Abstract
SlyD is a Ni(II)-binding protein that contributes to nickel homeostasis in Escherichia coli. The C-terminal domain of SlyD contains a rich variety of metal-binding amino acids, suggesting broader metal-binding capabilities, and previous work demonstrated that the protein can coordinate several types of first row transition metals. However, the binding of SlyD to metals other than Ni(II) has not been previously characterized. To further our understanding of the in vitro metal-binding activity of SlyD and how it correlates with the in vivo function of this protein, the interactions between SlyD and the series of biologically relevant transition metals Mn(II), Fe(II), Co(II), Cu(I) and Zn(II) were examined by using a combination of optical spectroscopy and mass spectrometry. SlyD binding to Mn(II) or to Fe(II) ions was not detected but the protein coordinates multiple ions of Co(II), Zn(II) and Cu(I) with appreciable affinities (KD ≤ nM), highlighting the promiscuous nature of this protein. The order of affinities of SlyD for the metals examined is Mn(II), Fe(II) < Co(II) < Ni(II) ~ Zn(II) ≪ Cu(I). Although the purified protein is unable to overcome the large thermodynamic preference for Cu(I) and exclude Zn(II) chelation in the presence of Ni(II), in vivo studies reveal a Ni(II)-specific function for the protein. Furthermore, these latter experiments support a specific role for SlyD as a [NiFe]-hydrogenase enzyme maturation factor. The implications of the divergence between the metal selectivity of SlyD in vitro and the specific activity in vivo are discussed.
Metal homeostasis is an essential yet complex process that is achieved through the action of a number of proteins often working in concert (1, 2). These factors ensure that metal-dependent proteins are paired with the correct metal ion(s) to produce biologically active complexes. The proteins involved in metal homeostasis include membrane importers/exporters, metal storage proteins that are responsible for safe sequestration, metallochaperones dedicated to targeted delivery of metal ions, as well as metal-sensing transcription factors, which together control the amount of a specific metal available in the cellular cytoplasm. The fidelity of these proteins is at the top of a hierarchy of metal-selective processes, and their selectivity is proposed to ultimately influence the metal occupancy of all other proteins that require a metal ion cofactor for function (2, 3). Therefore, a detailed understanding of the proteins involved in metal homeostasis and their ability to recognize and chelate a cognate metal partner is a prerequisite for deciphering how metal selectivity is achieved in the context of an entire cell.
Escherichia coli SlyD is an example of a protein that is involved in Ni(II) homeostasis. SlyD contributes to both nickel accumulation and energy metabolism in this organism because it participates in the Ni(II) insertion step during [NiFe]-hydrogenase metallocenter assembly (4, 5). NMR solution structures of E. coli SlyD revealed that the N-terminal region consists of two well-defined domains (6, 7). The FKBP (FK-506 binding protein) domain has a similar structure as other members of the FKBP family of peptidyl-prolyl isomerases (PPIases) and catalyzes the otherwise slow isomerisation of proline peptide bonds during protein folding (8, 9). The IF (insert in the flap) domain recognizes and binds to unfolded protein, contributing to the protein folding activity (10, 11). In addition, the final 50 residues at the C-terminus of SlyD correspond to the metal-binding domain (MBD) and include 28 potential metal-binding amino acids (6 Cys, 15 His, 2 Glu, 5 Asp) (Figure S1). This C-terminal domain remains unstructured according to NMR data and is variable between SlyD homologues (7, 12). A vital role for the MBD of SlyD was established upon truncation of the protein in E. coli, which resulted in compromised hydrogenase production (13). In support of the nickel-related in vivo function of SlyD, a recent analysis of the isolated protein demonstrated that it can bind up to 7 Ni(II) ions with an affinity in the nanomolar range (14). This in vitro investigation also revealed that SlyD coordinates Ni(II) through a non-cooperative mechanism and exists in a mixture of metalloforms at any given metal concentration. The ability of the unique MBD of SlyD to sequester a substantial amount of Ni(II) has been proposed to contribute to Ni(II) storage in E. coli and provide a source of Ni(II) for the hydrogenase enzymes (15).
Previous in vitro analysis revealed that in addition to Ni(II), SlyD can bind at least sub-stoichiometric amounts of other types of transition metals such as Zn(II), copper and cobalt (15). This finding is not surprising given the dense mix of different types of metal-binding amino acids in the MBD, which could feasibly meet the chemical coordination preferences of a range of metal ions. Thus, SlyD may contribute broadly to the homeostasis of other metal ions in E. coli. To test this hypothesis, we explored the metal-binding activity of SlyD to several biologically relevant first row transition metals, Mn(II), Fe(II), Co(II), Cu(I), and Zn(II) (16). Our results indicate that while SlyD does not bind Mn(II) or Fe(II) with appreciable affinity, it tightly binds to Co(II), Cu(I) and Zn(II) in addition to Ni(II). We also find that Ni(II) can replace Co(II) bound to SlyD, but that selectivity for Ni(II) is not observed in the presence of Cu(I) or Zn(II). In contrast, although SlyD did not exhibit selectivity towards Ni(II) in vitro, in vivo experiments reveal that SlyD can specifically influence the balance of nickel ions in E. coli under anaerobic conditions, while it had no effect on factors required for the homeostasis of other types of metals. This discrepancy is discussed in the context of the E. coli cell, and provides strong support for the assignment of SlyD as a dedicated nickel factor.
EXPERIMENTAL METHODS
Materials
The following metal salts, ZnSO4, [Cu(CH3CN)4]PF6, Ni(acetate)2, CoSO4, FeSO4 and MnCl2 were purchased from Aldrich as a minimum 99.9% pure. Except for copper, all metal solutions were prepared by dissolving the salts as purchased in Milli-Q water that was stirred in an anaerobic glovebox (O2 < 1 ppm) for at least 24 h to minimize oxygen content. Concentrations of metal stock solutions were verified by ICP-AES. The Cu(I) salts were re-crystallized as described previously to remove any trace amounts of Cu(II) and the resulting powder was dissolved in an Ar(g) saturated 30% (v/v) acetonitrile/water solution (17). The concentrations of the copper solutions were established by using bathocuproine sulfonate (Bcs) as described previously (18). Buffers for all metal assays were prepared with Milli-Q water that was then treated with Chelex-100 (Bio-Rad) and stirred in an anaerobic glovebox (O2 < 1 ppm) for at least 24 h prior to use. All titrations and assays were conducted at least in triplicate to ensure reproducibility. The metal chelators ethylene glycol tetraacetic acid (EGTA), bicinchoninate (Bca), Bcs, glycine and 4-(2-pyridylazo)resorcinol (PAR) were purchased from Sigma-Aldrich as a minimum 99% pure and the pHs of the stock solutions were adjusted to match that of the protein buffer.
Protein expression, purification and preparation
SlyD wildtype (WT) and triple mutant proteins were expressed and purified as mentioned previously (14). The triple mutant was prepared to investigate the role of the six cysteine residues, which occur in the SlyD MBD in pairs. All 3 sets of cysteines have been removed in this mutant, which has the following mutation/deletions: Cys to Ala mutation of residues 167,168,184,185, and removal of the last 4 residues (Δ193–196). Preparation of apo protein and determination of the reduction state of the WT SlyD via N-Ethyl maleimide modifications were carried out as described previously (14).
Metal binding via electronic absorption spectroscopy
Protein samples were buffer exchanged into 20 mM HEPES pH 7.5, 100 mM NaCl and 90 μL aliquots of apo SlyD were incubated with the appropriate metal concentrations as noted at 4 °C under anaerobic conditions. Samples were analyzed under aerobic conditions except for Fe(II) and Cu(I) titrations, which were measured in an anaerobic cuvette to minimize exposure to air. Due to significant background signals from Fe(II) and Cu(I) in HEPES buffer, metal-binding experiments for these two metals were conducted in 10 mM ammonium acetate, pH 7.5, that was bubbled with N2(g). Experiments involving Cu(I) and Bcs or Bca were conducted in 20 mM HEPES buffer pH 7.5, 100 mM NaCl and spectroscopic analysis was conducted under aerobic conditions.
Equilibrium dialysis
SlyD samples of 40 μM were dialyzed against an equal volume of 320 μM metal solutions at 4 °C overnight in an anaerobic glovebox using Microchambers of DIALYZER (Harvard apparatus). At the end of the equilibration period samples from the protein chamber (metal bound to protein + free metal) and the metal ion chamber (free metal) were measured in triplicate via a previously described HPLC method (19).
Metal binding via ESI-MS
Protein samples were buffer exchanged using 500 μL aliquots of 10 mM ammonium acetate, pH 7.5, in six dilution steps using 10K nanosep centrifugal devices (PALL Science). This volatile buffer is essential for acquiring clean MS data. Control experiments were conducted to ensure that results obtained when using ammonium acetate as a buffer were comparable to those obtained using 20 mM HEPES, pH 7.4, 100 mM NaCl, Direct metal titration results for Zn(II) were obtained by equilibrating the protein samples (10 μM each) with a known amount of ZnSO4. Titrations of SlyD with Cu(I) and Co(II) were obtained by incubating the protein samples (10 μM) with [Cu(MeCN)4]+ or CoSO4 in the presence of 100 μM DTT. Samples were allowed to equilibrate overnight at 4 °C in an anaerobic glovebox, followed by transfer to septum capped vials for MS analysis.
