Abstract
Modulating enzyme function with small molecule activators, as opposed to inhibitors, offers new opportunities for drug discovery and allosteric regulation. Previously, we identified a compound, called 1541, from a high through-put screen (HTS) that stimulates activation of a proenzyme, procaspase-3, to generate mature caspase-3. Here, we further investigate the mechanism of activation and report the surprising finding that 1541 self-assembles into nanofibrils, exceeding 1 μm in length. These particles are an unanticipated outcome from a HTS with properties distinct from standard globular protein aggregators. Moreover, 1541 nanofibrils function as a unique biocatalytic material, which activate procaspase-3 via induced proximity. These studies demonstrate a novel approach for proenzyme activation through binding to fibrils, which may mimic how procaspases are naturally processed on protein scaffolds.
Many proteases are expressed in dormant forms, known as zymogens, which are activated in response to diverse stimuli. Activation of these latent enzymes is often catalyzed by processing from upstream proteases and, in some cases, via auto-proteoylsis1. These transitions can be promoted by binding co-factors, by sequestering cellular inhibitors, or by interacting with signaling complexes2. Caspases, or cysteine aspartyl proteases, are expressed as such inactive precursors, or procaspases, which become activated in fate-determining transformations as diverse as cell death, innate immune responses, and differentiation3. Further mechanistic insight into the processes that facilitate procaspase activation will foster the design of novel chemical probes to study the sufficiency of caspases for these phenotypes4,5.
Procaspases are typically activated upon cleavage by upstream proteases or binding to protein scaffolds in response to intrinsic or extrinsic cellular signals6. For example, the apoptosome recruits procaspase-9, the death inducing signaling complex (DISC) interacts with procaspase-8, and the various inflammasome complexes associate with procaspase-1 to stimulate activity3,7. Procaspases bind to these signaling platforms, which triggers clustering, oligomerization and/or a conformation change, and proteolytic processing. Removal of an N-terminal prodomain and an additional cleavage to yield a large and small subunit generates the mature enzyme8. While procaspase-3 self-activation is normally restricted under physiological conditions, auto-proteolysis can occur9.
In an earlier investigation, we identified a synthetic small molecule, termed 1541, which promotes auto-activation of procaspase-310 (Chart 1). After a lag phase, the compound induced a burst in activity due to the formation of processed caspase-3. These results plus additional characterization suggested that the compounds worked through an allosteric mechanism to promote auto-proteolysis. In this study, we show that 1541 and related analogs spontaneously assemble into highly ordered nanofibrils. Procaspase-3 becomes immobilized on the surface of the fibrils and generates active caspase-3. It is conceivable that these “amyloid-like” fibrils mimic natural protein scaffolds for activating procaspases.
Chart 1.
Compound 1541 and Analogs.
Globular aggregates of small molecules have previously been described that inhibit enzyme activity11-13. These aggregates are readily identified by diagnostic experiments, including detergent sensitivity, β-lactamase inhibition, and sensitivity to bovine serum albumin (BSA)14-16. Furthermore, others have reported that detergent sensitive molecules in screening libraries can also promote enzyme activity17. We performed these diagnostic tests on 1541 with mixed results. For example, common detergents such as Triton or CHAPS did not disrupt procaspase-3 activation, and 1541 did not inhibit β-lactamase (Figure S1 and S2). Unexpectedly, the addition of BSA protected against procaspase-3 activation (Figure S3). This result alone is not definitive, since BSA contains hydrophobic patches that can bind soluble small molecules.
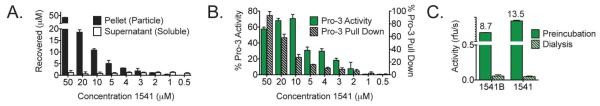
Due to these inconclusive results, we investigated the solubility of 1541 by centrifugation18. Surprisingly, 1541 pelleted from solution at 16,100-x g with a solubility constant (Ksp) between 1 and 2 μM, concentrations close to the AC50 of procaspase-3 activation by 1541 (Figure 1A and Figure S1). Interestingly, both active and inactive analogs pelleted upon centrifugation, suggesting that the particles may not necessarily be responsible for the activity observed (Figure S4).
Figure 1. 1541 forms particles that are required for binding and activation of procaspase-3.
