Abstract
Since class III unconventional myosins are motor proteins with an N-terminal kinase domain, it seems likely they play a role in both signaling and actin based transport. A growing body of evidence indicates that the motor functions of human class IIIA myosin, which has been implicated in progressive hearing loss, are modulated by intermolecular autophosphorylation. However, the phosphorylation sites have not been identified. We studied the kinase activity and phosphorylation sites of mouse class III myosins, mMyo3A and 3B, which are highly similar to their human orthologs. We demonstrate that the kinase domains of mMyo3A and 3B are active kinases, and that they have similar, if not identical, substrate specificities. We show that the kinase domains of these proteins autophosphorylate, and that they can phosphorylate sites within their myosin and tail domains. Using liquid chromatography-mass spectrometry, we identified phosphorylated sites in the kinase, myosin motor and tail domains of both mMyo3A and 3B. Most of the phosphorylated sites we identified and their consensus phosphorylation motifs are highly conserved among vertebrate class III myosins, including human class III myosins. Our findings are a major step toward understanding how the functions of class III myosins are regulated by phosphorylation.
Keywords: Unconventional myosin, Ste20 kinase, myosin phosphorylation, non-syndromic deafness (DFNB30), hair cells, photoreceptors
INTRODUCTION
The class III unconventional myosins of vertebrates and class XXI unconventional myosins of invertebrates are unique members of the myosin superfamily in that they have a kinase domain at their N-terminus (Odronitz et al., 2009). These kinase/myosins have been implicated in sensory functions; specifically in the photoreceptors of invertebrates and vertebrates (Montell and Rubin 1988; Li et al., 1998; Edwards and Battelle, 1987; Battelle et al., 1998; Cardasis et al., 2007; Dosé et al., 2003, 2004; Katti et al., 2009) and vestibular and cochlear hair cells of vertebrates (Walsh et al., 2002; 2011; Schneider et al., 2006).
Vertebrates express two kinase/myosins, myosin IIIA and IIIB (Myo3A and Myo3B), which are encoded by different genes. The functions of vertebrate class III myosins remain largely unknown, but mutations in MYO3A lead to nonsyndromic deafness (DFNB30) in humans (Walsh et al., 2002) and hearing loss in mice (Walsh et al., 2011). Surprisingly, these same mutations produce no detectable defects in photoreceptor or vestibular cell functions. A possible explanation is that Myo3B compensates for defects in 3A where they colocalize as they do in the photoreceptors of vertebrates (Katti et al., 2009; Lin-Jones et al., 2009).
The functions of kinase/myosins are of interest because of their importance for sensory cell function and survival. The kinase/myosins of vertebrates are molecular motors, and the motor and actin binding properties of human MYO3A are being studied in detail (Komaba et al., 2003; 2010; Kambara et al., 2006; Dosé et al., 2007; Quintero et al., 2010). All of the kinase/myosins studied so far are active serine/threonine kinases (Ng et al., 1996; Komaba et al., 2003; Kempler et al., 2007) belonging to the Sterile-20 (Ste20) family (Dan et al., 2001), and there is growing evidence that the functions of kinase/myosins are regulated by autophosphorylation. For example, the motor and actin binding properties of fish and human kinase/myosins are altered by deleting or inactivating the kinase domain (Erickson et al., 2003; Lin-Jones et al., 2004; Schneider et al., 2006; Dosé et al., 2007; Quintero et al., 2010; Komaba et al., 2010). In spite of the importance of phosphorylation for regulating the functions of kinase/myosins, none of the phosphorylation sites in vertebrate class III myosins has been identified.
This study focuses on mouse class III myosins, mMyo3A and 3B, which are highly homologous to the human proteins (Katti et al., 2009). We demonstrate here that the kinase domains of mMyo3A and 3B are active kinases which autophosphorylate and which have similar, if not identical, substrate specificities. We show that the kinase domain of these proteins phosphorylates sites within the myosin and tail domains, and we identified phosphorylated sites in the kinase, myosin motor and tail domains with mass spectrometry. Most of the phosphorylated sites identified, and their consensus phosphorylation motifs, are highly conserved among vertebrate class III myosins, including human class III myosins. Our findings are a major step toward understanding how the functions of class III myosins are regulated by phosphorylation.
MATERIALS AND METHODS
Reagents
Unless otherwise specified, reagents were purchased from Sigma-Aldrich (St. Louis, MO) or Fisher Scientific (Pittsburgh, PA).
Construction of recombinant baculovirus, expression, and purification of the mouse myosin III kinases
The mMyo3B kinase/transition domains were amplified from plasmid containing a cDNA clone encoding full length Myo3B (Katti et al., 2009); the mMyo3A kinase/transition domains were amplified from mouse testis cDNA (Clontech, Mountain View, CA). The sequences of the PCR primers used to amplify these domains are presented in Supplemental Table 1. The 5' PCR primer for the mMyo3B construct (mM3B K/T F1) was designed against a sequence located 5' of the coding region shown in Figure 1, and it included a BamHI restriction site along with a Kozak sequence and a start codon. The 3' reverse primer (mM3B K/T R1) contained a NotI restriction site. The amplified product was cloned into pCR4-TOPO, verified for the presence of the correct sequence then cloned as a BamHI-NotI fragment into pFASTBAC™1 transfer vector that had been modified to encode a FLAG-tag followed by a stop codon 3' of the NotI restriction site. The 5' PCR primer for the mMyo3A construct contained a Not1 restriction site along with a Kozak sequence and a start codon (Supplemental Table 1, mM3A K/T F1). The reverse primer (mM3A K/T R1) contained the sequence encoding the FLAG-tag followed by a HindIII restriction site. The PCR amplified product was cloned into pCR4-TOPO, verified for the presence of correct sequences, then cloned as a NotI-HindIII fragment into the pFASTBAC™1 baculovirus transfer vector which provided an in-frame stop codon (Invitrogen, Carlsbad, CA). The ends of the cloned fragments were sequenced with pFASTBAC™1 specific primers, and the open reading frames were confirmed. Recombinant baculovirus was made using the FastBac system (Invitrogen), and the mMyo3 kinases/transition domains were expressed in Sf9 cells.
Figure 1. Amino acid sequence alignment of full length mouse Myo3B (mMyo3B) with mouse Myo3A (mMyo3A) (Clustal X, Young, 1976) showing regions of particular interest for this study.
The amino acid numbering for each sequence is indicated at the right; black boxes indicate amino acids that are conserved between the two sequences. The kinase, myosin motor and tail domains are labeled and were predicted using the InterProScan program from EMBL-EBI. The loop2 actin binding region within the myosin domain and Mouse 3 tail homology domain I (3THDI) are indicated with dashed horizontal lines. The loop2 region was predicted from an alignment with chicken skeletal muscle myosin (Accession # P13538, Sellers, 1999). 3THDI was identified by Dosé et al. (2003) and is a region within the tail domains of class III myoisns that has relatively high amino acid identity among sequences. Sequences N-terminal to the vertical dashed line near the beginning of the motor domain include the kinase domain and most of the transition region between the kinase and myosin domains. These sequences were expressed in sf9 cells, served as the source of kinase activity and were tested as substrates for autophosphorylation. The sequences of mMyo3B below the colored solid lines were expressed in E. coli. and were tested as substrates for phosphorylation by the mMyo3 kinase domain. The sequence below the blue line includes the kinase activation region, that below the orange line includes the loop2 region and that below the green line includes THDI.. Red solid arrow heads indicate sites identified in this study that are phosphorylated by mMyo3 kinase; red open arrows indicate potential phosphorylation sites that could not be distinguished unambiguously from MS/MS fragment ions. Genbank Accession numbers: mMyo3A, AY10136; mMyo3B, NM_177376.
The expressed proteins were purified from sf9 cells using anti- FLAG- affinity column chromatography and the bound protein kinase was eluted using FLAG peptide dissolved in a buffer containing 0.5M NaCl (Wang et al., 2000; Kempler et al., 2007). The purity of the expressed kinase was assayed by staining the gels with SimplyBlue™ Safe Stain (Invitrogen). Purified protein was stored in 20–50µl drops by freezing over liquid nitrogen. Protein concentration was determined by separating the kinase by SDS PAGE together with a protein standard (ovalbumin), staining the gel with Coomassie Blue and quantifying the staining intensities of the kinase and the standard by ImageQuant software (GE HealthCare, Life Sciences, Pittsburgh, PA).
