Abstract
Cardioproteomics (Cardiovascular proteomics) is fast becoming an indispensible technique in deciphering changes in signaling pathways that occur in cardiovascular diseases (CVDs). The quality and availability of the instruments and bioinformatics software used for cardioproteomics continues to improve, and these techniques are now available to most cardiovascular researchers either directly or indirectly via university core centers. The heart and aorta are specialized tissues which present unique challenges to investigate. Currently, the diverse range of proteomic techniques available for cardiovascular research makes the choice of the best method or best combination of methods for the disease parameter(s) being investigated as important as the equipment used. This review focuses on proteomic techniques and their applications which have advanced our understanding of the signaling mechanisms involved in CVDs at the levels of protein complex/protein-protein interaction, post-translational modifications and signaling induced protein changes.
Keywords: Cardioproteomics, cardiovascular diseases, signaling pathway, proteomics, heart, mass spectrometry, drug signaling
Introduction
Cardiovascular diseases (CVDs) are a group of disorders of the heart and blood vessels. CVD is one of the leading causes of death in the world, especially in low- and middle-income countries. CVDs encompass many types of diseases which are subcharacterized into different types of cardiovascular disease in the heart or blood vessels (Table 1). About 17.1 million people died from CVDs in 2004, representing 29% of all global deaths, and an estimated 23.6 million people will die from CVDs each year by 2030 (World Health Organization 2001, www.who.int/mediacentre/factsheets/fs317/en). The American Heart Association's Heart Disease and Stroke Statistics suggest that CVD as the underlying cause of death accounted for 33.6% of all deaths in 2007, or 1 of every 3 deaths in the United States. The estimated direct and indirect cost of CVD in the United States for 2007 was $286.6 billion, which was a 5.8 percent increase over the previous year [1]. To prevent and control CVDs, significant efforts have been conducted to explore the pathogenesis of cardiovascular diseases. In general, CVDs result from the interplay between lifestyle risk factors, environmental stimuli and the inherent intracel-lular system. Therefore, the pathogenesis of cardiovascular diseases is complicated [1].
Table 1.
Main types of cardiovascular diseases
| Cardiovascular Diseases in the heart | Cardiovascular Diseases in the heart |
|---|---|
| Angina pectoris (angina) | -Aortic stenosis |
| -Stable angina | -Aortic regurgitation |
| -Unstable angina | -Tricuspid stenosis |
| -Variant angina (Prinzmetal's angina) | -Tricuspid regurgitation |
| Arrhythmias | Myocarditis |
| -Atrial fibrillation | Pericarditis |
| -Heart block | Rheumatic heart disease |
| -Premature atrial complex (PAC) | Sudden Cardiac Death |
| -Atrial flutter | Syncope |
| -Paroxysmal supraventricular tachycardia | Cardiac Tumor |
| -Wolff-Parkinson-White syndrome | Cardiovascular Diseases in the Blood Vessels |
| -Ventricular tachycardia | Aortic aneurysm |
| -Ventricular fibrillation | Aortitis |
| -Long QT syndrome | Arteriosclerosis |
| Cardiomyopathy | Atherosclerosis |
| -Dilated cardiomyopathy | Aortic dissection |
| -Hypertrophic cardiomyopathy | High blood pressure (hypertension) |
| -Restrictive cardiomyopathy | -Essential hypertension |
| Congestive heart failure | -Secondary hypertension |
| Congenital heart disease | -Malignant hypertension |
| -Atrial septal defect (ASD) | Stroke |
| -Ventricular septal defect (VSD) | Transient ischemic attack (TIA) |
| -Patent ductus arteriosus | Other problems in the arteries |
| -Pulmonic stenosis | -Atherosclerosis of the extremities |
| -Congenital aortic stenosis | (arteriosclerosis obliterans) |
| -Coarctation of aorta | -Arterial embolism |
| -Tetralogy of Fallot | -Acute arterial occlusion |
| -Tricuspid atresia | -Raynaud's phenomenon |
| -Truncus arteriosus | -Arteriovenous fistula |
| -Ebstein's anomaly of the tricuspid valve | -Vasculitis |
| -Transposition of the great vessels | -Thoracic outlet syndrome |
| Coronary artery disease (CAD) | Other problems in veins |
| Cor pulmonale | -Venous thrombosis |
| Diabetic cardiomyopathy | -Deep vein thrombosis (DVT) |
| Heart attack (myocardial infarction, MI) | -Thrombophlebitis |
| Heart valve disease: | -Varicose veins |
| -Mitral stenosis | -Spider veins |
| -Mitral valve regurgitation | Lymphedema |
| -Mitral valve prolapse |
Cardiovascular diseases which have been investigated by proteomics are underlined.
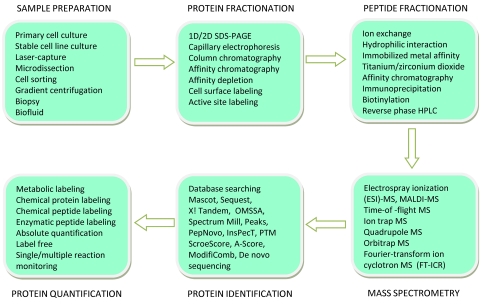
In recent years, the advancement of proteomic techniques has improved the methods available for investigating CVDs [2-5]. The application of proteomic methods to uncover the protein function and structure in normal or disease states in the cardiovascular field is called cardioproteomics. As with general proteomics, cardioproteomics can be subdivided into: 1) investigating protein function in different physiological or disease processes, called mechanistic studies, and 2) investigating proteins altered in response to different cardiovascular disease states for potential clinical use, called biomarker studies [2-7]. In general, the workflow of techniques used in a proteomic investigation includes sample preprocessing or/pre-fractionation, mass spectrometry (MS) analysis and data processing (Figure 1). In a recent review, the proteomics workflow, the general application of proteomics, and the difficulty of proteomics implementation were comprehensively detailed by Mallick et al., 2010 [8]. Sample pre-fractionation decreases the complexity of the sample for subsequent MS analysis. Depending on the aim(s) of the study, different methods of protein sample pre-fractionation are available, such as traditional one dimensional (1D) or two dimensional (2D) gel based protein separation, affinity chromatography/immune precipitation or multidimensional liquid chromatography. MS analysis takes the central stage of proteomics, and the development of other relevant proteomics techniques including sample pre-fraction and data analysis are centering on the development of MS. The peptide analysis methods of mass spectrometry are continually being improved with more advanced mass spectrometers and better MS analysis software. There are now numerous choices of mass spectrometers based on resolution, accuracy and precision. The most popular and common types of mass spectrometers used in proteomics are the MALDI-TOF/TOF-MS/MS, Q-TOF-MS/MS, and LTQ-Orbitrap MS. Each type of MS has its own advantages and disadvantages. The basic principles of MS based peptide sequencing, the mechanism of MS and the pros and cons of different MS are well reviewed [9, 10].
Figure 1.
Schematic of the typical Proteomic workflow for sample processing and mass spectrometry-based identification of a protein. Trypsin digestion (either in-gel or in-solution) occurs between protein and peptide fractionation.
Over the last few years a shift from protein identification to functional proteomics has occurred [11, 12]. The aim of functional proteomics is not only profiling the phenotype of certain tissues, cells, or organelles, but also to define protein function in different biological contexts to answer specific biological questions [13-18]. Cell signaling is a major research field of functional proteomics. Cell signaling pathways often govern basic cellular activities and coordinate cell actions; therefore, unraveling the nature of cellular signaling is crucial for a comprehensive understanding of normal biological processes and disease pathogenesis [19, 20]. The binding of a ligand to its receptor commonly triggers a cascade of reactions often involving protein-protein interactions, protein post-translational modifications (PTM) and signal-induced protein expression changes. Furthermore, signaling feedback loops usually help to regulate the triggered signaling, while crosstalk sometimes bridges diverse signaling pathways [20]. Most signaling pathways are not a linear cascade, but instead involve many complicated molecular interactions, which generally integrate into different signaling networks, making it difficult to uncover the full details of complex pathways [19, 20]. Functional proteomics has become one of the most successful methods to characterize these elaborate signaling systems which are difficult to determine by other biological methods [13, 16, 18]. Cardioproteomics, as one important research field of proteomics, has allowed us to dissect these signaling pathways, helping us to better understand the pathogenesis of cardiovascular diseases [3].
In this review, we summarize some new advancement in proteomics techniques and cardioproteomic applications for cardiovascular scientists. Although proteomics have been used in many fields of CVD research, in this review, we focus on the applications of these techniques in elucidating novel signaling pathways in cardiovascular research. The state-of-art integration of cardioproteomic tools and other techniques that enhance our ability to detect single functional molecules and signaling networks related to CVD is also discussed.
Cardioproteomic methods in discovering and identifying signaling pathways
Protein complex/protein-protein interaction
Protein complexes/protein-protein interactions are very important for linking chemical or physical stimuli to specific effecter molecules in dynamic signaling processes. Affinity purification coupled with mass spectrometry has enable researchers to determine the composition of protein complexes [8, 11, 12, 21]. Typical affinity purification utilizes affinity-tagged recombinant proteins to catch protein complexes from parallel samples. The more stringent and increasingly popular method for affinity purification is tandem affinity purification (TAP), which uses two or more different tags for two or more independent affinity purifications [22-24]. TAP allows highly purified complexes to be obtained in relatively short time periods reducing the nonspecific binding partners of the target protein. Utilizing specific antibodies to target proteins in protein complexes is an alternative method used to purify protein complexes [12, 21]. However, using specific antibodies requires highly precise antibodies and extensive optimization for obtaining optimal protein complexes. To explore the critical role of Ras interacting protein 1 (Rasip1) during normal blood vessel tubulogenesis, Xu et al., 2011, used two-step affinity purification and mass spectrometry to show that non-muscle myosin heavy-chain IIA (NMHCIIA) and a RhoA specific GAP, Arhgap29 interact with Rasip1 [25]. This group found that Rasip1 together with Arhgap29 suppresses RhoA signaling and dampens ROCK and nonmuscle myosin IIA activities in endothelial cells, which play critical roles in cell polarity formation, lumen morphogenesis, basal and apical adhesion and actomyosin contractility during blood vessel tubulogenesis. Using a TAP based approach, they were able to display a more detailed model of Rasip1 regulation of embryonic vascular tubulogenesis [25]. The advantages and disadvantages of the use of affinity chromatography and mass spectrometry to determine the composition of protein complexes have been well discussed [21, 22].