(A) The amount of 1541 in the pellet (filled black bars) and the supernatant (open bars) was analyzed after centrifugation. (B) (i) Samples of procaspase-3 (C163A) with varying concentrations of 1541 were centrifuged, and the amount of procaspase-3 in the pellet was determined (hatched bars). (ii) The activity of wild-type procaspase-3 with 1541 was determined (filled green bars). (C) In a preincubation control sample, procaspase-3 was added to 10 μM 1541 (or 1541B) inside a dialysis membrane (filled green bars). In the test sample, only procaspase-3 was inside the chamber with 1541 (or 1541B) outside the dialysis chamber (hatched green bars). After 12 h at 37°C, the activity of each sample was measured.
We next evaluated whether procaspase-3 could directly interact with the particulates. We used procaspase-3 (C163A), an inactive/ catalytically dead variant, to directly analyze binding without the complication of processing. Varying concentrations of 1541 and 1541B were added to 200 nM procaspase-3 (C163A). The solutions were immediately centrifuged, and the amounts of procaspase-3 in the pellet were assayed by gel electrophoresis and quantified by densitometry (Figure S4). We observed co-sedimentation of the C163A enzyme for both 1541 and 1541B at concentrations that correlate to their AC50s (Figure 1B and S5). Furthermore, enzyme sedimentation was not observed with the inactive analog, 1541D, even though compound sedimentation occurred. These data provide evidence that procaspase-3 binds to the high molecular weight particulates of the active compounds. While suggestive of a mechanism dependent on particles, both soluble small molecules and larger particles are present in the activation assay.
We performed a dialysis experiment to establish the relevant activating species. Procaspase-3 was placed inside a dialysis chamber with a 12-14 kDa molecular weight cutoff (approximately 2 nm radius of gyration), and 10 μM 1541 or 1541B was placed in buffer outside the membrane. After a 12-hour incubation, no activation or processing of the proenzyme was observed (Figure 1C and S6). In contrast, when the compounds were placed both inside and outside of the dialysis membrane, we saw dramatic activation and complete processing of procaspase-3. This suggested that particulates larger than roughly 4 nm were responsible for the activation effects.
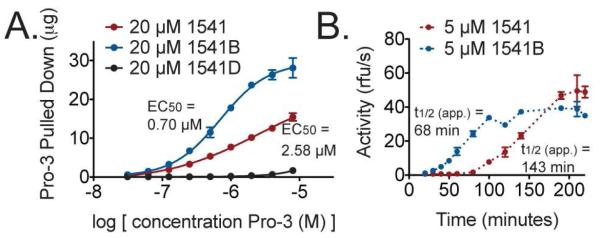
We subsequently analyzed if 1541 and 1541B particles exhibited saturation behavior to assess if the compounds provided a fixed number of binding sites to accommodate the enzyme or if they promoted nonspecific aggregation of procaspase-3. We set the concentration of the active analogs at 20 μM, ten-fold above their Ksp values, and titrated them with procaspase-3 (C163A) to directly investigate binding saturation. The mixtures were centrifuged and the amount of enzyme in the pellet was measured. The particulates from both compounds showed clear saturation behavior, yet with varying affinities for procaspase-3 (Figure 2A and S7). Furthermore, when monitoring the rates of procaspase-3 activation by 1541 and 1541B, we found that binding affinity and activation rate were coincident (Figure 2B). This suggests that procaspase-3 is immobilized by the particles with specific and saturable sites of interaction that impact activity.
Figure 2. 1541 and 1541B particles have different affinities for procaspase-3, which correlates to distinct rates of activation.
(A) 20 μM 1541, 1541B, or 1541D was incubated with a dilution series of procaspase-3 (C163A). After centrifugation, the pellet was examined for procaspase-3. (B) 5 μM 1541 or 1541B was added to wild-type procaspase-3, and initial rates were plotted over time. An apparent t1/2 value that incorporates the lag phase and the t1/2 was calculated for each compound. Notably, the distinct lag phases drive the difference in the apparent t1/2.
While the particles of 1541 and 1541B seemed to account for procaspase-3 activation, they were resistant to standard tests of small molecule aggregation. Thus, we determined whether compounds that are known to form aggregates could activate procaspase-311,16. A panel of six aggregators, well known for promiscuous enzyme inhibition, was incapable of activating procaspase-3 (Figure S8). We did see that one of the promiscuous compounds, Congo red, inhibited mature caspase-3. As expected, the inhibition of caspase-3 by the aggregator was detergent sensitive (Figure S8). This demonstrated a functional interaction between Congo red and caspase-3, however no activation of procaspase-3 was observed. In contrast, 1541 particles appear responsible for procaspase-3 activation and behave differently from standard aggregators. Due to the discrepancies in behavior, we sought to further characterize the physical properties of 1541 to establish if it exhibited novel properties of known colloidal aggregators or if it assembled into unique particles that promoted procaspase-3 activity.