ESI-MS instrumental parameters and data acquisition/processing
The mass spectra were acquired on a AB/Sciex QStarXL mass spectrometer equipped with an ion spray source in the positive ion mode and a Hot Source-Induced Desolvation (HSID) interface (Ionics Mass spectrometry Group Inc.). The HSID is an atmospheric pressure interface that is used for efficient desolvation of sample that is accomplished via energy transfer from the hot gas as the ions travel through multiple flow regions. The resulting ions are orthogonally introduced to the mass spectrometer and the interface is expected to lead to greater sensitivity and stability of the signal while operating similar to a conventional ion source. Ions were scanned from m/z 1300–3000 with accumulation of 1 s per spectrum with no interscan time delay and averaged for a 2–4 min period. The instrument parameters were set as follows: ion source temperature 200 °C, ion source gas 50.0 psi; curtain gas 50.0 psi; ion spray voltage 5000.0 V; declustering potential 60.0 V; focusing potential 60 V; collision gas 3.0; MCP (detector) 2200.0 V. The spectra were deconvoluted using the Bayesian protein reconstruction program over a mass range of 20000 – 22000 Da and a step mass of 1 Da, signal/noise ratio of 20 and the minimum intensity detected set to 1 % were used during reconstruction of the data. The addition of DTT to protein samples decreases the intensity of the signals detected and at metal concentrations at or above 2 equivalents of metal (i.e. 20 μM or higher) the signal is further suppressed due to high salt conditions. Therefore, samples containing DTT were acquired with similar instrument conditions listed above except the Q1 transmission was enhanced at m/z of 2500. The amounts of apo and holo SlyD species were calculated from the signal intensities of the reconstructed spectra assuming that all of the metalloforms are ionized to the same extent. Only the peaks corresponding to salt-free SlyD were used for calculating the metallation states of SlyD
Competition experiments for determining the KD
To determine the affinity of SlyD for Zn(II) a competition reaction was set up as noted in the equation below by adding increasing amounts of apo SlyD to separate aliquots of 400 μM PAR containing 8 μM Zn(II) in 20 mM HEPES pH 7.5, 100 mM NaCl.
Samples were equilibrated overnight at 4 °C and subsequently analyzed by electronic absorption spectroscopy at 500 nm. Titrations of 400 μM PAR with Zn(II) under identical conditions were used to calculate the amount of [Zn-PAR2] formed in the competition described above. As the pH of the solution greatly affects the formation constant as well as the molar absorption coefficient (ε) of the Zn-PAR2 complex, the metal-binding affinity of PAR under these particular buffer conditions was calibrated by using EGTA as described in detail by Zimmerman et al (18). Further details of the competition reactions are in the supplementary information (SI).
To determine the affinity of SlyD for Cu(I), apo SlyD was titrated into a solution of [Cu(Bcs)2]3− of defined molar ratio Bcs:Cu(I) ≥ 2.5. Transfer of Cu(I) from the chelator to protein or vice versa can be detected by the change in absorbance at 483 nm for Bcs (18). By systematically varying the concentration of the chelators and the protein, conditions that favoured competition were achieved and the resulting spectra were used for calculating the KD. For additional information see the SI.
To determine the affinity of SlyD for Co(II), separate aliquots of 35 μM Fura2 (Invitrogen) containing 8–12 μM Co(II) were titrated with apo SlyD. The resulting solutions were analyzed using electronic absorption spectroscopy at 368 nm and compared to titration of Fura2 with CoSO4 under identical conditions to establish the concentration of Co-Fura2 in each sample. An average KD was calculated using data from several replicates. For additional details see the SI.
Circular dichroism (CD) spectroscopy
Apo SlyD (60 μM) was buffer exchanged into 10 mM ammonium acetate, pH 7.5, using micro-centrifugal devices (MW cut-off of 10,000, Pall Nanosep) and incubated with known amounts of metal overnight at 4 °C in an anaerobic glovebox. Samples were loaded into a 0.1 cm cuvette and capped to minimize exposure to air. The spectra were collected on a Jasco J-710 spectropolarimeter by scanning 205–320 nm at room temperature. The final spectra obtained are averages of 5 scans collected by using a scan speed of 20 nm/min.
Cu(I)-binding stoichiometry using Bca
Apo protein samples were titrated into a solution of [Cu(Bca)2]3− of defined molar ratio L:Cu(I) ≥ 2.5, in which the ligand and copper concentrations were held constant. Transfer of Cu(I) from the chelator to protein or vice versa can be detected by monitoring the change in absorbance at 562 nm for Bca (18). By systematically varying the concentration of Bca and the protein, conditions that disfavoured competition were achieved.
Metal toxicity studies
In LB media
Wild type and ΔslyD strains from the BW25113 KEIO gene deletion collection were used in this assay (20). The growth rates of wild type and ΔslyD strains under different metal concentrations were assessed by measuring the OD600 over 24 h. The following metal salts were utilized to supplement the growth media; NiCl2, CoSO4, ZnSO4 and CuSO4. All cultures were maintained at 37 °C in an aerobic environment. The concentrations tested are summarized in Table S1.
In minimal media
All solutions used in this assay were treated with chelex-100 overnight to remove any trace metals. Metal stocks were prepared from Chelex-treated Milli-Q water. The M9 media (1x) (Sigma) was supplemented with 0.2 % glucose, 1 mM MgSO4 and 100 μM CaCl2 and sterile filtered (minimal media). Two wild-type bacterial strains were used in these toxicity studies: the above- mentioned BW25113 and MC4100, as well as the corresponding ΔslyD strains. The growth rates were monitored in M9 media by measuring the OD600 over 24 h. All cultures were maintained at 37 °C under aerobic conditions. The concentrations of metals tested are summarized in Table S1.
Growth conditions for quantitative real-time PCR (qRT-PCR)
Aerobic
All experiments were conducted using the bacterial strain MC4100 and the ΔslyD strain in the same background (5). Cultures were grown in minimal media at 37 °C with aerobic shaking at 250 rpm until the cultures reached exponential growth phase (approx. 16 h) followed by treatment with a known amount of metal for 30 min.
Anaerobic
For anaerobic growth experiments MC4100 wildtype and ΔslyD strains were used. The M9 salts (1x) were Chelex-treated overnight to remove any trace metal and then the following nutrients were added back to the media: 30 mM sodium formate, 0.5% glucose (v/v), 1 mM MgCl2, 100 nM CaCl2, 100 nM (NH4)2MoO4, 100 nM NaSeO3. Cultures were grown in a capped bottle without any headspace for 17 h at 37 °C allowing the cultures to reach a mid-log phase. These anaerobically-grown cultures were then divided into sterile Falcon tubes containing no metal or with 10 μM NiCl2 in an anaerobic glovebox. The small cultures were then capped and allowed to grow for an additional 30 min at 37 °C. For analysis of Zn(II) and Cu(II) under anaerobic conditions an identical procedure was followed, except the Chelex-treated M9 media was supplemented with only 0.2 % glucose (v/v), 1 mM MgCl2, 100 nM CaCl2.
RNA isolation and qRT-PCR
Total RNA was isolated by using the RNeasy Mini Kit (Qiagen) according to the manufacturers’ protocol for the mechanical disruption and purification of bacterial RNA. The QuantiTect Reverse Transcription Kit (Qiagen) was used for synthesis of cDNA from ~0.5–1 μg of RNA and to eliminate contaminating genomic DNA. qPCR reactions were performed on a LightCycler 480 Real-time PCR block (Roche) using SYBR green (Finnzymes) and primers specific for each transporter as listed in Table S2. holB mRNA, encoding for a DNA gyrase, was used as an internal control because the expression level of this protein is unaffected by metal supplementation (21).
RESULTS
Zn(II) binding to SlyD
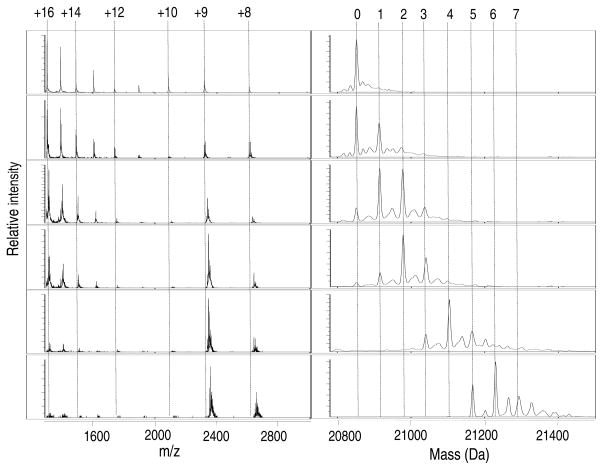
Zn(II) is one of the transition metals most avidly acquired by E. coli and the intracellular concentrations of this metal ion can reach up to ~0.1 mM (22). Furthermore, given the ligand composition of the SlyD MBD (Figure S1), which contains a distribution of cysteine and histidine residues, it was feasible that the softer Zn(II) ion would bind readily to SlyD. We first examined Zn(II) binding to SlyD in vitro by using mass spectrometry to analyze a titration experiment (Figure 1). The data reveal several metalloforms of the protein that differ in the number of Zn(II) ions bound at each titration point, indicating that SlyD contains multiple metal-binding sites. The existence of these stable, partially-metallated intermediates reflects a non-cooperative metallation mechanism similar to that of Ni(II) binding to SlyD (14, 23).
Figure 1.
ESI-MS of titration of SlyD with Zn(II). The addition of increasing amounts of Zn(II) to SlyD (10 μM) leads to concurrent filling of multiple metal sites indicating a non-cooperative metal-binding mechanism. From top to bottom the equivalents of Zn(II) added relative to protein concentration are: 0, 0.5, 1.5, 2, 4 and 6.3. The m/z spectra are shown on the left with the respective reconstructed spectra on the right. The numbers heading the dotted-lines indicate the number of Zn(II) ions bound to SlyD (right panels) and the charge states of the protein (left panels).
Recently, a change in charge state distribution has been recognized as a sensitive indicator of protein structure (24). Higher numbered charge states, with smaller m/z values, represent a greater surface area than lower numbered series and this can be interpreted as a more open structure. Changes in charge states observed during ESI-MS analysis as a function of metallation may indicate the formation of a structure that is folding around the metal-binding site (24). The m/z spectrum of apo SlyD reveals two different populations of SlyD in solution (Figures 1 and S2), with most of the protein carrying a higher charge (i.e. the +16 charge state is more intense than the signal for the +9 species) indicating that apo SlyD predominantly exists in a more unfolded population under these experimental conditions. Upon titration with Zn(II) the relative peak intensities for the species with the higher charge states decreases, suggesting a metal-induced conformational change that shifts the equilibrium to the more folded conformer.