Post-translational modification
Post-translational modification (PTM) of proteins is a common mechanism for controlling the behavior of a protein. More than 200 different types of PTMs are currently known and new ones are regularly being discovered [19, 20]. Some well-known PTMs include but are not limited to: phosphorylation, glycosylation, acetylation, methylation, and ubiquitylation. In addition, many proteins have more than one type of PTM [19, 20]. Therefore, PTMs significantly increase the possible cellular states of proteins, resulting in tremendous diversity, complexity and heterogeneity. It is very difficult for traditional molecular or biochemical methods to dissect the complicated PTMs that exist in most signaling pathways. PTM detection and analysis has been a major factor in the development of better mass spectrometers which have been continually improved to operate with greater sensitivity for detecting PTMs [13, 16, 18]. Current high resolution MSs are very powerful, because they enable the identification and quantification of proteins, determination of modified sites and extent of the PTMs under varying conditions [13, 16, 18].
In general, MS based methods for most PTM analyses are similar, but the most established method is utilized in the detection of protein phosphorylation. MS-based proteomics have become indispensable in studying protein phosphorylation [26]. Because of the relatively low abundance of most proteins containing PTMs in cells, the strategy of MS based protein phosphorylation identification includes first enriching the phosphorylated proteins or peptides, and then utilizing the enriched proteins or peptides for identification by MS. The enrichment of phosphorylated proteins or peptides can be achieved by methods involving chemical modification or by direct enrichment. The latter is often employed through resins (Immobilized Metal Ion Affinity Chromatography (iMAC Fe3+), titanium dioxide, strong cation-exchange column (SXC)), or specific antibodies (anti-phosphotyrosine antibody) [19, 20]. MS based identification of phosphorylated sites on proteins is not only high throughput, but also high-fidelity and high speed, which cannot be achieved by other biological or molecular methods. Thousands of phosphorylated proteins and their phosphorylation sites have been identified in many model organisms, from bacteria [27], yeast [28] and worms [29] to plants [30] and animals [31]. Over the last few years, many groups have contributed to cardiovascular phosphoproteomic analysis of cardiac myocytes [32], cardiac mitochondria [33, 34], 20S proteasomes [35] and myofilaments [36].
Quantitative methods
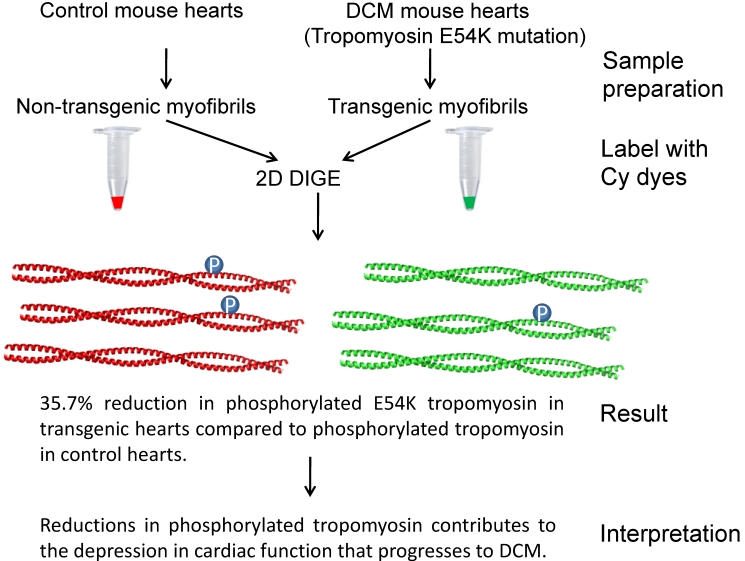
Quantitative methods are universally used to analyze global protein level changes in various biological processes [37-39]. Changes in protein expression levels are important in all diseases including cardiovascular diseases as changes in protein levels can affect the function, location and binding partners of the protein. Quantitative methods allow us to accurately determine how much protein changes. There are several studies using quantitative cardioproteomic techniques to explore CVDs (Table 2). In general, two main types of quantitative proteomic methods are utilized, 1) 2D gel based quantitative methods such as traditional 2D-SDS PAGE (polyacrylamide gel electrophoresis), and DIGE (Differential imaging of gel electrophoresis). While 2D gel based methods have disadvantages, including low dynamic range and inability to properly separate some membrane proteins, it is still widely used as a proteomics research tool because of its advantages. 2D gel based methods are directly observable, relatively inexpensive to perform, and protocols and software for 2D based methods are well developed. Results from 2D gel based research account for a significant proportion of current cardioproteomic studies, such as ventricular hypertrophy [40], pulmonary hypertension [41], hypertensive heart [42], arteriosclerosis [43], dilated cardiomyopathy [44], heart ischemia and ischemia-reperfusion [45], etc. In addition, there are dedicated applications using 2D gel based methods to resolve specific biological questions. A powerful use of 2D based applications is demonstrated in Figure 2. Transgenic mice expressing a mutant form of tropomyosin (E54K) that is associated with dilated cardiomyopathy was investigated by 2D phosphoproteomics [44]. Proteomics showed that decreased phosphorylation of tropomyosin may directly affect myofilament function and be part of the dilated car-diomyopathy signaling pathway in these transgenic mice. Using a rat model of myocardial infarction and 2D phosphoproteomics Dubois et al, 2011 found that phosphorylation of serine residue 208 of rat troponin T (residue 207 of human troponin T) is decreased in plasma and left ventricle of infarcted rat hearts [46]. Further evaluation of human plasma and ventricular samples using a phosphoantibody against serine 207 suggests that decreased phosphorylation of troponin T is a likely biomarker for left ventricular remodelling after myocardial infarction [46].
Table 2.
Summary of cardiovascular diseases investigated by proteomics and major findings
| Disease | Methods | Tissue/cell type or Subcellular fraction | Model | Findings | Ref. |
|---|---|---|---|---|---|
| Acute coronary syndrome | 2DE + MALDI-TOF/TOF + LC/MS/MS | Blood/monocytes | 25 human patients with non-ST-elevation acute coronary syndrome were randomized to receive atorvastatin 80 mg/dL (n = 14) or conventional treatment (n = 11) for two months. | Expression of 20 proteins was modified by intensive treatment with atorvastatin including protein disulfide isomerase ER60 (PDI), annexin 1, prohibitin, and HSP-70. | [93] |
| 2DE, DIGE, MALDIM S/MS | Plasma/pool remaining after depletion of high-abundance proteins. | Plasma from forty patients with acute coronary syndrome were compared with twenty healthy volunteers and 10 stable CAD patients. | Besides proteins previously identified as upregulated in plasma from patients with acute coronary syndrome, four other potential biomarkers were identified: alpha-1-B-glycoprotein, Hakata antigen, tetranectin, and tropomyosin 4 | [94] | |
| 2DE + MALDI-TOF/TOF | circulating platelets | Platelets from 18 patients with non-ST segment elevation acute coronary syndrome and 10 matched stable coronary artery disease patients were compared | 22 proteins differentially expressed including proteins involved in αllbβ3 and GPVI signaling. The number of differentially expressed proteins decreased at day 5 and further decreased 6 months after the acute event. | [77] | |
| iTRAQ + LC-MALDI-TOF/TOF | Cardiac/ventricular myocytes | Guinea pig ventricular myocytes were exposed to either 0 μM H2O2 for 5 min and then 10 units/ml catalase or 30 μM H2O2 for 5 min and then 10 units/ml catalase at 37 °C. | Altered expression of 35 proteins after transient exposure of myocytes to H2O2. Most protens altered were mitochondrial including malate dehydrogenase and cytochrome c oxidase subunit 2. | [95] | |
| Adenaline and Reactive Oxygen species on cardiomyocytes | 2DE + MALDI-TOF/TOF | Cardiac/myocytes and mitochondria from myocytes | Male rat cardiomyocytes under different conditions were compared: (i) control cells, with no exposure; (ii) cells incubated with adrenaline (ADR); (iii) cells exposed to ADR with XXO (xanthine with xanthine oxidase: a reactive oxygen species generating system); and (iv) cells exposed only to XXO. | Differential changes in myosin light chain-2, cytochrome c and voltage-dependent anion channel 1; redox regulation proteins (particular superoxide dismutase); energetic metabolism proteins (ATP synthase alpha chain and dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex); heat shock proteins. | [96] |
| Chronic model of type 1 diabetes | 2DE + MALDI-TOF | Cardiac/none | Hearts of 4-to 5-mo-old control and 0VE26 mice were compared. 0VE26 mice are a chronic model of type 1 diabetes | Altered expression of 20 identified proteins, of which 12 were mitochondrial and included aconitase 2 and ATP synthase, Fl complex, a. | [97] |
| Congestive heart failure | 2DE + DIGE/LC /MS/MS | Cardiac/Left atria myocytes | Dogs weighing 25-32 kg subjected to ventricular-tachypaced for 24hr (n=5) or 2 week periods (n=8). Sham-operated animals (instrumented but not paced) were used as controls (n=4 and 9 for 24hr and 2 week groups respectively). | Extensive changes (upregulation) in cardioprotective heat shock proteins, decreased antioxidant proteins (superoxide dismutase and peroxiredoxin), and desmin and filamin fragmentation in 2-week ventricular-tachypaced atrial cardiomyocytes. | [98] |
| Coronary atherosclerosis | 2DE, LC/MS/MS | Coronary arteries/none | 10 diseased and 7 normal human coronary arteries were compared. | Increased expression of ferritin light chain in diseased coronary arteries. | [99] |
| Dilated Cardiomyopathy (DCM) | Lable free LC/MS/MS | Cardiac/endomyocardial biopsies | Endomyocardial biopsies from 10 patients with inflammatory DCM as well as 7 controls with normal left ventricular function were compared. | 174 proteins were differentially expressed. The major changes in protein expression were observed for mitochondrial and cytoskeletal proteins. Deregulation of proteins of carbohydrate metabolism, the actin cytoskeleton, and extracellular matrix remodeling was observed in DCM samples. | [81] |