Expression of additional sequences tested as substrates for mMyo3A and 3B in E. coli
Sequences from mMyo3B were amplified by PCR from the following templates: activation region, from a bacmid containing the full-length sequence; loop2 region, from a full-length cDNA clone; tail domain from reverse transcribed total mouse retinal RNA. The 5' and 3' primers included an NdeI and a HindIII restriction site, respectively (Supplemental Table 1). The PCR products were cloned into pCR 4-TOPO (Invitrogen), sequenced in both directions to verify the correct sequence, inserted into pET 28a plasmid (Invitrogen) and expressed as His-tagged fusion proteins in Rosetta 2(DE3) E. coli cells (Novagen, Madison, WI). The kinase activation region expressed as an insoluble protein, therefore it was solubilized with 6 mol/l urea; the loop2 and tail proteins were soluble. Expressed proteins were enriched by standard Ni++ affinity chromatography in the absence or presence of urea as appropriate (PrepEaseR Histidine-tagged protein purification Midi kit – High Affinity, USB Corporation, Cleveland, OH). Before use, proteins purified in the absence of urea were dialyzed against 20 mmol/l TrisHCl, pH 7.5; for proteins purified in the presence of urea, 3 mol/l urea was added to the dialysate. Mutants of the loop2 region of Limulus Myo21, which were tested as substrates for the mMyo3 kinases, were expressed and purified as described by Kempler et al. (2007). Concentrations of the purified substrates were determined by their Coomassie Blue staining intensities on SDS PAGE gels as described above for the expressed kinase.
Protein phosphorylation assays
Incubation conditions for autophosphorylation of the kinase domains were as described in Kempler et al. (2007). The final concentration of ATP was 30 µmol/l containing 25 µCi [γ32P]ATP (PerkinElmer, Boston, MA). When the kinase domains were incubated with other substrates, they were preincubated for 1 hr with 30µmol/l non-radioactive ATP. Substrates and [γ32P] ATP were then added and the incubations continued for another 30 min. The concentrations of kinases and substrates used for individual experiments are indicated in the legends to the figures.
Protein phosphorylation assays were stopped by addition of SDS (sodium dodecyl sulfate) sample buffer (Laemmli, 1970) and sonication. Samples were separated by SDS PAGE (polyacrylamide gel electrophoresis), and protein phosphorylation was detected as Phosphorimages (Phosphorimager SI, GE HealthCare Biosciences, Pittsburgh, PA) of dried, Coomassie Blue-stained gels. Phosphorylation was quantified with ImageQuant software and normalized to protein concentration determined as described above from scans of the Coomassie Blue protein stains.
Phosphorylated amino acids were identified as described by Boyle et al. (1991). Briefly, components of an in vitro phosphorylation reaction were separated by SDS PAGE and blotted to PVDF (polyvinylidene fluoride membrane, Millipore Corp, Billerica, MA). The region of the blot containing the phosphorylated product of interest was located by autoradiography, cut out, hydrolyzed in acid, and the acid digest was separated in two dimensions on cellulose plates together with phosphoamino acid standards. The cellulose plates were stained with ninhydrin to visualize the locations of the phosphoamino acid standards. Phosphorylated amino acids were detected as Phosphorimages, and their relative levels of phosphorylation were quantified using ImageQuant.
Mass spectrometric analysis of phosphorylation sites in the kinase/transition domains of mMyo3A and 3B
The regions of mMyo3A and 3B encompassing the kinase/transition domains (Fig. 1) were expressed in sf9 cells, purified by FLAG affinity chromatography, and then purified further by one-dimensional SDS PAGE. Alternatively, affinity purified kinase/transition domains were incubated for 1hr in the presence of 30 µM non-radioactive ATP before gel purification. Gels were stained with Coomassie Blue to locate the bands of interest. These bands were cut from the gels and subjected to in-gel digestion. Tryptic peptides were run using liquid chromatography followed by tandem mass spectrometry (LC-MS/MS) as previously described (Alvares et al., 2009).
MS/MS spectra were extracted by Analyst version 1.1 (AB Sciex, Foster City, CA). Peptide sequencing and determination of the phosphorylated sites were performed using Mascot (Matrix Science, Boston, MA; version 2.0.01) to search enhanced product ion scan spectra against the mouse protein database assuming trypsin as the cleavage enzyme. Mascot was searched with both a parent and fragment ion mass tolerance of 0.30 Da. Iodoacetamide derivative of cysteine was specified in Mascot as a fixed modification. Deamidation of asparagine and glutamine, oxidation of methionine and phosphorylation of serine, threonine, and tyrosine were selected as variable modifications. Only charge states 2-4 were considered for sequencing. Scaffold (version Scaffold-01-06-03, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Each enhanced product ion scan spectrum was manually inspected to ensure acceptable b and y ion coverage and phosphorylation site identification.
Mass spectrometric analysis of phosphorylation sites in the loop2 region of the motor domain and tail domain of Myo3B
Solution-phase or gel bands containing the loop2 region or tail domain of mMyo3B were processed according to protocols used by the University of South Florida’s Center for Drug Discovery and Innovation (CDDI) Proteomics Facility. Online reversed phase LC-MS/MS analysis of tryptic digests was carried out on either a linear ion trap (LTQ XL, Thermo, San Jose, CA) or a hybrid linear ion trap-Orbitrap mass spectrometer (LTQ Orbitrap XL, Thermo), both equipped with a nanoelectrospray ionization source and operated with the Xcalibur (version 2.0) data acquisition software. Tryptic digests were loaded onto a 75 µm i.d. × 2 cm ProteoPep II capillary trap (New Objective, Woburn, MA) and desalted with 3% ACN, 1% acetic acid for 5 min prior to injection onto a 75 µm i.d. × 15 cm ProteoPep II analytical column (New Objective). A linear gradient to 40% ACN in 90 min at 250 nL/min was provided by a 2D Ultra nanoHPLC system (Eksigent, Dublin, CA) onto the analytical column. The standard data-dependent and neutral loss (NL)-driven MS3 modes of data acquisition were used for both the LTQ XL and LTQ Orbitrap XL analysis in which a m/z survey scan was performed followed by MS/MS analysis of the top five (standard) or three (NL-driven MS3) most intense precursor ions in each data dependent acquisition cycle. For LTQ Orbitrap XL analysis, full-scan mass spectra were acquired at 60,000 mass resolving power (at m/z 400) from m/z 350 to 1500 in parallel to linear ion trap MS/MS spectral acquisition using the automatic gain control mode of ion trapping. CID in the linear ion trap was performed using a 3.0-µ isolation width and 35% normalized collision energy with helium as the collision gas. For NL-driven MS3 data dependent acquisition, isolation and subsequent fragmentation of ions exhibiting a m/z 49, 32.67, or 24.5 difference (representing neutral loss of phosphorylation from +2, +3, or +4 precursor ions, respectively) from the precursor ion occurred if the neutral loss fragment ions were among the three most intense ions in the MS/MS spectra.
MS/MS spectra generated by data dependent acquisition via the LTQ XL or LTQ Orbitrap XL were extracted by BioWorks version 3.3 and searched against a current composite IPI mouse protein sequence database containing both forward and randomized sequences (v 3.54, 111,968 entries with randomized sequences) using the Mascot v 2.2 (Matrix Science, Boston, MA) search algorithm. Mascot was searched with a fragment ion mass tolerance of 0.6 Da and a parent ion tolerance of 2.5 Da (LTQ XL all spectra or LTQ Orbitrap XL MS3 spectra) or 10.0 ppm (LTQ Orbitrap XL full scan spectra) assuming the digestion enzyme trypsin with the possibility of one missed cleavage. Carbamidomethylation of cysteine was specified as a fixed modification while oxidation of methionine, and phosphorylation of serine and threonine were specified as variable modifications. For NL-driven MS3 data, extracted MS/MS/MS spectra were searched in a similar fashion; however, dehydrated serine or threonine (−18 Da) were included as variable modifications in the database search. Scaffold (version Scaffold_03_00_02) was employed to validate MS/MS-based peptide and protein identifications. In general, phosphopeptide identifications were accepted at a peptide probability value (Keller, 2002) that established a peptide identification false discovery rate of ≤1% (as calculated by Scaffold) in addition to manual validation of the phosphopeptide MS/MS spectra.