The charge distribution observed for the apo protein is slightly different than in our previous study, in which a higher abundance of lower charge states was observed (14). The experiments reported here were conducted on the same instrument under identical experimental conditions but following installation of an HSID interface. This interface results in desolvation at higher temperatures, which could affect the populations of protein conformations detected. Therefore, to allow for a comparison of data acquired under the same instrumental settings, Ni(II) coordination to SlyD was re-evaluated on the mass spectrometer equipped with the interface. As reported previously, the addition of Ni(II) caused a shift in the charge state envelopes to predominantly +8 and +9, indicating metal-induced folding similar to that caused by Zn(II) (Figure S2 and Ref.(14)). These new data support our previous findings and unambiguously show the noncooperative binding of up to 7 Ni(II) ions, as described above for Zn(II).
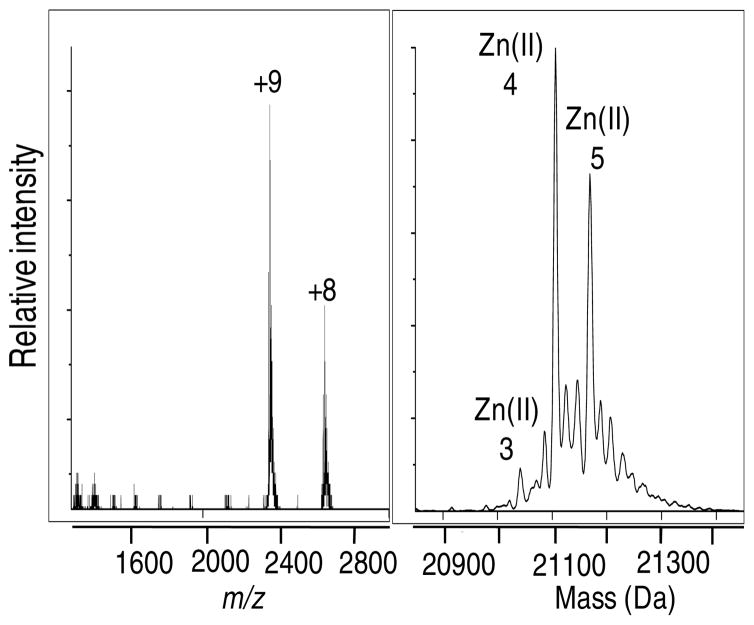
A definitive endpoint to the Zn(II) titration cannot be achieved by MS because the concentration of salt interferes with the quality of the data. Therefore, the total number of tight binding sites on SlyD was established by analyzing a protein sample that was treated with an 8-fold excess of Zn(II) overnight followed by gel-filtration chromatography to remove excess or loosely-bound Zn(II). The mass spectrum indicates the presence of a mixed population of metallated species with a maximum of five metal-binding sites and dominant peaks arising from the four and five Zn(II)-coordinated species (Figure 2). These results indicate that the affinity of additional zinc ions is sufficiently low such that they are lost on the column.
Figure 2.
Zn(II) stoichiometry of SlyD. The mass spectrum of a solution of SlyD (100 μM) incubated with 800 μM Zn(II) followed by gel filtration to remove any weak or non-specifically bound metal indicates a maximum capacity of 5 Zn(II) ions.
To estimate the affinity of SlyD for Zn(II), a competition assay was performed using the colorimetric indicator PAR. This chelator forms a 2:1 PAR to Zn(II) complex in solution that results in an increased absorbance at 500 nm (25). Titration of apo SlyD into a 400 μM PAR solution containing 8 μM Zn(II) results in a non-linear decrease in the signal at 500 nm, indicative of an effective competition between the protein and the ligand for Zn(II) (Figure S3). An identical titration with EGTA instead of protein was also conducted to calibrate the affinity of the Zn-PAR2 under the buffer conditions used, since the formation constant of this complex is affected by the pH (18). The resulting data from these two titrations were fitted to a model assuming a single binding event to obtain an apparent KD of 1.0 ± 0.4 × 10−10 M (Figure S3).
For comparison, the affinity of the Ni(II)-SlyD complex was also examined in more detail. Exploratory experiments indicated that the protein could compete with Fura2 (KD 2.4 × 10−8 M (26)) for Ni(II) coordination (data not shown), establishing a quantitative upper-limit to Ni(II)-SlyD complexes. Combining this result with the fact that Ni(II) can bind to SlyD concurrently with Zn(II) (see below) and compete with EGTA at low chelator concentrations (14), a KD(Ni(II)-SlyD) of ≤10−10 M can be estimated.
To determine whether the Zn(II)-induced structural changes of SlyD suggested by ESI-MS match those induced by Ni(II) binding, a Zn(II) titration was analyzed by using CD spectroscopy. Two minima are observed for the apo protein at 215 nm and 227 nm (Figure S4). Typical β-sheet structures give rise to the band at 215 nm, as would be expected for SlyD because both the PPIase and IF domains are predominantly composed of β-sheets (7), whereas the 227 nm band suggests the presence of β-turn type structures (27, 28). As SlyD is titrated with Zn(II) a gradual decrease in the molar ellipticity is observed in the far-UV region that reaches a plateau upon addition of 4 mol equiv of Zn(II). Although this trend is similar to that observed when SlyD chelates Ni(II), the overall degree of change in the molar ellipticity is significantly lower for Zn(II). Therefore, the CD spectra suggest that the Zn(II)-induced secondary structure for SlyD is likely different from that of Ni(II).
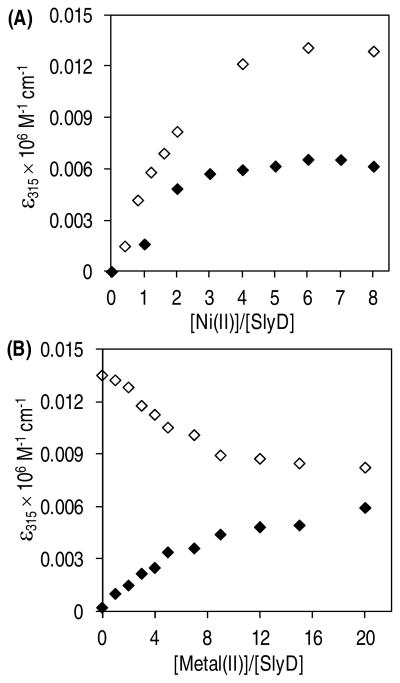
To assess the selectivity of SlyD for Zn(II) or Ni(II), a competition titration was conducted in which apo SlyD was incubated with equivalent amounts of Ni(II) and Zn(II) overnight. Binding of Zn(II) to the protein could be monitored indirectly because replacement of Ni(II) by Zn(II) results in a decrease in the ligand to metal charge transfer (LMCT) band of the Ni(II)-SlyD complex at 315 nm (14). A less intense signal is observed when apo SlyD is titrated with an equimolar solution of Ni(II) and Zn(II) in comparison to titration with Ni(II) alone (Figure 3A). Thus the data provide evidence for metal-binding sites on SlyD that preferentially sequester Zn(II) in the presence of Ni(II). In order to assess whether the order of metal addition affects the metallation state of SlyD, two competition titrations were carried out. SlyD was pre-incubated with 8 fold excess of Ni(II) or Zn(II) for 24 h followed by titration with the other metal and Ni(II) binding was monitored at 315 nm. A representative data set for each of the competition titrations that were allowed to equilibrate for 24 h after the addition of the second metal ion is shown in Figure 3B. Increasing the incubation period after addition of the second metal to 48 h or 72 h generated similar results, suggesting that 24 h is sufficient to reach a thermodynamic equilibrium (data not shown). The results clearly demonstrate that Ni(II) is able to replace a fraction of the Zn(II) bound to SlyD and vice versa. Furthermore, the two titration curves do not intersect at any of the metal concentrations tested, highlighting that the metallation state of SlyD is dependent on the order of metal addition when considering Ni(II) and Zn(II). This result is consistent with a model in which the binding of the first metal ion locks the protein in a specific conformation, such that only a few of the remining sites are then accessible for metal exchange reactions, as suggested by previous studies with nickel (14).
Figure 3.
Selectivity of SlyD for Ni(II) vs. Zn(II). (A) Addition of an equimolar solution of Ni(II) and Zn(II) to SlyD (filled diamonds) results in lower intensity at 315 nm when compared to a titration with equivalent amounts of Ni(II) alone (unfilled diamonds). The x-axis indicates the total amounts of Ni(II) added relative to apo SlyD (15 μM). (B) Change in absorbance at 315 nm as Ni(II) is added to a SlyD sample (15 μM) pre-incubated with 8 molar equivalents of Zn(II) (filled diamonds) or as Zn(II) is added to a SlyD sample pre-incubated with 8 molar equivalents of Ni(II) (unfilled diamonds).
Cu(I) binding to SlyD
Compared to zinc, the intracellular copper quota is lower in E. coli but still appreciable, reaching about 10 μM (22). As SlyD is a cytoplasmic protein we only characterized Cu(I) binding to the protein in detail (not Cu(II)) because this is likely the prevalent form of copper present in the reducing cytosolic environment (29). Cu(I) addition to SlyD is accompanied by a charge transfer band in the electronic absorption spectrum centered around 254 nm that reaches saturation at 5 equivalents of metal added (Figure S5). There is precedence for binding of electron rich Cu(I) to the softer thiolate ligands and the 254 nm signal is ascribed to a S → Cu(I) charge transfer band (30, 31). To verify this assignment, a similar titration of a SlyD mutant devoid of all Cys residues (referred to as the triple mutant because all three pairs of cysteines of SlyD have been removed (14)) was conducted. The addition of Cu(I) to the mutant SlyD failed to generate a change in the absorption spectrum (data not shown), suggesting that the LMCT bands are due to Cu(I) binding to SlyD via thiolate residues.