| 2DE + MALDITOF/TOF | Cardiac/none | Hearts from cows with bovine hereditary dilated cardiomyopathy were compared with nondiseased bovine hearts. | 24 proteins (including myoglobin) are of decreased abundance in diseased tissue, whilst 11 proteins are of increased abundance in the diseased state. | [100] | |
| 2DE + MALDITOF/TOF | Cardiac/affinit y purification of ubiquitinated proteins | 12 DCM, 9 ischemic (IHD) and 12 unused human donor hearts were compared. | All DCM hearts showed significantly higher expression of certain key enzymes of the ubiquitinproteasome pathway. | [101] | |
| 2DE + DIGE/LC/MS/MS | Cardiac/ventricular homogenates and myofibrils | Comparison of hearts from transgenic mice expressing a mutant tropomyosin (E54K) that is associated with dilated cardiomyopathy. | A significant (~ 40%) decrease in Tm phosphorylation in transgenic DCM hearts compared to nontransgenic mouse hearts. This suggests that altered phosphorylation may be a significant factor in the linkage of the E54K mutation to DCM. | [44] | |
| 2DE + DIGE/LC/MS/MS | Coronary arteries/none | Comparison of nondiseased coronary arteries from human heart transplant donors and patients with DCM with no evidence of coronary artery disease, to coronary arteries from patients with ischemic heart disease (IHD). | Hsp27 showed decreased abundance in ischemic vessels. The expression of cytoskeletal proteins, such as vimentin was significantly reduced, while transgelin and Tm showed significantly increased abundance in vessels with IHD. Together with western blotting data, the results suggest that phospho-Hsp27 protects against vascular disease possibly by stabilizing the actin cytoskeleton within endothelial and/or smooth muscle cells. | [102] | |
| Experimental alcoholic cardiomyopathy | iTRAQ/M ALDI TOF-TOF | Cardiac/ventricular nuclear, mitochondrial, sarcoplasmic and myofibrillar fractions | Male and female rats were maintained on either an alcohol-containing or alcohol-free diet for 18 wk and then compared. | Troponins were oppositely regulated by alcohol exposure in males (downregulated) vs. females (upregulated). Males consuming alcohol showed increased expression of proteins involved in oxidative phosphorylation (complexes I, III, IV, V) whereas females showed no change or decreased content. | [103] |
| Experimental Model of Type 1 Diabetes | iTRAQ + LC/MS/MS | Cardiac/mitochondria | Male rats treated with streptozotocin (60mg/kg) or saline and investigated after 120 days. | 65 proteins differed significantly between the groups: up-regulation of several enzymes involved in the oxidation of long-chain fatty acid, in combination with down-regulation of short-chain fatty acid catabolism. | [104] |
| in vitro stable isotope labeling, 2DE + MALDI-TOF | Cardiac/mitochondria | Diabetic rats and age-matched control rats were investigated at 1 or 4 weeks after STZ injection. | Up-regulation of the fatty acid β-oxidation. Down-regulation of protein levels for creatine kinase, voltage-dependent anion channel 1 (VDAC-1), HSP60, and Grp75. | [105] | |
| 2DE + iTRAQ + MALDI-TOF/TOF | Cardiac/subsarcolemm al mitochondria and interf ibrillar mitochondria | Male mice injected with streptozotocin (50mg/kg) or saline daily for five consecutive days after 6 h of fasting. Five weeks after hyperglycemia onset, animals were euthanized for further experimentation. | Interf ibrillar mitochondria were impacted by type 1 diabetes mellitus to a greater extent than subsarcolemmal mitochondria, including a decrease in abundance of fatty acid oxidation and electron transport chain proteins. Mitochondrial phosphate carrier and adenine nucleotide translocator were decreased in the diabetic interf ibrillar mitochondria | [106] | |
| Experimentaly induced cardiomyopathy | 2DE + MALDI-TOF | Cardiac/none | Comparison of WT, 4 wk (asymptomatic) and 9 wk old (severe cardiomyopathy) frataxin knockout mice. | Pronounced changes in protein expression profile in 9 wk-old KO mice with few changes in 4 wk-old KO mice. Frataxin KO mice showed decreased expression of components of the iron-dependent complex-l and -II of the mitochondrial electron transport chain, enzymes involved in ATP homeostasis (creatine kinase, adenylate kinase) and a variety of chaperones. The KO hearts exhibited increased expression of enzymes involved in the citric acid cycle, catabolism of branched-chain amino acids, ketone body utilization and pyruvate decarboxylation. | [107] |
| Experimental acute myocardial ischemia | isobaric tag labelling, iTRAQ, OFFGEL fractiona tion, LC/MS/MS | Cardiac/sarcomeric, nuclear, and cytoplasmic enriched fractions | Comparison of rat ventricular tissue from ischemic and non-ischemic regions of rat hearts induced by acute myocardial ischemia by ligatingthe left-anterior descending coronary artery in vivo for lhr without reperfusion. | 22 unique proteins in the sarcomeric enriched fraction had changed at least 20% including a decrease in ryanodine receptor 2 in ischemic regions. | [108] |
| Experimental ischemia-reperfusion injury | 2DE, LC/MS/MS | Cardiac/none | Isolated male rat hearts were perfused under aerobic conditions or subjected to ischemia-reperfusion in the presence or absence of the cardioprotective Rho kinase inhibitor, Y-27632. | Y-27632 treatment affected four proteins: lactate dehydrogenase and glyceraldehyde-3-phosphate dehydrogenase were significantly increased in the Y-27632 treated group, while creatine kinase and two different molecular fragments of ATP synthase were normalized to control levels by Y-27632. The cardioprotective effect of Y-27632 likely involves increased energy production. | [109] |
| Experimental hyperdynamic mouse hearts | 2DE, MALDI-TOF, LC-MS/MS | Cardiac/Ventricle | Ventricular proteins from phospholamban-KO and WT mice were compared. | Loss of phospholamban is associated with MLC-1 isoform switching and increased MLC-2v, HSP27 and aB-crystallin phosphorylation. | [110] |
| Left ventricular remodeling (LVR) | SELDI-TOF | Plasma/plasma albumin depleted | Human plasma samples from 93 patients (obtained on day 5 of hospitalization) were divided into three groups (no, low, or high remodeling) and compared. | Post-translational variants of the al-chain of haptoglobin were more elevated in remodeling patients. | [111] |
| 2DE + MALDI-TOF | Cardiac and plasma/left ventricle | Comparison of plasma from 10-week-old male rats which had myocardial infarction induced by left coronary ligation and plasma from 16 sham-operated rats. | 2D phosphoproteomics showed that troponin T phosphorylation was decreased in the left ventricle from rats with LVR. Western blotting with anti-phosphoserine residue 208 of troponin T showed that phosphorylation of this site was decreased in plasma and left ventricles from rats and humans. | [46] | |
| Hypertension | iTRAQ + LC/MS/MS | Cardiac/Mitochondria | 20-month-old spontaneously hypertensive rat (SHR) and Wistar-Kyoto controls were compared | 79 proteins were differentially expressed between groups. Changes in proteins involved in several metabolic pathways, chaperone and antioxidant systems. Multiple subunits of the oxidative phosphorylation complexes were increased (complexes 1, III and IV) or decreased (complexes II and V) in SHR heart mitochondria. | [112] |
| Myocardial Infarction | SELDI-TOFMS | Plasma/lmmu no-purification of cardiac troponin 1 forms from plasma | Cardiac Troponin 1 forms present in the human plasma from 64 patients with acute myocardial infarction | Several forms of cardiac troponin 1 were detected with intact and bis-phosphorylated troponin 1 mostly present by itself. | [113] |
| Reperfusion arrhythmias | isobaric tag labeling, iTRAQ, MALDI TOF-TOF | Cardiac/Left ventricular membrane enriched fraction | Rat myocardial ischemia reperfusion (IR) model was induced by 30 min coronary occlusion and 120 min reperfusion in the presence and absence of grape seed proanthocyanidin (GSPE) extract | 92 differentially expressed proteins. Na+/K+ ATPase al subunit was decreased in IR group while it was significantly increased in GSPE group compared to sham group. | [24] |
| Type 2 diabetes mellitus. | iTRAQ + MALDI TOF-TOF | Cardiac/subsarcolemm al mitochondria and interf ibrillar mitochondria | Pooled subsarcolemmal and interf ibrillar mitochondria subpopulations from 18wt old db/db and WT mouse hearts were compared | Inner mitochondrial membrane proteins and mitochondrial protein import machinery were predominantly decreased in diabetic mitochondria. Subsarcolemmal mitochondria from db/db showed greater differences than interfibrillar mitochondria when compared to their respective WT controls. | [114] |
Figure 2.
Schematic diagram showing a specific phosphorylation related cardiovascular signaling pathway involving tropomyosin which was partly resolved by cardioproteomics. A mutation in tropomyosin (E54K) is associated with dilated cardiomyopathy and this mutation was investigated using transgenic mice expressing this mutant protein [44]. Proteomics revealed that decreased phosphorylation of tropomyosin may directly affect myofilament function and be part of the dilated cardiomyopathy signaling pathway in E54K transgenic mice.
