Abstract
The cholesterol biosynthetic pathway produces not only sterols but also non-sterol mevalonate metabolites involved in isoprenoid synthesis. Mevalonate metabolites affect transcriptional and post-transcriptional events that in turn affect various biological processes including energy metabolism. In the present study, we examine whether mevalonate metabolites activate PPARγ (peroxisome-proliferator-activated receptor γ), a ligand-dependent transcription factor playing a central role in adipocyte differentiation. In the luciferase reporter assay using both GAL4 chimaera and full-length PPARγ systems, a mevalonate metabolite, FPP (farnesyl pyrophosphate), which is the precursor of almost all isoprenoids and is positioned at branch points leading to the synthesis of other longer-chain isoprenoids, activated PPARγ in a dose-dependent manner. FPP induced the in vitro binding of a co-activator, SRC-1 (steroid receptor co-activator-1), to GST (glutathione transferase)–PPARγ. Direct binding of FPP to PPARγ was also indicated by docking simulation studies. Moreover, the addition of FPP up-regulated the mRNA expression levels of PPARγ target genes during adipocyte differentiation induction. In the presence of lovastatin, an HMG-CoA (3-hydroxy-3-methylglutaryl-CoA) reductase inhibitor, both intracellular FPP levels and PPARγ-target gene expressions were decreased. In contrast, the increase in intracellular FPP level after the addition of zaragozic acid, a squalene synthase inhibitor, induced PPARγ-target gene expression. The addition of FPP and zaragozic acid promotes lipid accumulation during adipocyte differentiation. These findings indicated that FPP might function as an endogenous PPARγ agonist and regulate gene expression in adipocytes.
Keywords: adipocyte differentiation, farnesyl pyrophosphate (FPP), ligand, metabolic syndrome, mevalonate metabolite, peroxisome-proliferator-activated receptor γ (PPARγ)
Abbreviations: aP2, adipocyte fatty-acid-binding protein; CREB, cAMP-response-element-binding protein; CBP, CREB-binding protein; DMEM, Dulbecco's modified Eagle's medium; Fmoc, fluoren-9-ylmethoxycarbonyl; FPP, farnesyl pyrophosphate; FPTase, farnesyl pyrophosphate transferase; GGPP, geranylgeranyl pyrophosphate; GLUT4, glucose transporter 4; GST, glutathione transferase; 5HE, 5-hydroxyeicosapentaenoic acid; HMG-CoA, 3-hydroxy-3-methylglutaryl-CoA; IBMX, 1-methyl-3-isobutylxanthine; LC/MS, liquid chromatography MS; LPL, lipoprotein lipase; MD, molecular dynamics; Pio, pioglitazone; PPAR, peroxisome-proliferator-activated receptor; RMSD, root-mean-square displacement; RT–PCR, reverse transcription–PCR; RXR, retinoid X receptor; SCD, stearoyl-CoA desaturase; SRC-1, steroid receptor co-activator-1; SREBP, sterol-regulatory-element-binding protein; TAMRA, 6-carboxytetramethylrhodamine; WAT, white adipose tissue
INTRODUCTION
The pathophysiology of obesity and obesity-associated metabolic disorders including Type 2 diabetes mellitus, hypertension, hyperlipidaemia and cardiovascular disease is associated with abnormalities in endocrine signalling in WAT (white adipose tissue) [1,2]. The expansion of WAT during the development of obesity can occur through an increases in cell number and cell size [3]. Therefore the mechanisms controlling WAT development have been the focus of intense research.
The number of adipocytes is thought to increase as a result of the proliferation of pre-adipocytes and subsequent differentiation into mature adipocytes [4]. Adipocyte differentiation is characterized by marked changes in the pattern of gene expression that are achieved by sequential induction of various transcription factors [4]. PPARγ (peroxisome-proliferator-activated receptor γ), a member of the nuclear hormone receptor superfamily, plays a central role in the regulation of adipocyte differentiation [4]. PPARγ is a ligand-dependent transcription factor binding to the promoter of its target genes only as a heterodimer with RXR (retinoid X receptor) [4,5]. The thiazolidinediones, insulin sensitizers, promote the differentiation of pre-adipocytes by PPARγ activation [4] to increase the number of small adipocytes and to decrease the number of large adipocytes by increasing apoptosis [6], resulting in the improvement of metabolic disorders, such as insulin resistance. Although several naturally occurring compounds have been reported to activate PPARγ, the levels of these PPARγ ligands in tissue and plasma are frequently not precisely defined and, when so, are found at levels sometimes orders of magnitude lower than those required to activate specific α, γ or δ PPAR subtypes [7–9]. Thus the identification of bona fide high-affinity endogenous PPARγ ligands has been a controversial issue that, when resolved, will advance our understanding of PPARγ modulation and reveal new ways of intervention in diverse metabolic disorders and disease processes.
Non-sterol mevalonate metabolites involved in isoprenoid synthesis have until recently mainly been associated with the regulation of cell-cycle control and cytoskeletal organization [10]. Moreover, the mevalonate pathway in mammals has been shown to affect several nuclear hormone receptors (such as PPARα, PPARγ and liver X receptors) that regulate lipid and carbohydrate metabolism [11–14]. Previously, we have demonstrated that several isoprenoids contained in herbal and dietary plants function as PPAR ligands [15–19]. In the present paper, we report that FPP (farnesyl pyrophosphate), which is a mevalonate metabolite and the precursor of almost all isoprenoids, can activate PPARγ as an agonist and regulate the expression levels of PPARγ target genes during adipocyte differentiation. These findings suggest that FPP may function as an endogenous PPARγ ligand and regulate adipocyte functions.
MATERIALS AND METHODS
Materials
FPP was from Wako Pure Chemicals (Osaka, Japan). LG100268 and lovastatin were purchased from Santa Cruz Biotechnology and Calbiochem (San Diego, CA, U.S.A.). Unless otherwise indicated, all chemicals were purchased from Sigma or Nacalai Tesque (Kyoto, Japan) and were of guaranteed reagent grade or tissue culture grade.
Cell culture
3T3-L1 murine pre-adipocytes (from A.T.C.C., Manassas, VA, U.S.A.) were cultured in a growth medium, DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% (v/v) fetal bovine serum, 100 units/ml penicillin and 100 μg/ml streptomycin at 37 °C in 5% CO2. At 2 days after reaching confluence, the cells were incubated in a differentiation medium containing 0.25 μM dexamethazone, 10 μg/ml insulin and 0.5 mM IBMX (1-methyl-3-isobutylxanthine) in the growth medium with or without indicated compounds. After 48 h, the cells were harvested for mRNA expression level analysis. For the fatty acid synthesis activity assay and Oil Red O staining, 3T3-L1 cells, whose differentiation was induced as described above, were maintained for another 2–8 days in the growth medium containing insulin (5 μg/ml) with or without FPP or Pio (pioglitazone).
Monkey CV1 kidney cells and murine NIH 3T3 fibroblasts were cultured in the growth medium at 37 °C in 5% CO2.
Luciferase assay
A luciferase assay was performed as described previously [11–15]. Briefly, for the luciferase assay using GAL4 chimaera systems, we transfected p4xUASg-tk-luc (a reporter plasmid), pM-hPPARγ or pM-hRXRα (an expression plasmid for a chimaera protein for the GAL4 DNA-binding domain, and human PPARγ- or RXRα-ligand-binding domains) and pRL-CMV (an internal control for normalizing transfection efficiencies) into CV1 cells. For luciferase assays using a PPARγ full-length system, pDEST-hPPARγ (a human PPARγ expression vector), a reporter plasmid (p3xPPRE-tk-luc) and pRL-CMV were transfected into CV1 cells. Transfections into CV1 cells cultured on 100 mm dishes were performed using Lipofectamine™ (Invitrogen) according to the manufacturer's protocol. At 5 h after the transfections, the transfected cells were seeded on to 96-well plates with the medium containing each compound. After a 24 h incubation, a luciferase assay was performed using the dual-luciferase system (Promega) according to the manufacturer's protocol.
