Abstract
Broadly neutralizing antibodies (BNAbs) such as 2F5 are directed against the HIV-1 GP41 membrane proximal external region (MPER) and recognize well-defined linear core sequences. These epitopes can be engrafted onto protein scaffolds, serving as immunogens with high structural fidelity. Although antibodies that bind to the core gp41 epitope can be elicited, they lack neutralizing activity. To understand this paradox, we used biophysical methods to investigate 2F5 binding to the MPER in a membrane environment where it resides. Recognition is stepwise, through a paratope more extensive than core binding site contacts alone, and dynamically rearranging via an apparent CDRH3 scoop-like movement essential for MPER extraction from the viral membrane. Core epitope recognition on the virus requires induction of conformational changes in both the MPER and paratope. Hence, target neutralization through this lipid-embedded viral segment places stringent requirements on antibody combining-site plasticity.
A protective vaccine against HIV-1 requires the stimulation of a robust immune response to elicit broadly neutralizing antibodies (BNAbs)1. BNAbs are essential to prevent viral pathology, generally by inhibiting entry of the HIV retrovirus into host cells, thereby blocking viral replication as well as proviral integration into the human genome. The latter process establishes latent reservoirs of disease2. The trimeric envelope protein consisting of three protomers of GP120 non-covalently attached to GP41 is the only viral protein exposed on the virion surface. Entry of HIV-1 into human T lymphocytes is mediated first by attachment of its envelope GP120 subunit to the cellular receptor, CD4, followed by binding to the co-receptor (CCR5 or CXCR4)3. These interactions foster structural rearrangement of the membrane-anchored HIV-1 envelope GP41 subunit, subsequently leading to viral fusion with the host cell4,5. Therefore, antibody-mediated protection against HIV-1 must target accessible, functionally relevant and conserved spike epitopes.
Development of effective vaccines capable of eliciting BNAbs against HIV-1 has been extremely challenging. Failure to create an effective vaccine to control the global HIV-1 pandemic is a consequence of extensive mechanisms exploited by the virus to escape protective humoral immunity6–8. Through genetic sequence variability resulting from its error prone reverse transcriptase, HIV-1 has evolved into many subtypes with multiple quasispecies circulating at any time in each individual. In most HIV-1-infected patients, neutralizing antibody activities are strain specific6. However, a minority of chronically infected subjects develops antibodies capable of neutralizing diverse viral strains9. From the latter, monoclonal antibodies that are broadly neutralizing have been isolated and characterized in an effort to define potential targets for HIV-1 immunogen design. Among these antibodies, several (b12, 2G12, PG9, PG16 and VRC01) recognize conserved regions in GP120 while three others (2F5, 4E10 and Z13e1) bind to juxtaposed linear epitopes within the GP41 membrane proximal external region (MPER)10–16.
The MPER is a highly-conserved tryptophan-rich hydrophobic segment (residues 662–683), important for viral fusion17. This region lies at the base of the GP41 ectodomain, immediately proximal to the envelope’s transmembrane segment. Structural analysis of the HxB2 MPER suggests that the 2F5 and 4E10 epitopes are membrane-encrypted within an L-shaped MPER on the membrane surface18. The MPER is configured into two helical parts with different membrane orientations: a tilted N-terminal segment (residues 664–672) and a near-flat C-terminal helix (675–683), connected to one another via a flexible hinge. This bipartite arrangement is well suited to mediate fusion-related conformational change. In line with this observation, biochemical and structural analyses suggest possible interaction of 2F5 and 4E10 with phospholipids10,19–22. While there are no contacts between the tip of the unusually long heavy chain complementarity determining region-3 (CDRH3) and the MPER peptide segment observed in crystal structures, the CDRH3 is essential for neutralization by 2F5 as well as 4E1023–27. Elimination of the tip or key hydrophobic CDRH3 residues abrogates neutralizing activity of such 2F5 and 4E10 variants. As a consequence, it has been proposed that anti-MPER BNAbs associate initially with the viral membrane and subsequently capture the MPER in a two-step process28.