Fernando et al., 2005, used 2D-SDS-PAGE based kinase assays to compare the difference of heart kinase activities between a constitutively active mutant of mitogen-activated protein kinase kinase 6 (MKK6) transgenic mice and wild-type mice [47]. Heart lysates from MKK6 transgenic and wild-type mice were separated by isoelectric focusing (IEF), and then transferred to acrylamide gels containing kinase substrate. The reactions between kinase and substrate were detected using radioactive ATP, allowing the activity and location of kinases on the gel to be detected by autoradiography and the kinases subsequently identified by MS [47]. This group also used a similar method to identify substrates of MKK6 by separating IEF samples in a substrate-free second-dimension gel and incorporating a recombinant active MKK6 in the kinase incubation buffer. They found that the activity of MKK6, p38a, 5'-AMP activated kinase (AMPK), Rho associated kinase (RAK), and the serine/threonine kinase protein kinase N (PKN) was elevated in MKK6 transgenic mice, and some proteins including p38a, α-adducin, hsp90, elF4E, β-tubulin, and E1 ligase could be the substrates of MKK6. This approach allowed the determination of signaling pathways related to cardiomyocyte hypertrophy. In this study, they didn't compare protein expression levels but the activity of kinases, although they cannot exclude the effect on kinase activity levels due to changes in kinase expression levels [47].
2) The other main type of quantitative method used in cardioproteomics is shotgun based methods using stable isotope tagging [38, 48, 49] and label free methods [38, 50]. Since stable isotope tagging methods emerged, they have been well accepted for high-throughput determination of relative protein expression levels under different biological conditions [37, 38]. To introduce the analogue of the same peptide for the discrimination of mass spectrometry, different samples are labeled by light and heavy isotopes separately, resulting in a molecular weight shift between the same proteins in different samples. Several well developed stable isotope tagging methods are currently available including Isotope-Coded Affinity Tags (ICAT) [51, 52], Isobaric Tags for Relative and Absolute Quantitation (iTRAQ) [53], Isotope-Coded Protein Label (ICPL) [54], Tandem Mass Tags (TMT) [55], [180]-water labeling [56], Global Internal Standard Technology (GIST) [57] and Stable Isotope Labeling with Amino Acids in Cell Culture (SILAC) [58, 59]. All these methods except SILAC are in vitro methods used after protein extraction. Only SILAC labels proteins in vivo at the cell or animal level [58, 60]. Stable isotope methods generally achieve more high-throughput and effective data output than 2D gel based methods. However, these methods need to be combined with a high precision MS resulting in more extensive costs than 2D gel based methods. Of these stable isotope tagging methods, SILAC and iTRAQ are the most commonly used techniques for protein quantitation. SILAC introduces fewer quantitative errors compared to other isotopic labeling methods because of the nature of SILAC labeling. SILAC protein isotopic labels (heavy and light) occur intracellularly resulting in decreased artifacts which can be caused by sample fractionation or other manipulations following the mixing of light and heavy cells. As such, SILAC is more suitable for PTM and protein-protein interaction/protein complex analysis [16, 59, 61].
To get the bona fide interaction partners of target proteins, a method of protein complex purification is often combined with a quantitative method, such as ICAT [51], iTRAQ [61, 62] or SILAC [63, 64]. While these combinations of methods have not been significantly used in cardiovascular research for PTM analysis, the potential for the use of these methods is exemplified by successes in other proteomic fields. Olsen employed an integrated phosphoproteomic technology combining phosphopeptide enrichment, high-accuracy identification, and SILAC to determine time-dependent changes in phosphorylation dynamics after stimulating HeLa cells with epidermal growth factor (EGF) [65]. Selbach combined SILAC, RNA interference (RNAi), co-immunoprecipitation and mass-spectrometry analysis to detect cellular interaction partners of b-catenin and Cbl in mammalian cells [64]. This integrated approach significantly reduced non-specific interaction proteins, and was named as QUCIK [64].
Signaling in the understanding of the cellular processes that are responsible for the transition to disease phenotypes
Cardioproteomic approaches have been used to parse signaling molecules at the level of multiple protein complexes/protein interaction networks, PTMs and signaling affecting proteins. These studies often used focused-cardioproteomic techniques, allowing the function of the components of protein complexes investigated to be properly deciphered by straightforward follow-up experiments. The serine/threonine kinase, protein kinase C ε (PKCε), was found to play an important role by forming large multi-protein signaling complexes to accomplish signal transduction in protection against ischemic injury in the heart [66, 67]. In Ping's lab, they used 2DE and 1D SDS-PAGE followed by LC-MS/MS to identify purified PKCε complexes. A total of 93 proteins in PKCε complexes were reported in this study, including structural, signaling, stress-activated, metabolism-related, transcription- and translation-related proteins, which indicate PKCε could regulate multiple signaling pathways to integrate different functions against heart injury [66, 67]. Subsequently, they examined simultaneous association of PKCε and two of its binding partners, Akt and eNOS, in the regulation of NO production and cardiac protection. They found that PKCε could directly phosphorylate Akt and eNOS, and activation of PKCε increased phosphorylation of eNOS in a transgenic mouse heart. These investigations deepened our understanding of the function of the PKCε complex in heart injury protection [68].
Another investigation by Gomes et al., 2006 integrated glycerol gradient/ion-exchange chromatography, 1-DE, 2-DE, Blue native (BN)-PAGE, and LC/MS/MS to investigate the 20S and 26S proteasomes in murine hearts [69]. Proteasomes are very large protein complexes inside all eukaryotes and function mainly to degrade unneeded or damaged proteins. The cardiac 26S proteasome was found to contain alternatively spliced isoforms of Rpn10 as well as several kinases and phosphatases which directly interact with it. This study is an example of using multiple different techniques to obtain a more comprehensive complex analysis [69, 70]. One of the proteins found bound to the proteasome was protein phosphatase 2A (PP2A), which accounts for a large portion of serine/threonine phosphatase activity in cell signal transduction. Functional validation of the presence of PP2A interacting with the proteasome showed that this phosphatase not only interacts with the proteasome but endogenous PP2A in purified proteasomes was capable of regulating the activity of the proteasome, making PP2A a physiologically relevant interacting partner. Other physiologically relevant interacting partners of the cardiac proteasome include the kinases cAMP dependent protein kinase (PKA) and casein kinase II, which help to explain proteasome regulated local signaling and function in the heart [69, 71].
Identifying the types and locations of PTMs in cardiovascular signaling is currently only a small proportion of total cardioproteomic research. However, cardioproteomics have been used to examine several important types of PTMs in CVDs, such as phosphorylation [72], arginylation [73], oxidization [74] and S-nitrosylation [75]. Oxidative stress is the key factor for heart injury in heart ischemia-reperfusion. Chou et al., 2010 used hydrogen peroxide treated H9c2 rat cardiomyocytes as a model to determine changes in tyrosine phosphorylation signaling that may be induced during heart ischemia-reperfusion injury. They utilized anti-phosphotyrosine affinity purification with LC-MS/MS and 2D DIGE coupled with MALDI-TOF MS, and found that that the Src kinase may play an important role in oxidative stress-induced phosphorylation and cell damage in cardiomyocytes [72]. Post-translational arginylation mediated by arginyltransferase (ATE1) plays an important role in cardiovascular development. The arginylation reaction and the functioning of ATE1 remained poorly understood because of the lack of good biochemical models. Wang and colleagues took advantage of 2D SDS-PAGE combined with autoradiography to compare the arginylation functional difference of arginyltransferase (ATE1) isoforms. They incubated ATE1 isoforms and isotope labeled arginine in Ate1 KO cell extract. After 2D SDS-PAGE, the comparison of the difference in arginylated proteins was analyzed by radiography. They found the protein arginylation of the ATE1 isoforms is highly variable in vitro [73]. Tetsuro found that thioredoxin-1 (Trx1) regulates disulfide bond formation between two cysteine residues located in HDAC4 and its interaction protein DnaJb5 by reactive oxygen species (ROS) stimuli in cardiac myocytes [74]. They identified these oxidized cysteine residues located in the HDAC4 and DnaJb5 proteins by MS. In this study, proteomics was critical for the key results which were then further investigated in-depth [74]. Mutation of these cysteine residues in HDAC4 resulted in increased susceptibility to cardiac hypertrophy. Nitric Oxide (NO) is a very important signaling molecule in the cardiovascular system. NO-mediated S-nitrosylation was examined by 2-DE and the level of S-nitrosylation of 11 proteins was found to be significantly increased [75]. To explore the function of extracellular signal-regulated kinase 1/2 (ERK1/2) in cardiomyocytes exposed to ischemic hypoxia and reoxygenation, Mizukami et al., 2004 used 2DE and MS and found increased expression of a-enolase, a rate-limiting enzyme in the glycolytic pathway, in response to ischemic hypoxia [75]. The up-regulation of a-enolase could be inhibited by a MEK inhibitor, PD98059. They also showed that a-enolase could restore ATP levels and prevent cell death during ischemic hypoxia and reoxygenation in heart cells, suggesting that ERK1/2-α-enolase signaling pathway is important in ischemic hypoxia [76]. Platelet activation could be induced by the rupture of an unstable atherosclerotic plaque in acute coronary syndrome (ACS), which may lead to occlusion of an artery supplying a substantial part of the myocardium. In order to explore the functional change of platelets in ACS, Fernández and colleagues isolated platelets from patients with ACS and control patients and found that 14 proteins were differentially changed. These proteins were either signaling or cytoskeletal, and nine of them are known to participate in platelet activation by 2DE and Ingenuity pathway analysis (IPA) [77]. Other quantitative methods including ICAT, iTRAQ and label free proteomics have been used to investigate cardiovascular diseases [78-81].
Although there are many types of quantitative methods currently available, no one method is optimal for all investigations. A combination of different quantitative methods is usually a good choice for specific research goals. The results obtained by different methods are often partly complementary as well as independently beneficial. Taking advantage of these complementary methods often improves the result. Fu et al., 2009 used ICAT and iTRAQ to compare Trx1 induced proteins from the hearts of a cardiac specific Trx1-overexpressing transgenic mouse model. Using the results from ICAT and iTRAQ, they were able to reduce the false positive proteins and improved the accuracy of their protein list [80].