GST (glutathione transferase) pull-down assay
A GST pull-down assay was performed as described previously [12]. Briefly, for expression of the full-length PPARγ–GST fusion protein, the pDEST15 vector (Invitrogen) was used. Glutathione–Sepharose 4B (Amersham Biosciences) was washed extensively in binding buffer (10 mM Tris/HCl, pH 7.4, 1 mM EDTA, 150 mM NaCl, 1 mM dithiothreitol and 10% glycerol) containing an appropriate concentration of protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany) and resuspended in a volume of the binding buffer sufficient to generate a 50% slurry. This slurry was mixed with each cell lysate of Escherichia coli expressing GST or human PPARγ–GST and incubated at 4 °C overnight. The resin slurry was gently centrifuged, washed five times in the binding buffer to remove all unbound proteins and finally resuspended in 1 volume of the binding buffer. The GST pull-down experiments were conducted using 40 μl of GST-bound or GST–hPPARγ-bound, and unbound glutathione–Sepharose slurries and 20 μl of SRC-1-S2, a truncated SRC-1 (steroid receptor co-activator-1) protein (amino acids 623–770) produced by RTS500HY E. coli (Roche Diagnostics). After incubation for 2 h at 4 °C with continuous rotation, the samples were gently centrifuged and washed five times in the binding buffer. After the final wash, the resin was resuspended in 30 μl of SDS/PAGE sample buffer (Bio-Rad). Each sample was electrophoresed on a 7% denaturing gel. After blotting on to PVDF membranes, an enhanced chemiluminescence system (NEN Lifescience Products) was used to detect the SRC-1 protein binding to the PPARγ–GST protein. The density of protein bands was measured using the density analysis software NIH Image, as described previously [20].
Co-activator recruitment assay
The fluorescence polarization assay for SRC-1/PPARγ binding was performed as described previously [21]. The PPARbinding site (amino acids 623–770) of human SRC-1 was cloned into pROX-FL92 (Protein Express, Choshi, Japan). This expression vector enabled the addition of the FL tag of 12 amino acids [22] and the His tag to upstream and downstream of SRC-1 respectively. With an umber codon inserted during the addition of the FL tag and CloveDirect TAMRA (6-carboxytetramethylrhodamine; Protein Express), the pinpoint fluorescent labelling protein to which TAMRA-aminophenylalanine was introduced into the ninth amino acid was produced. By adding pROX-FL92-SRC-1 and CloveDirect TAMRA to the RST100 E. coli HY system (Roche Diagnostics), TAMRA-probed SRC-1 was expressed and purified. The mixture of 250 nM GST–PPARγ and 5 nM pinpoint fluorescent labelling SRC-1 (TAMRA–SRC-1) was added with 1–100 μM ligands. After a 1 h reaction, fluorescence polarization was monitored for 10 s, and the monitoring was repeated five times. Excitation and emission wavelengths were set at 555 and 580 nm respectively. By Scatchard analysis and Hill analysis, the dissociation constants of the PPARγ ligand and SRC-1 complex were estimated.
For the CBP [CREB (cAMP-response-element-binding protein)-binding protein]/PPARγ binding assay, we used a commercial ELISA kit (Fujikura Kasei, Tokyo, Japan).
Docking simulation study
From among the substrates of PPARγ complexes contained in the PDB, 5HE (5-hydroxyeicosapentaenoic acid) was selected on the basis of its similarity to FPP in terms of relevant factors, such as molecular length, molecular shape and the position of a polar group. The steric structure of the protein registered in PDB with the 2VV2 code was used for the molecular modelling of FPP in complex with PPARγ. In the model, the carboxylic acid of 5HE was substituted by phosphoric acid at the end of FPP; the OH group at the fifth carbon of 5HE was substituted by a second oxygen atom in phosphoric acid. The remaining part of the alkyl chain was aligned along the unsaturated hydrocarbon chain of 5HE. Using this model as an initial structure (Figure 3A), we carried out MD (molecular dynamics) simulation to examine the binding state of FPP. For comparison, the MD simulation of 5HE was also carried out to examine the binding stability of FPP. To construct the MD simulation system with the periodic boundary condition, each of these two complexes was placed in a cubic box (size of approximately 92 Å3; 1 Å=0.1 nm) filled with water molecules, and in order to neutralize this system, Na+ ions and Cl− ions were included, for which the program AMBER 9 [23] was used. The total number of water molecules was approximately 10100. We used the unified force-field model [24] of GAFF parameters [25] and RESP charges [26] for PPARγ with the modified topology file using an in-house program, GAFF parameters and AM1-BCC charges [27,28] for FPP and 5HE, and the TIP3P model [26] for water. To accelerate the calculation, a special-purpose parallel computer for non-bonded force sum, MD Server (http://www.nec.co.jp/press/en/0511/2901.html), was used. Temperature and pressure were controlled at 300 K and 1 atm respectively. The time step was 2 fs with the bond-length constraints for hydrogen atoms, and the simulation of each system was performed for 7500000 steps (corresponding to MD simulation of 15000 ps).
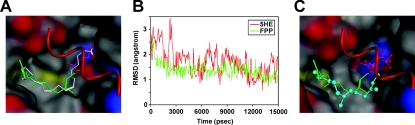
Figure 3. FPP can directly bind to PPARγ as an agonist in a docking simulation study.
(A) Modelling of the docking position of FPP in PPARγ protein by referring to the X-ray crystal structure of the 5HE–PPARγ complex. FPP (green carbon atoms) and 5HE (white carbon atoms) are represented as a stick model. The protein surface is shown in the colour of the atom. Colours: grey, carbon; red, oxygen; blue, nitrogen; yellow, sulfur. The α-helix by which the ligand-binding site is covered is represented by a red Cα-trace model. (B) RMSD of carbon atoms for FPP (green) and 5HE (red). Each complex structure during simulation is overlapped on the structure at 15000 ps with Cα root-mean-square fitting. (C) Difference between MD-simulated pose at 15000 ps (ball and stick model) and the initial pose of MD simulation (stick model).
Stable isotope-based fatty acid synthesis activity assay
Cells were washed with warm PBS, and the medium was replaced with glucose-free DMEM containing 4.5 mg/ml [13C6]-glucose. After a 48 h incubation, the medium was collected and diluted with methanol. Diluents were filtered using a 0.22 μm filter membrane, and filtrates were analysed using the LC/MS (liquid chromatography MS) system (Agilent Series 1100 LC system; Agilent Technologies, Waldbronn, Germany). Cells were dissolved in 5 ml of 20% KOH [ethanol/water (4:6, v/v)], hydrolysed for 60 min at 80 °C and then acidified with 6 ml of 5 M HCl. Fatty acids were extracted twice with 4 ml of diethyl ether, dried under N2 at 40 °C, dissolved in 5 ml of methanol and analysed using the LC/MS system.
Determination of FPP levels
Intracellular FPP levels were determined as described previously [29]. Briefly, NIH 3T3 cells cultured with each compound for 48 h in six-well plates were washed with PBS and collected. The cells were then extracted with butanol/75 mM ammonium hydroxide/ethanol (1:1.25:2.75, by vol.). After centrifugation, the supernatants were dried under reduced pressure. The resulting residue was then dissolved in 50 mM Tris/HCl assay buffer (pH 7.5) containing 5 mM dithiothreitol, 5 mM MgCl2, 10 μM ZnCl2 and 1.0% octyl-β-D-glucopyranoside. Dansylated-GCVLS peptides (Hayashi Kasei, Osaka, Japan) and FPTase (farnesyl pyrophosphate transferase) were added to the extracted solution, and the assay mixture was then incubated at 38 °C for 120 min. The reaction was terminated by the addition of acetonitrile and hydrogen chloride. FPP mass-reacting with the dansylated substrate peptide as catalysed by FPTase was separated and quantified by HPLC coupled with a fluorescence detector at λex=335 nm and λem=528 nm using a C18 reversed-phase analytical column.
Oil Red O staining
Cells were fixed with 10% formaldehyde/PBS and stained with Oil Red O solution (0.5% Oil Red O-isopropyl alcohol/water; 3:2, v/v). After staining, Oil Red O was extracted from cells with isopropyl alcohol and the attenuance at 490 nm was measured. Levels of Oil Red O staining were corrected for non-specific binding levels of the stain to untreated cells.
