Abstract
Exposure of Saccharomyces cerevisiae to alkaline pH provokes a stress condition that generates a compensatory reaction. In the present study we examined a possible role for the PKA (protein kinase A) pathway in this response. Phenotypic analysis revealed that mutations that activate the PKA pathway (ira1 ira2, bcy1) tend to cause sensitivity to alkaline pH, whereas its deactivation enhances tolerance to this stress. We observed that alkalinization causes a transient decrease in cAMP, the main regulator of the pathway. Alkaline pH causes rapid nuclear localization of the PKA-regulated Msn2 transcription factor which, together with Msn4, mediates a general stress response by binding with STRE (stress response element) sequences in many promoters. Consequently, a synthetic STRE–LacZ reporter shows a rapid induction in response to alkaline stress. A msn2 msn4 mutant is sensitive to alkaline pH, and transcriptomic analysis reveals that after 10 min of alkaline stress, the expression of many induced genes (47%) depends, at least in part, on the presence of Msn2 and Msn4. Taken together, these results demonstrate that inhibition of the PKA pathway by alkaline pH represents a substantial part of the adaptive response to this kind of stress and that this response involves Msn2/Msn4-mediated genome expression remodelling. However, the relevance of attenuation of PKA in high pH tolerance is probably not restricted to regulation of Msn2 function.
Keywords: alkaline stress, gene expression, Msn2, Msn4, protein kinase A (PKA), Saccharomyces cerevisiae, transcription factor
Abbreviations: CDRE, calcineurin-dependent response element; Cy3, indocarbocyanine; Cy5, indodicarbocyanine; GAP, GTPase activating proteins; GEF, guanine-nucleotide-exchange factor; GFP, green fluorescent protein; GO, Gene Ontology; PKA, protein kinase A; STRE, stress response element; TOR, target of rapamycin
INTRODUCTION
The regulation of the activity of the cAMP/PKA (protein kinase A) pathway plays a major role in the control of metabolism and cell proliferation in yeast cells, linked primarily to the available carbon source. In Saccharomyces cerevisiae, for instance, in response to a rapidly fermentable carbon source such as glucose the pathway activates the Cyr1 adenylate cyclase, which results in a transient increase in cAMP levels. PKA is a heterotetramer composed of two catalytic subunits and two regulatory subunits. The catalytic subunits can be encoded by three largely redundant genes (TPK1, TPK2 and TPK3), whereas the regulatory subunits are encoded by a single gene (BCY1). Binding of a second messenger cAMP to the regulatory subunits results in dissociation of the complex and activation of PKA. Restoration of cAMP levels is controlled by the low- and high-affinity phosphodiesterases encoded by PDE1 and PDE2 respectively, which hydrolyse cAMP to AMP. Sequentially, PKA affects diverse downstream targets often at the gene transcription level, including the stimulation of cell growth and cell cycle progression, up-regulation of glycolysis, down-regulation of gluconeogenesis, and mobilization of glycogen and trehalose [1–5].
PKA can be activated in response to glucose by two parallel signalling pathways. The first involves the Ras1 and Ras2 small GTPases, which are activated by glucose uptake and phosphorylation. The active (GTP-bound) Ras proteins increase the activity of the adenylate cyclase. In turn, the GDP/GTP exchange on the Ras proteins is controlled by the GEFs (guanine-nucleotide-exchange factors) Cdc25 and Sdc25 (S. cerevisiae homologue of Cdc25). The reverse process is accelerated by the Ira proteins (encoded by the IRA1 and IRA2 genes), which act as Ras GAPs (GTPase-activating proteins) and maintain Ras in the GDP-bound inactive state. The second pathway involves Gpr1 (a putative G-protein-coupled receptor) and its Gα protein Gpa2. Both pathways converge to activate adenylate cyclase, resulting in the generation of cAMP [1–3].
Activation of PKA has a major impact on gene expression. Consequently, several transcription factors are among the known PKA targets. Two of those are Msn2 and Msn4, which mediate the transcription of the so-called STRE (stress response element)-controlled genes [6–8]. STRE-regulated genes are involved in important processes such as carbohydrate metabolism and growth regulation, as well as in adaption to diverse types of stress, including heat, DNA damage, and oxidative and osmotic stresses [9–12]. Under growth-promoting conditions (growth on glucose with the absence of stress), Msn2 and Msn4 are phosphorylated and reside in the cytosol. Upon glucose exhaustion or other stress conditions [13], they become hypophosphorylated and translocate to the nucleus, where they induce expression of the STRE-controlled genes. PKA plays a very important role inhibiting nuclear import of Msn2/Msn4, either through direct phosphorylation of their nuclear localization signal [13–15] or indirectly via the protein kinases Yak1 and Rim15.
S. cerevisiae grows far better at acidic than neutral or alkaline pH and consequently even a modest alkalinization of the medium represents a stress situation that is able to trigger a compensatory multifactorial response (for a review see [16]). Alkaline stress activates diverse signalling pathways, including the Rim101/Nrg1 [17,18], the calcium/calcineurin [19–21] and the Wsc1/Pkc1/Slt2 MAPK (mitogen-activated protein kinase) pathways [22]. Alkalinization of the environment also disturbs nutrient homoeostasis, as deduced from its impact on iron/copper and phosphate uptake/utilization pathways [19,23]. Work in our laboratory in previous years has shown that alkaline pH stress also has a profound impact on the expression of genes encoding glucose uptake and metabolism-related proteins, finding that exposure to high pH would mimic a situation of glucose starvation [20,21]. Given the strong link between carbohydrate metabolism and the PKA pathway, we speculated that alkaline stress might involve changes in the activity of this pathway and specifically that the correct adaptation to high pH could entail its down-regulation. Indeed, in the present paper we showed that alkaline pH stress causes a transient decrease in cAMP levels and that change in the activity of the PKA pathway alters tolerance to alkaline pH. The results of the present paper also indicate that the adaptive response to high pH involves PKA-regulated Msn2/Msn4-mediated gene remodelling.
MATERIAL AND METHODS
Yeast strains and growth conditions
Yeast cells (the strains are listed in Table 1) were grown at 28 °C in YPD medium (10 g/l yeast extract, 20 g/l peptone and 20 g/l dextrose) or when carrying plasmids in synthetic complete medium [24] containing 2% glucose and lacking the appropriate selection requirements. Single kanMX deletion mutants on the BY4741 background, except MAR231, were generated in the context of the Saccharomyces Genome Deletion Project [25].
Table 1. Yeast strains used.
Except otherwise indicated, single BY4741-derived kanMX disruptans that are not listed here correspond to the systematic gene disruption project [25].