RESULTS
The amino acid sequence alignment of mMyo3A and 3B in Figure 1 highlights regions of the proteins of particular interest for this study. The kinase/transition region of mMyo3A and 3B were expressed in sf9 cells and served as the sources of kinase activity. These regions began with the initiation methionine of each protein and ended near the beginning of the motor domain as indicated by the dashed, vertical, line in Figure 1. Other regions of mMyo3B were expressed in E. coli and were tested as substrates for the kinases. These sequences are indicated below the solid, colored lines in Figure 1. The polypeptide below the blue line includes the activation region of the kinase domain, that below the orange line includes the loop2 region of the myosin domain, and that below the green line includes Myo3 tail homology domain I (3 THDI, Dosé et al., 2003). In addition, the entire tail domain of mMyo3B was tested as a substrate and analyzed by mass spectrometry. Red solid arrowheads in Figure 1 indicate amino acids identified by mass spectrometry as phosphorylation sites; red open arrows indicate additional possible phosphorylation sites (Supplemental Figures 1and 2).
Are mMyo3A and 3B active kinases?
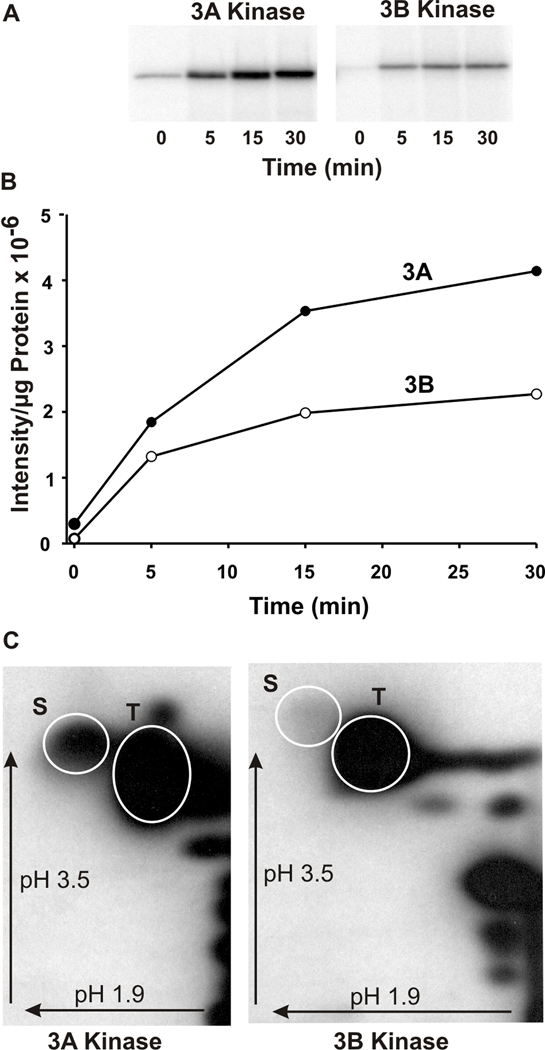
The kinase/transition region of both mMyo3A and 3B (amino acid 1–332, mMyo3B numbering) autophosphorylated in a time-dependent manner (Fig. 2A and 2B), therefore both are active kinases. Phosphoamino acid analyses of these regions revealed that both kinases autophosphorylated serine and threonine residues (Fig. 2C). In both proteins, threonines were more heavily phosphorylated than serines. 32P-labeled serines were clearly detected in mMyo3A. The incorporation of 32P-labeled onto serine residues in Myo3B was low when compared to that in mMyo3A, but it was seen consistently. Since proteins expressed in sf9 cells can become phosphorylated during expression, the significance of the differences in the relative incorporation of 32P onto serines and threonines in vitro is difficult to interpret without additional studies.
Figure 2. The kinase/transition domains of mMyo3A and 3B autophosphorylate on serine and threonine residues.
A. Phosphorimages of the autophosphorylation of mMyo3A and 3B with time. B. Plot of the intensities of the phosphorylated bands shown in A versus time per µg of protein incubated. Incubations contained 5.6µg mMyo3A (closed circles) and 2.4µg mMyo3B (open circles). C. Phosphorimages of phosphorylated amino acids in the kinase/transition domains of mMyo3A and 3B following a 1hr incubation in vitro. Incubations contained 10µmol/l and 8µmol/l of mMyo3A and 3B, respectively. Phosphorylated amino acids were separated by two dimensional high voltage thin layer electrophoresis using a pH 1.9 buffer in the first dimension and a pH 3.5 buffer in the second dimension. Arrows indicate the directions of electrophoretic migration. White circles show the locations of the phosphoserine (S) and phosphothreonine (T) amino acid standards.
We applied tandom mass spectrometry (MS/MS) to identify phosphorylated residues within the kinase/transition regions of recombinant mMyo3A and 3B directly after they were purified from sf9 cells. Four phosphopeptides were identified from mMyo3A; two from 3B (Supplemental Fig. 1). Each contained only one phosphorylated residue. This confirmed that both proteins are phosphorylated during expression.
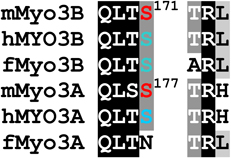
Two of the phosphorylated residues identified by MS/MS are conserved within the activation loops of the mMyo3A and 3B kinase domains (residues 169–178, mMyo3B numbering): a phosphothreonine located at the end of the activation loop (pT178 in Myo3B; pT184 in Myo3A) and a phosphoserine located within the activation loop (pS171 in Myo3B; pS177 in Myo3A) (Fig. 1 and Supplemental Fig. 1). Two additional phosphoserines (pS313 and pS332) were identified between the kinase and myosin domains of mMyo3A (the transition region) (Fig. 1 and Supplemental Fig. 1). Serines at these sites are not conserved in mMyo3B.
Since we identified the phosphorylated sites described above directly after purifying the protein from sf9 cells, it was not clear whether they were autophosphorylated or phosphorylated by some other kinase in sf9 cells. Two observations suggest the residues are autophosphorylated. First, when the purified kinase/transition regions were incubated in vitro with ATP prior to analysis by mass spectrometry, the same, and no additional, phosphorylated sites were identified. Second, results of the phosphoamino acid analyses of the kinase domains following autophosphorylation in vitro (Fig. 2C) showed a higher level of serine phosphorylation in mMyo3A compared to 3B, which is consistent with the MS/MS identification of three phosphoserines within the kinase/transition region of mMyo3A compared to one in 3B.
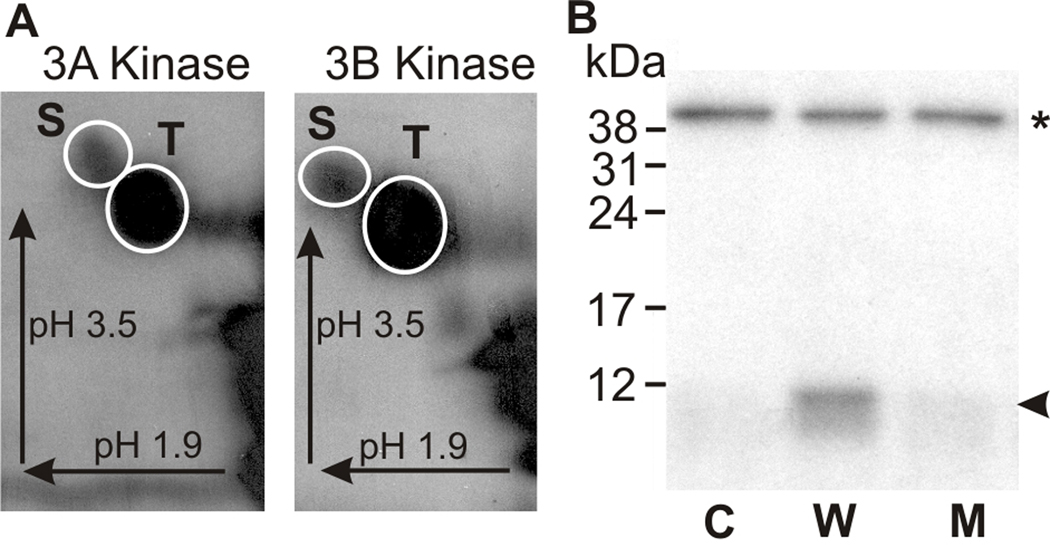
To test further whether both S171 and T178 are autophosphorylated, a polypeptide which includes the kinase activation loop of mMyo3B (the sequence below the blue line in Fig. 1) was expressed as a recombinant protein in E. coli and assayed as a substrate for mMyo3A and 3B kinase. The purified kinase activation region was incubated in vitro with mMyo3A or 3B kinase in the presence of [γ32P] ATP, separated from the kinases by SDS PAGE and blotted to PVDF membrane. Phosphorimages of these blots revealed that the activation region of mMyo3B was phosphorylated by both kinases, and phosphoamino acid analyses revealed that both kinases phosphorylated serines and threonines (Fig. 3A). Since proteins expressed in E. coli do not become phosphorylated as they are expressed, we were able to use the in vitro phosphorylation of the E. coli expressed kinase activation region to quantify the relative levels of serine and threonine phosphorylation. We found that threonines were preferred 5:1.