RNA preparation and real-time fluorescence monitoring RT–PCR (reverse transcription–PCR)
Total RNA was prepared from 3T3-L1 cells using Sepasol®-RNA I Super (Nacalai Tesuque) according to the manufacturer's protocol. Total RNA was reverse-transcribed using MMLV (Moloney-murine-leukaemia virus) reverse transcriptase (Promega) according to the manufacturer's instructions. To quantify mRNA expression, real-time RT–PCR was performed with a LightCycler system (Roche Diagnostics) using SYBR green fluorescence signals, as described previously [11–15,20]. The oligonucleotide primers of mouse 36B4 and adipogenic marker genes were designed using a PCR primer selection program in the website of the Virtual Genomic Center from the GenBank® Nucleotide Sequence Database. Primer sets used to measure the expression levels of 36B4, aP2 (adipocyte fatty-acid-binding protein), LPL (lipoprotein lipase) and adiponectin, have been described previously [11–15,20]. The primers used for the measurements of GLUT4 (glucose transporter 4) mRNA expression levels upstream and downstream were 5′-CGGATGCTATGGGTCCTTACG-3′ and 5′-TGAGATCTGGTCAAACGTCCG-3′ respectively. To compare mRNA expression levels among the samples, the copy numbers of all of the transcripts were divided by that of mouse 36B4, showing a constant expression level in adipocytes. All of the mRNA expression levels are presented here as the ratio relative to that of the control in each experiment.
Statistical analysis
The results are presented as means±S.E.M. and were statistically analysed by the unpaired Student's t test or the Welch t test when variances were heterogeneous. Differences were considered significant when P was <0.05.
RESULTS
FPP activates PPARγ in the luciferase reporter assay
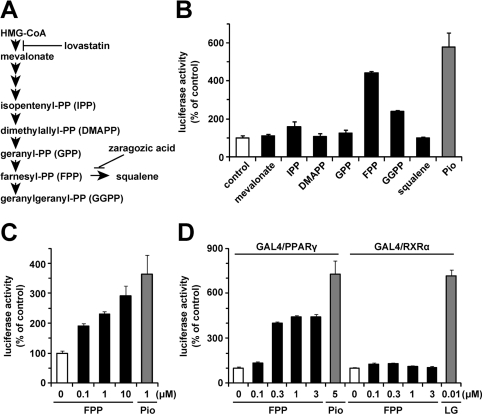
First, to examine whether mevalonate metabolites (Figure 1A) affect PPARγ ligand activity, we performed a luciferase reporter assay using the GAL4 chimaera system. In this assay system, Pio (1 μM), a synthetic PPARγ agonist, significantly enhanced luciferase activity (Figure 1B). We evaluated PPARγ ligand activity in the presence of various mevalonate metabolites at a concentration of 1 μM. As shown in Figure 1(B), in the presence of 1 μM FPP, luciferase activity was increased 4.4-fold compared with the vehicle control, suggesting that FPP increased PPARγ ligand activity. At this concentration, GGPP (geranylgeranyl pyrophosphate) also activated PPARγ, but the activation induced by GGPP was much weaker than that induced by FPP. Next, we investigated by luciferase reporter assay whether FPP activates full-length PPARγ. In this assay system, the addition of FPP also increased luciferase activity in a dose-dependent manner, suggesting that FPP activates full-length PPARγ (Figure 1C). Because the agonists of RXRs, the heterodimer partners of PPARγ, also activate full-length PPARγ [30], we evaluated the effect of FPP on RXRα ligand activity by luciferase reporter assay using the GAL4 chimaera system. As shown in Figure 1(D), FPP activated PPARγ in a dose-dependent manner, whereas FPP had no effect on RXRα ligand activity. These findings indicate that FPP increased PPARγ ligand activity followed by the activation of PPARγ.
Figure 1. FPP activates PPARγ as shown by luciferase reporter assay.
(A) Schematic illustration of the mevalonate pathway. The targets of lovastatin and zaragozic acid are also shown. (B–D) Effects of mevalonate metabolites (1 μM) or FPP on PPARγ or RXRα activity in a luciferase reporter assay using the GAL4/PPARγ chimaera system (B), full-length PPARγ system (C) or GAL4/PPARγ and GAL4/RXRα chimaera system (D). CV1 monkey kidney cells were transfected with pM-PPARγ, p4xUASg-tk-luc and pRL-CMV (B), pDEST-hPPARγ, p3xPPRE-tk-luc and pRL-CMV (C), or pM-PPARγ/pM-RXRα, p4xUASg-tk-luc and pRL-CMV (D). Cells were incubated in a medium with each mevalonate metabolite, Pio or LG100268 (LG; a synthetic RXR agonist) for another 24 h after the transfection. The activity of a vehicle control was set at 100% and the obtained relative luciferase activities are presented as the fold induction with respect to that in the vehicle control. All of the values are the means±S.E.M. for three or four tests.
FPP binds directly to PPARγ as an agonist
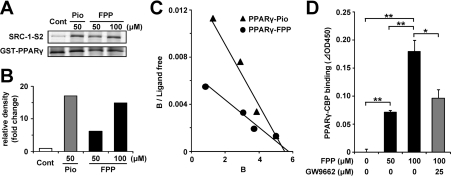
To confirm the direct effects of FPP on PPARγ activation, a GST pull-down assay was performed. A co-activator, SRC-1, interacts with PPAR proteins when the agonists of PPARs bind to the PPAR-ligand-binding domain [31]. Therefore the in vitro binding of SRC-1 to PPARs in the presence of a compound suggests that the compound binds to PPARs as an agonist. Under these conditions in the presence of 50 μM Pio, a His-tagged SRC-1 fragment, bound to PPARγ, is shown in Figure 2(A). FPP (100 μM) recruited the SRC-1 fragment on to PPARγ proteins as well as Pio. The densities of the bands are presented in Figure 2(B). The degrees of binding of SRC-1 to PPARγ in the presence of 50 μM Pio, and 50 and 100 μM FPP increased 17-, 6.1- and 15-fold respectively relative to that of the vehicle control. This suggests that FPP as an agonist of PPARγ can induce the binding of SRC-1 to PPARγ, which is important for the ligand-dependent activation of PPARγ. Next, to study co-activator recruitment to PPARγ kinetically, we performed an in vitro recruitment assay using pinpoint fluorescent labelling SRC-1 and GST–PPARγ. FPP enhanced the interaction between PPARγ and SRC-1 in a dose-dependent manner in a similar manner to Pio. As shown in Figure 2(C), the Scatchard plot shows a straight line. Bmax (Xint) might be slightly different however; we confirmed that Bmax (polarization) was approx. 260 mP at the maximum concentration in the case of all ligands, including troglitazone and rosiglitazone. The dissociation constants of FPP and Pio were estimated as 955 and 357 nM respectively. As GGPP also activated PPARγ weakly in the luciferase assay (Figure 1B), we performed a fluorescence polarization assay for SRC-1/PPARγ binding using GGPP. In that assay, we confirmed that GGPP did not induce an interaction between fluorescent SRC-1 and PPARγ. Moreover, we investigated another co-activator, CBP, for its recruitment to PPARγ. Similarly to SRC-1, CBP interacts with PPARγ when the agonists of PPARs bind to the PPAR-ligand-binding domain [32]. As is the case in SRC-1, FPP induced the interaction between CBP and PPARγ (Figure 2D), and this interaction was attenuated in the presence of GW9662, a synthetic PPARγ antagonist, suggesting that FPP binds to PPARγ specifically.
Figure 2. FPP induces the recruitment of co-activators to PPARγ in vitro.
(A) GST pull-down assay using full-length PPARγ protein. Recombinant SRC-1 fragment (SRC-1-S2) containing a PPAR-binding region was incubated with GST–human PPARγ and vehicle control, 50 μM Pio or 50 μM/100 μM FPP. After washing, the bound recombinant SRC-1 protein was detected by immunoblotting. Total GST–PPARγ protein was detected by Coomassie Brilliant Blue staining. The results are representative of three independent blots. (B) The densitometric analysis of immunoblotting membranes normalized by the amount of GST–PPARγ of (A) is shown. The density of the vehicle control was set at 1 and the relative densities were presented as the fold induction relative to that of the vehicle control. (C) Scatchard plot analysis of the binding of a PPARγ triangles ligand complex to the TAMRA–SRC-1 in the presence of pioglitazone (Pro; closed triangles) or FPP (closed circles). As the control experiment, TAMRA–SRC-1 was incubated together with GST protein. Ligand concentrations (in μM) at all data points are 0.15, 1.0, 2.0 and 3.8 for FPP and 0.1, 0.4, 1.2 and 3.8 for Pio from the left data point respectively. The results are representative of five independent blots. (D) The recruitment of CBP to PPARγ in the presence or absence of FPP (50 or 100 μM) and GW9662 (25 μM) was determined by ELISA. All the values are means±S.E.M. for three to five tests. *P<0.05, **P<0.01.