| Name | Relevant genotype | Source/reference |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ met15Δ ura3Δ | [25] |
| MAR231 | BY4741 bcy1::kanMX4 | The present paper |
| W303-1A | MATa ade2-1, can1-100, his3-112, leu2-3, trp1-1, ura3-1 | [52] |
| SC7 | W303-1A ira1::LEU2 | [42] |
| SC8 | W303-1A ira2::URA3 | [42] |
| PM903 | W303-1A ira1::LEU2 ira2::URA3 | [42] |
| WΔN1 | W303-1A cdc25::CDC25 aa907-1589/URA3 | [53] |
| WΔN2 | W303-1A cdc25::CDC25 aa1147-1589/URA3 | [53] |
| CCV35 | W303-1A pde1::kanMX4 | The present paper |
| PM942 | W303-1A pde2::URA3 | [27] |
| CCV36 | W303-1A pde1::kanMX4 pde2::URA3 | The present paper |
| MCY5278 | W303-1A msn2::kanMX4 msn4::hphMX4 | M. Carlson |
| CCV174 | W303-1A nrg1::nat1 | The present paper |
| CCV175 | W303-1A msn2::kanMX4 msn4::hphMX4 nrg1::nat1 | The present paper |
| tpk123 msn2 msn4 | W303-1A tpk1::URA3 tpk2::HIS3 tpk3::TRP1 msn2::HIS3 msn4::TRP1 | [14] |
| CCV37 | W303-1A snf1::LEU2 | The present paper |
| CCV38 | W303-1A msn2::kanMX4 msn4::hphMX4 snf1::LEU2 | The present paper |
| BY4742 | MATα his3Δ1 leu2Δ met15Δ ura3Δ | [25] |
| DC90 | BY4742 tpk1::kanMX::HIS3 tpk2w3 tpk3::LEU2 | J.M. Thevelein |
| DBY746 | MATa ura3-52 leu2-3112 his3-1 trp1-239 | D. Botstein |
| AGS66 | DBY746 URA3-STRE(7×)–lacZ | [29] |
The MAR231 strain was made by transforming BY4741 with a bcy1::kanMX4 cassette obtained from the KKY385 strain [26] by PCR amplification using the primers 5′-bcy1_disr (GAGGAGCATACGACTTCGGC) and 3′-bcy1_disr (CTGTCTTGTAGATCCTTTGG). The CCV35 and CCV36 strains were constructed as follows. A 1.6 kbp pde1::kanMX4 cassette was amplified from genomic DNA of the BY4741 pde1::kanMX4 strain with the oligonucleotides 5′-pde1 (CAAGGATCGTTACCCGGTA) and 3′-pde1 (GACTTATGTTGGGATAGGGG). The purified DNA fragment was used to transform the W303 or PM942 strains [27], to yield the CCV35 and CCV36 strains respectively. The CCV37 and CCV38 strains were obtained transforming W303-1A wild-type and MCY5278 strains with a SNF1::LEU2 disruption cassette [21] and the CCV174 and CCV175 strains were generated by transforming W303-1A and MCY5278 strains with the 2.1 kbp nrg1::nat1 cassette from the plasmid pBS-nrg1::nat1 as described previously [28]. The AGS66 strain contained an integrated STRE(7×)–lacZ reporter system at the URA3 locus [29].
Plasmids
The reporter plasmid pHXK1-lacZ was generated as follows. The HXK1 upstream DNA region containing −624 and +39 relative to the starting ATG was amplified by PCR with added BamHI/PstI restriction sites and cloned into the same sites of YEp357 [30]. The plasmid YCp50-RAS2Ala18Val19 [31] expressed the RAS2Ala18Val19 hyperactive Ras2 allele [32] from the centromeric YCp50. The plasmid pG2CT-112.2 expressed the constitutively active GPA2R273A allele from the episomal YEplac112 backbone [31]. The plasmid pAMS366 contains a tandem of four CDREs (calcineurin-dependent response elements) from the GSC2/FKS2 gene fused to the lacZ reporter [33]. pPHO84-LacZ contains a PHO84-LacZ reporter fusion as described in [19]. pKC201 contains the ENA1 promoter fused to LacZ [34,35].
Growth tests
The sensitivity of different yeast strains to alkaline pH was assayed by drop test on YPD plates containing 50 mM Taps adjusted with KOH at different pH values. Growth in liquid medium at high pH was performed in 5 ml cultures or in 96-well plates (250 μl), at 28 °C in YPD medium buffered with 50 mM Taps and adjusted with KOH at the pH values indicated below. Growth was monitored by measuring the absorbance (A) at 660 nm. BY4742 and DC90 cells at an initial A660 of 0.001 were grown for 17 and 24 h respectively. The strains prepared from the W303-1A genetic background were grown from an initial A660 of 0.01 for 17 h (wild-type and MCY5278 strains) and for 21 h (CCV37 and CCV38 strains).
Measurement of cAMP levels
Measurements were made as described in [36]. Briefly, BY4741 cells were grown to an A660 value of approximately 1.0 in YPD at 28 °C, and then the culture was centrifuged (5 min at 1220 g at room temperature) and resuspended in YPD containing 50 mM Taps (pH 8). Aliquots of 10 ml were filtered at the indicated times and extracted in the cold with 1 ml of 2 M perchloric acid. After neutralization with 1 ml of 1.8 M KOH and 0.4 M KHCO3 and centrifugation (5 min at 1220 g at 4 °C), the samples were purified with Amprep SAX minicolumns (Amersham), eluted with methanol/HCl and dried under vacuum. The assays were performed by a competitive binding method with the Amersham cAMP EIA (enzyme immunoassay) system. For calculation of yeast concentrations, one unit of absorbance at 660 nm was equivalent to 2.6 mg of wet weight/ml. In other experiments, the pH was raised to 8.2 by the addition of KOH (35 mM, final concentration). The control cells received the same concentration of KCl.
β-Galactosidase activity assay
Yeast cells were grown to saturation in the appropriate dropout medium and then inoculated into YPD medium (or YP plus 4% glucose, where indicated) at pH 5.5. Growth was resumed until A660 was 0.5–0.7 and the cultures were centrifuged at room temperature for 5 min at 1620 g. The cells were resuspended in YPD or YP 4% glucose where indicated (no induction) or YPD (or YPD 4% glucose) plus 50 mM Taps adjusted to pH 8.0 (alkaline stress), and growth was resumed for the indicated times. In all cases, β-galactosidase activity was measured as described previously [37].
Microscopy techniques
For the Msn2 subcellular localization experiments, the indicated strains were transformed with the plasmid pMSN2-GFP (given by Professor Francisco Estruch, University of Valencia, Valencia, Spain), a YCplac111-based vector that contains a C-terminal Msn2–GFP (green fluorescent protein) fusion [13]. The cells were grown in YPD or YP 2% ethanol until an A660 value of 0.8–1.0 was reached. The cultures (5 ml) were treated by either the addition of 100 μl of 1 M KCl (control cells, pH 5.5) or of 100 μl of 1 M KOH (alkaline stress, pH 8.0). Samples (500 μl) were taken at the appropriate times and fixed for 5 min by adding 30 μl of 37% formaldehyde. The cells were harvested, washed three times with PBS and concentrated 10-fold before visualization. In all cases the cells were visualized with a fluorescein filter using a Nikon Eclipse E800 fluorescence microscope (magnification, 1000×). Digital images were captured with an ORCA-ER 4742-80 camera (Hamamatsu) using Wasabi software. Intracellular distribution of Msn2–GFP was quantified by scoring at least in 200 cells per sample into one of three possible categories: cytoplasmic (fluorescence in the cytoplasm only), nuclear/cytoplasmic (fluorescence in the cytoplasm and nucleus) and nuclear (fluorescence in the nucleus only).
RNA purification, cDNA synthesis and DNA microarray experiments
For RNA purification, 50 ml of yeast culture (strains W303-1A and MCY5278) was grown at 28 °C in YPD medium until an A660 value of 0.6–0.8 was reached, and then KOH or KCl was added from a concentrated stock solution (1 M) to reach a final concentration of 20 mM (pH 8.05 and pH 5.5 respectively). Yeast cells were harvested by filtration after 10 and 30 min and washed with ice-cold water; dried cells were kept at −80 °C until RNA purification was performed. Total RNA was purified using a RiboPure-Yeast kit (Ambion) following the manufacturer's instructions. RNA quality was assessed by denaturing 0.8% agarose gel electrophoresis, and RNA quantification was performed by measuring absorbance at 260 nm in a BioPhotometer (Eppendorf). Transcriptional analyses were performed using DNA microarrays (printed at the Universitat Autònoma de Barcelona Genomics Facility) containing PCR-amplified fragments from 6014 S. cerevisiae open reading frames [20,38]. Fluorescent Cy3- and Cy5-labelled cDNA probes were prepared from 8 μg of purified total RNA by the indirect dUTP labelling method using a CyScribe post-labelling kit (Amersham Biosciences).