Deciphering of drug related signaling
The use of drugs to treat cardiovascular diseases is an important field of cardiovascular research. The increasing number of patients with cardiovascular diseases amplifies the need for more effective drugs to get better therapeutic efficacy and reduce the cost of treatment for patients. Drug discovery requires not only new drugs but also redesigned drugs. How diseases and infections are controlled at the molecular and physiological level is the basic knowledge required for drug disign. However, comprehensively deciphering the mechanism(s) of a drug's action is still a major challenge for researchers. Recent research on signaling processes has laid the foundation for fundamental understanding of cellular biology and has begun to influence choices made in drug discovery, including drug screens, drug design, drug modification and drug optimization [53, 82]. Most drugs bind to intracellular targets and elicit a series of dynamic signaling changes in cells, which will lead to many positive and negative side effects on the cells during the treatment process [53, 82]. Knowing the drug elicited signaling pathway is important to understand the action and mechanism of these drugs. Mass spectrometry has made important contributions to the understanding of drug related signaling [83-88]. Most drugs derived from small molecular compounds are ideal drug discovery candidates for human diseases. The application of proteomics in drug discovery is increasing and has driven chemical proteomic developments that integrate synthetic organic chemistry, cell biology, biochemistry and mass spectrometry [83-88]. Chemical proteomics focus mainly on the finding of specific drug targets based on small molecular compound probes [85-88]. Nearly half of the drug targets fall into just six gene families: serine/threonine and tyrosine protein kinases, G-protein-coupled receptors (GPCRs), zinc metallopeptidases, serine proteases, nuclear hormone receptors and phosphodiesterases [82].
The interaction of drugs on these targets typically leads to target relevant signaling changes. Many model applications of drug target discovery have been carried out, especially in anti-cancer drug discovery. For example, more than 20 different protein kinases and various other cellular proteins were identified as putative gefitinib (epidermal growth factor receptor (EGFR)-directed kinase inhibitor) targets in HeLa cells through affinity chromatography and MS identification [89]. Overall, the strategy for drug target identification using MS is similar to the identification method used for protein-protein interaction studies. To get more specific drug targets, quantitative proteomic methods can be combined with the above mentioned strategies. Bantscheff et al., 2007 used kinobeads bound to multiple kinase inhibitors combined with iTRAQ and found new drug target proteins. They were also able to profile the effect of a drug on BCR-ABL-RasGAP-MAPK1 signaling pathways [90].
Amid the applications of proteomics in the research of drug related signaling, the integration of quantitative methods and PTM analysis methods is beneficial for exploring pharmaceutical effects on target cells or tissue. Currently there are only a few applications of proteomics in drug research for the treatment of cardiovascular diseases. Salvianolic acid B (SB) is the most abundant and bioactive component of the herb Danshen, which is popularly used in China. It possesses considerable protective effects against cardiovascular disorders such as angina pectoris, myocardial infarction, and stroke. But its direct target proteins and downstream signal -related proteins remain unknown [91]. Feng et al., 2011 predicted that EGFR may be one of the ligands of SB. They employed 2DE to compare the effect of SB on H9c2 cardiac cells undergoing stimulated ischemic-reperfusion (IR) injury, and found 9 signaling-related proteins involved in a network together with EGFR through direct interaction [91]. Atenolol, a β1-selective drug, can significantly block β1 adreno-receptor activity. In order to investigate the difference between R- and S-enantiomers of atenolol on vascular smooth muscle cells, they employed an iTRAQ-coupled 2D LC-MS/MS approach. Their data showed some molecular evidence on the metabolic effect and possible link of calcium-binding proteins with treatment of hypertension associated with atenolol [92]. Since CVD is the major cause of death due to disease worldwide, cardiovascular drug discovery will continue and cardioproteomics will be a major player in drug discovery research.
Future prospects
With continued improvements in proteomic methods, exploration of cardiovascular signaling pathways will be a central point in future cardioproteomics applications. Although many advanced proteomic techniques are currently available, utilization of cardioproteomics is currently still limited in CVD signaling research, especially in CVD related drug signaling. This is partially due to the specific characteristics of heart tissue. 2DE based approaches still account for a significant proportion of cardioproteomics (Table 2). Most shotgun based proteomic techniques are still labor intensive and costly for many cardiovascular researchers. To some extent, 2DE based and shotgun based techniques are complementary for resolving some biological questions. For optimal results it is important to choose the strategy most suitable to the study goals.
The most advanced proteomic techniques are not always the best choice for some studies. In addition, the most advanced techniques still are unable to routinely detect some PTMs. It is still difficult to analyze glycosylation, ubiquitinylation and very transient PTM events by MS based PTM detection. The strategy of combining different approaches is encouraged so as to make the most of the unique advantages of different techniques in resolving biological problems. For most biologists, it is important to discuss their goals with proteomic experts to help find the most suitable strategy to achieve their research aims. Although proteomics has been applied to various fields of life science as a tool for biological research, it is not one size-fits-all, and has certain limitations to answering some biological questions. Proteomics has a disadvantage in that it is unable to detect the functional details of identified proteins in signaling pathways. The integration of proteomic methods with other biological and molecular methods is very important to unraveling functional details of certain proteins.
During the process of charting signaling pathways, cardioproteomic techniques are often used either on a restrictive scale or large scale. Both restrictive and global characteristics of proteomic investigations help to decipher the signaling pathways in different CVDs. The data produced by proteomics are usually large compared to other biological studies. Most proteomic labs are unable to fully analyze the proteomic data to achieve the functional and phenotypic assessment in different models. Dissecting the specific function of these signaling modules could be achieved by collaboration between proteomics laboratories and laboratories that specialize with functional studies. Because the heart and aorta are specialized tissues, protein extraction approaches should be developed or optimized for CVD signaling research. Although there are some challenges ahead for the widespread application of cardioproteomics in the future, it will continue to be a powerful tool for signaling pathway detection, especially for protein complex and PTMs in CVD relevant research. As Table 1 shows, relatively few cardiovascular diseases have been investigated using proteomic techniques compared to the total number of different cardiovascular diseases. Both signaling in CVDs and signaling of CVD related drugs are important research fields in the near future.
Acknowledgments
The authors' work was supported by NIH grant HL096819 (AVG), American Heart Association grant 0835335N (AVG) and partly supported by a HHMI basic science research training program (SD).
References
- 1.Roger VL, Go AS, Lloyd-Jones DM, Adams RJ, Berry JD, Brown TM, Carnethon MR, Dai S, de Simone G, Ford ES, Fox CS, Fullerton HJ, Gillespie C, Greenlund KJ, Hailpern SM, Heit JA, Ho PM, Howard VJ, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Makuc DM, Marcus GM, Marelli A, Matchar DB, McDermott MM, Meigs JB, Moy CS, Mozaffarian D, Mussolino ME, Nichol G, Paynter NP, Rosamond WD, Sorlie PD, Stafford RS, Turan TN, Turner MB, Wong ND, Wylie-Rosett J. Heart disease and stroke statistics-2011 update: a report from the American Heart Association. Circulation. 2011;123:e18–e209. doi: 10.1161/CIR.0b013e3182009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arrell DK, Neverova I, Van Eyk JE. Cardiovascular proteomics: evolution and potential. Circ Res. 2001;88:763–773. doi: 10.1161/hh0801.090193. [DOI] [PubMed] [Google Scholar]
- 3.Ping P. Identification of novel signaling complexes by functional proteomics. Circ Res. 2003;93:595–603. doi: 10.1161/01.RES.0000093221.98213.E0. [DOI] [PubMed] [Google Scholar]