The X-ray crystal structure of the 5HE–PPARγ complex (PDB code 2VV2) and the model structure of the FPP–PPARγ complex shown in Figure 3(A) were subjected to MD simulation for 15000 ps to examine the binding stability of FPP. The RMSD (root-mean-square displacement) of carbon atoms for 5HE and FPP for 15000 ps, which is calculated with respect to the atomic coordinates at 15000 ps, is shown in Figure 3(B). After the start of MD simulation, structural changes and positional displacements were induced in the initial structure for both 5HE and FPP. The structure of 5HE reached a steady state and position in approx. 5000 ps, whereas that of FPP reached a steady state and position in approx. 3000 ps. After the elapse of these times, both 5HE and FPP retained structural stability for the subsequent 10000 ps in the presence of thermal fluctuation alone. For both 5HE–PPARγ and FPP–PPARγ complexes, the steady-state Cα RMSD was constant at approx. 1.1 Å. On the basis of these findings, the FPP–PPARγ complex shows the same level of stability as the 5HE–PPARγ complex on the time scale of this MD simulation. In Figure 3(C), the binding state of the initial model is compared with that of the MD-simulated model at 15000 ps. The positions of the phosphate group are the same in both models, whereas the structures of the carbon chain are slightly different. These findings indicate that FPP can directly bind to PPARγ as its agonist.
The addition of FPP up-regulated PPARγ target genes during induction of adipocyte differentiation
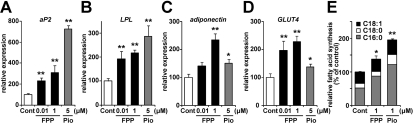
To elucidate whether FPP actually induces PPARγ target gene expression during the induction of adipocyte differentiation, we evaluated mRNA expression levels using 3T3-L1 pre-adipocytes. Adipocyte differentiation was induced by the addition of a standard adipogenic mixture of dexamethasone, insulin and IBMX and an incubation for 48 h. FPP or Pio (a synthetic PPARγ agonist) was also added with this standard adipogenic mixture, and after 48 h, mRNA expression levels were measured. In the presence of 5 μM Pio, well-known PPARγ target genes, such as aP2, LPL and adiponectin, in adipocytes were induced 7.3-, 2.8- and 1.5-fold respectively (Figures 4A–4C). The addition of FPP also increased the mRNA expression levels of these genes in a dose-dependent manner (Figures 4A–4C). Treatment with 1 μM FPP resulted in 3.1-, 2.2- and 2.4-fold increases in aP2, LPL and adiponectin levels respectively. Moreover, the expression levels of GLUT4, which encodes the insulin-sensitive GLUT induced by PPARγ activation [33], were increased by both FPP and Pio (Figure 4D). These findings indicate that FPP treatment enhances PPARγ target gene expression and promotes adipocyte differentiation.
Figure 4. The addition of FPP up-regulates PPARγ target genes and promotes de novo fatty acid synthesis.
(A–D) Expression levels of adipogenic marker genes, such as aP2 (A), LPL (B), adiponectin (C) and GLUT4 (D), in 3T3-L1 cells treated with or without FPP. 3T3-L1 cells were induced to differentiate with or without 0.01 μM/1 μM FPP or 5 μM Pio for 48 h. Total RNA was isolated and analysed by real-time monitoring RT–PCR. mRNA expression levels of each gene were normalized to the expression levels of the ribosomal 36B4 gene. The expression level of cells treated with the vehicle control is set at 100% and relative expression levels are presented as the fold inductions over the vehicle control. (E) Rate of de novo fatty acid synthesis from glucose after chronic FPP treatment during adipocyte differentiation. 3T3-L1 cells were induced to differentiate and maintained with or without 1 μM FPP or 1 μM Pio for 10 days. Cells were analysed using [13C6]-glucose and the LC/MS system as described in the Materials and methods section. All of the values are means±S.E.M. for five or six tests. *P<0.05, **P<0.01 compared with vehicle controls.
Next, we estimated the rate of de novo fatty acid synthesis from glucose after chronic FPP treatment using [13C6]-glucose and the LC/MS system. In the case of FPP addition to the medium throughout the differentiation period (day 0–10), total fatty acid (palmitate, stearate and oleate) synthesis, particularly palimitate synthesis, was significantly up-regulated compared with vehicle administration (Figure 4E). A similar effect was observed in the case of Pio treatment. These findings indicate that FPP treatment enhances PPARγ target gene expression and de novo fatty acid synthesis, leading to the promotion of adipocyte differentiation.
Endogenous FPP might regulate PPARγ activity during induction of adipocyte differentiation
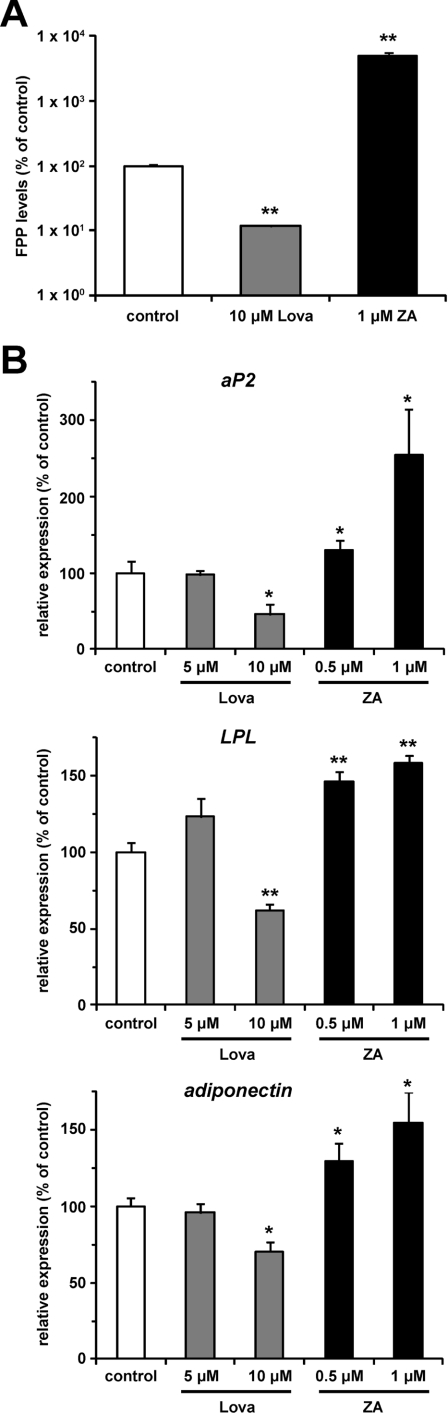
To assess whether FPP produced endogenously affects adipocyte differentiation, we measured the aP2 expression level in the presence of inhibitors of the mevalonate pathway. We used two inhibitors, lovastatin for HMG-CoA (3-hydroxy-3-methylglutaryl-CoA) reductase catalysing the conversion of HMG-CoA into mevalonate, and zaragozic acid for squalene synthase catalysing the conversion of FPP into squalene (Figure 1A). As previously reported [29], in the presence of lovastatin, the intracellular FPP level decreased, whereas it increased in the presence of zaragozic acid in NIH 3T3 fibroblasts, which can differentiate to adipocytes by the ectopic expression of PPARγ [34] (Figure 5A). The intracellular FPP level decreased by 88.4% in the presence of 10 μM lovastatin. On the other hand, 1 μM zaragozic acid induced a 49-fold increase in the intracellular FPP level. We then treated 3T3-L1 cells with these inhibitors during the induction of adipocyte differentiation, and measured the mRNA expression level of PPARγ target genes (aP2, LPL and adiponectin). As shown in Figure 5(B), the addition of 10 μM lovastatin decreased the mRNA expression levels of PPARγ target genes (54%, 38% and 29% decreases in aP2, LPL and adiponectin expression levels respectively). Conversely, these gene expression levels were increased by zaragozic acid in a dose-dependent manner; 1 μM zaragozic acid increased aP2, LPL and adiponectin mRNA expression levels by 2.5-, 1.6- and 1.5-fold respectively. These findings indicate that endogenous FPP can regulate PPARγ activity during the induction of adipocyte differentiation.
Figure 5. Endogenous FPP might regulate PPARγ activity during induction of adipocyte differentiation.