Pre-hybridization, hybridization and washes were carried out as recommended by The Institute for Genomic Research with minor modifications. Briefly, prehybridizations of the DNA microarrays were carried out at 42 °C for 1 h in a solution containing 5× SSC (1× SSC is 0.15 M NaCl/0.015 M sodium citrate), 0.1% SDS and 1% BSA. For hybridization, dried Cy3 (indocarbocyanine)- and Cy5 (indodicarbocyanine)-labelled probes were resuspended in 35 μl of hybridization solution (50% formamide, 5× SSC and 0.1% SDS) each and mixed. Salmon sperm DNA (5 μg) was added to the mix before denaturation for 3 min at 95 °C. DNA microarrays were hybridized in an ArrayBooster hybridization station (Sunergia Group) for 14 h at 42 °C. The scanner ScanArray 4000 (Packard Instrument) was used to obtain the Cy3 and Cy5 images with a resolution of 10 μm. The fluorescent intensity of the spots was measured and processed using the GenePix Pro 6.0 software (Molecular Devices). Spots with either a diameter smaller than 120 μm or fluorescence intensity for Cy3 and Cy5 lower than 150 units were not considered for further analysis.
For each condition assayed, two independent experiments were performed and dye swapping was carried out for each experiment. Microarray data was deposited at the GEO (Gene Expression Omnibus; accession number GSE27925). The results from different experiments were combined, and the mean was calculated. A given gene was considered to be induced or repressed when the mean of the ratios (alkaline stress compared with no stress) was >2.0 or <0.50 respectively. Software from the GEPAS server (http://gepas.bioinfo.cipf.es/) was used to carry out clustering and other data analyses [39]. According to the expression of these genes in the msn2 msn4 strain, different levels of dependence on these transcription factors were defined. Thus genes showing a mutant/wild-type ratio of 0.67>X>0.50 were considered ‘weakly dependent’, those with a ratio 0.50>X>0.25 were ranked as ‘strongly dependent’ (SD) and those with a ratio ≤0.25 were defined as ‘totally dependent’. Likewise, genes induced more than 2.5-fold in wild-type cells and considered not induced (i.e. the ratio of alkaline stress/no stress) <1.3) in msn2 msn4 cells were also considered as totally dependent. This score is the same employed in our previous report on calcineurin dependence of high pH response [20].
RESULTS
Manipulation of upstream elements of the PKA pathway affects growth at alkaline pH
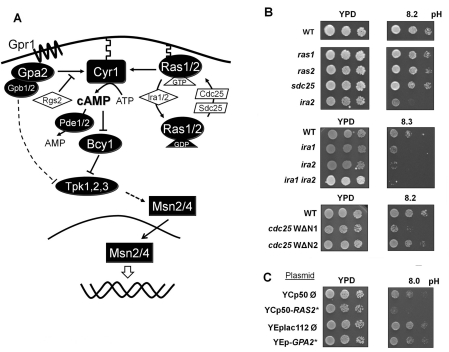
Our starting hypothesis was that adaptation to high pH stress should entail a decrease in the PKA pathway activity. Therefore we speculated that mutations increasing the activity of the pathway should result in a decreased tolerance to alkaline pH. We first tested strains lacking components of the Gpr1/Gpa2-sensing pathway such as gpr1, gpa2, rgs2 (lacking a GAP of Gpa2), gpb1 or gpb2 (lacking the multistep regulator of cAMP/PKA signalling) (Figure 1A). However, none of these strains showed an altered tolerance to alkaline pH (results not shown). In contrast, deletion of ira1 or ira2, the GAPs of Ras1 and Ras2, resulted in increased sensitivity. This phenotype was enhanced in the double ira1 ira2 mutant (Figure 1B). Conversely, expression of the Cdc25 WΔN1 allele, which lacks the N-terminal domain of Cdc25 and elicits constitutive activation of Ras proteins, also results in poor growth at high pH (Figure 1B). All of these manipulations lead to increased Ras activity and hyperactivation of the PKA pathway. In contrast, the effect on high pH tolerance of the expression of the Cdc25 WΔN2 allele which does not lead to an increase in PKA activity was barely noticeable. These results were confirmed by expression of the hyperactive Ras allele RAS2Ala18Val19 from a centromeric plasmid [31]. Among other phenotypes, expression of this allele decreased heat-shock tolerance. The expression of the RAS2Ala18Val19 allele decreased alkaline pH tolerance (Figure 1C). In contrast, expression of a constitutively active form of Gpa2 (pG2CT-112.2) constructed by replacing the arginine residue at position 273 with an alanine residue [31] did not alter high pH tolerance.
Figure 1. Effect of mutations in the upstream components of the PKA pathway on alkaline pH tolerance.
(A) A simplified schematic depiction of the PKA signalling pathway. See the Introduction section for additional information. (B) Upper panels: three dilutions of cultures of wild-type BY4741 cells and the isogenic kanMX disruption derivatives ras1, ras2, sds25 and ira2 mutants were spotted on to YPD plates adjusted at the indicated pH values. Middle panels: wild-type strains W303-1A and its ira1::LEU2 (strain SC7), ira2::URA3 (SC8) and ira1::LEU2 ira2::URA3 (PM903) derivatives were spotted as above. Lower panels: wild-type strains W303-1A and isogenic cells expressing the WΔN1 or WΔN2 alleles of CDC25 were spotted. All plates were incubated for 3 days. (C) Wild-type strain W303-1A was transformed with the indicated plasmids. Positive clones were grown overnight in synthetic selective medium and spotted on to YPD plates adjusted at the indicated pH and growth was monitored after 2 days. YCp50-RAS2* expresses a hyperactive allele (Ras2Ala18Val19) of the Ras protein. YEp-GPA2* generates a constitutively active Gpa2R273A version of the protein.
Alkaline pH stress transiently decreases the levels of cAMP
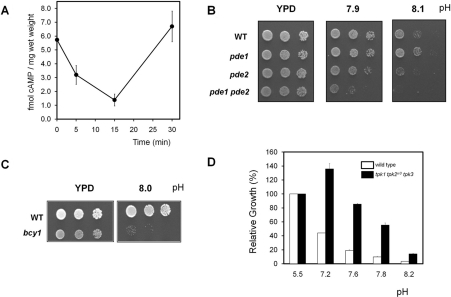
cAMP levels determine the interaction between the regulatory subunit (Bcy1) and the catalytic subunits (Tpk) of PKA and are therefore crucial for the activity of the kinase. We have measured the levels of cAMP after shifting the cells from pH 5.5 to medium buffered to pH 8.0 and found that this treatment drastically decreased the concentration of the second messenger cAMP in the first 5–15 min, followed by a recovery to the initial levels after 30 min of stress (Figure 2A). A similar result was obtained when KOH was added directly to the exponentially growing cultures to raise the pH. In this case the decrease in cAMP levels was apparent even only 2 min after stress (results not shown). Raising the pH of the liquid medium up to 8.2 did not result in detectable cell lysis, as determined by microscopic examination, viability counting or release of alkaline phosphatase activity to the medium.
Figure 2. Changes in PKA pathway activity influences high pH tolerance.
(A) Wild-type BY4741 cells were subjected to alkaline pH stress as described in the Materials and methods section and cultures were processed for cAMP determination. Results are means±S.E.M of at least four independent experiments. (B) Wild-type (WT) W303-1A cells and their derivatives pde1 (CCV35), pde2 (PM942) and pde1 pde2 (CCV36) were spotted on to YPD plates adjusted at the indicated pH values. Growth was monitored after 2 days. (C) The BY4741 strain (WT) and its bcy1 derivative (MAR231) were grown on YPD plates adjusted at the indicated pH for 3 days. (D) Wild-type strain BY4742 (open bars) and its derivative DC90 (tpk1 tpk2w3 tpk3, closed bars) were grown on liquid YPD medium as described in the Materials and methods section. Growth is represented as the percentage over the same strain at initial pH 5.5. Results are the average±S.E.M. from two independent experiments.
The results of the present paper were consistent with the possibility that a drop in cAMP levels leading to down-regulation of PKA activity could be an adaptive strategy to confront high pH stress. Therefore we considered that mutations resulting directly in increased cAMP levels should be deleterious for cells subjected to alkaline pH stress. The PDE1 and PDE2 genes encode the yeast cAMP phosphodiesterase activity, which degrades cAMP to AMP. When the single mutants are grown on alkaline-pH plates, a growth defect can be observed for the pde2 mutant (lacking the high-affinity isoform) that is exacerbated in the double pde1 pde2 mutant strain (Figure 2B).