Extensive structural and biochemical information on the 2F5 BNAb and its sequential core epitope have guided design of immunogens to elicit antibodies capable of inhibiting viral infection [29 and references therein]. For example, epitope-engrafted scaffold immunogens configure the 2F5 core epitope (ELDKWA) in a manner precisely mimicking that of the 2F5-bound epitope as verified by X-ray crystallography, yet scaffold-elicited antibodies like 11F10 lack detectable neutralizing activity30. To understand this paradox, we performed nuclear magnetic resonance (NMR), electron paramagnetic resonance (EPR) and hydrogen-deuterium exchange mass spectrometry (HX-MS) studies to assess the manner in which 2F5 binds to the MPER in a membrane environment. Our results show that the 2F5 paratope interacts extensively with the MPER segment and surrounding lipid in addition to the core epitope. Moreover, 2F5 induces substantial conformational changes in the hinge and N-terminal helical segment in a process fostering extraction of lipid-buried core residues and is dependent upon the CDRH3 loop whose lipid interaction per se is negligible in the absence of MPER binding. The rigidification of the CDRH3 by an internal hydrogen bond network and proline cluster in conjunction with hydrophobic residues at its tip allow for the antibody to function as an extraction scoop. In addition to explaining why core 2F5 epitope recognition itself is insufficient to mediate viral neutralization, our findings suggest that antibody can function as an atomic tool mediating structural rearrangement. The findings extend the notion of antibody motion from that required to recognize a single state (i.e. induced fit) to that required to recognize two states (membrane-embedded epitope and extracted epitope). These data offer new ideas to approach MPER-directed BNAb generation.
RESULTS
2F5 binding to membrane-embedded MPER is affected by hydrophobic residues in the CDRH3 tip
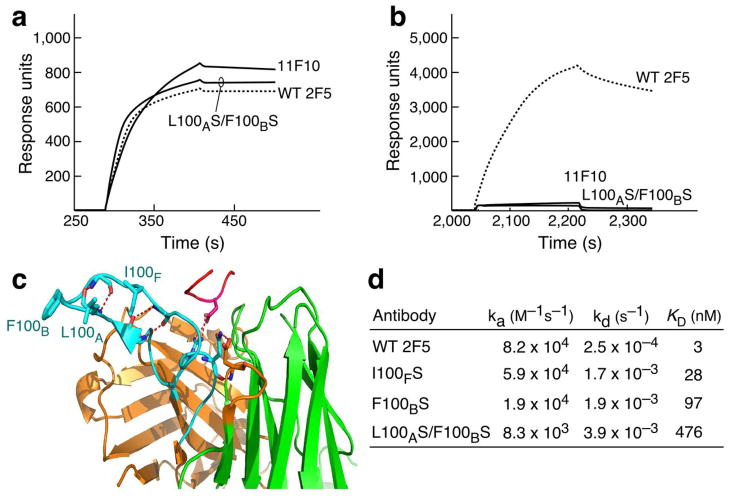
To investigate why an antibody with high affinity for target epitope recognition lacks functional anti-viral activity, the binding mode of the non-neutralizing 11F10 mAb, was compared to that of the 2F5 BNAb. Consistent with structure studies, comparable 11F10 and 2F5 binding measured by BIAcore was observed to an MPER peptide in the absence of lipid (Fig. 1a). Surprisingly, 11F10 was incapable of interacting with the MPER when the peptide was arrayed on liposomes (Fig. 1b). The substantial length of the 2F5 CDRH3 loop (i.e. 22 residues) and its hydrophobic character (Fig. 1c) are thought to be important for mediating membrane interactions and 2F5 BNAb activity. Pointedly, 11F10 lacks these characteristics. Furthermore, the double serine mutations in 2F5 at residue L100A and F100B of CDRH3 (2F5m L100AS F100BS) completely abrogate neutralizing activity, whereas 2F5m I100FS and 2F5m F100BS, reduce potency by approximately 2 orders of magnitude, relative to wild-type (wt) 2F5 mAb in IC50 viral neutralization assays26. While the affinity of 2F5 mutants for the MPER peptide in the absence of lipid is not affected by the CDRH3 tip mutations (Fig. 1a), consistent with Ofek et al.26, the 2F5m L100AS F100BS binding to the MPER segment is substantially reduced when configured on an HIV virion mimic membrane surface (Fig. 1b). As shown in Fig. 1d and representative sensorgrams (see Supplementary Fig. 1), a 2 order of magnitude reduced binding KD (476 nM) was observed for 2F5m L100AS F100BS compared to that of wt 2F5 (3.1 nM). 2F5m I100FS bound to the MPER segment less well than wt 2F5 with a 28 nM KD and 2F5m F100BS manifests a decrease in KD to 97 nM.