As with centrifugation experiments, dynamic light scattering (DLS) studies demonstrated the presence of particles at room temperature for 10 μM 1541 and 1541B. Remarkably, these particles showed characteristics distinct from standard aggregators (Table 1 and Figure S9). Common properties of aggregators have been previously published, with 3′,3″,5′,5″-tetraiodophenolpthalein (TIPT) shown here for direct comparison15,19. The intensity of scattered light for 1541 and 1541B was approximately 10 to 20-fold lower than for TIPT (270.2, 170.1, and 3903.5 kilocounts per second (kcps), respectively). Conversely, the radii of 1541 and 1541B particles determined by DLS were over 10-fold greater, 1112.8 nm and 910.7 nm, compared to TIPT, 71.1 nm. The inactive analog, 1541D, has properties similar to TIPT with an intensity of 3317.0 kcps and a radius of 164.1 nm. These results suggest that 1541 and 1541B form particles that are different from 1541D and other standard colloidal aggregates.
Table 1.
Unique properties of 1541 and related analogs compared to standard aggregators.
| Sample | Temp. (°C) | DLS Conc. (μM) | DLS Intensity (kcps) | Radius (nm) | Ksp (μM)† | AC50 (μM)‡ |
|---|---|---|---|---|---|---|
| Control | 25 | - | 19.7 ± 1.3 | 1.0 ± 0.3 | no particles | NA |
| Control | 37 | - | 20.8 ± 1.2 | 1.3 ± 0.3 | no particles | NA |
| 1541 | 25 | 10 | 270.2 ± 77.9 | 1112.8 ± 86.6 | 1.3 ± 0.3 | >50 |
| 1541 | 37 | 10 | 17.5 ± 1.4 | 0.5 ± 0.1 | 10 ± 5 | 3.0 ± 0.6 |
| 1541B | 25 | 10 | 170.1 ± 44.7 | 910.7 ± 66.7 | 1.6 ± 0.4 | 3.1 ± 0.5§ |
| 1541B | 37 | 10 | 15.8 ± 0.9 | 0.4 ± 0.1 | 20 ± 5 | 1.8 ± 0.1 |
| 1541D | 25 | 10 | 3317.0 ± 854.0* | 164.1 ± 7.7 | 7.5 ± 0.5 | NA |
| 1541D | 37 | 10 | 3071.9 ± 465.1* | 186.4 ± 21.6 | 7.5 ± 0.5 | NA |
| TIPT | 25 | 50 | 3903.5 ± 1423.0 | 71.1 ± 8.6 | - | NA |
| TIPT | 37 | 50 | 3933.2 ± 1369.0 | 76.7 ± 7.8 | - | NA |
1541D samples were run at 25% laser power.
Ksp values reported are from particle flow cytometry.
Activity measurements were taken at 8 h.
Maximum percent activation for 1541B at 25°C is 8% compared to 60% at 37°C. NA = no activation.
The distinct properties of 1541 and 1541B were further characterized by particle flow cytometry18. Notably, the Ksp determined for 1541 and 1541B closely matched the AC50’s for self-activation of procaspase-3 (Table 1). Particle count increased linearly with concentration until reaching a critical particle count at 4 μM for 1541 and at 5 μM for 1541B (Figure S10). Increasing concentration beyond this point only generated larger particle sizes but not particle numbers. Once nucleated, 1541 and 1541B particles appear to favor recruitment of additional small molecules to grow in size. Conversely, TIPT and 1541D appear to maintain a constant size but increase particle count with increasing concentration (Figure S10). These results further depict unique features of 1541 and 1541B compared to standard aggregators.
Since the procaspase-3 self-activation assay is performed at 37°C, we studied the properties of 1541 and 1541B particles after agitation at this temperature. Interestingly, the intensity of scattered light as well as the radii of 1541 and 1541B particles decreased to buffer values for these conditions (Table 1). Since the particle flow cytometer uses light scattering to detect particles, a shift in Ksp was similarly observed for both 1541 and 1541B at 37°C to 10 and 20 μM, respectively. Nonetheless, incubation and centrifugation of 1541 and 1541B at 37°C still resulted in pelleting of the compound (Figure S11). These results indicate that the particles were still present at increased temperatures, but significantly altered as to become undetectable by light scattering. Notably, temperature had minimal impact on the properties of both TIPT and 1541D.