Lack of Bcy1, the regulatory subunit of PKA, leads to constitutive PKA activity, which results in a variety of phenotypes (impaired growth on different carbon sources, temperature sensitivity etc.; see [2] and references therein). Although the quantification of the effect is difficult because of the relatively poor growth of the bcy1 mutant even under standard conditions, we can show that this strain exhibits a dramatic alkaline pH-sensitive phenotype (Figure 2C). The same effect was observed in the W303-1A background (results not shown). Similarly, we tested whether a decrease in PKA activity could improve growth at high pH. Since complete lack of PKA activity is not compatible with survival, for this experiment we resorted to a strain lacking TPK1 and TPK3 and carrying an attenuated allele of TPK2 (tpk2w3) as the sole source of PKA activity. As shown in Figure 2(D), the relative growth of this strain in alkaline pH medium compared with non-restrictive conditions is higher than the wild-type strain. In essence, the results presented so far show that yeast cells respond to the alkalinization of their medium with a sharp decrease in the intracellular level of cAMP. The observation that when PKA activity is attenuated cells grow better at high pH, whereas any situation resulting in increased PKA activity makes cells more sensitive to this condition, supports the notion that down-regulating PKA activity contributes to adaptation to alkaline pH stress.
Alkaline pH stress promotes entry of Msn2 in the nucleus and STRE-dependent promoter activation
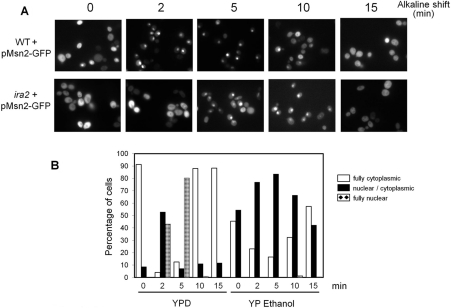
Nuclear localization of Msn2, a transcription factor that together with Msn4 regulates the general stress response of S. cerevisiae, is controlled by PKA activity [13,14,40]. We considered that alkaline pH-triggered down-regulation of the PKA activity could result in the translocation of Msn2 to the nucleus and that this transition might have a relevant effect in the pH-induced adaptive response. To test this possibility, an Msn2–GFP fusion was introduced into wild-type cells and the cultures subjected to alkaline treatment. As shown in Figures 3(A) and 3(B), exposure to high pH (8.0) triggered an almost immediate entry of Msn2 into the nucleus, which reached a maximum after 5 min (Figure 3B). Nuclear localization of Msn2 was transient and after 10–15 min the transcription factor was again mostly cytosolic. Interestingly, when the same experiment was carried out in a strain lacking the IRA2 gene (sensitive to alkaline pH, see Figure 1B), entry of Msn2 into the nucleus was somewhat delayed and nuclear localization was a less general effect than in the wild-type strain. Nuclear translocation of Msn2 was also evaluated in cells subjected to alkaline stress (pH 8.0) growing in the presence of ethanol as a carbon source (Figure 3B). Under these conditions, Msn2 showed both cytoplasmic and nuclear/cytoplasmic distribution in unstressed cells. Interestingly, exposure of cells to alkalinization resulted in only a modest increase in the percentage of cells showing nuclear/cytoplasmic distribution, but very few cells with only nuclear Msn2. Therefore subcellular distribution of Msn2 in response to alkaline stress is affected by the carbon source in which cells are grown.
Figure 3. Alkaline pH triggers nuclear entry of the Msn2 transcription factor.
Cultures of wild-type BY4741 (WT) and its ira2::kanMX derivative (right-hand panels) were transformed with plasmid pMsn2-GFP and subjected to pH stress (8.0) on YPD medium. Samples were collected at the specified times and fixed for fluorescence microscopy. (B) BY4741 cells carrying the pMsn2-GFP construct were grown to exponential phase (A660 of 0.8) on YPD medium or YP medium containing ethanol (2%) as the carbon source, shifted to pH 8.0. The subcellular distribution of the Msn2–GFP fusion was monitored for at least 200 cells per time point. Open bars denote full cytoplasmic, closed bars denote nuclear/cytoplasmic and crossed bars denote full nuclear localization. The results shown correspond to a representative experiment. Three independent experiments were performed with similar results.
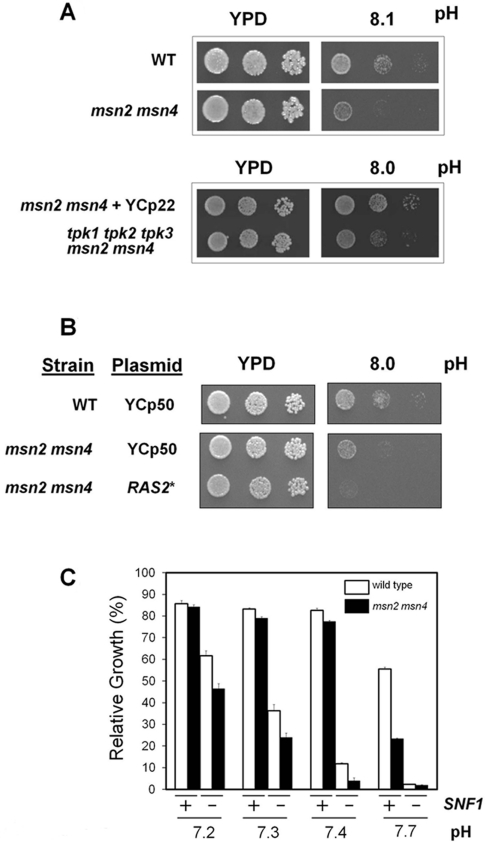
We then investigated whether the Msn2 and Msn4 transcription factors are necessary for a normal tolerance to high pH stress. As shown in Figure 4(A), lack of these transcription factors somewhat reduces tolerance to high pH, suggesting that they are components of the adaptive response to alkalinization. Remarkably, additional deletion of all three PKA catalytic subunits (which is feasible in the absence of the transcription factors) yields cells that are even more sensitive than the msn2 msn4 mutant. It should be noted that in this case the msn2 msn4 strain was transformed with a centromeric plasmid which contains a TRP1 marker. The introduction of this marker in the reference strain was necessary for accurate comparison because the tpk msn2,4 strain was constructed using the TRP1 gene as a marker, and it has been reported that the absence of this gene somewhat decreases tolerance to high pH [41]. These results suggested that, besides its role in Msn2,4 function, regulation of PKA activity may exert additional effects on high pH tolerance. This notion was reinforced by the results shown in Figure 4(B). We observed that expression of the hyperactive form of RAS2 further decreased high pH tolerance even in the absence of Msn2/Msn4 transcription factors, suggesting again that not all of the effects relevant for alkaline tolerance mediated by PKA are based necessarily on the activation of Msn2/Msn4. On the other hand, cells lacking the Snf1 kinase are sensitive to alkaline pH and Snf1 has been reported to repress Msn2 function. Therefore we considered whether the sensitive phenotype of snf1 cells could be due to deregulation of Msn2 function. As shown in Figure 4(C), snf1 cells are more sensitive than the msn2 msn4 mutant to alkaline pH. In addition, deletion of msn2 msn4 further increases sensitivity to alkaline pH of the snf1 mutant. Therefore deregulation of Msn2/Msn4 is not the basis of the snf1 alkali-sensitive phenotype.
Figure 4. Effect of the lack of Msn2 and Msn4 transcription factors on high pH tolerance.