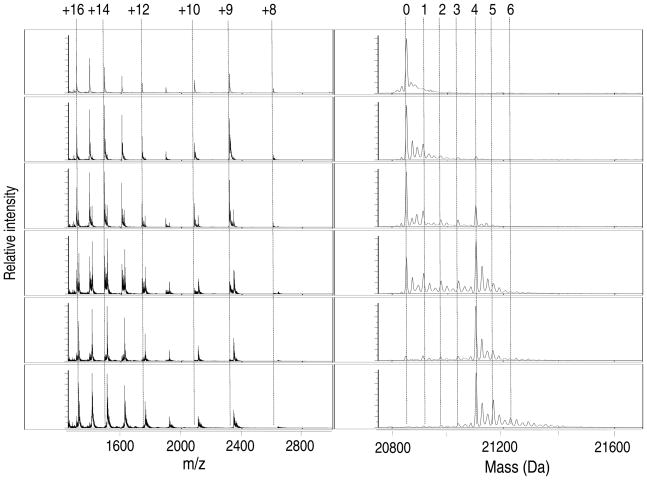
To gain an understanding of the Cu(I)-binding mechanism of SlyD, copper titrations were analyzed via ESI-MS. Although all possible precautions were taken to ensure anaerobic conditions, the addition of Cu(I) resulted in the appearance of protein dimers that were accompanied by a decrease in the protein mass by 6 Da (data not shown), suggesting oxidation of the protein. Titration of SlyD with Cu(II) followed by MS analysis revealed similar results, so further analysis with Cu(II) was not performed. Instead, copper titrations of SlyD were re-evaluated in the presence of 100 μM DTT. Inclusion of the reducing agent did indeed prevent the formation of protein dimers and the accompanying loss of mass (Figure 4 and data not shown), but it significantly reduced the ion signal detected, resulting in spectra with poor resolution (data not shown). Therefore, to improve the quality of the spectra, Cu(I) titrations containing DTT in the buffer were acquired with the ion signal enhanced at a m/z of 2500 (Figure 4). Upon titration with less than 4 molar equivalents of Cu(I), the predominant species observed correspond to apo SlyD, a small amount of SlyD with a single copper ion bound, and SlyD with 4 Cu(I) ions bound. Assuming that all metal-SlyD species are ionized to the same extent, this metallation pattern implies that binding of a single Cu(I) ion to the protein facilitates the binding of the 2nd, 3rd and the 4th Cu(I) ion through a partially cooperative mechanism, unlike Ni(II) and Zn(II). However, it is not possible to rule out this pattern of metallation is due to the decreased stability of the lower metallated Cu(I) species (i.e. < 4 Cu(I)) in the MS. The addition of 6 molar equivalents of Cu(I) results in the appearance of metalloforms with more than 4 Cu(I) ions bound, but a definitive saturation point could not be obtained through this direct titration method as the higher concentrations of salt interfere with the quality of the MS data. It should be noted that significant changes in the population of charge states are not detected when Cu(I) is added. This observation is due to the above mentioned enhancement of the signal at m/z 2500, because Ni(II) titrations analyzed under these instrument settings did not reveal the expected shift in the m/z spectra (Figure S2 and data not shown).
Figure 4.
ESI-MS of titration of SlyD with Cu(I) in the presence of 100 μM DTT. The addition of increasing amounts of Cu(I) to SlyD (10 μM) leads to the appearance of several Cu(I)-SlyD species. From top to bottom the equivalents of Cu(I) added relative to protein concentrations are: 0, 0.3, 0.8, 2.3, 3.9, and 6. The m/z spectra are shown on the left with the respective reconstructed spectra on the right. The numbers heading the dotted lines indicate the number of Cu(I) ions bound to SlyD (right panels) and the charge states of the protein (left panels). The most intense peaks correspond to apo, 1 Cu(I) and 4 Cu(I)-bound SlyD species (at Cu(I) equivalents < 4).
Given that ESI-MS could not be used to determine the Cu(I) stoichiometry, coupled with the fact that Cu(I) binding to SlyD was analyzed in the presence of DTT, which has a considerable affinity for Cu(I) (32), the stoichiometry of the metal complex was re-evaluated by using the small molecule chelator Bca. This copper probe forms an air-stable [Cu(Bca)2]3− complex with a formation constant of β2 = 1017.2 and gives rise to a charge transfer band in the visible region with a maximum at 562 nm (ε = 7900 M−1 cm−1) (18). Titration of apo wild-type protein into a solution of 40 μM Cu(I) and 100 μM Bca (or 300 μM, data not shown) resulted in a linear decrease in the signal at 562 nm, indicating non-competitive conditions (Figure S6 inset). A linear fit of the data reveals that 10 μM SlyD is sufficient to remove the 40 μM Cu(I) bound to Bca, thereby confirming that the protein binds 4 equivalents of Cu(I) (Figure S6). To determine the KCu(I) of SlyD, competitive conditions were achieved by using the Bcs chelator. Similar to Bca, Bcs also coordinates Cu(I) to form an air-stable [Cu(Bcs)2]3−, albeit with a higher formation constant (β2 = 1019.8), which can be detected by a charge transfer band with a maximum at 483 nm (ε = 13000 M−1cm−1) (18). Titrations of protein into a solution containing constant concentrations of Bcs and copper at 300 μM and 40 μM, respectively (Figure S7), were fitted assuming a single binding scheme to obtain an apparent KD of 1.5 ± 0.3 × 10−17 M for Cu(I) binding to SlyD.
To evaluate the metal selectivity of SlyD with respect to Cu(I), a second series of experiments were conducted using the weaker copper probe, Bca. To establish whether the reaction described in equation (A) can occur, 10 μM SlyD was first incubated with 50 μM Cu(I) for 1 h, followed by addition of 200 μM Bca. Then 80 μM of Ni(II) or Zn(II) was added and allowed to equilibrate for 24 h.
| (A) |
As expected, the addition of Bca to the SlyD sample leads to an increase in the absorption at 562 nm corresponding to a concentration of 10 μM [CuI(Bca)2]3− (data not shown). However, neither excess Ni(II) nor Zn (II) affected the signal at 562 nm, demonstrating that the forward reaction in (A) did not occur under these experimental conditions (data not shown). Control experiments were conducted with Bca and Ni(II) or Zn(II) alone to ensure that these metals do not alter the spectrum of Bca. To determine if the reverse reaction could be achieved, a 10 μM protein sample was pre-incubated with 8 equivalents of Ni(II) or Zn(II) for 24 h, followed by addition of 5 equivalents of Cu(I) and 200 μM Bca and equilibration for an additional 24 h. The resulting signal at 562 nm corresponded to only 10 μM [CuI(Bca)2]3− suggesting that 40 μM Cu(I) was bound by SlyD (data not shown). Therefore, these results support tight binding of 4 Cu(I) ions even in the presence of Ni(II) or Zn(II). This preferential binding of Cu(I) corroborates the binding constants measured for the different metals thus far, where KD(Cu(I)-SlyD) < KD (Ni(II)-SlyD), KD(Zn-SlyD).
Co(II) binding to SlyD
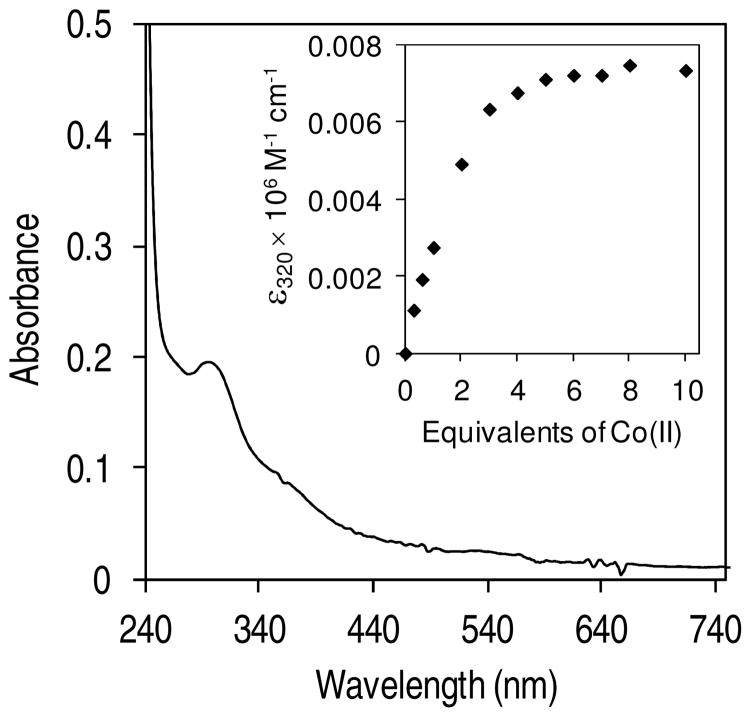
Although cobalt is not widely used by E. coli, this transition metal ion can still be found in low abundance in the cytosol of this bacteria (16). In addition, Co(II) was of particular interest to us because two other proteins involved in nickel homeostasis, RcnR and RcnA, also contribute to cobalt homeostasis in E. coli (33–35). To determine whether SlyD could coordinate Co(II), a direct cobalt titration of apo SlyD was analyzed via electronic absorption spectroscopy (Figure 5). The addition of Co(II) leads to an increase in the absorption at 280 nm accompanied by a broad shoulder in the 300–390 nm region that saturates upon addition of 3–4 equivalents of the metal (Figure 5 inset). The spectrum of the Co(II)-saturated SlyD sample reveals an LMCT band at 293 nm (εc = 7800 M−1 cm−1) and a broader charge transfer band centred around 360 nm (εc = 3418 M−1 cm−1) (the linear increase in absorption upon addition of Co(II) was used to determine the molar absorptivity of the metal-protein complex). These electronic absorption bands are attributed to LMCT from thiolates to the metal centre (33, 36), an assignment supported by the lack of such a CT band when Co(II) is added to the triple mutant (data not shown), and the intensity of these near UV bands suggests that all six Cys residues contribute to cobalt coordination.