Figure 1. 2F5, 2F5m and 11F10 antibody binding to the MPER peptide in solution vs on a lipid membrane.
(a) Antibody binding to the biotin-MPER peptide attached to streptavidin on the CM5 surface chip by BIAcore. (b) Antibody binding to HIV virion mimic liposome-MPER complex attached to L1 chip surface. The peptide sequence biotin-GGG-QQEKNEQELLELDKWASLWN was used for the binding analysis in solution and with QQEKNEQELLELDKWASLWNWFNI for the binding analysis in membrane environment. (c) Location of mutation residues in the CDRH3 loop (light blue) of 2F5 in the complex with MPER peptide (pink) taken from PDB ITJI19. Note that in long CDRH3, residues between 100 and 101 are numbered with appended letters (i.e. 100, 100A, 100B, etc). (d) BIAcore binding constants of 2F5 and 2F5m variants to the MPER peptide segments on the surface of DOPC-DOPG membrane linked to an L1 chip. Given that no inverse correlation between dissociation rate and various contact time21 ranging from 30 sec to 15 min was shown under our experimental conditions (see Supplementary Fig. 1b, c), binding data are fit to a 1:1 Langmuir model.
Interaction of 2F5 with MPER in membrane protects CDRH3 and L2 from deuterium exchange
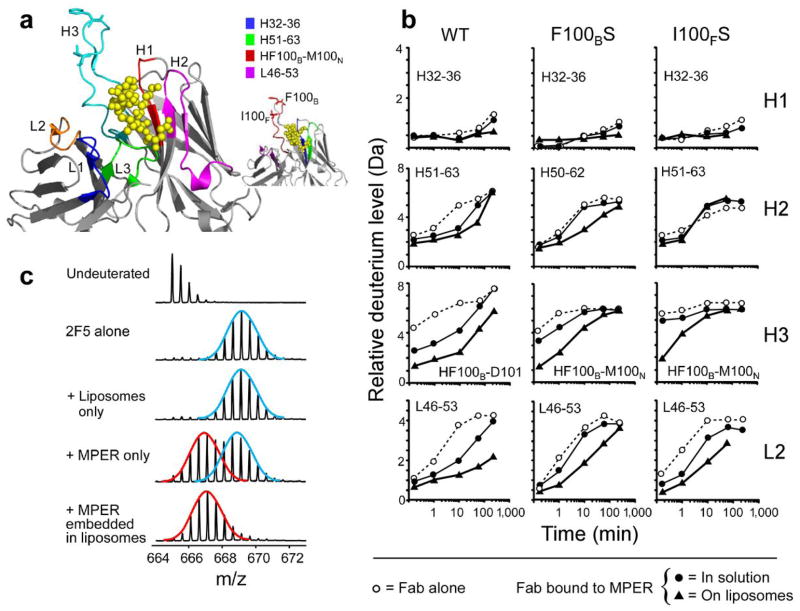
Despite absence of contact with the MPER peptide in a crystal structure (Fig. 1c), the apex of CDRH3 may play an indirect but key functional role in mediating 2F5 binding to its core epitope. To examine this possibility, we employed HX-MS, to assess conformational changes31. After solvent deuterons exchanged for various time periods (15 seconds to 4h) with backbone amide hydrogens at rates dependent on hydrogen bonding and solvent accessibility, the quenched protein was digested with pepsin. Deuterium incorporation into each pepsin fragment was analyzed by electrospray mass spectrometry32 (over 40 in total, see Supplementary Fig. 2). Key peptides and their relation to each of the CDR regions are shown in Fig. 2a. Exchange into the 2F5 Fab alone provided baseline data for the protein in solution to be compared with the antibody fragment complexed to MPER in the presence and absence of liposomes. The same experiments for wt 2F5 Fab were repeated for 2F5m F100BS and 2F5m I100FS Fabs and data compared. As a control, 2F5 Fab exchange was also measured in liposomes lacking the MPER peptide.