(A) Upper panels: wild-type W303-1A alone (WT) was plated along with strain MCY5278, a W303-1A-derived strain lacking both MSN2 and MSN4 genes. Lower panels: the MCY5278 strain was transformed with centromeric plasmid YCp22 (which carries a TRP1 gene marker, see the main text for an explanation) and plated with the equivalent strain but lacking all three TPK genes [14]. Dilutions of the cultures were grown for 2 days at the indicated pH. (B) The W303-1A strain (WT) and the msn2 msn4 derivative transformed with the empty plasmid YCp50 or the same plasmid carrying a hyperactive RAS2Ala18Val19 allele (RAS2*). Dilutions of the cultures were spotted on to YPD plates at the indicated pH values and growth was monitored after 2 days. (C) The W303-1A strain (open bars) and its msn2 msn4 derivative (closed bars) carrying a wild-type allele of SNF1 (+) or a snf1 deletion (−) were grown at the indicated pH values. Growth is denoted as the percentage compared with the same strains cultured at pH 5.5. Results are means±S.E.M. from two independent experiments performed in triplicate.
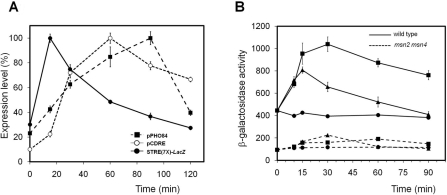
Because Msn2 is known to bind with STRE elements in response to glucose shortage and other forms of stress, we tested the activation of a synthetic promoter containing a tandem of seven STRE elements fused to the LacZ reporter (the AGS66 strain). As shown in Figure 5(A), this promoter exhibited a very fast activation with a peak of activity 15 min after moving the cells to a pH 8.0 environment. This activation was much faster than that observed when the LacZ gene is expressed from a PHO84 promoter, which is considered a gene with a relatively late response to high pH stress [19] and it was completely abolished in a msn2 msn4 strain (results not shown). Remarkably, the response was also faster than that obtained from a synthetic promoter based in a cluster of four copies of the CDRE motif from the calcineurin-responsive GSC2/FKS2 gene. It must be noted that the calcineurin/Crz1-mediated response is considered to be an early response to alkaline pH stress [19,20]. We also tested the response to alkaline pH of the HXK1 gene which encodes hexokinase, a gene with predicted STRE elements in its promoter [10] and known to be induced by high temperature (37 °C) and oxidative stress in a msn2 msn4-dependent way [12]. As shown in Figure 5(B), expression of the HXK1 promoter induced by high pH stress was as fast as the one induced by exposure to 37 °C, although the level of β-galactosidase activity declined faster (probably due to yeast-promoted acidification of the medium). Remarkably, in the msn2 msn4 strain the basal activity of the promoter, as well as the response to both stress conditions, is dramatically reduced. However, a small increase can still be detected after 30 min of exposure to high pH stress, suggesting an Msn2/Msn4-independent input to the promoter, which could be attributed to activation of calcineurin as this gene was demonstrated to be induced by calcium as well as by high pH [21].
Figure 5. Alkaline pH stress induces STRE-mediated transcriptional activation.
(A) The wild-type strain DBY746 was transformed with episomal plasmids pPHO84-LacZ (pPHO84) and pAMS366 (pCDRE). These strains, together with strain AGS66 [carrying a STRE(7×)–LacZ reporter system integrated at the URA3 locus] were grown and subjected to high pH stress (pH 8.0). β-Galactosidase activity was measured as described and it is represented as the percentage over the maximum value for each strain. (B) The wild-type strain W303-1A (continuous lines) and its msn2 msn4 derivative (broken lines) were transformed with the HXK1–LacZ reporter. Cultures grown on YP medium (plus 4% glucose) were subjected to high pH (8.0, ▲) or high temperature (37 °C, ■) and samples taken at different times for β-galactosidase activity measurements. Non-induced cultures are denoted by circles. Results are means±S.E.M. from six independent experiments.
Relevance of Msn2/Msn4 transcription factors in the transcriptional response to high pH stress
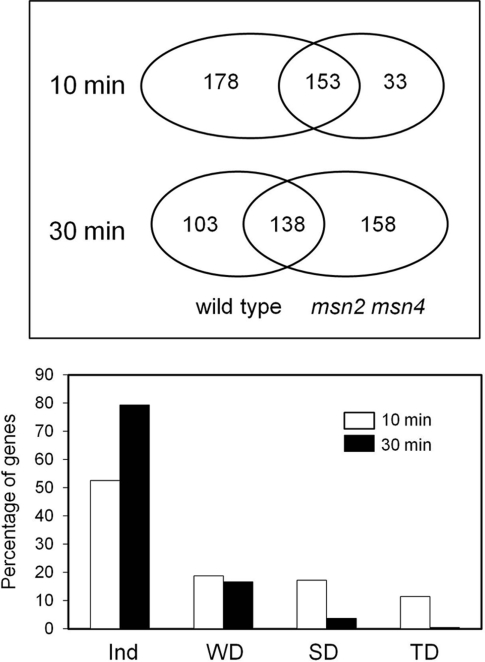
Since these results indicated that Msn2/Msn4 could be responsible for the response of certain genes to alkaline pH stress, we considered it necessary to evaluate to what extent this pathway is responsible for the remodelling of gene expression observed upon exposure to high pH. To this end, DNA microarray analysis was conducted using wild-type and msn2 msn4 strains subjected to pH 8.0 for 10 and 30 min. After 10 min of exposure to high pH, in the wild-type cells 331 genes had a magnitude of induced expression at least 2-fold greater, whereas only 186 genes found in the msn2 msn4 cells strain (from a total number of 3463 genes with valid data; Figure 6 and Supplementary Table S1 at http://www.BiochemJ.org/bj/438/bj4380523add.htm).
Figure 6. Msn2/Msn4 dependence of the transcriptional response to alkaline stress.
Upper panel: number of genes induced in the wild-type and the msn2 msn4 strains after 10 or 30 min of alkaline stress deduced from the DNA microarray analysis. Lower panel: evaluation of the Msn2/Msn4 level of dependence of the transcriptional response to high pH after 10 min (open bars) and 30 min (closed bars). Ind, Independent; WD, weakly dependent; SD, strongly dependent; TD, totally dependent.
GO (Gene Ontology) analysis of the genes induced in the wild-type strain yielded, as expected, an excess of genes related to carbohydrate metabolism (P<7.74×10−9), in particular to trehalose (P<5.05×10−8) and glucose (P<1.42×10−6) and glycogen (P<1.61×10−3) metabolism. Analysis of the msn2 msn4 dependence for induction revealed that 157 genes required the presence of the transcription factors for full induction (Figure 6). Although dependence was limited in some cases (62 genes were rated as weakly dependent), the majority of the short-term induced genes were strongly (57) or totally (38) dependent of the presence of Msn2 and Msn4. GO analysis of the 95 genes defined as strongly or totally dependent revealed a strong excess of genes encoding proteins involved in trehalose (P<9.40×10−9) and glycogen (P<1.46×10−7) metabolism. A similar analysis of genes showing weak dependence provided no distinctive profile.
Exposure of cells to pH 8.0 for 30 min resulted in 241 genes induced in the wild-type strain, whereas 296 genes increased expression at least 2-fold in the msn2 msn4 mutant (Figure 6). In this case, the level of dependence of the transcription factors was much lower (only 50 genes). In addition, the vast majority (40) were weakly dependent; nine were ranked as strongly dependent and only one as totally dependent. GO analysis of the set of strongly dependent plus totally dependent genes did not produce any specific profile. Therefore Msn2 and Msn4 transcription factors are responsible for the induction of a substantial subset of the early alkali-responsive genes. Analysis of repressed genes showed 146 and 279 genes repressed at least 0.5-fold in the wild-type strain 10 and 30 min after a shift to high pH respectively. The absence of msn2 msn4 did not result in activation of any of these genes.