- 4.Agnetti G, Husberg C, Van Eyk JE. Divide and conquer: the application of organelle proteomics to heart failure. Circ Res. 2011;108:512–526. doi: 10.1161/CIRCRESAHA.110.226910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ping P, Chan DW, Srinivas P. Advancing cardiovascular biology and medicine via proteomics: Opportunities and present challenges of cardiovascular proteomics. Circulation. 2010;121:2326–2328. doi: 10.1161/CIRCULATIONAHA.110.949230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards AV, White MY, Cordwell SJ. The role of proteomics in clinical cardiovascular biomarker discovery. Mol Cell Proteomics. 2008;7:1824–1837. doi: 10.1074/mcp.R800007-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Arab S, Gramolini AO, Ping P, Kislinger T, Stanley B, van Eyk J, Ouzounian M, MacLennan DH, Emili A, Liu PP. Cardiovascular proteomics: tools to develop novel biomarkers and potential applications. J Am Coll Cardiol. 2006;48:1733–1741. doi: 10.1016/j.jacc.2006.06.063. [DOI] [PubMed] [Google Scholar]
- 8.Mallick P, Kuster B. Proteomics: a pragmatic perspective. Nat Biotechnol. 2010;28:695–709. doi: 10.1038/nbt.1658. [DOI] [PubMed] [Google Scholar]
- 9.Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 10.Steen H, Mann M. The ABC's (and XYZ's) of peptide sequencing. Nat Rev Mol Cell Biol. 2004;5:699–711. doi: 10.1038/nrm1468. [DOI] [PubMed] [Google Scholar]
- 11.Colland F, Jacq X, Trouplin V, Mougin C, Groizeleau C, Hamburger A, Meil A, Wojcik J, Legrain P, Gauthier JM. Functional proteomics mapping of a human signaling pathway. Genome Res. 2004;14:1324–1332. doi: 10.1101/gr.2334104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kocher T, Superti-Furga G. Mass spectrometry-based functional proteomics: from molecular machines to protein networks. Nat Methods. 2007;4:807–815. doi: 10.1038/nmeth1093. [DOI] [PubMed] [Google Scholar]
- 13.Jensen ON. Modification-specific proteomics: characterization of post-translational modifications by mass spectrometry. Curr Opin Chem Biol. 2004;8:33–41. doi: 10.1016/j.cbpa.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 14.Major MB, Camp ND, Berndt JD, Yi X, Goldenberg SJ, Hubbert C, Biechele TL, Gingras AC, Zheng N, Maccoss MJ, Angers S, Moon RT. Wilms tumor suppressor WTX negatively regulates WNT/beta-catenin signaling. Science. 2007;316:1043–1046. doi: 10.1126/science/1141515. [DOI] [PubMed] [Google Scholar]
- 15.Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G, Drewes G, Gavin AC, Jackson DB, Joberty G, Neubauer G, Rick J, Kuster B, Superti-Furga G. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 16.Choudhary C, Mann M. Decoding signalling networks by mass spectrometry-based proteomics. Nat Rev Mol Cell Biol. 2010;11:427–439. doi: 10.1038/nrm2900. [DOI] [PubMed] [Google Scholar]
- 17.Jensen ON. Interpreting the protein language using proteomics. Nat Rev Mol Cell Biol. 2006;7:391–403. doi: 10.1038/nrm1939. [DOI] [PubMed] [Google Scholar]
- 18.Witze ES, Old WM, Resing KA, Ahn NG. Mapping protein post-translational modifications with mass spectrometry. Nat Methods. 2007;4:798–806. doi: 10.1038/nmeth1100. [DOI] [PubMed] [Google Scholar]
- 19.Kholodenko BN. Cell-signalling dynamics in time and space. Nat Rev Mol Cell Biol. 2006;7:165–176. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brandman O, Meyer T. Feedback loops shape cellular signals in space and time. Science. 2008;322:390–395. doi: 10.1126/science.1160617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gingras AC, Aebersold R, Raught B. Advances in protein complex analysis using mass spectrometry. J Physiol. 2005;563:11–21. doi: 10.1113/jphysiol.2004.080440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gingras AC, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes using mass spectrometry. Nat Rev Mol Cell Biol. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- 23.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 24.Graumann J, Dunipace LA, Seol JH, McDonald WH, Yates JR, 3rd, Wold BJ, Deshaies RJ. Applicability of tandem affinity purification Mud-PIT to pathway proteomics in yeast. Mol Cell Proteomics. 2004;3:226–237. doi: 10.1074/mcp.M300099-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Xu K, Sacharidou A, Fu S, Chong DC, Skaug B, Chen ZJ, Davis GE, Cleaver O. Blood vessel tubulogenesis requires Rasip1 regulation of GTPase signaling. Dev Cell. 2011;20:526–539. doi: 10.1016/j.devcel.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Preisinger C, von Kriegsheim A, Matallanas D, Kolch W. Proteomics and phosphoproteomics for the mapping of cellular signalling networks. Proteomics. 2008;8:4402–4415. doi: 10.1002/pmic.200800136. [DOI] [PubMed] [Google Scholar]
- 27.Soufi B, Jers C, Hansen ME, Petranovic D, Mijakovic I. Insights from site-specific phospho-proteomics in bacteria. Biochim Biophys Acta. 2008;1784:186–192. doi: 10.1016/j.bbapap.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 28.Gruhler A, Olsen JV, Mohammed S, Mortensen P, Faergeman NJ, Mann M, Jensen ON. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol Cell Proteomics. 2005;4:310–327. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Zielinska DF, Gnad F, Jedrusik-Bode M, Wisniewski JR, Mann M. Caenorhabditis elegans has a phosphoproteome atypical for metazoans that is enriched in developmental and sex determination proteins. Journal of proteome research. 2009;8:4039–4049. doi: 10.1021/pr900384k. [DOI] [PubMed] [Google Scholar]
- 30.Komatsu S, Tanaka N. Rice proteome analysis: a step toward functional analysis of the rice genome. Proteomics. 2005;5:938–949. doi: 10.1002/pmic.200401040. [DOI] [PubMed] [Google Scholar]
- 31.Ballif BA, Villen J, Beausoleil SA, Schwartz D, Gygi SP. Phosphoproteomic analysis of the developing mouse brain. Mol Cell Proteomics. 2004;3:1093–1101. doi: 10.1074/mcp.M400085-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Chu G, Egnaczyk GF, Zhao W, Jo SH, Fan GC, Maggio JE, Xiao RP, Kranias EG. Phosphoproteome analysis of cardiomyocytes subjected to beta-adrenergic stimulation: identification and characterization of a cardiac heat shock protein p20. Circ Res. 2004;94:184–193. doi: 10.1161/01.RES.0000107198.90218.21. [DOI] [PubMed] [Google Scholar]
- 33.Deng N, Zhang J, Zong C, Wang Y, Lu H, Yang P, Wang W, Young GW, Korge P, Lotz C, Doran P, Liem DA, Apweiler R, Weiss JN, Duan H, Ping P. Phosphoproteome analysis reveals regulatory sites in major pathways of cardiac mitochondria. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.000117. MHO 000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng J, Zhu M, Schaub MC, Gehrig P, Roschitzki B, Lucchinetti E, Zaugg M. Phosphoproteome analysis of isoflurane-protected heart mitochondria: phosphorylation of adenine nucleotide translocator-1 on Tyr194 regulates mitochondrial function. Cardiovasc Res. 2008;80:20–29. doi: 10.1093/cvr/cvn161. [DOI] [PubMed] [Google Scholar]
- 35.Lu H, Zong C, Wang Y, Young GW, Deng N, Souda P, Li X, Whitelegge J, Drews O, Yang PY, Ping P. Revealing the dynamics of the 20 S proteasome phosphoproteome: a combined CID and electron transfer dissociation approach. Mol Cell Proteomics. 2008;7:2073–2089. doi: 10.1074/mcp.M800064-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan C, Sheng Q, Tang H, Li Y, Zeng R, Solaro RJ. Quantitative comparison of sarcomeric phosphoproteomes of neonatal and adult rat hearts. Am J Physiol Heart Circ Physiol. 2008;295:H647–656. doi: 10.1152/ajpheart.00357.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 38.Elliott MH, Smith DS, Parker CE, Borchers C. Current trends in quantitative proteomics. J Mass Spectrom. 2009;44:1637–1660. doi: 10.1002/jms.1692. [DOI] [PubMed] [Google Scholar]
- 39.Wilm M. Quantitative proteomics in biological research. Proteomics. 2009;9:4590–4605. doi: 10.1002/pmic.200900299. [DOI] [PubMed] [Google Scholar]
- 40.Gallego-Delgado J, Lazaro A, Osende Jl, Barderas MG, Duran MC, Vivanco F, Egido J. Comparison of the protein profile of established and regressed hypertension-induced left ventricular hypertrophy. Journal of proteome research. 2006;5:404–413. doi: 10.1021/pr0503275. [DOI] [PubMed] [Google Scholar]
- 41.Kwapiszewska G, Wygrecka M, Marsh LM, Schmitt S, Trosser R, Wilhelm J, Helmus K, Eul B, Zakrzewicz A, Ghofrani HA, Schermuly RT, Bohle RM, Grimminger F, Seeger W, Eickelberg O, Fink L, Weissmann N. Fhl-1, a new key protein in pulmonary hypertension. Circulation. 2008;118:1183–1194. doi: 10.1161/CIRCULATIONAHA.107.761916. [DOI] [PubMed] [Google Scholar]
- 42.Arias-Loza PA, Hu K, Dienesch C, Mehlich AM, Konig S, Jazbutyte V, Neyses L, Hegele-Hartung C, Heinrich Fritzemeier K, Pelzer T. Both estrogen receptor subtypes, alpha and beta, attenuate cardiovascular remodeling in aldosterone salt-treated rats. Hypertension. 2007;50:432–438. doi: 10.1161/HYPERTENSIONAHA.106.084798. [DOI] [PubMed] [Google Scholar]
- 43.Huang ZY, Yang PY, Almofti MR, Yu YL, Rui YC. Comparative analysis of the proteome of left ventricular heart of arteriosclerosis in rat. Life Sci. 2004;75:3103–3115. doi: 10.1016/j.lfs.2004.04.054. [DOI] [PubMed] [Google Scholar]