(A) Intracellular FPP levels in NIH 3T3 cells treated with or without lovastatin (Lova) or zaragozic acid (ZA). NIH 3T3 cells treated with vehicle control, 10 μM Lova or 1 μM ZA for 48 h were extracted with butanol/75 mM ammonium hydroxide/ethanol (1:1.25:2.75, by vol.) and the intracellular FPP level was determined as described in the Materials and methods section. All of the values are means±S.E.M. for three tests. (B) mRNA expression levels of aP2, LPL and adiponectin in 3T3-L1 cells treated with or without Lova or ZA. 3T3-L1 cells were induced to differentiate with vehicle control, Lova (5 or 10 μM) or ZA (0.5 or 1 μM) for 48 h. mRNA expression levels were determined as described in Figure 4. All the values are means±S.E.M. for four to six tests. *P<0.05, **P<0.01 compared with vehicle controls.
FPP promotes lipid accumulation during adipocyte differentiation
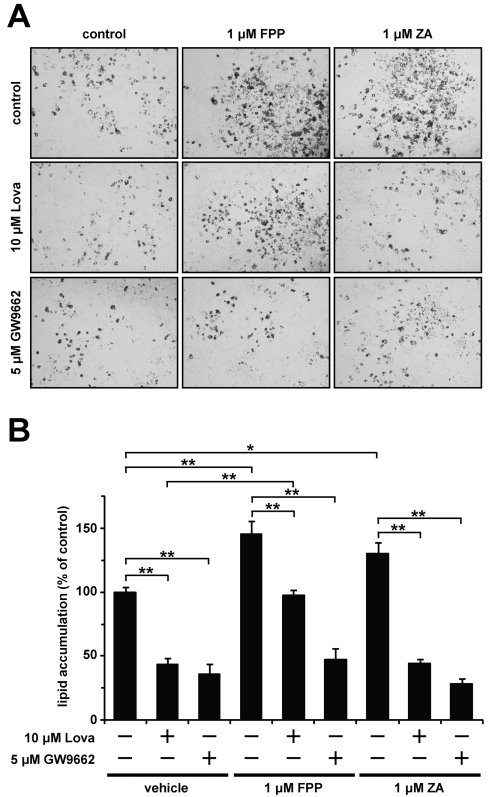
Finally, to investigate the effect of FPP on lipid accumulation during adipocyte differentiation, we performed Oil Red O staining. The addition of 1 μM FPP and zaragozic acid enhanced lipid accumulation during adipocyte differentiation (1.4- and 1.3-fold increases respectively; Figures 6A and 6B). This enhancement of lipid accumulation was cancelled in the presence of the PPARγ antagonist GW9662. Moreover, the lovastatin-induced decrease in lipid accumulation level was rescued by adding FPP, but not zaragozic acid. These findings suggest that FPP promotes lipid accumulation during adipocyte differentiation through PPARγ activation.
Figure 6. FPP promotes lipid accumulation during adipocyte differentiation.
(A, B) 3T3-L1 cells were induced to differentiate with or without the indicated compounds for 48 h. The cells were incubated with or without the indicated compounds for an additional 2 days. The cells were fixed with formalin, and stained with Oil Red O. Microscopy views of representative 3T3-L1 cells (the original magnification is ×100) (A). Oil Red O was extracted from the cells with isopropyl alcohol and attenuance was measured at 490 nm (B). The levels of Oil Red O staining were corrected for the levels of non-specific binding levels of the stain to untreated cells. The values are means±S.E.M. for four tests. *P<0.05, **P<0.01.
DISCUSSION
Isoprenoids make up a large group of essential molecules involved in diverse cellular processes including energy metabolism [10–14]. In all metazoan organisms, isoprenoids are produced via the mevalonate pathway. Thus, in the present study, we assessed the effects of various mevalonate metabolites on PPARγ activity, which plays an important role in adipocyte differentiation. We have reported that one of the mevalonate metabolites, FPP, which is the precursor of almost all isoprenoids and is positioned at branch points leading to the synthesis of other longer-chain isoprenoids [35], enhances the expression levels of adipogenic genes, such as aP2, LPL, adiponectin and GLUT4, suggesting that the addition of FPP promoted adipocyte differentiation.
FPP plays an important role as the substrate of protein farnesylation reactions catalysed by FPTase [36]. Members of the Ras GTPase family are major substrates of post-translational modification by farnesylation, a process essential for their proper membrane localization and activation [37]. Previously, Klemm et al. [38] suggested that protein farnesylation is an essential process in adipocyte differentiation. Thus the increase in farnesylated protein level induced by FPP treatment might also contribute to the promoting effect of FPP on adipocyte differentiation. In our preliminary study, we tried the experiment using an inhibitor of farnesyltransferase (α-hydroxy farnesyl phosphonic acid) to elucidate the contribution of protein farnesylation. This inhibitor (1 μM) significantly inhibited the mRNA expression level of an adipogenic marker gene (aP2) during adipocyte differentiation, and this inhibitory effect was rescued by FPP treatment (1 μM). Under these conditions, it is suggested that the farnesyltransferase reaction hardly proceeds, because the binding ability of α-hydroxy farnesyl phosphonic acid to farnesyltransferase is 10-fold stronger than that of FPP [39]. These results might indicate that FPP induces adipogenic marker genes without affecting protein farnesylation. Besides serving as the substrate of post-translational modification, FPP has been reported to have several bioactivities by itself [40,41]. A direct interaction between FPP and PPARγ was determined by a co-activator recruitment assay and docking simulation studies. Many novel ligands were studied by these methods as well [42–44]. Compared with Pio or troglitazone, which are potent PPARγ agonists, the Kd of FPP was larger. However, the Kd of FPP seems to be smaller than those of fatty acids such as linoleic acid, which are known to be PPARγ endogenous ligands [44]. As the ability of the polar heads of ligands to form hydrogen bonds with PPARγ is important [44], the phosphoric acid of FPP may play a role in forming hydrogen bonds with PPARγ strongly similarly to carbonic acid of fatty acids. On the other hand, an hydrophobic interaction between the tails of ligands and pockets of PPARγ is also required. Indomethacin, which has a short chain, or palmitic acid, which has a flexible chain, loses entropy, resulting in a weak interaction. It is suggested that FPP has a suitable length of chains, but flexibility may slightly decrease the ability to bind the hydrophobic pockets of PPARγ. Indeed, both glucose and lipid metabolisms of adipocytes were activated by adding FPP. Therefore, at least partially, FPP seems to promote adipocyte differentiation via the activation of PPARγ as an agonist, although further studies are required to clarify the detailed mechanisms.
In the present study, Pio and FPP vary in their relative size of effect (FPP>Pio for some genes, FPP<Pio for others), and palmitate accumulation is observed rather than oleate in the case of FPP. The molecular mechanism of the agonist-dependent transactivation of PPARγ is said to be not uniform but dependent on the agonist type and specific cofactors. It has been reported that Fmoc (fluoren-9-ylmethoxycarbonyl)-L-leucine acts as a selective PPARγ ligand and potently improves insulin resistance despite a weak induction of the mRNA expression of PPARγ target genes in WAT [45]. In that paper, Fmoc-L-leucine interacts with the ligand-binding site of PPARγ in a different manner from thiazolidinediones to result in the recruitment of different cofactors. This recruitment of different cofactors was shown between troglitazone and 15-deoxy-Δ12,14-prostaglandin J2 in spite of the equal potential in the transactivation of PPARγ [46]. Moreover, Camp et al. [47] reported that troglitazone, but not rosiglitazone, may behave as a partial agonist under certain physiological circumstances and as a full agonist in others. As for SCD (stearoyl-CoA desaturase), rosiglitazone has been reported to enhance SCD expression [48], whereas troglitazone and Pio have been reported to down-regulate SCD expression [49]. Thus it is likely that the increase in PPARγ-mediated gene expression levels in WAT is not always paralleled by amelioration of metabolic disorders, and that the agonist-dependent cofactor recruitment to PPARγ might be important for determining target genes. Further studies are required to elucidate the characterization of FPP as a PPARγ agonist.