Figure 5.
Co(II) binding to SlyD. Difference spectrum generated by subtracting apo SlyD (25 μM) from a protein sample incubated with 7 equivalents of Co(II). Inset: Plot of molar absorptivity at 320 nm vs. equivalents of Co(II) indicates saturation around 3–4 equivalents of Co(II) added.
To evaluate Co(II) binding in more depth, ESI-MS was used. Similar to the copper titrations described above, the addition of cobalt ions led to a concomitant decrease in the mass of the protein by 6 Da and the appearance of dimer species (data not shown), indicating oxidation of the protein. Therefore, titrations were carried out in the presence of 100 μM DTT. Mass spectrometry data revealed that similar to the Ni(II) and Zn(II) titrations, Co(II) binding by the protein occurred through a non-cooperative mechanism that produced mixtures of metallated species at all Co(II) concentrations tested (Figure S8). At a given amount of Co(II), lower stoichiometries of Co(II)-SlyD species were observed for samples containing DTT than samples without DTT, indicating that the reducing agent was competing with SlyD for cobalt ion binding (data not shown). Therefore, the stoichiometry of the protein for Co(II) was established through equilibrium dialysis, which indicated an average of 4 Co(II) ions bound per monomer, a capacity analogous to that of Ni(II) (Table S3).
To determine the affinity of SlyD for Co(II), competition experiments with the fluorescence indicator Fura2 were performed. Although Fura2 was originally designed as a molecular sensor for Ca(II), this chelator has also been used for determining the affinity of several proteins for various transition metals including Co(II) (36, 37). The chelator forms a 1:1 complex with Co(II) that can be detected by the decrease in absorbance at 368 nm. Competitive conditions required for determining the dissociation constant of SlyD for cobalt ions were achieved by titrating apo protein into a constant concentration of [Co(II)-Fura2], leading to an increase in the signal at 368 nm (indicative of a decreasing [Co(II)-Fura2] in solution) (Figure S9). To determine the affinity, data with a fractional saturation between 0.8–0.2 (with respect to the Fura2 complex) from several replicates were compiled to obtain an average KD (SlyD-Co(II)) of 4 ± 1 × 10−9 M. It should be noted that we used the KD of 8.64 × 10−9 M reported for the [Co(II)-Fura2] complex at pH 7.0 (38), because the affinity of this chelator for metal is constant near neutral pH (39).
To establish the metal selectivity of SlyD for Co(II) versus Ni(II), equilibrium dialysis was used. Metal analysis of samples equilibrated with 8-fold amounts of both Co(II) and Ni(II) revealed that the protein was exclusively bound to Ni(II) with an average stoichiometry of 4.4 (Table S3). Given that SlyD binds an average of ~4 Ni(II) ions when equilibrated with Ni(II) alone (Table S3 and (14)) these data indicate that SlyD selectively coordinates Ni(II) to its’ full capacity in the presence of Co(II). Similarly, titration of Zn(II) into a sample of SlyD pre-incubated with 8-fold excess of Co(II) caused the LMCT band at 320 nm to disappear completely at 6 equivalents of Zn(II) added, indicating effective replacement of Co(II) with Zn(II) at metal sites composed of Cys ligands (Figure S10).
Fe(II) binding to SlyD
The high iron quota of E. coli, ~ 0.1 mM (16) and the propensity of this metal to coordinate to Cys residues suggested that Fe(II) binding to SlyD was also likely. However, electronic absorption spectroscopy of SlyD titrated with Fe(II) did not reveal any changes in the spectrum of apo SlyD (data not shown). Given that Fe(II) binding to thiolate ligands is expected to generate LMCT or MLCT bands in the visible or UV regions of the absorption spectrum (40), this result suggests that SlyD does not bind Fe(II) under the experimental conditions used. In support of this conclusion, Ni(II) binding to SlyD was unaffected by pre-incubation with 10 molar equivalents of Fe(II) (Figure S11).
Mn(II) binding to SlyD
Considering the large number of carboxylate and His residues present in SlyD, Mn(II) is another metal ion that could be coordinated by this protein. The addition of Mn(II) to SlyD did not produce any change in the electronic absorption profile of the protein (data not shown), and equilibrium dialysis revealed only a small fraction of the protein bound to the metal (Table S3). Furthermore, the presence of Mn(II) did not affect Ni(II) binding to SlyD monitored either by electronic absorption spectroscopy (data not shown) or metal analysis (Table S3).
Susceptibility of ΔslyD E. coli to external metal concentrations
The metal-binding data described above demonstrate that SlyD can coordinate several different types of metal ions in vitro with considerable affinity, so it was feasible that SlyD plays a role in minimizing the effects of metals in E. coli by functioning as a general metal buffer/detoxifier and a metal-ion sink. To test this hypothesis, the growth rates of wild type and ΔslyD strains in either LB or minimal media supplemented with different metals (Table S1) were examined by monitoring optical density. The amounts of metals used were sufficient to inhibit growth of the bacteria, but no significant differences in the growth rates were observed for the ΔslyD strain compared to wildtype strain at any of the metal concentrations tested (Figure S12 and data not shown) suggesting that SlyD does not confer metal resistance against cobalt, nickel, copper or zinc in E. coli under aerobic growth conditions.
Impact of the slyD deletion on transcriptional metalloregulators
To assess the role of SlyD in metal homeostasis, we examined the transcription of several metal transporters in E. coli by using quantitative PCR (qRT-PCR). The expression of these proteins is tightly regulated by metal-sensing transcription factors that gauge the available concentrations of a particular metal with extreme selectivity (41). Thus, a comparison between the mRNA levels of wild type and ΔslyD strains should enable us to determine if there is an impact of SlyD on a given metal homeostasis pathway. SlyD appears to be constitutively expressed in E. coli in the presence or absence of oxygen (5), a variable that might modulate the metal requirements of the bacteria, so we examined the effects of SlyD under both conditions. Given that SlyD is connected to nickel biochemistry as an accessory protein for Ni(II) delivery during biosynthesis of the [NiFe]-hydrogenase enzymes (5), we hypothesized that SlyD could lead to perturbation of the nickel ion balance in E. coli. Furthermore, because SlyD can bind multiple Zn(II) or Cu(I) ions with affinities equal or greater than that for Ni(II), we also examined the contribution of SlyD in the homeostasis of these metals.
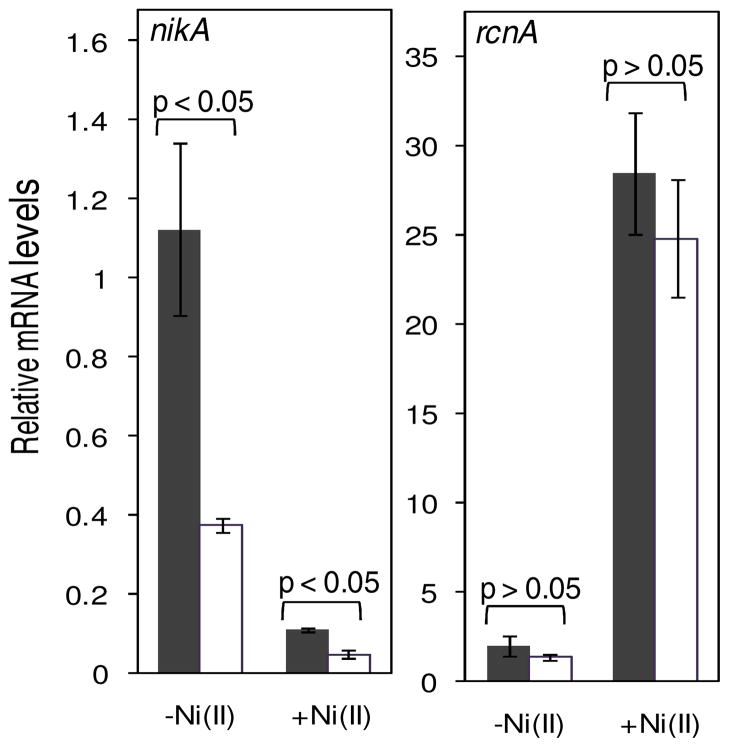
While a specific Ni(II) uptake system is not known to be expressed under aerobic conditions, Ni(II) can still gain access to the cytoplasm in the presence of oxygen through non-specific metal importers such as CorA or other unidentified means (1). Repression of the gene encoding the Ni(II) exporter RcnA by the metalloregulator RcnR is alleviated in response to nickel, an effect observed in the presence or absence of oxygen (42). As expected, the expression of rcnA was induced when aerobically-grown bacteria were treated with 10 μM Ni(II), but we observed no difference in the transcript levels in the ΔslyD strain compared to wild type cells (Figure S13). We next examined the transcript levels of rcnA in E. coli grown anaerobically with or without 10 μM Ni(II). Under these conditions, we also examined the expression levels of the nik operon, which encodes the NikABCDE Ni(II) importer that is up-regulated during anaerobic growth (43), when presumably it is pulling in Ni(II) required for the hydrogenase enzymes, and repressed by the transcription factor NikR in response to Ni(II) (44, 45). While a substantial induction in rcnA expression was observed upon metal supplementation of the growth media, the changes in mRNA levels in the wild type and ΔslyD strains were comparable, indicating that SlyD does not modulate the nickel export pathway (Figure 6). In contrast, without Ni(II) supplementation of the media a ~3 fold decrease in the nikA mRNA levels was detected in the ΔslyD strain when compared to wild type strain (Figure 6). Upon addition of 10 μM Ni(II) a ~10 fold reduction in the expression of nikA was observed for the wild type strain (Figure 6). Ni(II) also caused a decrease in nikA transcription in the ΔslyD strain, such that the levels were about half that of the wild-type cells grown under the same conditions. These diminished nikA transcript levels in the ΔslyD strain indicate an impact of SlyD on the Ni(II) uptake pathway.