Figure 3. The kinase activation region of mMyo3B is phosphorylated by mMyo3A and 3B kinase on both serine and threonine residues.
The kinase activation region of mMyo3B used as substrate in these assays is indicated below the blue line in Figure 1. The polypeptide was expressed in E. coli and thus not post-translationally modified during expression. A. Phosphorimages of phosphorylated amino acids released from the mMyo3B kinase activation region phosphorylated by mMyo3A or 3B kinase as indicated. Incubations contained 10 µmol/Lsubstrate and 2.5 µmol/L kinase and continued for 1hr. Phosphorylated amino acids were separated in two dimensions by high voltage thin layer electrophoresis as described in the legend to Figure 2. Arrows indicate the directions of electrophoretic migration. White circles show locations of the phosphoserine (S) and phosphothreonine (T) amino acid standards. Both serines and threonines were phosphorylated. B. Autoradiograph of an SDS-PAGE gel examining the phosphorylation of the wild-type and mutated kinase activation region of mMyo3B by the full length mMyo3B kinase domain. Incubations contained 6µmol/L substrate and 0.15 µmol/L kinase. The full length kinase domain migrates at about 40 kDa (asterisk); the kinase activation region migrates at about 10 kDa (arrowhead). The full length kinase domain was incubated with no substrate (C, control), with the wild-type sequence of the kinase activation region (W); and with a mutated kinase activation region (M) in which the two phosphorylated residues identified by mass spectrometry, S171 and T178, were converted to alanines. The incubation in B was for 5 min.
To confirm that S171 and T178 were the phosphorylated residues, we mutated both to alanines, expressed the mutated sequence in E. coli and tested whether the mutated protein could be phosphorylated by mMyo3A or 3B kinase. Figure 3B shows that the mutated sequence was not phosphorylated by either kinase. These results, taken together with the MS/MS data (Supplemental Figures 1and 2), provide direct evidence for the autophosphorylation of both S171 and T178 in mMyo3B with T178 being preferred. Both residues are conserved and become phosphorylated in mMyo3A; therefore both are probably autophosphorylated in mMyo3A.
Do mMyo3A and 3B have similar substrate specificities?
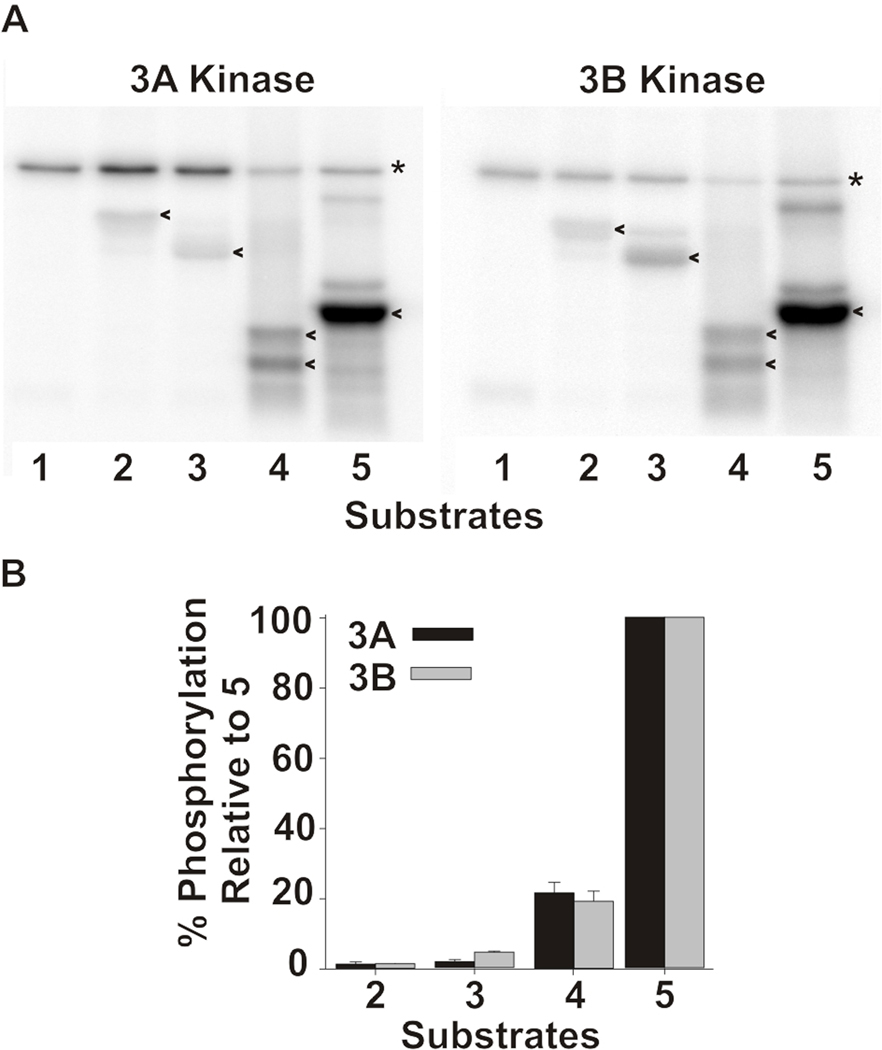
To compare the substrate specificities of mMyo3A and 3B we tested their ability to phosphorylate a number of different proteins (Fig. 4) and heterologously expressed polypeptides (Fig. 5). The sequences of the polypeptides are based on the sequence of the loop2 region of Limulus kinase/myosin. The wild-type sequence contains three predicted phosphorylation sites, two predicted PKA sites and a predicted PKC site (Fig. 5A). The wild-type sequence was mutated such that each substrate tested contained only one predicted phosphorylation site. The mMyo3A and 3B kinases showed similar specificities for the proteins assayed (Fig. 4) and for all three sites in the polypeptides (Fig. 5B). These finding suggest that mMyo3A and 3B have similar, if not identical substrate specificities.
Figure 4. mMyo3A and 3B kinases show similar specificities for different protein substrates.
A. Phosphorimages of dried SDS-PAGE gels showing the incorporation of 32P from γ32P ATP into the kinase domains (asterisk)(0.4µmol/L) and different protein substrates (20 µmol/L)(arrow heads). Substrates assayed were: 1. none. 2. α casein. 3. β casein. 4. histone. 5. myelin basic protein. B. Intensities of the phosphorylated substrate bands were quantified with ImageQuant in three separate experiments. The levels of phosphorylation in each experiment were normalized to the level of phosphorylation observed with the substrate in lane 5 and expressed as the mean % of that phosphorylation ± the standard error of the means. The substrates are as listed in A.
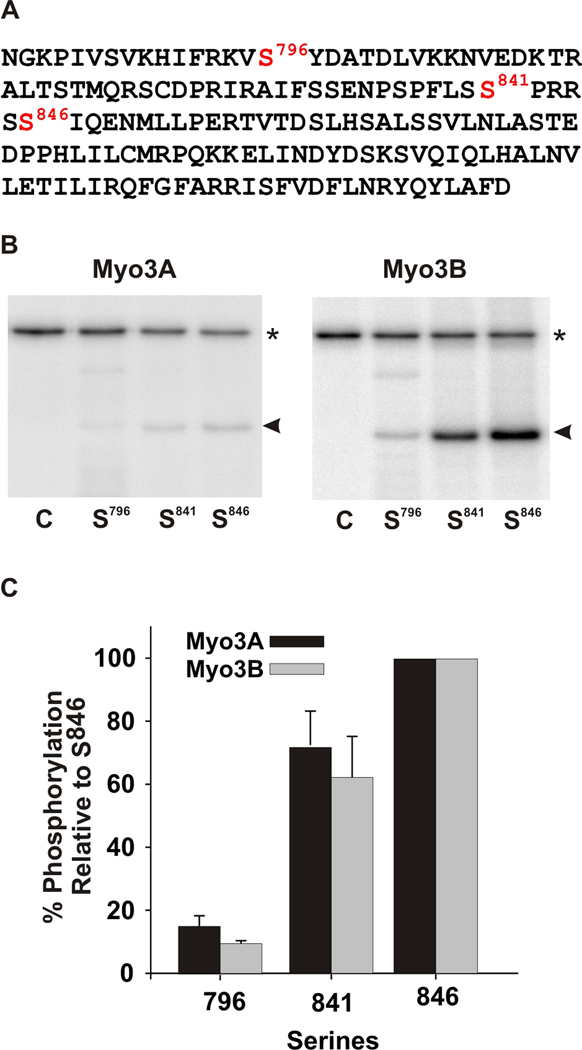
Figure 5. mMyo3A and 3B kinases show similar specificities for predicted PKA and PKC phosphorylation sites in test polypeptide substrates.
