry. Annu Rev Biophys. 2010;39:291–308. doi: 10.1146/annurev.biophys.093008.131339. [Review of proteomic mass spectrometry quantitation techniques.] [DOI] [PubMed] [Google Scholar]
- 13 *.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci U S A. 2003;100(12):6940–5. doi: 10.1073/pnas.0832254100. [PMCID: 165809 Initial publication showing isotope dilution method for peptide quantitation using selective reaction monitoring mass spectrometry, coined “absolute quantification” or AQUA method.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14 **.Lewandowska-Gnatowska E, Johnston ML, Antoine W, Szczegielniak J, Muszynska G, Miernyk JA. Using multiplex-staining to study changes in the maize leaf phosphoproteome in response to mechanical wounding. Phytochemistry. 2011 doi: 10.1016/j.phytochem.2011.01.030. [Two-dimensional gel electrophoresis was done on maize leaf proteins following mechanical wounding. Phosphorylation staining identified differentially modified proteins in the sample. In total, 21 proteins were identified, although magnitude of phosphorylation changes were not reported.] [DOI] [PubMed] [Google Scholar]
- 15 **.Chen Y, Hoehenwarter W, Weckwerth W. Comparative analysis of phytohormone-responsive phosphoproteins in Arabidopsis thaliana using TiO2-phosphopeptide enrichment and mass accuracy precursor alignment. Plant J. 2010;63(1):1–17. doi: 10.1111/j.1365-313X.2010.04218.x. [Label-free quantitative analysis of Arabidopsis cell culture treated with five individual hormones over a time-course. Resulted in the identification of 152 differentially modified peptides. Novel phosphopeptides were identified for abscisic acid, as well as a Gα subunit tyrosine phosphorylation that appeared to be a common response to multiple hormones.] [DOI] [PubMed] [Google Scholar]
- 16 **.Zorb C, Schmitt S, Muhling KH. Proteomic changes in maize roots after short-term adjustment to saline growth conditions. Proteomics. 2010;10(24):4441–9. doi: 10.1002/pmic.201000231. [Utilized a 2D gel electrophoresis method with phosphate specific dyes to identify more than 2-fold phosphorylation level changes in sixteen proteins from maize roots following a one-hour salt treatment. Proteins were identified by peptide mass fingerprinting following in gel extraction.] [DOI] [PubMed] [Google Scholar]
- 17 **.Kline KG, Barrett-Wilt GA, Sussman MR. In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc Natl Acad Sci U S A. 2010;107(36):15986–91. doi: 10.1073/pnas.1007879107. [PMCID: 2936636 Use of reciprocal metabolic labeling strategy for the precise quantitation of phosphorylation changes in Arabidopsis proteins following exposure to abscisic acid. Known ABA-responsive proteins and kinases were identified as having significantly altered phosphorylation levels, along with multiple novel candidates not previously shown to be involved in ABA signalling.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18 **.Haruta M, Burch HL, Nelson RB, Barrett-Wilt G, Kline KG, Mohsin SB, et al. Molecular characterization of mutant Arabidopsis plants with reduced plasma membrane proton pump activity. J Biol Chem. 2010;285(23):17918–29. doi: 10.1074/jbc.M110.101733. [PMCID: 2878554. First publication using online phosphopeptide enrichment and SRM-MS for the determination of phosphorylation stoichiometry in a biological sample. Demonstrated compensation for the loss of individual protein isoforms was occuring at the level of phosphorylation stoichiometry.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19 *.Huttlin EL, Hegeman AD, Harms AC, Sussman MR. Comparison of full versus partial metabolic labeling for quantitative proteomics analysis in Arabidopsis thaliana. Mol Cell Proteomics. 2007;6(5):860–81. doi: 10.1074/mcp.M600347-MCP200. [First publication showing the ability to isotopically label hydroponically grown Arabidopsis to >98% incorporation with 15N enriched salts. Additionally this paper introduces a method by which partially metabolically labeled tissue can be utilized for quantitative proteomic studies.] [DOI] [PubMed] [Google Scholar]