Figure 6.
Expression of Ni(II) transporters in response to Ni(II). The mRNA levels from anaerobically-cultured bacteria grown in minimal media optimized for hydrogenase expression without (−Ni(II)) or with 10 μM Ni(II) (+Ni(II)) were measured. The amounts of mRNA observed in wildtype and slyD strains are shown in black and clear bars respectively. The observed transcripts levels are relative to that of the DNA gyrase, holB. The p-value (based on student t-test) of < 0.05 indicates that the values are significantly different and p-values > 0.05 are considered not to be significantly different. Data are an average of three replicates.
To test whether SlyD contributes to nickel homeostasis specifically even in the presence of other metals, nikA and rcnA transcript levels were monitored in anaerobically-grown bacteria treated for 30 min with 10 μM Ni(II) as well as 400 μM Zn(II), a concentration high enough to elicit a response in zinc homeostasis (see below). The amounts of mRNA of the two transporters followed a similar trend to that observed when the bacteria were treated with Ni(II) alone (Figure S14), indicating that SlyD contributes to nickel pathways even in the presence of excess zinc.
To determine whether SlyD can influence zinc homeostasis, the transcription of znuA, which encodes a subunit of the major inducible high-affinity Zn(II) uptake ABC transporter system, and zntA, which encodes a Zn(II) exporter from the P1-type ATPase family (2), was monitored. The expression of these transporters is controlled by the metalloregulators Zur and ZntR, respectively (22). The concentration of Zn(II) used was chosen based on the toxicity experiments, which revealed that 400 μM Zn(II) results in slower growth (as opposed to a lethal concentration leading to no growth). Furthermore, several previous investigations assessing the genome-wide transcriptional response of E. coli to zinc demonstrated that [Zn(II)] >100 μM is sufficient to observe a change in the transcript levels of the above-mentioned transporters, albeit under different media compositions (46, 47). While the addition of Zn(II) led to an increase in zntA transcription and a decrease in znuA transcription, as expected, we observed no significant difference between wild type and ΔslyD strains grown in the presence or absence of oxygen (Figures S13 and S15).
We next tested the involvement of SlyD in copper homeostasis by examining copA and cusF transcripts upon addition of 150 μM Cu(II) to the media in the presence of molecular oxygen. While the copper sensing transcription factor CueR regulates the expression of copA, cusF is under the control of the CusRS system (48). Similar to Zn(II), this particular copper concentration was selected based on our toxicity studies that indicated 150 μM is sufficient to reduce the growth rate of the bacteria. In addition, previous investigations on copper toxicity in E. coli by Outten et al. revealed that CuSO4 above 100 μM is sufficient to cause extreme stress levels (48). At these high copper concentrations the primary copper exporter CopA as well as the CusCFBA system, which exports copper across both membranes, are necessary to render copper tolerance to the bacteria (48). While the expression of both exporters increases in the presence of copper, copA transcripts are induced to a higher level compared to cusF in the presence of oxygen (Figure S13). In contrast, in anaerobically grown bacteria a more dramatic increase in mRNA levels was detected for cusF relative to copA (Figure S15), confirming the differential expression of the two exporters in response to molecular oxygen noted previously (48). However, the mRNA levels were similar in the wildtype and ΔslyD strains, indicating that the lack of SlyD did not lead to perturbation of copper homeostasis in E. coli (Figure S13 and S15).
DISCUSSION AND CONCLUSIONS
In this investigation, the in vitro metal-binding activities of E. coli SlyD and the impact of this protein on other metal homeostasis factors in vivo were examined. Among the metals tested, SlyD is capable of binding Zn(II), Cu(I), and Co(II) in vitro, as well as Ni(II), whereas Mn(II) and Fe(II) do not bind appreciably to isolated SlyD. Based on the in vitro studies, the order of preference of SlyD for metal ions is Mn(II), Fe(II) < Co(II) < Ni(II), Zn(II) ≪ Cu(I). This series resembles the relative affinities observed for small molecule chelators for divalent metals, commonly termed the Irving-Williams series (49). From these results it is evident that the strength of metal binding is dictated by the properties of the metals rather than the protein and that SlyD is able to meet the coordination preferences of many of these metal ions. The fact that the sequence of SlyD does not impose selective metal binding on the protein is not surprising given the number and diversity of potential metal-binding residues in the MBD of SlyD (Figure S1), allowing the protein to coordinate multiple ions of many of the first row transition metals. In addition to the non-discriminating primary structure, the lack of specificity is likely also due to the highly malleable conformation of the MBD, which is substantiated by the fact that the structure of this region cannot be resolved from NMR measurements even upon addition of 1 equivalent of Ni(II) (6).
Given the promiscuous and extensive metal-binding capabilities of SlyD it is reasonable to predict that it would contribute to the distribution of metals other than nickel in vivo. Deletion of slyD did not lead to a significant change in the transcription profiles of the copper or zinc transporters, suggesting that although the protein cannot preferentially coordinate Ni(II) over the other metals in vitro, its activity is specific for Ni(II) in vivo. Furthermore, the results from the RT-PCR and the toxicity studies clearly indicate that although the protein is capable of tightly coordinating multiple ions of several types of transition metals, SlyD does not function as a general metal detoxifier or buffer like some other Ni(II) storage factors such as Hpn or Hpn-like proteins (50, 51). However, it is possible that SlyD may play a back-up role in metal sequestration/management when other metal transporters are severely compromised or overwhelmed, a hypothesis that remains to be tested.
When E. coli were grown in the presence of excess Zn(II) along with Ni(II), the influence of SlyD on the nikA mRNA levels was similar to that observed upon supplementation with Ni(II) alone. Thus, even toxic levels of Zn(II) that activate the cognate metalloregulators do not interfere with the impact of SlyD on nickel homeostasis. This result raises the question as to how SlyD in vivo is able to overcome the preference for Zn(II) over Ni(II). One possible explanation involves metal-induced allostery (2, 41). In this event, the binding of the “correct metal” is exploited to drive changes in the secondary/tertiary/quaternary structure of the protein and/or dynamics (i.e. protein-protein interactions) that ultimately result in a biological function. For example, the coordination preferences of Ni(II) and Zn(II) are sufficiently different such that, even though the charge states observed in the ESI-MS data suggest similar overall changes in structure, it is likely that the two metals result in distinct protein folds, as indicated by the CD spectra. However, it is also clear that the effective intracellular concentrations of metal ions are tightly controlled within a cell. In E. coli, copper homeostasis is regulated by the copper sensor CueR, which has an estimated affinity for copper in the zeptomolar range (10−21 M) (52). Similarly, the E. coli Zn(II) sensors Zur and ZntR bind their cognate metal with KD ~ 10−15 M (22). Such tight metal affinities of the regulators imply that all cellular copper and zinc is tightly bound and buffered within the cytosol. In support of this model, although Zn(II) can disrupt the Ni(II)-binding capacity of SlyD in vitro, the lack of an impact of Zn(II) on the ability of SlyD to function in nickel homeostasis suggests a restricted availability of cytoplasmic zinc even when the bacteria are exposed to toxic zinc levels.
The differential impact of ΔslyD on the transcript levels of the Ni(II) importer nikA and the exporter rcnA provides several insights into the complex nickel homeostasis pathways in E. coli. Under anoxic conditions the Nik transporter is expressed to establish a supply of nickel that meets the demands of the [NiFe]-hydrogenase enzymes (1, 53). Deletion of slyD caused a down-regulation of nikA transcription, with or without Ni(II) supplementation of the media. This result is consistent with the previously reported observation that a ΔslyD strains of E. coli has diminished uptake of 63Ni(II) under anaerobic conditions (5). The lower amounts of nikA imply a reduced Ni(II) capacity in the cytoplasm such that there is more nickel available to activate NikR. Deletion of SlyD would reduce the number of Ni(II)-binding sites and thus the ability of E. coli to sequester the Ni(II) imported by the Nik transporter. In addition, the ΔslyD strain has significantly less hydrogenase maturation (5), which would alleviate the demand for nickel ions. However, whether the decrease in active hydrogenase is a consequence of the lower nikA levels, as opposed to a cause, is not clear. An analogous connection was observed upon treatment of E. coli with formate (53), which activates hydrogenase production and decreases repression of the nik promoter.
In contrast to the affect on NikR activity, deletion of slyD did not affect rcnA expression under any of the conditions investigated, indicating that SlyD does not influence the nickel-exporting pathway or the activity of the rcnA metalloregulator RcnR. In the absence of Ni(II) supplementation, the distinct effects of ΔslyD on the activities of NikR and RcnR could be explained by the higher affinity of NikR for Ni(II) (33, 44), such that it is more responsive than RcnR to changes in low levels of available metal. However, upon Ni(II) supplementation a marked change in rcnA expression is observed, indicating that sufficient Ni(II) enters the cell to cause de-repression by RcnR, but the slyD deletion still has no impact. These results support a model in which nickel is compartmentalized in the E. coli cytoplasm, resulting in two distinct pools of Ni(II) that are maintained by the transcription factors RcnR and NikR (42). Furthermore, it appears that the role of SlyD is constrained to the nickel that is directed towards the hydrogenase biosynthetic pathway. The assignment of SlyD as a dedicated metallochaperone is in agreement with the observation that SlyD does not influence the sensitivity of E. coli to nickel toxicity, suggesting that the Ni(II) capacity of the protein is reserved for the maturation of the hydrogenase enzymes.