A. Wild-type sequence of the test polypeptide, which is the sequence of the loop2 region of the kinase/myosin expressed in Limulus photoreceptors. The sequence contains one predicted PKC phosphorylation site (S841) and two predicted PKA sites (S796 and S846). This sequence was mutated to produce three different substrates in which only one of the three predicted phosphorylation sites was present. The other two serines were converted to alanines. Mutated sequences were expressed in E. coli, purified and incubated together with the mMyo3A and 3B kinases. Kinases were present at 0.4µmol/L; substrates at (1.6 µmol/L). B. Phosphorimages of SDS-PAGE gels which separated the kinases (asterisk) from the substrates (arrow head). The serine available for phosphorylation is indicated below each lane. The control lane (C) contained no substrate. C. Quantification of the phosphorylation of the substrates in three different experiments relative to the phosphorylation of the substrate containing S846 and expressed as the mean % of this phosphorylation ± the standard error of the means.
Can the kinase domains of mMyo3A and 3B autophosphorylate other regions of the protein?
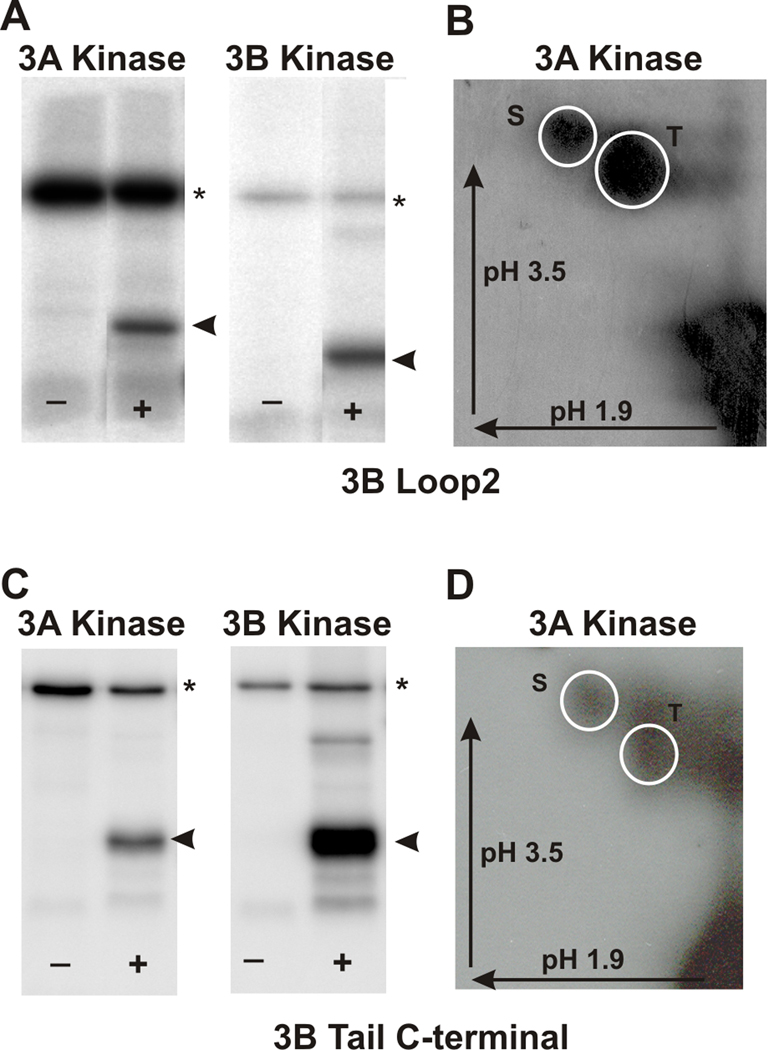
We tested whether the mMyo3A and 3B kinases could phosphorylate the loop2 region of the myosin motor domain of mMyo3B and its tail domain (Fig. 1). Loop2 of the myosin motor domain is of particular interest because it is a target for autophosphorylation in the Limulus kinase/myosin (Kempler et al., 2007), and because it is important for determining myosin motor functions (see Discussion). Different regions of the tail domain are thought to target the myosin to particular subcellular compartments (Porter et al., 1992) and to bind to cargo proteins and to actin (see Discussion). Figures 6A and C show that both the loop2 region and C-terminus of the tail domain are phosphorylated by mMyo3A and 3B kinases. The loop2 region was phosphorylated on both serine and threonine residues with threonines preferred 2:1 (Fig. 6B; the C-terminus of the tail domain was phosphorylated equally on serine and threonine residues (Fig. 6D).
Figure 6. mMyo3A and 3B kinases phosphorylate the loop2 region of the myosin motor domain and the C-terminal tail of Myo3B on both serines and threonines.
The loop2 and C-terminal tail regions of mMyo3B were expressed as His-tagged proteins in E. coli, purified and used as substrates for mMyo3A and 3B kinases. A and C. Phosphorimages of SDS-PAGE gels which separated the kinases (asterisk) from the substrates (arrow head). The kinase used is indicated at the top of each pair of Phosphorimages. (−) lanes contained the kinase and no added substrate. (+) lanes contained the kinase and substrate. B and D. Phosphorimages of phosphorylated amino acids from the loop2 region of Myo3B (B) and the C-terminal of the tail of Myo3B (D). Both were phosphorylated by Myo3A kinase. Identical results were obtained following phosphorylation with the 3B kinase. The phosphoamino acids were separated by two-dimensional high voltage thin layer electrophoresis as described in the legend to Figure 2. The arrows indicate the directions of electrophoretic migration. The white circles indicate the locations of the phosphoserine (S) and phosphothreonine (T) standards.
Phosphorylated residues in the loop2 and tail domains of Myo3B were identified by MS/MS (Supplemental Fig. 2). A polypeptide that includes loop2 (the region below the orange line in Figure 1) the full-length tail domain and the C-terminus of the tail domain (the region below the green line in Figure. 1) were expressed in E. coli and incubated separately in vitro with non-radioactive ATP and mMyo3A kinase. Solutions or gel bands from these reactions were processed as described in Methods.
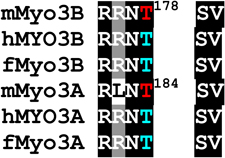
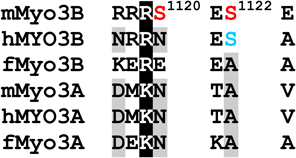
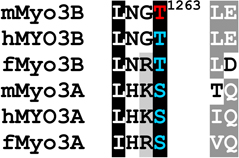
Phosphorylated residues in the loop2 region were identified as pS887, which is within loop2 (Fig. 1 and Supplemental Fig. 2A), and pT935, which is close to the C-terminal end of loop2 (Fig. 1 and Supplemental Fig. 2B and C). A phosphopeptide identified near the N-terminus of the tail domain contains two potential phosphorylation sites, S1120 and S1122, but insufficient fragment ions were obtained to identify the site unequivocally (Fig. 1 and Supplemental Fig. 2D). A phosphorylation site within the C-terminal of the tail domain was identified as pT1263 (Fig. 1 and Supplemental Fig. 2E). It is located within 3THDI, one of two more highly conserved regions within the tail domains of class III myosins (Dosé et al., 2003).
Standard data-dependent acquisition was able to identify a majority of phosphorylation sites in the loop2 and tail domains. However, for the phosphopeptide from the C-terminal of the tail domain, predominant neutral loss of H3PO4 (98 Da) upon CID fragmentation produced minimal fragment ions, which were inadequate to identify the sequence from a database search. Accordingly, neutral loss-driven MS3 was employed. This approach resulted in the identification of T1263 from the tryptic peptide MLSSPEDTMYYNQLNGpTLEYQGSQR (Supplemental Fig. 2E) and validated phosphorylation sites identified from standard data-dependent acquisition (for example Supplemental Fig. 2C).