- 44.Warren CM, Arteaga GM, Rajan S, Ahmed RP, Wieczorek DF, Solaro RJ. Use of 2-D DIGE analysis reveals altered phosphorylation in a tropomyosin mutant (Glu54Lys) linked to dilated cardiomyopathy. Proteomics. 2008;8:100–105. doi: 10.1002/pmic.200700772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakai J, Ishikawa H, Satoh H, Yamamoto S, Kojima S, Kanaoka M. Two-dimensional differential gel electrophoresis of rat heart proteins in ischemia and ischemia-reperfusion. Methods Mol Biol. 2007;357:33–43. doi: 10.1385/1-59745-214-9:33. [DOI] [PubMed] [Google Scholar]
- 46.Dubois E, Richard V, Mulder P, Lamblin N, Drobecq H, Henry JP, Amouyel P, Thuillez C, Bauters C, Pinet F. Decreased serine207 phosphorylation of troponin T as a biomarker for left ventricular remodelling after myocardial infarction. Eur Heart J. 2011;32:115–123. doi: 10.1093/eurheartj/ehq108. [DOI] [PubMed] [Google Scholar]
- 47.Fernando P, Deng W, Pekalska B, DeRepentigny Y, Kothary R, Kelly JF, Megeney LA. Active kinase proteome screening reveals novel signal complexity in cardiomyopathy. Mol Cell Proteomics. 2005;4:673–682. doi: 10.1074/mcp.M400200-MCP200. [DOI] [PubMed] [Google Scholar]
- 48.Beynon RJ, Pratt JM. Metabolic labeling of proteins for proteomics. Mol Cell Proteomics. 2005;4:857–872. doi: 10.1074/mcp.R400010-MCP200. [DOI] [PubMed] [Google Scholar]
- 49.Gouw JW, Krijgsveld J, Heck AJ. Quantitative proteomics by metabolic labeling of model organisms. Mol Cell Proteomics. 2010;9:11–24. doi: 10.1074/mcp.R900001-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiener MC, Sachs JR, Deyanova EG, Yates NA. Differential mass spectrometry: a label-free LC-MS method for finding significant differences in complex peptide and protein mixtures. Anal Chem. 2004;76:6085–6096. doi: 10.1021/ac0493875. [DOI] [PubMed] [Google Scholar]
- 51.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 52.Han DK, Eng J, Zhou H, Aebersold R. Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nat Biotechnol. 2001;19:946–951. doi: 10.1038/nbt1001-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 54.Schmidt A, Kellermann J, Lottspeich F. A novel strategy for quantitative proteomics using isotope-coded protein labels. Proteomics. 2005;5:4–15. doi: 10.1002/pmic.200400873. [DOI] [PubMed] [Google Scholar]
- 55.Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 56.Wang YK, Ma Z, Quinn DF, Fu EW. Inverse 180 labeling mass spectrometry for the rapid identification of marker/target proteins. Anal Chem. 2001;73:3742–3750. doi: 10.1021/ac010043d. [DOI] [PubMed] [Google Scholar]
- 57.Chakraborty A, Regnier FE. Global internal standard technology for comparative proteomics. J Chromatogr A. 2002;949:173–184. doi: 10.1016/s0021-9673(02)00047-x. [DOI] [PubMed] [Google Scholar]
- 58.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 59.Ibarrola N, Kalume DE, Gronborg M, Iwahori A, Pandey A. A proteomic approach for quantitation of phosphorylation using stable isotope labeling in cell culture. Anal Chem. 2003;75:6043–6049. doi: 10.1021/ac034931f. [DOI] [PubMed] [Google Scholar]
- 60.Kruger M, Moser M, Ussar S, Thievessen I, Luber CA, Forner F, Schmidt S, Zanivan S, Fassler R, Mann M. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell. 2008;134:353–364. doi: 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 61.Corvey C, Koetter P, Beckhaus T, Hack J, Hofmann S, Hampel M, Stein T, Karas M, Entian KD. Carbon Source-dependent assembly of the Snf1p kinase complex in Candida albicans. J Biol Chem. 2005;280:25323–25330. doi: 10.1074/jbc.M503719200. [DOI] [PubMed] [Google Scholar]
- 62.Zieske LR. A perspective on the use of iTRAQ reagent technology for protein complex and profiling studies. J Exp Bot. 2006;57:1501–1508. doi: 10.1093/jxb/erj168. [DOI] [PubMed] [Google Scholar]
- 63.Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol. 2006;7:952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 64.Selbach M, Mann M. Protein interaction screening by quantitative immunoprecipitation combined with knockdown (QUICK) Nat Methods. 2006;3:981–983. doi: 10.1038/nmeth972. [DOI] [PubMed] [Google Scholar]
- 65.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 66.Edmondson RD, Vondriska TM, Biederman KJ, Zhang J, Jones RC, Zheng Y, Allen DL, Xiu JX, Cardwell EM, Pisano MR, Ping P. Protein kinase C epsilon signaling complexes include metabolism- and transcription/translation-related proteins: complimentary separation techniques with LC/MS/MS. Mol Cell Proteomics. 2002;1:421–433. doi: 10.1074/mcp.m100036-mcp200. [DOI] [PubMed] [Google Scholar]
- 67.Ping P, Zhang J, Pierce WM, Jr, Bolli R. Functional proteomic analysis of protein kinase C epsilon signaling complexes in the normal heart and during cardioprotection. Circ Res. 2001;88:59–62. doi: 10.1161/01.res.88.1.59. [DOI] [PubMed] [Google Scholar]
- 68.Zhang J, Baines CP, Zong C, Cardwell EM, Wang G, Vondriska TM, Ping P. Functional proteo-mic analysis of a three-tier PKCepsilon-Akt-eNOS signaling module in cardiac protection. Am J Physiol Heart Circ Physiol. 2005;288:H954–961. doi: 10.1152/ajpheart.00756.2004. [DOI] [PubMed] [Google Scholar]
- 69.Gomes AV, Zong C, Edmondson RD, Li X, Stefani E, Zhang J, Jones RC, Thyparambil S, Wang GW, Qiao X, Bardag-Gorce F, Ping P. Mapping the murine cardiac 26S proteasome complexes. Circ Res. 2006;99:362–371. doi: 10.1161/01.RES.0000237386.98506.f7. [DOI] [PubMed] [Google Scholar]
- 70.Zong C, Gomes AV, Drews O, Li X, Young GW, Berhane B, Qiao X, French SW, Bardag-Gorce F, Ping P. Regulation of murine cardiac 20S proteasomes: role of associating partners. Circ Res. 2006;99:372–380. doi: 10.1161/01.RES.0000237389.40000.02. [DOI] [PubMed] [Google Scholar]
- 71.Zhou XW, Mudannayake M, Green M, Gigena MS, Wang G, Shen RF, Rogers TB. Proteomic studies of PP2A-B56gamma1 phosphatase complexes reveal phosphorylation-regulated partners in cardiac local signaling. Journal of proteome research. 2007;6:3433–3442. doi: 10.1021/pr060619l. [DOI] [PubMed] [Google Scholar]
- 72.Chou HC, Chen YW, Lee TR, Wu FS, Chan HT, Lyu PC, Timms JF, Chan HL. Proteomics study of oxidative stress and Src kinase inhibition in H9C2 cardiomyocytes: a cell model of heart ischemia-reperfusion injury and treatment. Free Radic Biol Med. 2010;49:96–108. doi: 10.1016/j.freeradbiomed.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 73.Wang J, Han X, Saha S, Xu T, Rai R, Zhang F, Wolf YI, Wolfson A, Yates JR, 3rd, Kashina A. Arginyltransferase is an ATP-independentself -regulating enzyme that forms distinct functional complexes in vivo. Chem Biol. 2011;18:121–130. doi: 10.1016/j.chembiol.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ago T, Liu T, Zhai P, Chen W, Li H, Molkentin JD, Vatner SF, Sadoshima J. A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy. Cell. 2008;133:978–993. doi: 10.1016/j.cell.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 75.Chen SC, Huang B, Liu YC, Shyu KG, Lin PY, Wang DL. Acute hypoxia enhances proteins' S-nitrosylation in endothelial cells. Biochem Biophys ResCommun. 2008;377:1274–1278. doi: 10.1016/j.bbrc.2008.10.144. [DOI] [PubMed] [Google Scholar]
- 76.Mizukami Y, Iwamatsu A, Aki T, Kimura M, Nakamura K, Nao T, Okusa T, Matsuzaki M, Yoshida K, Kobayashi S. ERK1/2 regulates intracellular ATP levels through alpha-enolase expression in cardiomyocytes exposed to ischemic hypoxia and reoxygenation. J Biol Chem. 2004;279:50120–50131. doi: 10.1074/jbc.M402299200. [DOI] [PubMed] [Google Scholar]
- 77.Parguina AF, Grigorian-Shamajian L, Agra RM, Teijeira-Fernandez E, Rosa I, Alonso J, Vinuela-Roldan JE, Seoane A, Gonzalez-Juanatey JR, Garcia A. Proteins involved in platelet signaling are differentially regulated in acute coronary syndrome: a proteomic study. PLoS One. 2010;5:e13404. doi: 10.1371/journal.pone.0013404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grant JE, Bradshaw AD, Schwacke JH, Baicu CF, Zile MR, Schey KL. Quantification of protein expression changes in the aging left ventricle of Rattus norvegicus. Journal of proteome research. 2009;8:4252–4263. doi: 10.1021/pr900297f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Isserlin R, Merico D, Alikhani-Koupaei R, Gramolini A, Bader GD, Emili A. Pathway analysis of dilated cardiomyopathy using global proteomic profiling and enrichment maps. Proteomics. 2010;10:1316–1327. doi: 10.1002/pmic.200900412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fu C, Wu C, Liu T, Ago T, Zhai P, Sadoshima J, Li H. Elucidation of thioredoxin target protein networks in mouse. Mol Cell Proteomics. 2009;8:1674–1687. doi: 10.1074/mcp.M800580-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hammer E, Goritzka M, Ameling S, Darm K, Steil L, Klingel K, Trimpert C, Herda LR, Dorr M, Kroemer HK, Kandolf R, Staudt A, Felix SB, Volker U. Characterization of the human myocardial proteome in inflammatory dilated cardiomyopathy by label-free quantitative shotgun proteomics of heart biopsies. Journal of proteome research. 2011;10:2161–2171. doi: 10.1021/pr1008042. [DOI] [PubMed] [Google Scholar]
- 82.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 83.Dandapani S, Marcaurelle LA. Grand challenge commentary: Accessing new chemical space for ‘undruggable’ targets. Nat Chem Biol. 2010;6:861–863. doi: 10.1038/nchembio.479. [DOI] [PubMed] [Google Scholar]