As previously reported [50], an HMG-CoA reductase inhibitor, lovastatin, inhibited the induction of PPARγ target genes during adipocyte differentiation induction. Conversely, a squalene synthase inhibitor, zaragozic acid, promoted it. These findings indicate that mevalonate metabolites such as FPP are important for the regulation of adipocyte differentiation induction. As is the case for most anabolic pathways, isoprenoid biosynthesis involving the mevalonate pathway is regulated tightly in order to allow a constant production of various isoprenoid molecules and to avoid overaccumulation of toxic intermediates or products, such as cholesterol [51]. The feedback regulation of isoprenoid biosynthesis by cholesterol is achieved predominantly through the repression of transcription of genes that govern the synthesis of cholesterol, such as HMG-CoA reductase and HMG-CoA synthase [51]. This transcriptional regulation is performed by SREBPs (sterol-regulatory-element-binding proteins), which are transcription factors under the conditions of cholesterol starvation [51]. There is substantial evidence that most, if not all, enzymes of isoprenoid and cholesterol biosynthesis are under co-ordinated regulation by SREBPs, since the overexpression of these proteins in mice induces the expression of all enzymes involved in isoprenoid and cholesterol biosyntheses studied [52]. In the obese Zucker rat adipocytes, the expression level of the nuclear-active form of SREBP2 was found to be elevated compared with that in lean controls [53]. In agreement with that paper, in vivo experiments showed that cholesterol synthesis was found to be enhanced in the adipose tissue from obese rodents and humans [54]. In adipocyte differentiation, SREBP1 plays a key role [55] and the overexpression of SREBP1 in 3T3-L1-generated ligands for PPARγ, suggesting that PPARγ ligands are generated during isoprenoid and cholesterol biosyntheses in adipocytes [56]. In our preliminary study, the expression level of FPP synthase, which catalyses the conversion from geranyl pyrophosphate into FPP, was increased in parallel with adipocyte differentiation. These findings indicate that the isoprenoid and cholesterol biosynthesis pathways are flexibly regulated during adipocyte differentiation. Therefore there is a possibility that a regulatory mechanism for them is important for adipocyte differentiation. Although further studies are required, these papers raise the possibility that FPP stimulates PPARγ as an endogenously produced agonist.
In the present study, lovastatin treatment inhibited adipocyte differentiation. Although there have been a number of clinical reports indicating that statins affect insulin sensitivity, this is far from being an accepted fact in the field, with an almost equal number of reports arguing against a direct impact on any parameters with respect to carbohydrate metabolism [57–60]. Much of the variability of observations is likely to be due to differences in the type of statin used, the duration of treatment and/or differences in patient populations. Even if statins inhibit PPARγ activity in vivo, they do not necessarily decrease insulin-sensitivity, because not only the pharmacological activation of PPARγ induced by thiazolidinediones, but also the reduction in PPARγ activity prevents insulin resistance [61–64]. Thus, depending on certain conditions, the inhibitory effects of statins on adipocyte differentiation might be involved in the effects of statins on insulin sensitivity.
In conclusion, the present study indicates that FPP serves as an agonist of PPARγ in cultured adipocytes, and it might function as an endogenous regulator of adipocyte differentiation. The circulating FPP level in feeding dogs appears to be higher than that in fasting dogs, raising the possibility that it is affected by whole-body nutritional conditions [65]. Because the adipocyte differentiation process is closely related to obesity and obesity-associated metabolic disorders, FPP might be implicated in the pathogenesis of these metabolic diseases.
AUTHOR CONTRIBUTION
Tsuyoshi Goto, Hiroyuki Nagai, Nobuyuki Takahashi and Teruo Kawada led the design and overall implementation of the trial. Tsuyoshi Goto, Hiroyuki Nagai and Hiroh Miyagawa wrote the initial draft of the paper in consultation with Shogo Ebisu, Takahiro Hohsaka, Shigeru Murakami, Nobuyuki Takahashi and Teruo Kawada. Tsuyoshi Goto, Hiroyuki Nagai, Kahori Egawa, Young-Il Kim, Sota Kato, Aki Taimatsu, Tomoya Sakamoto and Hiroh Miyagawa were responsible for laboratory analyses. All authors contributed to the interpretation of data, and have seen and approved the final paper.
ACKNOWLEDGEMENTS
We thank Y. Tada and S. Shinotoh for technical assistance and secretarial support.
FUNDING
This work was mainly supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan [grant numbers 22780816, 22228001, 22380075]; and by The Uehara Memorial Foundation.
References
- 1.Kadowaki T., Yamauchi T., Kubota N., Hara K., Ueki K., Tobe K. Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J. Clin. Invest. 2006;116:1784–1792. doi: 10.1172/JCI29126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsuzawa Y. The metabolic syndrome and adipocytokines. FEBS Lett. 2006;580:2917–2921. doi: 10.1016/j.febslet.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 3.Sakai T., Sakaue H., Nakamura T., Okada M., Matsuki Y., Watanabe E., Hiramatsu R., Nakayama K., Nakayama K. I., Kasuga M. Skp2 controls adipocyte proliferation during the development of obesity. J. Biol. Chem. 2007;282:2038–2046. doi: 10.1074/jbc.M608144200. [DOI] [PubMed] [Google Scholar]
- 4.Spiegelman B. M., Flier J. S. Adipogenesis and obesity: rounding out the big picture. Cell. 1996;87:377–389. doi: 10.1016/s0092-8674(00)81359-8. [DOI] [PubMed] [Google Scholar]
- 5.Glass C. K. Going nuclear in metabolic and cardiovascular disease. J. Clin. Invest. 2006;116:556–560. doi: 10.1172/JCI27913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamauchi T., Kamon J., Waki H., Murakami K., Motojima K., Komeda K., Ide T., Kubota N., Terauchi Y., Tobe K., et al. The mechanisms by which both heterozygous peroxisome proliferator-activated receptor gamma (PPARgamma) deficiency and PPARgamma agonist improve insulin resistance. J. Biol. Chem. 2001;276:41245–41254. doi: 10.1074/jbc.M103241200. [DOI] [PubMed] [Google Scholar]
- 7.Bell-Parikh L. C., Ide T., Lawson J. A., McNamara P., Reilly M., FitzGerald G. A. Biosynthesis of 15-deoxy-delta12,14-PGJ2 and the ligation of PPARgamma. J. Clin. Invest. 2003;112:945–955. doi: 10.1172/JCI18012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosen E. D., Spiegelman B. M. PPARgamma: a nuclear regulator of metabolism, differentiation, and cell growth. J. Biol. Chem. 2001;276:37731–37734. doi: 10.1074/jbc.R100034200. [DOI] [PubMed] [Google Scholar]
- 9.Tzameli I., Fang H., Ollero M., Shi H., Hamm J. K., Kievit P., Hollenberg A. N., Flier J. S. Regulated production of a peroxisome proliferator-activated receptor-gamma ligand during an early phase of adipocyte differentiation in 3T3-L1 adipocytes. J. Biol. Chem. 2004;279:36093–36102. doi: 10.1074/jbc.M405346200. [DOI] [PubMed] [Google Scholar]
- 10.Edwards P. A., Ericsson J. Sterols and isoprenoids: signaling molecules derived from the cholesterol biosynthetic pathway. Annu. Rev. Biochem. 1999;68:157–185. doi: 10.1146/annurev.biochem.68.1.157. [DOI] [PubMed] [Google Scholar]