MS/MS analyses did not reveal a phosphorylated serine within the C-terminal of the tail domain, although the two-dimensional analysis of phosphoamino acids indicates a serine in this region becomes phosphorylated (Fig. 6D). This suggests we have not identified all sites that can be phosphorylated in this region.
DISCUSSION
We show here that mMyo3A and 3B are active kinases with similar, if not identical, substrate specificities, and we identify with mass spectrometry autophosphorylation sites within their kinase, myosin motor and tail domains. This is the first direct identification of autophosphorylation sites within any vertebrate class III myosin. Many of the sites identified are highly conserved among vertebrate class III myosins, including those from humans, and located in regions of known functional significance. Many sites phosphorylated by mouse class III myosin kinases are also predicted sites for phosphorylation by cyclic AMP-dependent protein kinase (PKA) or protein kinase C (PKC). This suggests that class III myosins can be phosphorylated and regulated by second messenger activated kinases as well as autophosphorylation.
Substrate specificities
The substrate specificities of mMyo3A and 3B were similar when tested against a number of different proteins (Fig. 4) and three different predicted phosphorylation sites in the same polypeptide (Fig. 5). In the polypeptide substrate, the preferred phosphorylation site for both kinases was S846 (Fig. 5), a predicted PKA site with a basic amino acid located at the third position on the N-terminal side of the phosphorylation site (P−3) (Yaffe et al., 2001; Fujii et al., 2004; Zhu et al., 2005). This site is phosphorylated by PKA (Kempler et al., 2007). S796 in the same polypeptide is also a predicted PKA site, but it is a poorer substrate than S846 for PKA (Kempler et al., 2007), mMyo3A and 3B (Fig. 5). Interestingly, mMyo3A and 3B phosphorylate S841 in the polypeptide nearly as well as S846 (Fig. 5). S841 is a predicted PKC phosphorylation site with a neighboring hydrophobic amino acid at its C-terminal side (P+1) and a basic amino acid at P+2 (Fujii et al., 2004; Nishikawa et al., 1997). This site is not phosphorylated by PKA (Kempler et al., 2007). These findings suggest that mMyo3A and 3B can target both PKA- and PKC-like sites.
We anticipated mMyo3A and 3B kinases would have similar substrate specificities because their kinase domains show high sequence identity, and because they have an identical pattern of acid residues at sites thought to be critical for determining the substrate specificity of kinases. This pattern includes an acidic residue that aligns with E128 in mouse PKA (E104and D118 in Myo3B and 3A, respectively). An acidic residue at this site is thought to confer strong preference among kinases for a basic amino acid at P−3 (Yaffe et al., 2001; Fujii et al., 2004). In this respect mMyo3A and 3B differ from most other Ste20 kinases (Zhu et al., 2005). However, like other Ste20 kinases, mMyo3A and 3B have a ny pattern of acidic residues at sites within their substrate binding pocket, a pattern thought to strongly influence substrate specificity (Zhu et al., 2005). Both kinases lack an acidic residue that aligns with E171 in mouse PKA (N148 and N162 in mMyo3B and 3A, respectively) and have an acidic residue that aligns with E231 in mouse PKA (E216 and E230 in mMyo3B and 3A, respectively). The three critical acidic residues in the kinase domains of mouse class III myosins described above are highly conserved among all vertebrate class III myosins; therefore the specificity of vertebrate class III myosins from fish to man is probably similar to that described here for the mouse proteins.
Our analysis of sites autophosphorylated in mMyo3A and 3B provides further support to the idea that their kinase domains target PKA- and PKC-like sites. Several sites have a basic residue at P−3, which is a PKA-like motif (Table 1). These include threonines which our two dimensional amino acid analyses suggest are preferred autophosphorylation sites within the activation region of the kinase domains (pT178 and pT184 in mMyo3B and 3A, respectively) and the preferred phosphorylation site in the loop2 region of mMyo3B (pT935). In addition, each of the two possible phosphoserines identified in the tail domain of mMyo3B has a basic residue at P−3 (pS1120 and S1122). Several other autophosphorylated residues have a hydrophobic amino acid at P+1 or a basic amino acid at P+2, common motifs for PKC phosphorylation. These include pS171 and pS177 in the kinase domains of mMyo3B and 3A, respectively, pS332 in the transition region of mMyo3A, pS887 within loop2 of the mMyo3B motor domain and pT1263 in the 3THDI of mMyo3B. Taken together, our findings show that mouse class III myosins, and probably other vertebrate class III myosins, target serines and threonines with a basic residue at P−3 or P+2, or with a hydrophobic residue at P+1.
Table 1.
Alignments of mouse, human and fish class III myosins in those regions where mMyo3B contains a phosphorylated residue.
Regions are as described in Figure 1
Alignments of sequences from the database were performed in ClustalW. Amino acids highlighted in black are conserved in all six sequences, those in dark gray are conserved in five of the six sequences; those in light gray are conserved in at least four of the six sequences Amino acids in red were identified as autophosphorylation sites in this study; those in blue are predicted autophosphorylation sites. Superscript numbers indicate the position of the autophosphorylated residue in the sequence of mMyo3B or 3A. See Figure 1. Accession numbers: mMyo3A, AY101368; mMyo3B, NM_177376; hMYO3B (human), AF369908; hMYO3A, BC119811; fMyo3B (fish, bass), AF512506; fMyo3A, AF003249.
Phosphorylated sites identified in mMyo3B and their consensus phosphorylation motifs are conserved in other vertebrate class III myosins and positioned to influence class III myosin function
A ClustalW alignment of many vertebrate class III myosins revealed that most of the phosphorylation sites we identified in mouse class III myosins are highly conserved together with their phosphorylation motifs. Regions of interest from representative sequences are shown in Table 1.
Kinase domain
The major autophosphorylation site in the kinase domain of mouse class III myosins (pT178 in mMyo3B) is highly conserved in all vertebrate class III myosins, and in each sequence, a basic residue is located at P−3. This conserved threonine at the C-terminal end of the kinase activation loop aligns with the primary phosphorylation site in other Ste20 kinases, which is required for kinase activation (Glantschnig et al., 2002; Delpire, 2009). Komaba et al. (2003) showed that human MYO3A kinase activity is activated by autophosphorylation, but the autophosphorylation sites responsible were not identified. Our findings suggest that vertebrate class III myosins are activated by autophosphorylation of threonines aligning with pT178 in mMyo3B.
pS171 in mMyo3B and pS177 in mMyo3A are probably secondary autophosphorylation sites because they are much less heavily autophosphorylated than threonines (Fig. 3A). This site, together with a basic residue at P+2, is conserved in mammalian class IIIA and IIIB myosins, fish class IIIB myosins, but not fish class IIIA myosins. Secondary phosphorylation sites have also been identified in other Ste20 kinases (Glantschnig et al., 2002), but the functional consequences of phosphorylating these sites are not yet known.
Loop2 region of the myosin domain
Loop2 in the motor domain of myosins is a region of low amino acid conservation among different myosins. Nevertheless, there is conservation in this region among different vertebrate class IIIB myosins. Importantly for this study, each has a serine that aligns with pS887 in mMyo3B, and this serine is followed by a hydrophobic residue at P+1. Thus, all vertebrate class IIIB myosins may autophosphorylate this site. This site is not conserved in class IIIA myosins.
In contrast to sequences within loop2, sequences at the C-terminal end of loop2 are highly conserved among all class III myosins. In particular, a threonine that aligns with pT935 in mMyo3B, the major phosphorylation site in the loop2 region of mMyo3B, is highly conserved and each has a basic residue at P−3 and a hydrophobic amino acid at P+1. Thus, this site is probably a major autophosphorylation site in all vertebrate class III myosins. Consistent with our findings, Komaba et al. (2003) located an autophosphorylation site in human MYO3A to a peptide from the C-terminal end of the myosin domain which includes a threonine aligning with pT935 in mMyo3B.