Although deletion of the genes for the Nik importer abrogates hydrogenase activity (43), it does not affect the response of NikR to exogenous Ni(II), suggesting that metallochaperones sequester Ni(II) for hydrogenase maturation at the transporter (42, 53). Given that disruption of slyD impacts the activity of NikR as well as hydrogenase maturation, it is likely that its role is not just as a source of nickel for the hydrogenase enzymes. SlyD forms a complex with the hydrogenase accessory protein HypB (5), so it is possible that it contributes to the biosynthetic pathway through this partnership. How it influences the communication between the hydrogenase-directed pool of Ni(II) and management of this pool by NikR will be the subject of future studies.
Similar to copper and zinc, nickel concentrations are thought to be tightly buffered within E. coli (42). Then how is SlyD able to bind Ni(II) in vivo and impact the homeostasis of this metal? NikR has two different Ni(II)-binding sites, with affinities of 10−12 and 10−7 M (1). Ni(II) binding to both sites allows the regulator to bind to its recognition sequence in vitro with a tighter affinity than when only a single site is occupied by Ni(II) (44, 54), suggesting two levels of gene regulation. If one were to follow the above argument presented for Cu(I) and Zn(II) ion regulation then SlyD, which chelates Ni(II) with a ~10−10 M affinity, would not be able to compete with the high-affinity metal sites of NikR. However, it is possible that the protein is able to sequester enough metal for storage and/or transfer before the low-affinity site of NikR is filled and transcription of the nickel importer is completely turned off. On the other hand, if SlyD can interact in some fashion with the Nik transporter there may be no such competition. Ni(II) transfer from the Nik transporter to SlyD, either directly or indirectly, would circumvent the thermodynamically-dictated metal affinities of competing proteins, and ensure a population of protein loaded with nickel through kinetic control.
In conclusion, SlyD is a remarkable protein with a number of sites that will accept a variety of transition metals, but Ni(II) is the target metal in vivo. Utilizing metallochaperones for metal delivery and compartmentalization is a means to overcome the thermodynamic preferences of proteins for metals. Further experiments are currently underway to determine the details of how SlyD contributes to nickel trafficking to the hydrogenase enzyme in E. coli.
Supplementary Material
Table 1.
Affinities of metal binding to SlyD.§
| Metal | KMe (M) |
|---|---|
| Co(II) | 4 ± 1 × 10−9a |
| Ni(II) | ≤ 10−10b |
| Cu(I) | 1.5 ± 0.3 × 10−17c |
| Zn(II) | 1.0 ± 0.1 × 10−10d |
The data were treated by assuming binding of a single metal ion to obtain a macroscopic KD.
Determined via competition with Fura2.
Estimated from competition with PAR, Fura2 and Zn-SlyD.
Determined by competition with Bcs.
Calculated from competition experiments with PAR.
Acknowledgments
We thank Prof. B. J. Andrews (University of Toronto) for her generous support in providing materials and instrumentation for gathering qRT-PCR data. We also thank Prof. A. Böck for the donation of the MC4100 strains and Prof A. Emili for the donation of the BW25113 strains.
ABBREVIATIONS
- Bcs
Bathocuproine sulfonate
- Bca
Bicinchoninate E. coli, Escherichia coli
- ESI-MS
Electrospray ionization mass spectrometry
- EGTA
Ethylene glycol tetraacetic acid
- HSID
Hot source-induced desolvation
- MBD
metal-binding domain
- PAR
4-(2-pyridylazo)resorcinol
- WT
wildtype
Footnotes
This work was supported in part by funding from the Canadian Institutes of Health Research (DBZ), the Natural Sciences and Engineering Research Council (NSERC) of Canada (MJS) and the Canada Research Chairs program. We also thank NSERC for financial support through Graduate Scholarships for DEKS and HK.
Supporting Information Available. Experimental details of the competition titrations with PAR, Bcs and Fura2. Tables S1–S3 containing a summary of metal concentrations used for metal toxicity studies, a list of primers used for qRT-PCR analysis and summary of Mn(II), Co(II) and Ni(II) stoichiometries of SlyD; Figure S2 shows representative spectra of a Ni(II) titrations analyzed via ESI-MS; Figures S3–S4 shows a graphical representation of the PAR competition assay used for determining the average KD (Zn(II)-SlyD) and a comparison of CD spectra for Ni(II) and Zn(II) bound SlyD. Figure S5 is a representative data set for a Cu(I) titration of SlyD analyzed via electronic absorption spectroscopy. Figure S6–S7 are competition titrations with Bca and Bcs used for determining copper stoichiometry and affinity, respectively. Figure S8–S10 are a graphical representation of Co(II) titrations analyzed via ESI-MS, competition titrations with Fura2 for determining average KD (Co(II)-SlyD) and titration of a cobalt saturated SlyD sample with Zn(II). Figure S11 is spectroscopy data obtained for titration of Ni(II) compared with a similar titration carried out in the presence of excess Fe(II). Figure S12 depicts representative growth curves of WT and ΔslyD strains conducted in minimal media supplemented with various amounts of metal. Figure S13 is relative mRNA levels measured for various metal transporters under aerobic conditions. Figure S14–S15 are relative mRNA levels measured for various metal transporters under anaerobic conditions. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Li Y, Zamble DB. Nickel homeostasis and nickel regulation: an overview. Chem Rev. 2009;109:4617–4643. doi: 10.1021/cr900010n. [DOI] [PubMed] [Google Scholar]
- 2.Waldron KJ, Robinson NJ. How do bacterial cells ensure that metalloproteins get the correct metal? Nat Rev Microbiol. 2009;7:25–35. doi: 10.1038/nrmicro2057. [DOI] [PubMed] [Google Scholar]
- 3.Robinson NJ. A more discerning zinc exporter. Nat Chem Biol. 2007;3:692–693. doi: 10.1038/nchembio1107-692. [DOI] [PubMed] [Google Scholar]
- 4.Leach MR, Zamble DB. Metallocenter assembly of the hydrogenase enzymes. Curr Opin Chem Biol. 2007;11:159–165. doi: 10.1016/j.cbpa.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 5.Zhang JW, Butland G, Greenblatt JF, Emili A, Zamble DB. A role for SlyD in the Escherichia coli hydrogenase biosynthetic pathway. J Biol Chem. 2005;280:4360–4366. doi: 10.1074/jbc.M411799200. [DOI] [PubMed] [Google Scholar]
- 6.Martino L, He Y, Hands-Taylor KL, Valentine ER, Kelly G, Giancola C, Conte MR. The interaction of the Escherichia coli protein SlyD with nickel ions illuminates the mechanism of regulation of its peptidyl-prolyl isomerase activity. FEBS J. 2009;276:4529–4544. doi: 10.1111/j.1742-4658.2009.07159.x. [DOI] [PubMed] [Google Scholar]
- 7.Weininger U, Haupt C, Schweimer K, Graubner W, Kovermann M, Bruser T, Scholz C, Schaarschmidt P, Zoldak G, Schmid FX, Balbach J. NMR solution structure of SlyD from Escherichia coli: spatial separation of prolyl isomerase and chaperone function. J Mol Biol. 2009;387:295–305. doi: 10.1016/j.jmb.2009.01.034. [DOI] [PubMed] [Google Scholar]
- 8.Knappe TA, Eckert B, Schaarschmidt P, Scholz C, Schmid FX. Insertion of a chaperone domain converts FKBP12 into a powerful catalyst of protein folding. J Mol Biol. 2007;368:1458–1468. doi: 10.1016/j.jmb.2007.02.097. [DOI] [PubMed] [Google Scholar]
- 9.Scholz C, Eckert B, Hagn F, Schaarschmidt P, Balbach J, Schmid FX. SlyD proteins from different species exhibit high prolyl isomerase and chaperone activities. Biochemistry. 2006;45:20–33. doi: 10.1021/bi051922n. [DOI] [PubMed] [Google Scholar]
- 10.Jakob RP, Zoldak G, Aumuller T, Schmid FX. Chaperone domains convert prolyl isomerases into generic catalysts of protein folding. Proc Natl Acad Sci U S A. 2009;106:20282–20287. doi: 10.1073/pnas.0909544106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zoldak G, Schmid FX. Cooperation of the prolyl isomerase and chaperone activities of the protein folding catalyst SlyD. J Mol Biol. 2011;406:176–194. doi: 10.1016/j.jmb.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 12.Erdmann F, Fischer G. The nickel-regulated peptidyl prolyl cis/trans isomerase SlyD. In: Sigel A, Sigel H, Sigel RKO, editors. Metal Ions in Life Sciences. John Wiley and Sons; 2007. pp. 501–518. [Google Scholar]
- 13.Leach MR, Zhang JW, Zamble DB. The role of complex formation between the Escherichia coli hydrogenase accessory factors HypB and SlyD. J Biol Chem. 2007;282:16177–16186. doi: 10.1074/jbc.M610834200. [DOI] [PubMed] [Google Scholar]
- 14.Kaluarachchi H, Sutherland DE, Young A, Pickering IJ, Stillman MJ, Zamble DB. The Ni(II)-Binding Properties of the Metallochaperone SlyD. J Am Chem Soc. 2009;131:18489–18500. doi: 10.1021/ja9081765. [DOI] [PubMed] [Google Scholar]
- 15.Wulfing C, Lombardero J, Pluckthun A. An Escherichia coli protein consisting of a domain homologous to FK506-binding proteins (FKBP) and a new metal binding motif. J Biol Chem. 1994;269:2895–2901. [PubMed] [Google Scholar]