- 11.Martin G., Duez H., Blanquart C., Berezowski V., Poulain P., Fruchart J. C., Najib-Fruchart J., Glineur C., Staels B. Statin-induced inhibition of the Rho-signaling pathway activates PPARalpha and induces HDL apoA-I. J. Clin. Invest. 2001;107:1423–1432. doi: 10.1172/JCI10852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forman B. M., Ruan B., Chen J., Schroepfer G. J., Jr, Evans R. M. The orphan nuclear receptor LXRalpha is positively and negatively regulated by distinct products of mevalonate metabolism. Proc. Natl. Acad. Sci. U.S.A. 1997;94:10588–10593. doi: 10.1073/pnas.94.20.10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Argmann C. A., Edwards J. Y., Sawyez C. G., O'Neil C. H., Hegele R. A., Pickering J. G., Huff M. W. Regulation of macrophage cholesterol efflux through hydroxymethylglutaryl-CoA reductase inhibition: a role for RhoA in ABCA1-mediated cholesterol efflux. J. Biol. Chem. 2005;280:22212–22221. doi: 10.1074/jbc.M502761200. [DOI] [PubMed] [Google Scholar]
- 14.Yano M., Matsumura T., Senokuchi T., Ishii N., Murata Y., Taketa K., Motoshima H., Taguchi T., Sonoda K., Kukidome D., et al. Statins activate peroxisome proliferator-activated receptor gamma through extracellular signal-regulated kinase 1/2 and p38 mitogen-activated protein kinase-dependent cyclooxygenase-2 expression in macrophages. Circ. Res. 2007;100:1442–1451. doi: 10.1161/01.RES.0000268411.49545.9c. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi N., Kawada T., Goto T., Yamamoto T., Taimatsu A., Matsui N., Kimura K., Saito M., Hosokawa M., Miyashita K., et al. Dual action of isoprenols from herbal medicines on both PPARgamma and PPARalpha in 3T3-L1 adipocytes and HepG2 hepatocytes. FEBS Lett. 2002;514:315–322. doi: 10.1016/s0014-5793(02)02390-6. [DOI] [PubMed] [Google Scholar]
- 16.Goto T., Takahashi N., Kato S., Egawa K., Ebisu S., Moriyama T., Fushiki T., Kawada T. Phytol directly activates peroxisome proliferator-activated receptor alpha (PPARalpha) and regulates gene expression involved in lipid metabolism in PPARalpha-expressing HepG2 hepatocytes. Biochem. Biophys. Res. Commun. 2005;337:440–445. doi: 10.1016/j.bbrc.2005.09.077. [DOI] [PubMed] [Google Scholar]
- 17.Kuroyanagi K., Kang M. S., Goto T., Hirai S., Ohyama K., Kusudo T., Yu R., Yano M., Sasaki T., Takahashi N., et al. Citrus auraptene acts as an agonist for PPARs and enhances adiponectin production and MCP-1 reduction in 3T3-L1 adipocytes. Biochem. Biophys. Res. Commun. 2008;366:219–225. doi: 10.1016/j.bbrc.2007.11.119. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi N., Kawada T., Goto T., Kim C. S., Taimatsu A., Egawa K., Yamamoto T., Jisaka M., Nishimura K., Yokota K., et al. Abietic acid activates peroxisome proliferator-activated receptor-gamma (PPARgamma) in RAW264.7 macrophages and 3T3-L1 adipocytes to regulate gene expression involved in inflammation and lipid metabolism. FEBS Lett. 2003;550:190–194. doi: 10.1016/s0014-5793(03)00859-7. [DOI] [PubMed] [Google Scholar]
- 19.Park J. Y., Kawada T., Han I. S., Kim B. S., Goto T., Takahashi N., Fushiki T., Kurata T., Yu R. Capsaicin inhibits the production of tumor necrosis factor alpha by LPS-stimulated murine macrophages, RAW 264.7: a PPARgamma ligand-like action as a novel mechanism. FEBS Lett. 2004;572:266–270. doi: 10.1016/j.febslet.2004.06.084. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi N., Kawada T., Yamamoto T., Goto T., Taimatsu A., Aoki N., Kawasaki H., Taira K., Yokoyama K. K., Kamei Y., et al. Overexpression and ribozyme-mediated targeting of transcriptional coactivators CREB-binding protein and p300 revealed their indispensable roles in adipocyte differentiation through the regulation of peroxisome proliferator-activated receptor gamma. J. Biol. Chem. 2002;277:16906–16912. doi: 10.1074/jbc.M200585200. [DOI] [PubMed] [Google Scholar]
- 21.Abe R., Shiraga K., Ebisu S., Takagi H., Hohsaka T. Incorporation of fluorescent non-natural amino acids into N-terminal tag of proteins in cell-free translation and its dependence on position and neighboring codons. J. Biosci. Bioeng. 2010;110:32–38. doi: 10.1016/j.jbiosc.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 22.Nagai H., Ebisu S., Abe R., Goto T., Takahashi N., Hosaka T., Kawada T. Development of a novel PPARγ ligand screening system using pinpoint fluorescence-probed protein. Biosci. Biotechnol. Biochem. 2011;75:337–341. doi: 10.1271/bbb.100810. [DOI] [PubMed] [Google Scholar]
- 23.Case D. A., Darden T. A., Cheatham T. E., III, Simmerling C. L., Wang J., Duke R. E., Luo R., Merz K. M., Pearlman D. A., Crowley M., et al. AMBER 9 Users' Manual. San Francisco, CA: University of California; 2006. [Google Scholar]
- 24.Fujitani H., Tanida Y., Matsuura A. Massively parallel computation of absolute binding free energy with well-equilibrated states. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2009;79:021914. doi: 10.1103/PhysRevE.79.021914. [DOI] [PubMed] [Google Scholar]
- 25.Wang J., Wolf R. M., Caldwell J. W., Kollamn P. A., Case D. A. Development and testing of a general Amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 26.Jorgensen W. L., Chandrasekhar J., Madura J., Klein M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 27.Jakalian A., Bush B. L., Jack D. B., Bayly C. I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: I. Method. J. Comput. Chem. 2000;21:132–146. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 28.Jakalian A., Jack D. B., Bayly C. I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 2002;23:1623–1641. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 29.Tong H., Holstein S. A., Hohl R. J. Simultaneous determination of farnesyl and geranylgeranyl pyrophosphate levels in cultured cells. Anal. Biochem. 2005;336:51–59. doi: 10.1016/j.ab.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 30.Leibowitz M. D., Ardecky R. J., Boehm M. F., Broderick C. L., Carfagna M. A., Crombie D. L., D'Arrigo J., Etgen G. J., Faul M. M., Grese T. A., et al. Biological characterization of a heterodimer-selective retinoid X receptor modulator: potential benefits for the treatment of type 2 diabetes. Endocrinology. 2006;147:1044–1053. doi: 10.1210/en.2005-0690. [DOI] [PubMed] [Google Scholar]
- 31.DiRenzo J., Söderstrom M., Kurokawa R., Ogliastro M. H., Ricote M., Ingrey S., Hörlein A., Rosenfeld M. G., Glass C. K. Peroxisome proliferator-activated receptors and retinoic acid receptors differentially control the interactions of retinoid X receptor heterodimers with ligands, coactivators, and corepressors. Mol. Cell. Biol. 1997;17:2166–2176. doi: 10.1128/mcb.17.4.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gelman L., Zhou G., Fajas L., Raspé E., Fruchart J. C., Auwerx J. p300 interacts with the N- and C-terminal part of PPARgamma2 in a ligand-independent and -dependent manner, respectively. J. Biol. Chem. 1999;274:7681–7688. doi: 10.1074/jbc.274.12.7681. [DOI] [PubMed] [Google Scholar]
- 33.Armoni M., Kritz N., Harel C., Bar-Yoseph F., Chen H., Quon M. J., Karnieli E. Peroxisome proliferator-activated receptor-gamma represses GLUT4 promoter activity in primary adipocytes, and rosiglitazone alleviates this effect. J. Biol. Chem. 2003;278:30614–30623. doi: 10.1074/jbc.M304654200. [DOI] [PubMed] [Google Scholar]
- 34.Tontonoz P., Hu E., Spiegelman B. M. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 35.Houten S. M., Frenkel J., Waterham H. R. Isoprenoid biosynthesis in hereditary periodic fever syndromes and inflammation. Cell. Mol. Life Sci. 2003;60:1118–1134. doi: 10.1007/s00018-003-2296-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang F. L., Casey P. J. Protein prenylation: molecular mechanisms and functional consequencesProtein prenylation: molecular mechanisms and functional consequences. Annu. Rev. Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 37.Bar-Sagi D., Hall A. Ras and Rho GTPases: a family reunion. Cell. 2000;103:227–238. doi: 10.1016/s0092-8674(00)00115-x. [DOI] [PubMed] [Google Scholar]
- 38.Klemm D. J., Leitner J. W., Watson P., Nesterova A., Reusch J. E., Goalstone M. L., Draznin B. Insulin-induced adipocyte differentiation. Activation of CREB rescues adipogenesis from the arrest caused by inhibition of prenylation. J. Biol. Chem. 2001;276:28430–28435. doi: 10.1074/jbc.M103382200. [DOI] [PubMed] [Google Scholar]