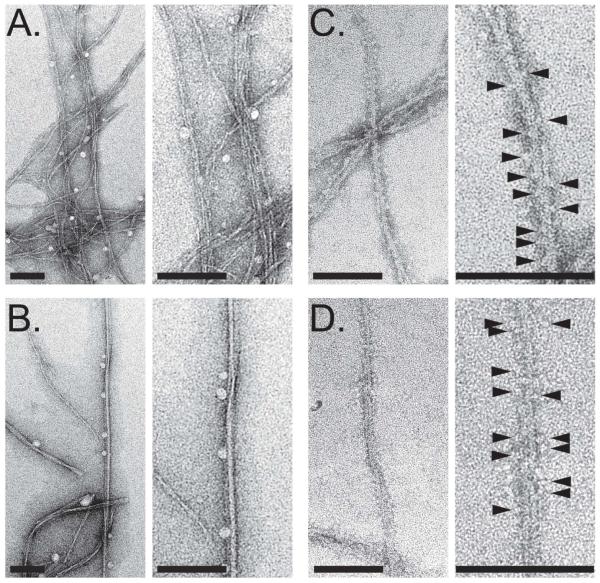
Given the strong evidence that the particles of 1541 and 1541B were distinct from promiscuous inhibitors, we investigated the molecular structure of 1541 compared to a colloidal aggregator, TIPT, by transmission electron microscopy (TEM). TEM images of TIPT showed the typical globular structures of known aggregators (Figure S12). In dramatic contrast, 1541 produced long thin fibrils extending to over 1 μm in length (Figure 3). The fibrils tended to cluster into braided bundles, but single strands appeared as thin as 2.6 nm, perhaps only a few molecules thick (Figure S12). At 37°C, 1541 tended to form thicker fibrils and typically appeared as individual strands, rather than the larger tangles observed at room temperature. Consistent with the tangles breaking apart at higher temperatures, we observed delayed centrifugation of 1541 particles after 1 h incubation at 37°C compared to room temperature (Figure S13). Disrupting the clusters at 37°C may allow a greater extent of the fibril surface to be accessible to the procaspase to facilitate activation, however, additional experiments are necessary to elucidate temperature effects.
Figure 3. Transmission electron micrographs of 1541 nanofibrils with and without procaspase-3.
(A) Negatively stained 1541 at 25°C shows bundles of very thin and flexible fibrils. (B) In contrast, 1541 at 37°C consists mainly of larger, less flexible fibrils. (C) 1541 fibrils at 25°C decorated with procaspase-3; and (D) 1541 fibrils at 37°C decorated with procaspase-3. In (C) and (D) the surface of the fibrils appears decorated with procaspase-3 molecules (arrow heads). Magnification bars = 100 nm.
We subsequently examined if binding of procaspase-3 to 1541 fibrils could be observed by TEM. We incubated 50 μM 1541 with 500 nM procaspase-3 (C163A) at 37°C and also at 25°C. At both temperatures, the edges of the nanofibrils no longer looked crisp, but appeared to be decorated with protein particles, suggesting that procaspase-3 was lining the length of the fibrils (Figure 3 and S12). The nanofibrils also appeared to be wider, consistent with the enzyme binding to the surface.
Our results show that 1541 and 1541B spontaneously assemble into nanofibrils that can activate procaspase-3. These particles have distinct properties from standard aggregators identified in HTS, which typically lead to enzyme inhibition11. We propose that these fibrils act as a scaffold to concentrate procaspase-3 where it can be processed by other enzymes in close proximity (Figure 4). In this regard, these synthetic fibrils mimic protein scaffolds, such as the inflammosomes, the apoptosome, and the DISC, which facilitate procaspase activation3,7,20. Alternatively, the fibrils may alter the conformation of the proenzyme to promote intramolecular processing. Further studies are underway to distinguish between cis- versus trans-activation promoted by the fibrils.
Figure 4. Model of procaspase-3 activation by 1541 nanofibrils.
1541 and analogs spontaneously self-assemble into nanofibrils. The fibrils bind directly to procaspase-3 to promote increased local concentration of the enzyme. Upon recruitment to the fibrils, procaspase-3 is activated to generate mature caspase-3.