Figure 2. Hydrogen exchange mass spectrometry of 2F5 and 2F5m variants.
(a) Location of CDR loops, displayed on the crystal structure of 2F5 (PDB 1TJG)19. CDRH1 (red), CDRH2 (pink), CDRH3 (cyan), CDR L1 (blue), CDRL2 (orange), CDRL3 (green). The atoms of MPER peptide in the crystal structure are shown as yellow balls. The F100B and I100F amino acids in CDRH3 are shown. The inset shows the same structure with peptides used in deuterium exchange curves shown in panel b. (b) Representative deuterium exchange curves for important regions of 2F5, F100BS and I100FS for protein alone (open circles), protein bound to MPER in solution (solid circles) and protein bound to MPER embedded in liposomes (solid triangles). The residues of each peptide are indicated and their position described in the inset of Panel A. The s.d. of each data point is, at most, ± 0.25 Da. (c) Mass spectra of peptide 100BFGVPIARGPVNAM100N from the CDRH3 after 10 minutes in deuterated buffer. Idealized distributions in the bimodal pattern are indicated by the red (lower mass, more protected) and blue (upper mass, less protected) lines.
For the majority of peptides derived from wt and 2F5m Fabs, there was very little change in exchange upon binding to MPER-embedded in liposomes (Fig. 2b, top row; Supplementary Fig. 3–5). Likewise, exchange into the several CDR regions that contact the MPER in the crystal structure (H1, H2) was not affected differentially by the MPER in solution versus lipid-embedded. Although CDRL3 also interacts with the MPER, no peptides covering CDRL3 could be located in the HX-MS experiments (see Supplementary Fig. 2). However, there were reproducible differences in wt 2F5 Fab HX in the CDRH3 and the CDRL2 (Fig. 2b, left column). The observed differences were well outside the error of deuterium level determination (±0.25 Da). In these regions, the amount of deuterium incorporated into 2F5 was lowest in the presence of MPER embedded in liposomes, indicating that 2F5 is protected from exchange therein as a result of binding to MPER in the liposome.
Hydrogen exchange into the mutant 2F5m F100BS and 2F5m I100FS Fabs was compared to that of wt 2F5 Fab (Fig 2b). Exchange into each of the mutant Fabs was not dramatically altered in the absence of MPER, but obvious differences were found between wt and mutant Fabs when bound to MPER with and without lipid. For example, protection from exchange in CDRH3 and CDRL2 as a result of binding MPER in solution was reduced in both mutants relative to wt. The shape of the deuterium incorporation lines was also different between wt and mutants in that MPER-bound 2F5m became deuterated more quickly than wt 2F5. Based on the raw mass spectra, these changes were the result of redistribution of the population of multiple conformations.