- 39.Gibbs J. B., Pompliano D.L, Mosser S. D., Rands E., Lingham R. B., Singh S. B., Scolnick E. M., Kohl N. E., Oliff A. Selective inhibition of farnesyl-protein transferase blocks ras processing in vivo. J. Biol. Chem. 1993;268:7617–7620. [PubMed] [Google Scholar]
- 40.Oh D. Y., Yoon J. M., Moon M. J., Hwang J. I., Choe H., Lee J. Y., Kim J. I., Kim S., Rhim H., O'Dell D. K., et al. Identification of farnesyl pyrophosphate and N-arachidonylglycine as endogenous ligands for GPR92. J. Biol. Chem. 2006;283:21054–21064. doi: 10.1074/jbc.M708908200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murthy S., Tong H., Hohl R. J. Regulation of fatty acid synthesis by farnesyl pyrophosphate. J. Biol. Chem. 2005;280:41793–41804. doi: 10.1074/jbc.M504101200. [DOI] [PubMed] [Google Scholar]
- 42.Lehmann J. M., Moore L. B., Smith-Oliver T. A., Wilkison W. O., Willson T. M., Kliewer S. A. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma) J. Biol. Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- 43.Ferry G., Bruneau V., Beauverger P., Goussard M., Rodriguez M., Lamamy V., Dromaint S., Canet E., Galizzi J. P., Boutin J. A. Binding of prostaglandins to human PPARgamma: tool assessment and new natural ligands. Eur. J. Pharmacol. 2001;417:77–89. doi: 10.1016/s0014-2999(01)00907-4. [DOI] [PubMed] [Google Scholar]
- 44.Murakami K., Ide T., Nakazawa T., Okazaki T., Mochizuki T., Kadowaki T. Fatty-acyl-CoA thioesters inhibit recruitment of steroid receptor co-activator 1 to a and c isoforms of peroxisome-proliferator-activated receptors by competing with agonists. Biochem. J. 2001;353:231–238. doi: 10.1042/0264-6021:3530231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rocchi S., Picard F., Vamecq J., Gelman L., Potier N., Zeyer D., Dubuquoy L., Bac P., Champy M. F., Plunket K. D., et al. A unique PPARgamma ligand with potent insulin-sensitizing yet weak adipogenic activity. Mol. Cell. 2001;8:737–747. doi: 10.1016/s1097-2765(01)00353-7. [DOI] [PubMed] [Google Scholar]
- 46.Kodera Y., Takeyama K., Murayama A., Suzawa M., Masuhiro Y., Kato S. Ligand type-specific interactions of peroxisome proliferator-activated receptor gamma with transcriptional coactivators. J. Biol. Chem. 2000;275:33201–33204. doi: 10.1074/jbc.C000517200. [DOI] [PubMed] [Google Scholar]
- 47.Camp H. S., Li O., Wise S. C., Hong Y. H., Frankowski C. L., Shen X., Vanbogelen R., Leff T. Differential activation of peroxisome proliferator-activated receptor-gamma by troglitazone and rosiglitazone. Diabetes. 2000;49:539–547. doi: 10.2337/diabetes.49.4.539. [DOI] [PubMed] [Google Scholar]
- 48.Risérus U., Tan G. D., Fielding B. A., Neville M. J., Currie J., Savage D. B., Chatterjee V. K., Frayn K. N., O'Rahilly S., Karpe F. Rosiglitazone increases indexes of stearoyl-CoA desaturase activity in humans: link to insulin sensitization and the role of dominant-negative mutation in peroxisome proliferator-activated receptor-gamma. Diabetes. 2005;54:1379–1384. doi: 10.2337/diabetes.54.5.1379. [DOI] [PubMed] [Google Scholar]
- 49.Kurebayashi S., Hirose T., Miyashita Y., Kasayama S., Kishimoto T. Thiazolidinediones downregulate stearoyl-CoA desaturase 1 gene expression in 3T3-L1 adipocytes. Diabetes. 1997;46:2115–2118. doi: 10.2337/diab.46.12.2115. [DOI] [PubMed] [Google Scholar]
- 50.Nicholson A. C., Hajjar D. P., Zhou X., He W., Gotto A. M., Jr, Han J. Anti-adipogenic action of pitavastatin occurs through the coordinate regulation of PPARgamma and Pref-1 expression. Br. J. Pharmacol. 2007;151:807–815. doi: 10.1038/sj.bjp.0707250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goldstein J. L., Brown M. S. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 52.Sakakura Y., Shimano H., Sone H., Takahashi A., Inoue N., Toyoshima H., Suzuki S., Yamada N. Sterol regulatory element-binding proteins induce an entire pathway of cholesterol synthesis. Biochem. Biophys. Res. Commun. 2001;286:176–183. doi: 10.1006/bbrc.2001.5375. [DOI] [PubMed] [Google Scholar]
- 53.Boizard M., Le Liepvre X., Lemarchand P., Foufelle F., Ferré P., Dugail I. Obesity-related overexpression of fatty-acid synthase gene in adipose tissue involves sterol regulatory element-binding protein transcription factors. J. Biol. Chem. 1998;273:29164–29171. doi: 10.1074/jbc.273.44.29164. [DOI] [PubMed] [Google Scholar]
- 54.Le Lay S, Ferré P., Dugail I. Adipocyte cholesterol balance in obesity. Biochem. Soc. Trans. 2004;32:103–106. doi: 10.1042/bst0320103. [DOI] [PubMed] [Google Scholar]
- 55.Kim J. B., Spiegelman B. M. ADD1/SREBP1 promotes adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes Dev. 1996;10:1096–1107. doi: 10.1101/gad.10.9.1096. [DOI] [PubMed] [Google Scholar]
- 56.Kim J. B., Wright H. M., Wright M., Spiegelman B. M. ADD1/SREBP1 activates PPARgamma through the production of endogenous ligand. Proc. Natl. Acad. Sci. U.S.A. 1998;95:4333–4337. doi: 10.1073/pnas.95.8.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Freeman D. J., Norrie J., Sattar N., Neely R. D., Cobbe S. M., Ford I., Isles C., Lorimer A. R., Macfarlane P. W., McKillop J. H., et al. Pravastatin and the development of diabetes mellitus: evidence for a protective treatment effect in the West of Scotland Coronary Prevention Study. Circulation. 2001;103:357–362. doi: 10.1161/01.cir.103.3.357. [DOI] [PubMed] [Google Scholar]
- 58.Sever P. S., Dahlöf B., Poulter N. R., Wedel H., Beevers G., Caulfield M., Collins R., Kjeldsen S. E., Kristinsson A., McInnes G. T. Prevention of coronary and stroke events with atorvastatin in hypertensive patients who have average or lower-than-average cholesterol concentrations, in the Anglo-Scandinavian Cardiac Outcomes Trial–Lipid Lowering Arm (ASCOT-LLA): a multicentre randomised controlled trial. Lancet. 2003;361:1149–1158. doi: 10.1016/S0140-6736(03)12948-0. [DOI] [PubMed] [Google Scholar]
- 59.Diabetes Atorvastin Lipid Intervention (DALI) Study Group. The effect of aggressive versus standard lipid lowering by atorvastatin on diabetic dyslipidemia: the DALI study: a double-blind, randomized, placebo-controlled trial in patients with type 2 diabetes and diabetic dyslipidemia. Diabetes Care. 2001;24:1335–1341. doi: 10.2337/diacare.24.8.1335. [DOI] [PubMed] [Google Scholar]
- 60.Nakata M., Nagasaka S., Kusaka I., Matsuoka H., Ishibashi S., Yada T. Effects of statins on the adipocyte maturation and expression of glucose transporter 4 (SLC2A4): implications in glycaemic control. Diabetologia. 2006;49:1881–1892. doi: 10.1007/s00125-006-0269-5. [DOI] [PubMed] [Google Scholar]
- 61.Altshuler D., Hirschhorn J. N., Klannemark M., Lindgren C. M., Vohl M. C., Nemesh J., Lane C. R., Schaffner S. F., Bolk S., Brewer C., et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat. Genet. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 62.Yamauchi T., Waki H., Kamon J., Murakami K., Motojima K., Komeda K., Miki H., Kubota N., Terauchi Y., Tsuchida A., et al. Inhibition of RXR and PPARgamma ameliorates diet-induced obesity and type 2 diabetes. J. Clin. Invest. 2001;108:1001–1013. doi: 10.1172/JCI12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kubota N., Terauchi Y., Miki H., Tamemoto H., Yamauchi T., Komeda K., Satoh S., Nakano R., Ishii C., Sugiyama T., et al. PPAR gamma mediates high-fat diet-induced adipocyte hypertrophy and insulin resistance. Mol. Cell. 1999;4:597–609. doi: 10.1016/s1097-2765(00)80210-5. [DOI] [PubMed] [Google Scholar]
- 64.Miles P. D., Barak Y., He W., Evans R. M., Olefsky J. M. Improved insulin-sensitivity in mice heterozygous for PPAR-gamma deficiency. J. Clin. Invest. 2000;105:287–292. doi: 10.1172/JCI8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saisho Y., Morimoto A., Umeda T. Determination ofFPP in dog and human plasma by high-performance liquid chromatography with fluorescence detection. Anal. Biochem. 1997;252:89–95. doi: 10.1006/abio.1997.2314. [DOI] [PubMed] [Google Scholar]