Intriguingly, 1541 particles appear structurally similar to the fibrous β-sheet aggregates formed by β-amyloid proteins (Aβ). Aβ proteins have been shown to facilitate proenzyme activation, such as the conversion of prekallikrein to kallikrein of the plasma kinin-forming cascade as well as the conversion of plasminogen into the active protease plasmin by the tissue-type plasminogen activator (tPA)21,22. Furthermore, several cellular proteins assemble into an amyloid fold, such as fibrin and Pmel17, to facilitate natural processes, including proenzyme activation23-25. Filamentous structures within the cell have even been shown to directly associate with caspase-326. In this regard, 1541 fibrils appear to mimic endogenous and possibly disease-related fibrous structures on which the procaspases can become concentrated and activated.
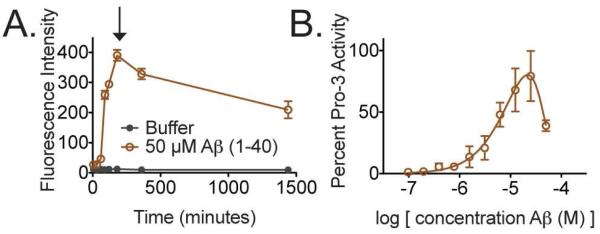
To evaluate if such physiologically relevant fibrils could interact with procaspase-3, we generated fibrils from Aβ (1-40) by agitation of the peptide at 37°C in a caspase activity buffer (Figure 5A). The Aβ fibrils were subsequently serially diluted and incubated with procaspase-3. Similar to our small molecule fibrils, addition of these peptide fibrils to procaspase-3 stimulated activation (Figure 5B). This result has potential implications for Alzheimer’s disease where aggregates of Aβ have been linked to caspase-dependent neurotoxicity27. Moreover, we previously described the apoptotic activity induced by 1541 and 1541B10. We are now exploring how these nanofibrils promote cell death. Such a mechanism is certainly intriguing and non-standard from a drug discovery perspective.
Figure 5. Aβ fibrils activate procaspase-3.
(A) Aβ peptide (1-40) was agitated at 37°C in a caspase activity buffer to form fibrils. Fibril formation was monitored over time by an increase in thioflavin T fluorescence. (B) Aβ peptide (1-40) samples were taken at 3 h (black arrow), serially diluted, and incubated with procaspase-3 for 6 h. Percent activity is shown.
For bioprocessing applications, enzyme immobilization on nanostructures offers an exciting alternative to traditional approaches for manipulating enzyme functionality, such as genetic and chemical engineering28,29. 1541 represents a first-in-class, self-assembling, small-molecule nanofibril, which acts as a catalyst for procaspase-3 activation. Future studies aim to elucidate the specific structural properties of the molecule that drive fibril assembly as well as those that facilitate procaspase-3 activation. It may be possible to use this scaffold for the design of other proenzyme activators or to discover novel fibril-forming small molecules that activate proenzymes.
Supplementary Material
ACKNOWLEDGMENT
We thank S. Mahrus, N. Agard, and J. Porter for invaluable advice, guidance, and support. We acknowledge M. Bennett and H. Rodriguez at Calithera for advice and discussions. We thank A. Doak, B. Feng, S. Prusiner, and B. Shoichet for helpful discussions on promiscuous enzyme inhibitors and for use of their instruments. We thank D. Hostetter for providing granzyme B. We thank the Wells lab, the Small Molecule Discovery Center at UCSF, and W. F. DeGrado for suggestions. This research was supported by grants from the National Institutes of Health (R01 CA136779, P01 AG02132) as well as the National Cancer Institute Postdoctoral Fellowship (F32 CA119641-03) (to D.W.W.), the ARCS Foundation Award (to J.A.Z.), and the Schleroderma Research Foundation Evnin-Wright Fellowship (to J.A.Z.).