- 84.Kruse U, Bantscheff M, Drewes G, Hopf C. Chemical and pathway proteomics: powerful tools for oncology drug discovery and personalized health care. Mol Cell Proteomics. 2008;7:1887–1901. doi: 10.1074/mcp.R800006-MCP200. [DOI] [PubMed] [Google Scholar]
- 85.Rix U, Superti-Furga G. Target profiling of small molecules by chemical proteomics. Nat Chem Biol. 2009;5:616–624. doi: 10.1038/nchembio.216. [DOI] [PubMed] [Google Scholar]
- 86.Kritzer JA. Grand challenge commentary: Beyond discovery: probes that see, grab and poke. Nat Chem Biol. 2010;6:868–870. doi: 10.1038/nchembio.469. [DOI] [PubMed] [Google Scholar]
- 87.Inglese J, Johnson RL, Simeonov A, Xia M, Zheng W, Austin CP, Auld DS. High-throughput screening assays for the identification of chemical probes. Nat Chem Biol. 2007;3:466–479. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- 88.Wierzba K, Muroi M, Osada H. Proteomics accelerating the identification of the target molecule of bioactive small molecules. Curr Opin Chem Biol. 2011;15:57–65. doi: 10.1016/j.cbpa.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 89.Brehmer D, Greff Z, Godl K, Blencke S, Kurtenbach A, Weber M, Muller S, Klebl B, Cotten M, Keri G, Wissing J, Daub H. Cellular targets of gefitinib. Cancer Res. 2005;65:379–382. [PubMed] [Google Scholar]
- 90.Bantscheff M, Eberhard D, Abraham Y, Bastuck S, Boesche M, Hobson S, Mathieson T, Perrin J, Raida M, Rau C, Reader V, Sweetman G, Bauer A, Bouwmeester T, Hopf C, Kruse U, Neubauer G, Ramsden N, Rick J, Kuster B, Drewes G. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat Biotechnol. 2007;25:1035–1044. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]
- 91.Feng LX, Jing CJ, Tang KL, Tao L, Cao ZW, Wu WY, Guan SH, Jiang BH, Yang M, Liu X, Guo DA. Clarifying the signal network of salvianolic acid B using proteomic assay and bioinformatic analysis. Proteomics. 2011;11:1473–1485. doi: 10.1002/pmic.201000482. [DOI] [PubMed] [Google Scholar]
- 92.Sui J, Zhang J, Tan TL, Ching CB, Chen WN. Comparative proteomics analysis of vascular smooth muscle cells incubated with S- and R- enantiomers of atenolol using iTRAQ-coupled two-dimensional LC-MS/MS. Mol Cell Proteomics. 2008;7:1007–1018. doi: 10.1074/mcp.M700485-MCP200. [DOI] [PubMed] [Google Scholar]
- 93.Barderas MG, Tunon J, Darde VM, De la Cuesta F, Jimenez-Nacher JJ, Tarin N, Lopez-Bescos L, Egido J, Vivanco F. Atorvastatin modifies the protein profile of circulating human monocytes after an acute coronary syndrome. Proteomics. 2009;9:1982–1993. doi: 10.1002/pmic.200700583. [DOI] [PubMed] [Google Scholar]
- 94.Darde VM, de la Cuesta F, Dones FG, Alvarez-Llamas G, Barderas MG, Vivanco F. Analysis of the plasma proteome associated with acute coronary syndrome: does a permanent protein signature exist in the plasma of ACS patients? Journal of proteome research. 2010;9:4420–4432. doi: 10.1021/pr1002017. [DOI] [PubMed] [Google Scholar]
- 95.Seenarain V, Viola HM, Ravenscroft G, Casey TM, Lipscombe RJ, Ingley E, Laing NG, Bringans SD, Hool LC. Evidence of altered guinea pig ventricular cardiomyocyte protein expression and growth in response to a 5 min in vitro exposure to H(2)0(2) Journal of proteome research. 2010;9:1985–1994. doi: 10.1021/pr9011393. [DOI] [PubMed] [Google Scholar]
- 96.Costa VM, Silva R, Tavares LC, Vitorino R, Amado F, Carvalho F, Bastos Mde L, Carvalho M, Carvalho RA, Remiao F. Adrenaline and reactive oxygen species elicit proteome and energetic metabolism modifications in freshly isolated rat cardiomyocytes. Toxicology. 2009;260:84–96. doi: 10.1016/j.tox.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 97.Shen X, Zheng S, Thongboonkerd V, Xu M, Pierce WM, Jr, Klein JB, Epstein PN. Cardiac mitochondrial damage and biogenesis in a chronic model of type 1 diabetes. Am J Physiol Endocrinol Metab. 2004;287:E896–905. doi: 10.1152/ajpendo.00047.2004. [DOI] [PubMed] [Google Scholar]
- 98.De Souza AI, Cardin S, Wait R, Chung YL, Vijayakumar M, Maguy A, Camm AJ, Nattel S. Proteomic and metabolomic analysis of atrial profibrillatory remodelling in congestive heart failure. J Mol Cell Cardiol. 2010;49:851–863. doi: 10.1016/j.yjmcc.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 99.You SA, Archacki SR, Angheloiu G, Moravec CS, Rao S, Kinter M, Topol EJ, Wang Q. Proteomic approach to coronary atherosclerosis shows ferritin light chain as a significant marker: evidence consistent with iron hypothesis in atherosclerosis. Physiological genomics. 2003;13:25–30. doi: 10.1152/physiolgenomics.00124.2002. [DOI] [PubMed] [Google Scholar]
- 100.Weekes J, Wheeler CH, Yan JX, Weil J, Eschenhagen T, Scholtysik G, Dunn MJ. Bovine dilated cardiomyopathy: proteomic analysis of an animal model of human dilated cardiomyopathy. Electrophoresis. 1999;20:898–906. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<898::AID-ELPS898>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 101.Weekes J, Morrison K, Mullen A, Wait R, Barton P, Dunn MJ. Hyperubiquitination of proteins in dilated cardiomyopathy. Proteomics. 2003;3:208–216. doi: 10.1002/pmic.200390029. [DOI] [PubMed] [Google Scholar]
- 102.Robinson AA, Dunn MJ, McCormack A, dos Remedios C, Rose ML. Protective effect of phosphorylated Hsp27 in coronary arteries through actin stabilization. J Mol Cell Cardiol. 2010;49:370–379. doi: 10.1016/j.yjmcc.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 103.Fogle RL, Hollenbeak CS, Stanley BA, Vary TC, Kimball SR, Lynch CJ. Functional proteomic analysis reveals sex-dependent differences in structural and energy-producing myocardial proteins in rat model of alcoholic cardiomyopathy. Physiological genomics. 2011;43:346–356. doi: 10.1152/physiolgenomics.00203.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jullig M, Hickey AJ, Middleditch MJ, Crossman DJ, Lee SC, Cooper GJ. Characterization of proteomic changes in cardiac mitochondria in streptozotocin-diabetic rats using iTRAQ isobaric tags. Proteomics Clin Appl. 2007;1:565–576. doi: 10.1002/prca.200600831. [DOI] [PubMed] [Google Scholar]
- 105.Turko IV, Murad F. Quantitative protein profiling in heart mitochondria from diabetic rats. J Biol Chem. 2003;278:35844–35849. doi: 10.1074/jbc.M303139200. [DOI] [PubMed] [Google Scholar]
- 106.Baseler WA, Dabkowski ER, Williamson CL, Croston TL, Thapa D, Powell MJ, Razunguzwa TT, Hollander JM. Proteomic alterations of distinct mitochondrial subpopulations in the type 1 diabetic heart: contribution of protein import dysfunction. Am J Physiol Regul Integr Comp Physiol. 2011;300:R186–200. doi: 10.1152/ajpregu.00423.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sutak R, Xu X, Whitnall M, Kashem MA, Vyoral D, Richardson DR. Proteomic analysis of hearts from frataxin knockout mice: marked rearrangement of energy metabolism, a response to cellular stress and altered expression of proteins involved in cell structure, motility and metabolism. Proteomics. 2008;8:1731–1741. doi: 10.1002/pmic.200701049. [DOI] [PubMed] [Google Scholar]
- 108.Warren CM, Geenen DL, Helseth DL, Jr, Xu H, Solaro RJ. Sub-proteomic fractionation, iTRAQ, and OFFGEL-LC-MS/MS approaches to cardiac proteomics. J Proteomics. 2010;73:1551–1561. doi: 10.1016/j.jprot.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cadete VJ, Sawicka J, Polewicz D, Doroszko A, Wozniak M, Sawicki G. Effect of the Rho kinase inhibitor Y-27632 on the proteome of hearts with ischemia-reperfusion injury. Proteomics. 2010;10:4377–4385. doi: 10.1002/pmic.201000393. [DOI] [PubMed] [Google Scholar]
- 110.Chu G, Kerr JP, Mitton B, Egnaczyk GF, Vazquez JA, Shen M, Kilby GW, Stevenson TI, Maggio JE, Vockley J, Rapundalo ST, Kranias EG. Proteomic analysis of hyperdynamic mouse hearts with enhanced sarcoplasmic reticulum calcium cycling. FASEB J. 2004;18:1725–1727. doi: 10.1096/fj.04-2025fje. [DOI] [PubMed] [Google Scholar]
- 111.Pinet F, Beseme O, Cieniewski-Bernard C, Drobecq H, Jourdain S, Lamblin N, Amouyel P, Bauters C. Predicting left ventricular remodeling after a first myocardial infarction by plasma proteome analysis. Proteomics. 2008;8:1798–1808. doi: 10.1002/pmic.200700781. [DOI] [PubMed] [Google Scholar]
- 112.Jullig M, Hickey AJ, Chai CC, Skea GL, Middleditch MJ, Costa S, Choong SY, Philips AR, Cooper GJ. Is the failing heart out of fuel or a worn engine running rich? A study of mitochondria in old spontaneously hypertensive rats. Proteomics. 2008;8:2556–2572. doi: 10.1002/pmic.200700977. [DOI] [PubMed] [Google Scholar]
- 113.Peronnet E, Becquart L, Poirier F, Cubizolles M, Choquet-Kastylevsky G, Jolivet-Reynaud C. SELDI-TOF MS analysis of the Cardiac Troponin I forms present in plasma from patients with myocardial infarction. Proteomics. 2006;6:6288–6299. doi: 10.1002/pmic.200600158. [DOI] [PubMed] [Google Scholar]
- 114.Dabkowski ER, Baseler WA, Williamson CL, Powell M, Razunguzwa TT, Frisbee JC, Hollander JM. Mitochondrial dysfunction in the type 2 diabetic heart is associated with alterations in spatially distinct mitochondrial proteomes. Am J Physiol Heart Circ Physiol. 2010;299:H529–540. doi: 10.1152/ajpheart.00267.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]