Phosphorylation of sites within and near loop2 could profoundly affect class III myosin motor functions. Vertebrate class III myosins are molecular motors directed toward the plus ends of actin filaments (Komaba et al., 2003; Erickson et al., 2003; Lin-Jones et al., 2004), and a variety of biochemical and cell biological studies of human MYO3A indicate that the phosphorylation of sites in the motor domain reduces motor ATPase activity and actin binding affinity (Kambara et al., 2006; Dosé et al., 2007; 2008; Salles et al., 2009; Komaba et al., 2010; Quintero et al., 2010). Loop2, and particularly its net charge, strongly influences the enzymatic and actin binding properties of myosins (Furch et al., 1998; Uyeda et al., 1994; Yengo and Sweeney, 2004). The phosphorylation of sites we identified within and at the C-terminal end of loop2 in mouse class III myosins should strongly influence the net charge in this region, and thus may play a major role in regulating myosin motor and actin binding properties. Our results also suggest that phosphorylating the loop2 regions of class IIIA and IIIB myosins will have different functional consequences since IIIB myosins have two phosphorylation sites while IIIA myosins have one.
Autophosphorylation of sites within and near loop2 of the myosin domain may be a characteristic of kinase/myosins from diverse species. For example, even the distantly related kinase/myosin from Limulus autophosphorylates a site within loop2 of its myosin domain (S846 from Fig. 5) (Kempler et al., 2007).
Tail domain
Sequences within the tail domains of class III myosin are diverse and therefore difficult to align. Note that the ClustalW alignment of the tail domains mMyo3A and 3B in Figure 1 is different from that obtained when many vertebrate class IIIA and IIIB sequences were included in the alignment (Table 1). This sequence diversity probably reflects differences in function that are still mostly unknown. Based on the alignment shown in Table 1, S1120, one of the two potential phosphorylation sites near the N-terminal end of the tail domain of mMyo3B, is not conserved in other vertebrate class III myosins. Therefore, if this site is phosphorylated, the functional effects will be unique to mMyo3B. On the other hand, a serine or threonine aligning with S1122 in mMyo3B with a basic residue is located at P−3, are highly conserved in mammalian class IIIB myosins but not in class IIIA myosins or in class IIIB myosins of non-mammalian vertebrates. Phosphorylation at this site may regulate a function common to mammalian class IIIB myosins.
Although the overall sequence homology among tail domains of different class III myosins is low, two regions of relatively higher homology have been identified: 3THDI (Fig.1), which is present in all vertebrate class III myosins and 3THDII, which is present only in class IIIA myosins (Dosé et al., 2003). Specific functions have been ascribed to these domains. 3THDI binds to the actin bundling protein epsin 1 (Salles et al., 2009); 3THDII contains an important actin binding motif which binds actin in vivo and in vitro (Erickson et al., 2003).
An autophosphorylation site in mMyo3B, pT1263, is located within 3THDI (Fig. 1). This site aligns with either a serine or threonine in other vertebrate class III myosins, and in each of these sequences, except for mMyo3A, the site is followed by a hydrophobic residue at P+1. Thus this site may be autophosphorylated in all class III myosins, with the possible exception of mMyo3A. In 3THDI of human MYO3A, the serine aligning with pT1263 is located only two residues away on the C-terminal side of the epsin 1 binding region (Salles et al., 2009); thus phosphorylation of this site may influence interactions between class III myosins and epsin 1, or other proteins.
mMyo3B lacks 3THDII, therefore we have no direct evidence for a phosphorylation site in this region. However, 3THDII contains a highly conserved serine with a basic residue at P−3 located only four residues away on the C-terminal side of its actin binding site. Based on the substrate specificity for vertebrate class III kinases suggested by this study, this serine is an excellent candidate site for autophosphorylation that could regulate actin-Myo3A interactions.
Phosphorylation of class IIIA and IIIB myosins by one another and by second messenger regulated kinases
Autophosphorylation of kinase/myosins is intermolecular (Kempler et al., 2007; Quintero et al., 2010), and our results indicate that the substrate specificities of Myo3A and 3B kinases are similar, if not identical, to one another. Therefore, where these proteins co-localize, they may phosphorylate one another. Myo3A and 3B co-localize in inner segments of mouse rod and cone photoreceptors (Katti et al., 2009) and in inner segments and calycal processes of fish cone photoreceptors (Lin-Jones et al., 2009). Many of the autophosphorylation sites we identified are also predicted PKA or PKC phosphorylation sites. This suggests that class III myosins can also be phosphorylated and regulated by second messenger dependent kinases. A precedent for this suggestion is the finding that autophosphorylation sites in the Limulus kinase/myosin are also phosphorylated by PKA (Kempler et al., 2007; Cardasis et al., 2007).
In summary, in this study we show that mMyo3A and 3B are active kinases, provide an initial characterization of the substrate specificity of these kinases, and identify autophosphorylation sites. Many of the phosphorylation sites in mouse class III myosins are located at important functional regions and are highly conserved in other vertebrate class III myosins. Our findings suggest new testable hypotheses regarding the role of phosphorylation in the regulation of class III myosin function.
Supplementary Material
ACKNOWLEDGEMENTS
Funding sources. NIH-EY03575 to B.B, NSF-IOS 0517273 to B-A. B., the University of Florida’s Proteomics Core Facility, the University of South Florida Center for Drug Discovery and Innovation Proteomics Facility and Whitney Laboratory internal funds. We thank Lynn Milstead and Joy Nguyen Schade for assistance with the preparation of figures, and Dr. Helena Cardasis for comments on an earlier version of this manuscript.
Abbreviations
- ACN
acetonitrile
- CID
collision induced dissociation
- IDA
information dependent acquisition
- PAGE
polyacrylamide gel electrophoresis
- PKA
cyclic AMP dependent protein kinase
- PKC
protein kinase C
- PVDF
polyvinylidene fluoride
- LC
liquid chromatography
- MS/MS
tandom mass spectrometry
- MS3
multiple stage mass spectrometry
- Myo3A
myosin IIIA
- Myo3B
myosin IIIB
- NL
neutral loss
- SDS
sodium dodecyl sulfate
- Ste20 kinase
Sterile-20 kinase
- TFA
trifluoroacetic acid
- 3THDI
myosin III tail homology domain I
REFERENCES
- Alvarez S, Wilson GH, Chen S. Determination of in vivo disulfide-bonded proteins in Arabidopsis. J. Chromatogr. B. 2009;877:101–104. doi: 10.1016/j.jchromb.2008.11.027. [DOI] [PubMed] [Google Scholar]
- Battelle BA, Andrews AW, Calman BG, Sellers JR, Greenberg RM, Smith WC. A myosin III from Limulus eyes is a clock-regulated phosphoprotein. J. Neurosci. 1998;18:4548–4559. doi: 10.1523/JNEUROSCI.18-12-04548.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biemann K. Mass-spectrometry of peptides and proteins. Ann. Rev. Biochem. 1992;61:977–1010. doi: 10.1146/annurev.bi.61.070192.004553. [DOI] [PubMed] [Google Scholar]
- Boyle WJ, Vandergeer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by 2-dimensional separation on thin-layer cellulose plates. Meth. Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- Cardasis HL, Stevens SM, McClung S, Kempler KE, Powell DH, Eyler JR, Battelle BA. The actin-binding interface of a myosin III is phosphorylated in vivo in response to signals from a circadian clock. Biochemistry. 2007;46:13907–13919. doi: 10.1021/bi701409f. [DOI] [PubMed] [Google Scholar]
- Dan I, Watanabe NM, Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–230. doi: 10.1016/s0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- Delpire E. The mammalian family of sterile 20p-like protein kinases. Eur. J. Physiol. 2009;458:953–967. doi: 10.1007/s00424-009-0674-y. [DOI] [PubMed] [Google Scholar]