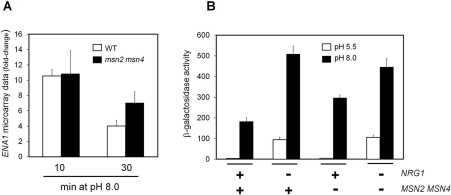
The ENA1 gene encodes a Na+-ATPase that has been reported repeatedly to be strongly induced upon alkaline pH stimulation. Interestingly, our microarray results indicate that deletion of MSN2/MSN4 does not affect the induction of this ATPase gene in the short-term (10 min), but results in higher-than-normal expression after 30 min of stress (Figure 7A). In an attempt to identify a possible cause for this behaviour we searched our microarray raw data for changes in the expression of genes encoding known regulatory components of ENA1. We observed that after 10 min after alkalinization of the medium, the expression of NRG1, a known repressor of ENA1 expression, was essentially identical in the wild-type and msn2 msn4 cells. However, after 30 min the expression of NRG1 was decreased by 37% in the msn2 msn4 strain compared with the wild-type. To explore a possible influence of this change on the response of ENA1, we evaluated the expression from the ATPase gene promoter in wild-type and msn2 msn4 cells deleted for NRG1 by means of a LacZ reporter. As it can be observed (Figure 7B), high-pH-dependent expression from the ENA1 promoter is stronger in the msn2 msn4 mutant than in the wild-type strain, thus confirming the microarray results. Moreover, the induction observed upon alkaline pH stimulation in the nrg1 strain is virtually identical with that observed in the msn2 msn4 nrg1 triple mutant, suggesting that changes in the Nrg1 levels could be at the basis of the abnormal response of ENA1 in the msn2 msn4 mutant.
Figure 7. Dependence of ENA1 expression on the presence of Msn2/Msn4.
(A) Fold-change of ENA1 expression deduced from microarray data for W303-1A wild-type cells (WT, empty bars) and MCY5278 (msn2 msn4, filled bars) exposed to pH 8.0 for the indicated times. Results are means±S.E.M. from four microarray experiments. (B) Strains with the indicated genotype (+, wild type allele; -, deletion mutant) were transformed with plasmid pKC201, which bears a LacZ fusion of the ENA1 promoter. Exponential cultures were resuspended in medium buffered at pH 5.5 (empty bars) or pH 8.0 (filled bars) for 1 h and β-galactosidase activity was measured. Results are means±S.E.M. from six to nine independent experiments.
DISCUSSION
Exposure to alkaline stress triggers a set of responses that are known to be mediated by different signalling pathways, such as the calcineurin/Crz1, the Rim101 and the Slt2 pathways [16]. PKA mediates a major signalling pathway that is crucial for linking stress responses and cell proliferation. Therefore the major goal of the present study was to evaluate the relevance of the PKA pathway in the adaptive response to high pH stress. We observed that mutations leading to activation of PKA resulted in decreased alkaline pH tolerance, whereas those that lead to inhibition of the pathway yield cells with increased tolerance. Our results also show that exposure to high pH provokes a sharp and transient decrease in the cAMP levels, which may lead to down-regulation of PKA activity. In this regard, a previous study reported a transitory increase of cAMP levels as a result of sudden acidification of the cytosol [42]. It is worth noting that these authors noticed that Gpa2 was not required for the stimulation of cAMP accumulation in response to intracellular acidification. Remarkably, we observe that only alterations in the small G-proteins Ras1 and Ras2 branch of the pathway do result in changes in tolerance to alkaline pH, whereas mutations in diverse genes involved in the Gpr1/Gpa2 glucose sensor system do not produce phenotypic effects. This suggests that the observed changes are due to alteration of intracellular glucose metabolism and not to defects in the extracellular glucose detection system, and reinforce the notion that changes in cytosolic pH may result in fluctuations in cAMP levels and consequent regulation of PKA activity. In the case of sudden alkalinization of the medium, the sharp decrease in cAMP levels would inhibit PKA activity and allow cells to adapt to the stress.
The results of the present study show that alkalinization of the medium triggers a very fast entry of the Msn2 transcription factor to the nucleus (2–5 min). This time frame is very similar to that observed when cells are shifted to a low-glucose medium [13,14]. Interestingly, the fact that we observe only a very modest response in ethanol-grown cells suggested that nuclear transition of Msn2 is related to alkaline stress-induced alterations in glucose signalling. We also show that Msn2 and Msn4 are important for mediating a substantial part of the transcriptional response induced by alkalinization of the medium. It is conceivable that this response constitutes one of the factors that allow normal tolerance to the stress, since the msn2 msn4 mutant is sensitive to high pH (Figure 4A). Moreover, our results suggest that regulation of Msn2 function in response to high pH is the result of inhibition of PKA. It is known that intracellular localization of Msn2 can be regulated not only by PKA, but also by the Snf1 kinase [43,44] and the TOR (target of rapamycin) pathway [44,45], thus raising the possibility that these two pathways could contribute to the alkaline pH-triggered entry of Msn2 into the nucleus. In fact, activation of the Snf1 pathway has been previously linked to high pH stress [21,28,46]. However, we observe that mutation of SNF1 yields cells even more sensitive to alkaline pH than those lacking msn2 msn4, and that deletion of both factors in a snf1 background further decreases tolerance. Furthermore, we have observed that, within the time-span shown in Figure 3(A), entry into the nucleus of Msn2 is not affected by the lack of the protein kinase Snf1 after alkaline treatment (results not shown). Finally, comparison of the dependence level for Msn2,4 of the short-term transcriptional response to high pH described in the present paper with the dependence on Snf1, evaluated in a parallel project (A. Casamayor, A. Ruiz, R. Serrano, M. Platara, J. Ferrer-Dalmau and J. Ariño, unpublished work), indicates low overlap (only 34 of the 157 genes reported as Msn2,4-dependent in the present study are found to exhibit some degree of dependence for the Snf1 kinase). As an example, most of the genes encoding enzymes involved in trehalose metabolism whose expression is increased by alkalinization are Msn2,4-dependent, but not Snf1-dependent. Therefore the results of the present study do not support the notion that activation of Msn2/4 by high pH is mediated by the Snf1 kinase.
The PKA and TOR pathways illustrate diverse ways of functional interaction in the regulation of cell growth, showing some functional overlap [47]. However, a role of the TOR pathway in regulation of Msn2 function in response to high pH is questionable because, with the exception of the ure2 strain (a very pleiotropic strain), mutants in genes of the pathway do not show altered tolerance to high pH nor does alkalinization trigger increased expression of well-established readouts for inhibitors of the TOR pathway, such as GAP1, GLN1 or GDH1 (results not shown). Two protein kinases, Yak1 and Rim15, have been reported to modulate Msn2 function in response to PKA activation (see [5] for a review). However, yak1 and rim15 mutant strains are not sensitive to high pH (results not shown) and furthermore it has been proposed that Yak1 regulates Msn2 function by an unknown mechanism that does not implicate the control of Msn2 subcellular localization. Therefore the most probable scenario is that Msn2 entry into the nucleus after high pH stress is caused by inhibition of PKA activity.
Comparison of the time sequence of the events described in our work is rather coherent, with a very fast decrease in cAMP levels and subsequent entry of Msn2 in the nucleus, followed by a massive, fast (10 min) and transient activation of Msn2/Msn4-dependent genes, as deduced by DNA microarray transcriptomic analysis. It is remarkable that, after 30 min of high pH stress, the transcriptional response becomes largely msn2 msn4 independent. In fact, our results (Figure 5A) suggest that Msn2/Msn4 may be responsible for the first chronological set of transcriptional responses to alkaline stress, even a faster response than that mediated by the calcineurin/Crz1 pathway, which were currently defined as early responses [19,20].