Footnotes
Supporting Information Placeholder
ASSOCIATED CONTENT Supporting Information. Supplemental figures and detailed experimental procedures. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Kassell B, Kay J. Science. 1973;180:1022. doi: 10.1126/science.180.4090.1022. [DOI] [PubMed] [Google Scholar]
- (2).Turk B. Nat. Rev. Drug discovery. 2006;5:785. doi: 10.1038/nrd2092. [DOI] [PubMed] [Google Scholar]
- (3).Li J, Yuan J. Oncogene. 2008;27:6194. doi: 10.1038/onc.2008.297. [DOI] [PubMed] [Google Scholar]
- (4).Shen A. Mol. Biosyst. 2010;6:1431. doi: 10.1039/c003913f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Zorn JA, Wells JA. Nat. Chem. Biol. 2010;6:179. doi: 10.1038/nchembio.318. [DOI] [PubMed] [Google Scholar]
- (6).Salvesen GS, Riedl SJ. Adv. Exp. Med. Biol. 2008;615:13. doi: 10.1007/978-1-4020-6554-5_2. [DOI] [PubMed] [Google Scholar]
- (7).Schroder K, Tschopp J. Cell. 2010;140:821. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- (8).Stennicke HR, Salvesen GS. Biochim. Biophys. Acta. 2000;1477:299. doi: 10.1016/s0167-4838(99)00281-2. [DOI] [PubMed] [Google Scholar]
- (9).Roy S, Bayly CI, Gareau Y, Houtzager VM, Kargman S, Keen SL, Rowland K, Seiden IM, Thornberry NA, Nicholson DW. Proc. Natl. Acad. Sci. USA. 2001;98:6132. doi: 10.1073/pnas.111085198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Wolan DW, Zorn JA, Gray DC, Wells JA. Science. 2009;326:853. doi: 10.1126/science.1177585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).McGovern SL, Helfand BT, Feng B, Shoichet BK. J. Med. Chem. 2003;46:4265. doi: 10.1021/jm030266r. [DOI] [PubMed] [Google Scholar]
- (12).Ryan AJ, Gray NM, Lowe PN, Chung C-W. J. Med. Chem. 2003;46:3448. doi: 10.1021/jm0340896. [DOI] [PubMed] [Google Scholar]
- (13).Reddie KG, Roberts DR, Dore TM. J. Med. Chem. 2006;49:4857. doi: 10.1021/jm060115z. [DOI] [PubMed] [Google Scholar]
- (14).Feng BY, Simeonov A, Jadhav A, Babaoglu K, Inglese J, Shoichet BK, Austin CP. J. Med. Chem. 2007;50:2385. doi: 10.1021/jm061317y. [DOI] [PubMed] [Google Scholar]
- (15).Coan KED, Shoichet BK. Mol. Biosyst. 2007;3:208. doi: 10.1039/b616314a. [DOI] [PubMed] [Google Scholar]
- (16).Seidler J, McGovern SL, Doman TN, Shoichet BK. J. Med. Chem. 2003;46:4477. doi: 10.1021/jm030191r. [DOI] [PubMed] [Google Scholar]
- (17).Goode DR, Totten RK, Heeres JT, Hergenrother PJ. J. Med. Chem. 2008;51:2346. doi: 10.1021/jm701583b. [DOI] [PubMed] [Google Scholar]
- (18).Coan KED, Shoichet BK. J. Am. Chem. Soc. 2008;130:9606. doi: 10.1021/ja802977h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Doak AK, Wille H, Prusiner SB, Shoichet BK. J. Med. Chem. 2010;53:4259. doi: 10.1021/jm100254w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Yuan S, Yu X, Asara JM, Heuser JE, Ludtke SJ, Akey CW. Structure. 2011;19:1084. doi: 10.1016/j.str.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Shibayama Y, Joseph K, Nakazawa Y, Ghebreihiwet B, Peerschke EI, Kaplan AP. Clin. Immunol. 1999;90:89. doi: 10.1006/clim.1998.4621. [DOI] [PubMed] [Google Scholar]
- (22).Kingston IB, Castro MJ, Anderson S. Nat. Med. 1995;1:138. doi: 10.1038/nm0295-138. [DOI] [PubMed] [Google Scholar]
- (23).Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. PLoS Biol. 2006;4:e6. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Kranenburg O, Bouma B, Kroon-Batenburg LMJ, Reijerkerk A, Wu Y-P, Voest EE, Gebbink MFBG. Curr. Biol. 2002;12:1833. doi: 10.1016/s0960-9822(02)01224-1. [DOI] [PubMed] [Google Scholar]
- (25).Fowler DM, Koulov AV, Balch WE, Kelly JW. Trends Biochem. Sci. 2007;32:217. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- (26).Dinsdale D, Lee JC, Dewson G, Cohen GM, Peter ME. Am. J. Pathol. 2004;164:395. doi: 10.1016/S0002-9440(10)63130-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Wellington CL, Hayden MR. Clin. Genet. 2000;57:1. doi: 10.1034/j.1399-0004.2000.570101.x. [DOI] [PubMed] [Google Scholar]
- (28).Kim J, Grate J, Wang P. Chem. Eng. Sci. 2006 [Google Scholar]
- (29).Ge J, Lu D, Liu Z, Liu Z. Biochem. Eng. J. 2009 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.