- Dosé AC, Hillman DW, Wong C, Sohlberg L, Lin-Jones J, Burnside B. Myo3A, one of two class III myosin genes expressed in vertebrate retina, is localized to the calycal processes of rod and cone photoreceptors and is expressed in the sacculus. Mol. Biol. Cell. 2003;14:1058–1073. doi: 10.1091/mbc.E02-06-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosé AC, Lin-Jones J, Burnside B. Myosin III in photoreceptors: What does it do? In: Williams DS, editor. Cell Biology and Disease of the Outer Retina. New Jersey: World Scientific Press; 2004. pp. 133–162. [Google Scholar]
- Dosé AC, Ananthanarayanan S, Moore JE, Burnside B, Yengo CM. Kinetic mechanism of human myosin IIIA. J. Biol. Chem. 2007;282:216–231. doi: 10.1074/jbc.M605964200. [DOI] [PubMed] [Google Scholar]
- Dosé AC, Ananthanarayanan S, Moore JE, Corsa AC, Burnside B, Yengo CM. The kinase domain alters the kinetic properties of the myosin IIIA motor. Biochemistry. 2008;47:2485–2496. doi: 10.1021/bi7021574. [DOI] [PubMed] [Google Scholar]
- Edwards S, Battelle B. Octopamine- and cyclic AMP-stimulated phosphorylation of a protein in Limulus ventral and lateral eyes. J Neurosc. 1987;7:2811–2820. doi: 10.1523/JNEUROSCI.07-09-02811.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson FL, Corsa AC, Dosé AC, Burnside B. Localization of a class III myosin to filopodia tips in transfected HeLa cells requires an actin-binding site in its tail domain. Mol. Biol. Cell. 2003;14:4173–4180. doi: 10.1091/mbc.E02-10-0656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii K, Zhu GZ, Liu Y, Hallam J, Chen L, Herrero J, Shaw S. Kinase peptide specificity: Improved determination and relevance to protein phosphorylation. Proc. Natl. Acad. Sci., USA. 2004;101:13744–13749. doi: 10.1073/pnas.0401881101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furch M, Geeves MA, Manstein DJ. Modulation of actin affinity and actomyosin adenosine triphosphatase by charge changes in the myosin motor domain. Biochemistry. 1998;37:6317–6326. doi: 10.1021/bi972851y. [DOI] [PubMed] [Google Scholar]
- Glantschnig H, Rodan GA, Reszka AA. Mapping of MST1 kinase sites of phosphorylation - Activation and autophosphorylation. J.Biol. Chem. 2002;277:42987–42996. doi: 10.1074/jbc.M208538200. [DOI] [PubMed] [Google Scholar]
- Kambara T, Komaba S, Ikebe M. Human myosin III is a motor having an extremely high affinity for actin. J. Biol. Chem. 2006;281:37291–37301. doi: 10.1074/jbc.M603823200. [DOI] [PubMed] [Google Scholar]
- Katti C, Dalal JS, Dosé AC, Burnside B, Battelle B-A. Cloning and distribution of myosin 3B in the mouse retina: Differential distribution in cone outer segments. Exp. Eye Res. 2009;89:224–237. doi: 10.1016/j.exer.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A. Empirical statistical model to estimate the accuracy of peptide identification made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Kempler K, Toth J, Yamashita R, Mapel G, Robinson K, Cardasis H, Stevens S, Sellers JR, Battelle B-A. Loop 2 of limulus myosin III is phosphorylated by protein kinase A and autophosphorylation. Biochemistry. 2007;46:4280–4293. doi: 10.1021/bi062112u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komaba S, Inoue A, Maruta S, Hosoya H, Ikebe M. Determination of human myosin III as a motor protein having a protein kinase activity. J.Biol. Chem. 2003;278:21352–21360. doi: 10.1074/jbc.M300757200. [DOI] [PubMed] [Google Scholar]
- Komaba S, Watanabe S, Umeki N, Sato O, Ikebe M. Effect of phosphorylation in the motor domain of human Myosin IIIA on its ATP hydrolysis cycle. Biochemistry. 2010;49:3695–3702. doi: 10.1021/bi902211w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during assembly of head of bacteriophage-T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Li HS, Porter JA, Montell C. Requirement for the NINAC kinase/myosin for stable termination of the visual cascade. J.Neurosci. 1998;18:9601–9606. doi: 10.1523/JNEUROSCI.18-23-09601.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Jones J, Parker E, Wu M, Dosé A, Burnside B. Myosin 3A transgene expression produces abnormal actin filament bundles in transgenic Xenopus laevis rod photoreceptors. J. Cell Sci. 2004;117:5825–5834. doi: 10.1242/jcs.01512. [DOI] [PubMed] [Google Scholar]
- Lin-Jones J, Sohlberg L, Dosé A, Breckler J, Hillman D, Burnside B. Identification and localization of myosin superfamily members in fish retina and retinal pigmented epithelium. J. Comp. Neurol. 2009;513:209–223. doi: 10.1002/cne.21958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C, Rubin GM. The Drosophila ninaC locus encodes two photoreceptor cell specific proteins with domains homologous to protein kinases and the myosin heavy chain head. Cell. 1988;52:757–772. doi: 10.1016/0092-8674(88)90413-8. [DOI] [PubMed] [Google Scholar]
- Ng KP, Kambara T, Matsuura M, Burke M, Ikebe M. Identification of myosin III as a protein kinase. Biochemistry. 1996;35:9392–9399. doi: 10.1021/bi960181a. [DOI] [PubMed] [Google Scholar]
- Nishikawa K, Toker A, Johannes FJ, Zhou SY, Cantley LC. Determination of the specific substrate sequence motifs of protein kinase C isozymes. J. Biol.Chem. 1997;272:952–960. doi: 10.1074/jbc.272.2.952. [DOI] [PubMed] [Google Scholar]
- Odronitz F, Becker S, Kollmar M. Reconstructing the phylogeny of 21 completely sequenced arthropod species based on their motor proteins. BMC Genomics. 2009;10 doi: 10.1186/1471-2164-10-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter JA, Hicks JL, Williams DS, Montell C. Differential localizations of and requirements for the two Drosophila ninaC kinase/myosins in photoreceptor cells. J. Cell Biol. 1992;116:683–693. doi: 10.1083/jcb.116.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero OA, Moore JE, Unrath WC, Manor U, Salles FT, Grati M, Kachar B, Yengo CM. Intermolecular Autophosphorylation Regulates Myosin IIIa Activity and Localization in Parallel Actin Bundles. J. Biol. Chem. 2010;285:35770–35782. doi: 10.1074/jbc.M110.144360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salles FT, Merritt RC, Manor U, Dougherty GW, Sousa AD, Moore JE, Yengo CM, Dosé AC, Kachar B. Myosin IIIa boosts elongation of stereocilia by transporting espin 1 to the plus ends of actin filaments. Nat. Cell Biol. 2009;11:443–U180. doi: 10.1038/ncb1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider ME, Dosé AC, Salles FT, Chang WS, Erickson FL, Burnside B, Kachar B. A new compartment at stereocilia tips defined by spatial and temporal patterns of myosin IIIa expression. J. Neurosci. 2006;26:10243–10252. doi: 10.1523/JNEUROSCI.2812-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeda TQP, Ruppel KM, Spudich JA. Enzymatic-activities correlate with chimeric substitutions at the actin-binding face of myosin. Nature. 1994;368:567–569. doi: 10.1038/368567a0. [DOI] [PubMed] [Google Scholar]
- Walsh T, Walsh V, Vreugde S, Hertzano R, Shahin H, Haika S, Lee MK, Kanaan M, King MC, Avraham KB. From flies' eyes to our ears: Mutations in a human class IIImyosin cause progressive nonsyndromic hearing loss DFNB30. Proc. Natl. Acad. Sci. U.S.A. 2002;99:7518–7523. doi: 10.1073/pnas.102091699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh VL, Raviv D, Dror AA, Shahin H, Walsh T, Kanaan MN, Avraham KB, King MC. A mouse model for human hearing loss DFNB30 due to loss of function of myosin IIIA. Mamm. Genome. 2011;22:170–177. doi: 10.1007/s00335-010-9310-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Harvey EV, Conti MA, Wei DF, Sellers JR. A conserved negatively charged amino acid modulates function in human nonmuscle myosin IIA. Biochemistry. 2000;39:5555–5560. doi: 10.1021/bi000133x. [DOI] [PubMed] [Google Scholar]
- Yaffe MB, Leparc GG, Lai J, Obata T, Volinia S, Cantley LC. A motif-based profile scanning approach for genome-wide prediction of signaling pathways. Nat. Biotech. 2001;19:348–353. doi: 10.1038/86737. [DOI] [PubMed] [Google Scholar]
- Yengo CM, Sweeney HL. Functional role of loop 2 in myosin V. Biochemistry. 2004;43:2605–2612. doi: 10.1021/bi035510v. [DOI] [PubMed] [Google Scholar]
- Zhu GZ, Fujii K, Liu Y, Codrea V, Herrero J, Shaw S. A single pair of acidic residues in the kinase major groove mediates strong substrate preference for P-2 or P-5 arginine in the AGC, CAMK, and STE kinase families. J. Biol. Chem. 2005;280:36372–36379. doi: 10.1074/jbc.M505031200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.