It has been widely documented that expression of the ENA1 Na+-ATPase is potently induced by alkaline pH (see [48] and references therein). An interesting observation derived from our microarray results is that the increase in ENA1 expression is virtually identical in wild-type and msn2 msn4 cells at short time frames (10 min), but is further enhanced by the lack of Msn2/Msn4 at 30 min. Interestingly, we observed previously a similar effect by using a LacZ translational fusion of the ENA1 promoter and a different msn2 msn4 mutant strain [19], although at that moment it was not further characterized. Our observations suggest that lack of Msn2/4 results in either activation of a positive regulatory element for ENA1 expression or removal of a negative regulator. It is well known that Nrg1 acts as a repressor of ENA1 expression by directly binding to the gene promoter, thus participating in alkaline pH signalling [18,49,50]. We observe in the present paper that NRG1 levels decrease after 30 min of high pH stress and that alkaline pH induction of ENA1 is not further increased by mutation of msn2 msn4 in a nrg1 deletion background. This suggests that the enhanced expression of the ATPase gene in the msn2 msn4 mutant can be due to a decrease in the amount of the Nrg1 repressor. In this regard, it must be noted that the remarkable effect of lack of RIM101 in the expression of the ATPase gene [18,19,28], is mediated fully by a rather small change (2.8-fold) in NRG1 mRNA levels [18].
The results of the present study also suggest involvement of the PKA pathway in high pH tolerance in way that would be independent of the Msn2/Msn4 transcription factors. This is based in the combination of tpk and msn2,4 mutations and in the observation that hyperactivation of RAS2 in the msn2 msn4 mutant still increases high pH sensitivity (Figure 4B). A possible explanation could be based in a co-operative role for the PKA and calcineurin pathways in the adaptive response to high pH stress. In this regard, it was reported that PKA can phosphorylate Crz1, promoting its exit from the nucleus and thus opposing the action of calcineurin [26]. Therefore inhibition of PKA may result in further enhancement of calcineurin/Crz1-mediated responses. A further step of complexity may exist in the interaction between the calcineurin and the PKA pathways, as it was recently reported that Crz1 may exert a destabilization effect on Msn2 levels upon calcium treatment [51]. In this context, it is conceivable that entry of Crz1 in the nucleus, following activation of calcineurin by high pH may serve, in addition to activate calcineurin/Crz1-responsive promoters, as a negative-feedback system to avoid persistent activation of Msn2/Msn4-responsive genes. This regulatory system may contribute to the transient Msn2/4-dependent transcriptional effect observed in the present study.
Online data
AUTHOR CONTRIBUTION
Carlos Casado performed most of the experiments. Asier González and Amparo Ruiz performed cAMP measurements and constructed some strains, and contributed to the design of the experiments. Maria Platara contributed with diverse lacZ reporter assays. Joaquín Ariño conceived the project, supervised its development and wrote the paper.
ACKNOWLEDGEMENTS
We thank Raquel Serrano and Antonio Casamayor for support. The excellent technical assistance of Montserrat Robledo and Anna Vilalta is acknowledged. We thank Francisco Estruch, Martha Cyert, Jeanne Hirsch, Johan Thevelein, Christoph Schüller, Marian Carlson, Renata Tisi and Enzo Martegani for constructs, strains and advice.
FUNDING
This work was supported by the Ministry of Science and Innovation, Spain and FEDER [grant numbers BFU2008-04188-C03-01, GEN2006-27748-C2-1-E/SYS (SysMo) and EUI2009-04147 (SysMo2) to (J.A.)]. J.A. was the recipient of an ‘Ajut 2009SGR-1091’ (Generalitat de Catalunya). C.C. was supported by a predoctoral fellowship from the Spanish Ministry of Science and Technology.
References
- 1.Thevelein J. M., de Winde J. H. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 1999;33:904–918. doi: 10.1046/j.1365-2958.1999.01538.x. [DOI] [PubMed] [Google Scholar]
- 2.Santangelo G. M. Glucose signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2006;70:253–282. doi: 10.1128/MMBR.70.1.253-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gancedo J. M. The early steps of glucose signalling in yeast. FEMS Microbiol. Rev. 2008;32:673–704. doi: 10.1111/j.1574-6976.2008.00117.x. [DOI] [PubMed] [Google Scholar]
- 4.Tamaki H. Glucose-stimulated cAMP-protein kinase A pathway in yeast Saccharomyces cerevisiae. J. Biosci. Bioeng. 2007;104:245–250. doi: 10.1263/jbb.104.245. [DOI] [PubMed] [Google Scholar]
- 5.Smets B., Ghillebert R., De Snijder P., Binda M., Swinnen E., De Virgilio C., Winderickx J. Life in the midst of scarcity: adaptations to nutrient availability in Saccharomyces cerevisiae. Curr. Genet. 2010;56:1–32. doi: 10.1007/s00294-009-0287-1. [DOI] [PubMed] [Google Scholar]
- 6.Estruch F., Carlson M. Two homologous zinc finger genes identified by multicopy suppression in a SNF1 protein kinase mutant of Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:3872–3881. doi: 10.1128/mcb.13.7.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez-Pastor M. T., Marchler G., Schuller C., Marchler-Bauer A., Ruis H., Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE) EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- 8.Schmitt A. P., McEntee K. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 1996;93:5777–5782. doi: 10.1073/pnas.93.12.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mai B., Breeden L. Xbp1, a stress-induced transcriptional repressor of the Saccharomyces cerevisiae Swi4/Mbp1 family. Mol. Cell. Biol. 1997;17:6491–6501. doi: 10.1128/mcb.17.11.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moskvina E., Schuller C., Maurer C. T., Mager W. H., Ruis H. A search in the genome of Saccharomyces cerevisiae for genes regulated via stress response elements. Yeast. 1998;14:1041–1050. doi: 10.1002/(SICI)1097-0061(199808)14:11<1041::AID-YEA296>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 11.Smith A., Ward M. P., Garrett S. Yeast PKA represses Msn2p/Msn4pdependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 1998;17:3556–3564. doi: 10.1093/emboj/17.13.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., Storz G., Botstein D., Brown P. O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorner W., Durchschlag E., Martinez-Pastor M. T., Estruch F., Ammerer G., Hamilton B., Ruis H., Schuller C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorner W., Durchschlag E., Wolf J., Brown E. L., Ammerer G., Ruis H., Schuller C. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 2002;21:135–144. doi: 10.1093/emboj/21.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garreau H., Hasan R. N., Renault G., Estruch F., Boy-Marcotte E., Jacquet M. Hyperphosphorylation of Msn2p and Msn4p in response to heat shock and the diauxic shift is inhibited by cAMP in Saccharomyces cerevisiae. Microbiology. 2000;146:2113–2120. doi: 10.1099/00221287-146-9-2113. [DOI] [PubMed] [Google Scholar]
- 16.Arino J. Integrative responses to high pH stress in S. cerevisiae. OMICS. 2010;14:517–523. doi: 10.1089/omi.2010.0044. [DOI] [PubMed] [Google Scholar]
- 17.Lamb T. M., Xu W., Diamond A., Mitchell A. P. Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J. Biol. Chem. 2001;276:1850–1856. doi: 10.1074/jbc.M008381200. [DOI] [PubMed] [Google Scholar]
- 18.Lamb T. M., Mitchell A. P. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 2003;23:677–686. doi: 10.1128/MCB.23.2.677-686.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Serrano R., Ruiz A., Bernal D., Chambers J. R., Arino J. The transcriptional response to alkaline pH in Saccharomyces cerevisiae: evidence for calcium-mediated signalling. Mol. Microbiol. 2002;46:1319–1333. doi: 10.1046/j.1365-2958.2002.03246.x. [DOI] [PubMed] [Google Scholar]
- 20.Viladevall L., Serrano R., Ruiz A., Domenech G., Giraldo J., Barcelo A., Arino J. Characterization of the calcium-mediated response to alkaline stress in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:43614–43624. doi: 10.1074/jbc.M403606200. [DOI] [PubMed] [Google Scholar]