- 16.Finney LA, O’Halloran TV. Transition metal speciation in the cell: Insights from the chemistry of metal ion receptors. Science. 2003;300:931–936. doi: 10.1126/science.1085049. [DOI] [PubMed] [Google Scholar]
- 17.Salgado MT, Bacher KL, Stillman MJ. Probing the structural change in the alpha and beta domains of copper- and silver-substituted metallothionein by emission spectroscopy and electrospray ionization mass spectrometry. J Biol Inorg Chem. 2007;12:294–312. doi: 10.1007/s00775-006-0187-5. [DOI] [PubMed] [Google Scholar]
- 18.Zimmermann M, Clarke O, Gulbis JM, Keizer DW, Jarvis RS, Cobbett CS, Hinds MG, Xiao ZG, Wedd AG. Metal Binding Affinities of Arabidopsis Zinc and Copper Transporters: Selectivities Match the Relative, but Not the Absolute, Affinities of their Amino-Terminal Domains. Biochemistry. 2009;48:11640–11654. doi: 10.1021/bi901573b. [DOI] [PubMed] [Google Scholar]
- 19.Atanassova A, Lam R, Zamble DB. A high-performance liquid chromatography method for determining transition metal content in proteins. Anal Biochem. 2004;335:103–111. doi: 10.1016/j.ab.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 20.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graham AI, Hunt S, Stokes SL, Bramall N, Bunch J, Cox AG, McLeod CW, Poole RK. Severe Zinc Depletion of Escherichia coli ROLES FOR HIGH AFFINITY ZINC BINDING BY ZinT, ZINC TRANSPORT AND ZINC-INDEPENDENT PROTEINS. J Biol Chem. 2009;284:18377–18389. doi: 10.1074/jbc.M109.001503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Outten CE, O’Halloran TV. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 23.Ngu TT, Stillman MJ. Metal-binding mechanisms in metallothioneins. Dalton Trans. 2009:5425–5433. doi: 10.1039/b902008j. [DOI] [PubMed] [Google Scholar]
- 24.Kaltashov IA, Abzalimov RR. Do ionic charges in ESI MS provide useful information on macromolecular structure? J Am Soc Mass Spectrom. 2008;19:1239–1246. doi: 10.1016/j.jasms.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka M, Funahash S, Shirai K. Kinetics of Ligand Substitution Reaction of Zinc(2)-4-(2-Pyridylazo)Resorcinol Complex with (Ethylene Glycol)Bis(2-Aminoethyl Ether)-N,N,N′,N′-Tetraacetic Acid. Inorg Chem. 1968;7:573–578. [Google Scholar]
- 26.McNulty TJ, Taylor CW. Extracellular heavy-metal ions stimulate Ca2+ mobilization in hepatocytes. Biochem J. 1999;339:555–561. [PMC free article] [PubMed] [Google Scholar]
- 27.Hottenrott S, Schumann T, Pluckthun A, Fischer G, Rahfeld JU. The Escherichia coli SlyD is a metal ion-regulated peptidyl-prolyl cis/trans-isomerase. J Biol Chem. 1997;272:15697–15701. doi: 10.1074/jbc.272.25.15697. [DOI] [PubMed] [Google Scholar]
- 28.Perczel A, Hollosi M, Fasman GD. Circular Dichroism and the Conformational Analysis of Biomolecules. 1996. [Google Scholar]
- 29.Rensing C, Grass G. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol Rev. 2003;27:197–213. doi: 10.1016/S0168-6445(03)00049-4. [DOI] [PubMed] [Google Scholar]
- 30.Cobine PA, George GN, Jones CE, Wickramasinghe WA, Solioz M, Dameron CT. Copper transfer from the Cu(I) chaperone, CopZ, to the repressor, Zn(II)CopY: Metal coordination environments and protein interactions. Biochemistry. 2002;41:5822–5829. doi: 10.1021/bi025515c. [DOI] [PubMed] [Google Scholar]
- 31.Badarau A, Firbank SJ, McCarthy AA, Banfield MJ, Dennison C. Visualizing the Metal-Binding Versatility of Copper Trafficking Sites. Biochemistry. 2010;49:7798–7810. doi: 10.1021/bi101064w. [DOI] [PubMed] [Google Scholar]
- 32.Xiao Z, Brose J, Schimo S, Ackland SM, La Fontaine S, Wedd AG. Unification of the Copper(I) Binding Affinities of the Metallo-chaperones Atx1, Atox1, and Related Proteins: DETECTION PROBES AND AFFINITY STANDARDS. J Biol Chem. 2011;286:11047–11055. doi: 10.1074/jbc.M110.213074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwig JS, Leitch S, Herbst RW, Maroney MJ, Chivers PT. Ni(II) and Co(II) sensing by Escherichia coli RcnR. J Am Chem Soc. 2008;130:7592–7606. doi: 10.1021/ja710067d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koch D, Nies DH, Grass G. The RcnRA (YohLM) system of Escherichia coli: a connection between nickel, cobalt and iron homeostasis. Biometals. 2007;20:759–771. doi: 10.1007/s10534-006-9039-6. [DOI] [PubMed] [Google Scholar]
- 35.Rodrigue A, Effantin G, Mandrand-Berthelot MA. Identification of rcnA (yohM), a nickel and cobalt resistance gene in Escherichia coli. J Bacteriol. 2005;187:2912–2916. doi: 10.1128/JB.187.8.2912-2916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang SC, Dias AV, Bloom SL, Zamble DB. Selectivity of metal binding and metal-induced stability of Escherichia coli NikR. Biochemistry. 2004;43:10018–10028. doi: 10.1021/bi049405c. [DOI] [PubMed] [Google Scholar]
- 37.Golynskiy MV, Gunderson WA, Hendrich MP, Cohen SM. Metal binding studies and EPR spectroscopy of the manganese transport regulator MntR. Biochemistry. 2006;45:15359–15372. doi: 10.1021/bi0607406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwan CY, Putney JW. Uptake and Intracellular Sequestration of Divalent-Cations in Resting and Methacholine-Stimulated Mouse Lacrimal Acinar-Cells - Dissociation by Sr-2+ and Ba-2+ of Agonist-Stimulated Divalent-Cation Entry from the Refilling of the Agonist-Sensitive Intracellular Pool. J Biol Chem. 1990;265:678–684. [PubMed] [Google Scholar]
- 39.Grynkiewicz G, Poenie M, Tsien RY. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- 40.Morleo A, Bonomi F, Iametti S, Huang VW, Kurtz DM., Jr Iron–nucleated folding of a metalloprotein in high urea: resolution of metal binding and protein folding events. Biochemistry. 2010;49:6627–6634. doi: 10.1021/bi100630t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giedroc DP, Arunkumar AI. Metal sensor proteins: nature’s metalloregulated allosteric switches. Dalton Trans. 2007;29:3107–3120. doi: 10.1039/b706769k. [DOI] [PubMed] [Google Scholar]
- 42.Iwig JS, Rowe JL, Chivers PT. Nickel homeostasis in Escherichia coli - the rcnR-rcnA efflux pathway and its linkage to NikR function. Mol Microbiol. 2006;62:252–262. doi: 10.1111/j.1365-2958.2006.05369.x. [DOI] [PubMed] [Google Scholar]
- 43.Wu LF, Mandrand-Berthelot MA, Waugh R, Edmonds CJ, Holt SE, Boxer DH. Nickel deficiency gives rise to the defective hydrogenase phenotype of hydC and fnr mutants in Escherichia coli. Mol Microbiol. 1989;3:1709–1718. doi: 10.1111/j.1365-2958.1989.tb00156.x. [DOI] [PubMed] [Google Scholar]
- 44.Chivers PT, Sauer RT. Regulation of high affinity nickel uptake in bacteria. Ni2+-Dependent interaction of NikR with wild-type and mutant operator sites. J Biol Chem. 2000;275:19735–19741. doi: 10.1074/jbc.M002232200. [DOI] [PubMed] [Google Scholar]
- 45.De Pina K, Desjardin V, Mandrand-Berthelot MA, Giordano G, Wu LF. Isolation and characterization of the nikR gene encoding a nickel-responsive regulator in Escherichia coli. J Bacteriol. 1999;181:670–674. doi: 10.1128/jb.181.2.670-674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee LJ, Barrett JA, Poole RK. Genome-wide transcriptional response of chemostat-cultured Escherichia coli to zinc. J Bacteriol. 2005;187:1124–1134. doi: 10.1128/JB.187.3.1124-1134.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamamoto K, Ishihama A. Transcriptional response of Escherichia coli to external zinc. J Bacteriol. 2005;187:6333–6340. doi: 10.1128/JB.187.18.6333-6340.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Outten FW, Huffman DL, Hale JA, O’Halloran TV. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J Biol Chem. 2001;276:30670–30677. doi: 10.1074/jbc.M104122200. [DOI] [PubMed] [Google Scholar]
- 49.Irving H, Williams RJP. Order of Stability of Metal Complexes. Nature. 1948;162:746–747. [Google Scholar]
- 50.Ge R, Zhang Y, Sun X, Watt RM, He QY, Huang JD, Wilcox DE, Sun H. Thermodynamic and kinetic aspects of metal binding to the histidine-rich protein, Hpn. J Am Chem Soc. 2006;128:11330–11331. doi: 10.1021/ja062589t. [DOI] [PubMed] [Google Scholar]
- 51.Zeng YB, Yang N, Sun H. Metal-binding properties of an hpn-like histidine-rich protein. Chemistry. 2011;17:5852–5860. doi: 10.1002/chem.201100279. [DOI] [PubMed] [Google Scholar]
- 52.Changela A, Chen K, Xue Y, Holschen J, Outten CE, O’Halloran TV, Mondragon A. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science. 2003;301:1383–1387. doi: 10.1126/science.1085950. [DOI] [PubMed] [Google Scholar]
- 53.Rowe JL, Starnes GL, Chivers PT. Complex transcriptional control links NikABCDE-dependent nickel transport with hydrogenase expression in Escherichia coli. J Bacteriol. 2005;187:6317–6323. doi: 10.1128/JB.187.18.6317-6323.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bloom SL, Zamble DB. Metal-selective DNA-binding response of Escherichia coli NikR. Biochemistry. 2004;43:10029–10038. doi: 10.1021/bi049404k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.