- 21.Ruiz A., Serrano R., Arino J. Direct regulation of genes involved in glucose utilization by the calcium/calcineurin pathway. J. Biol. Chem. 2008;283:13923–13933. doi: 10.1074/jbc.M708683200. [DOI] [PubMed] [Google Scholar]
- 22.Serrano R., Martin H., Casamayor A., Arino J. Signaling alkaline pH stress in the yeast Saccharomyces cerevisiae through the Wsc1 cell surface sensor and the Slt2 MAPK pathway. J. Biol. Chem. 2006;281:39785–39795. doi: 10.1074/jbc.M604497200. [DOI] [PubMed] [Google Scholar]
- 23.Serrano R., Bernal D., Simon E., Arino J. Copper and iron are the limiting factors for growth of the yeast Saccharomyces cerevisiae in an alkaline environment. J. Biol. Chem. 2004;279:19698–19704. doi: 10.1074/jbc.M313746200. [DOI] [PubMed] [Google Scholar]
- 24.Adams A., Gottschling D. E., Kaiser C. A., Stearns T. Methods in Yeast Genetics. New York: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- 25.Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J. D., Bussey H., et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 26.Kafadar K. A., Cyert M. S. Integration of stress responses: modulation of calcineurin signaling in Saccharomyces cerevisiae by protein kinase A. Eukaryot. Cell. 2004;3:1147–1153. doi: 10.1128/EC.3.5.1147-1153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma P., Wera S., Van D. P., Thevelein J. M. The PDE1-encoded low-affinity phosphodiesterase in the yeast Saccharomyces cerevisiae has a specific function in controlling agonist-induced cAMP signaling. Mol. Biol. Cell. 1999;10:91–104. doi: 10.1091/mbc.10.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Platara M., Ruiz A., Serrano R., Palomino A., Moreno F., Arino J. The transcriptional response of the yeast Na+-ATPase ENA1 gene to alkaline stress involves three main signaling pathways. J. Biol. Chem. 2006;281:36632–36642. doi: 10.1074/jbc.M606483200. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalez A., Ruiz A., Casamayor A., Arino J. Normal function of the yeast TOR pathway requires the type 2C protein phosphatase Ptc1. Mol. Cell Biol. 2009;29:2876–2888. doi: 10.1128/MCB.01740-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myers A. M., Tzagoloff A., Kinney D. M., Lusty C. J. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene. 1986;45:299–310. doi: 10.1016/0378-1119(86)90028-4. [DOI] [PubMed] [Google Scholar]
- 31.Xue Y., Batlle M., Hirsch J. P. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Galpha subunit and functions in a Ras-independent pathway. EMBO J. 1998;17:1996–2007. doi: 10.1093/emboj/17.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toda T., Uno I., Ishikawa T., Powers S., Kataoka T., Broek D., Cameron S., Broach J., Matsumoto K., Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985;40:27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- 33.Stathopoulos A. M., Cyert M. S. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 1997;11:3432–3444. doi: 10.1101/gad.11.24.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alepuz P. M., Cunningham K. W., Estruch F. Glucose repression affects ion homeostasis in yeast through the regulation of the stress-activated ENA1 gene. Mol. Microbiol. 1997;26:91–98. doi: 10.1046/j.1365-2958.1997.5531917.x. [DOI] [PubMed] [Google Scholar]
- 35.Cunningham K. W., Fink G. R. Calcineurin inhibits VCX1-dependent H+/Ca2+ exchange and induces Ca2+ ATPases in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:2226–2237. doi: 10.1128/mcb.16.5.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marquez J. A., Serrano R. Multiple transduction pathways regulate the sodium-extrusion gene PMR2/ENA1 during salt stress in yeast. FEBS Lett. 1996;382:89–92. doi: 10.1016/0014-5793(96)00157-3. [DOI] [PubMed] [Google Scholar]
- 37.Reynolds A., Lundblad V., Dorris D., Keaveney M. Yeast vectors and assays for expression of cloned genes. In: Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K., editors. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1997. pp. 13.6.1–13.6.6. [DOI] [PubMed] [Google Scholar]
- 38.Alberola T. M., Garcia-Martinez J., Antunez O., Viladevall L., Barcelo A., Arino J., Perez-Ortin J. E. A new set of DNA macrochips for the yeast Saccharomyces cerevisiae: features and uses. Int. Microbiol. 2004;7:199–206. [PubMed] [Google Scholar]
- 39.Herrero J., Al Shahrour F., Diaz-Uriarte R., Mateos A., Vaquerizas J. M., Santoyo J., Dopazo J. GEPAS: a web-based resource for microarray gene expression data analysis. Nucleic Acids Res. 2003;31:3461–3467. doi: 10.1093/nar/gkg591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jacquet M., Renault G., Lallet S., De M. J., Goldbeter A. Oscillatory nucleocytoplasmic shuttling of the general stress response transcriptional activators Msn2 and Msn4 in Saccharomyces cerevisiae. J. Cell Biol. 2003;161:497–505. doi: 10.1083/jcb.200303030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gonzalez A., Larroy C., Biosca J. A., Arino J. Use of the TRP1 auxotrophic marker for gene disruption and phenotypic analysis in yeast: a note of warning. FEMS Yeast Res. 2008;8:2–5. doi: 10.1111/j.1567-1364.2007.00315.x. [DOI] [PubMed] [Google Scholar]
- 42.Colombo S., Ma P., Cauwenberg L., Winderickx J., Crauwels M., Teunissen A., Nauwelaers D., de Winde J. H., Gorwa M. F., Colavizza D., Thevelein J. M. Involvement of distinct G-proteins, Gpa2 and Ras, in glucose- and intracellular acidification-induced cAMP signalling in the yeast Saccharomyces cerevisiae. EMBO J. 1998;17:3326–3341. doi: 10.1093/emboj/17.12.3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Wever V., Reiter W., Ballarini A., Ammerer G., Brocard C. A dual role for PP1 in shaping the Msn2-dependent transcriptional response to glucose starvation. EMBO J. 2005;24:4115–4123. doi: 10.1038/sj.emboj.7600871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mayordomo I., Estruch F., Sanz P. Convergence of the target of rapamycin and the Snf1 protein kinase pathways in the regulation of the subcellular localization of Msn2, a transcriptional activator of STRE (Stress Response Element)-regulated genes. J. Biol. Chem. 2002;277:35650–35656. doi: 10.1074/jbc.M204198200. [DOI] [PubMed] [Google Scholar]
- 45.Beck T., Hall M. N. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 46.Hong S. P., Carlson M. Regulation of snf1 protein kinase in response to environmental stress. J. Biol. Chem. 2007;282:16838–16845. doi: 10.1074/jbc.M700146200. [DOI] [PubMed] [Google Scholar]
- 47.Chen J. C., Powers T. Coordinate regulation of multiple and distinct biosynthetic pathways by TOR and PKA kinases in S. cerevisiae. Curr. Genet. 2006;49:281–293. doi: 10.1007/s00294-005-0055-9. [DOI] [PubMed] [Google Scholar]
- 48.Ruiz A., Arino J. Function and regulation of the Saccharomyces cerevisiae ENA sodium ATPase system. Eukaryot. Cell. 2007;6:2175–2183. doi: 10.1128/EC.00337-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vyas V. K., Berkey C. D., Miyao T., Carlson M. Repressors Nrg1 and Nrg2 regulate a set of stress-responsive genes in Saccharomyces cerevisiae. Eukaryot. Cell. 2005;4:1882–1891. doi: 10.1128/EC.4.11.1882-1891.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vyas V. K., Kuchin S., Carlson M. Interaction of the repressors Nrg1 and Nrg2 with the Snf1 protein kinase in Saccharomyces cerevisiae. Genetics. 2001;158:563–572. doi: 10.1093/genetics/158.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takatsume Y., Ohdate T., Maeta K., Nomura W., Izawa S., Inoue Y. Calcineurin/Crz1 destabilizes Msn2 and Msn4 in the nucleus in response to Ca2+ in Saccharomyces cerevisiae. Biochem. J. 2010;427:275–287. doi: 10.1042/BJ20091334. [DOI] [PubMed] [Google Scholar]
- 52.Thomas B. J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 53.Belotti F., Tisi R., Martegani E. The N-terminal region of the Saccharomyces cerevisiae RasGEF Cdc25 is required for nutrient-dependent cell-size regulation. Microbiology. 2006;152:1231–1242. doi: 10.1099/mic.0.28683-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.