HX-MS defines a single 2F5 CDRH3 conformation in the presence of lipid-embedded MPER
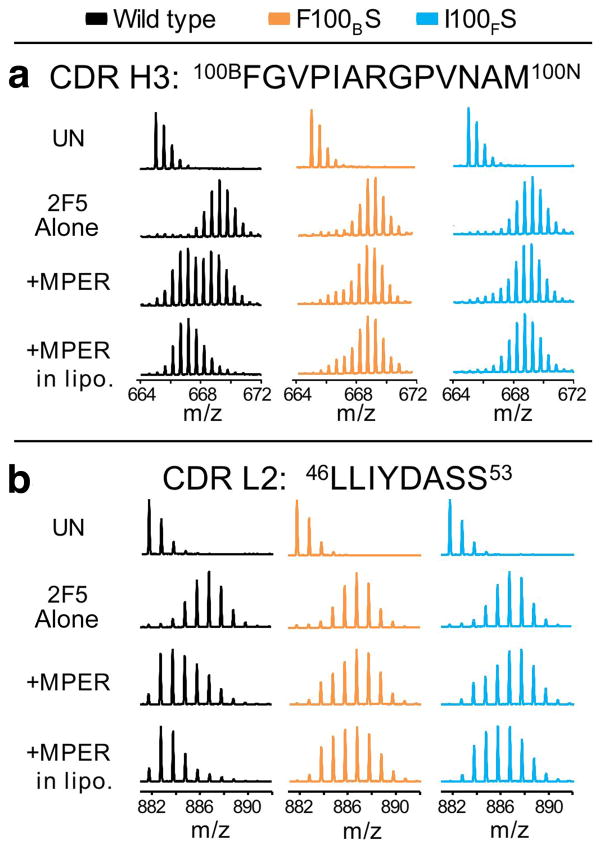
In addition to protection from exchange, when 2F5 bound to the MPER in solution, the mass spectra indicated that the antibody adopted multiple conformers, in contrast to a single conformational population found for unligated 2F5. The regions where multiple conformational forms exist cover the C-terminal half (residues F100B-D101) of the CDRH3 and the N-terminal portion (residues 46–49) of the CDRL2 identified by the unique isotope distributions characteristic of multiple conformation in these peptides. An example of the spectra is shown in Fig. 2c for the CDRH3 peptide (see also Fig. 3 for spectra of L2). Multiple populations distinguish themselves in these experiments by the appearance of bimodal isotope patterns which may be due to EX1 kinetics33,34. One population represents a more exposed form of the protein, hence incorporating more deuterium to yield a higher mass (Fig. 2c blue line). In contrast, the other population is more protected with less deuterium incorporated and therefore has a lower mass (Fig. 2c, red line). These populations interconvert due to protein “breathing” or dynamics but because the deuterium labeling reaction is unidirectional (~95% (v/v) D2O solution), all molecular species eventually reach the higher mass, i.e. more deuterated form, with the rate of conversion between the two forms correlating with the change in the population distribution. A less protected form was found for the CDRH3 of unbound 2F5 (2F5 alone) while multiple populations were found for the soluble MPER-bound form (+ MPER only). This suggests that MPER binding slowed the 2F5 protein dynamics, requiring more time to reach the upper distribution. The 10 minute time point is shown in Figs. 2–3 for purposes of illustration, but the distributions were characterized for all time points. Interestingly, in the presence of lipid (+MPER embedded in liposomes), there was only one protected form as evidenced by the single isotope distribution at lower mass (Fig. 2c) and the decreased amount of exchange found in the incorporation graphs (Fig. 2b, triangle symbols in wt Fab column). MPER embedded in liposomes, therefore, had the greatest ability to prevent protein breathing and dynamics in wt 2F5 Fab, presumably due to the influence of the lipids in altering the motions in the Fab, particularly the CDRH3, and to a lesser extent the CDRL2.
Figure 3. Mass spectra showing the loss of protection from deuterium exchange in 2F5m F100BS and I100FS for peptides from (a) the CDRH3 and (b) the CDRL2 after 10 min in deuterated buffer.
For each protein, four conditions are shown: undeuterated form, UN; 2F5 alone; 2F5 in the presence of MPER in solution (+MPER); and 2F5 in the presence of liposomes containing embedded MPER (+MPER in lipo). The data shown in panel a for wt 2F5 are the same as the data shown in Fig. 2c. The bimodal, multiple population phenomena can also be observed at the other exchange timepoints (Fig 2b).
In contrast to the wt 2F5 Fab, in the CDRH3 and CDRL2 regions of the 2F5m F100BS and 2F5m I100FS Fabs there was no bimodal pattern after 10 min of labeling. Instead, an unprotected population resembling the unbound protein was observed (Fig. 3). There was a bimodal pattern earlier in the time course of exchange but the conversion rate between the protected and unprotected forms was much more rapid for the mutants than for the wild-type, which is also shown by the rapid increase in the relative deuterium levels in the kinetic analysis (Fig. 2b). Maximal deuteration was reached much more quickly than the wt 2F5 as seen by comparing the line shapes of 2F5 bound to MPER in liposomes (triangles) in Fig. 2b (H3 and L2). By the 10 min time point shown in Fig. 2, both mutants were heavily deuterated without protection as in wt 2F5. Thus, mutation at the tip of the CDRH3 loop accelerates deuteration and dynamic conversion between conformations, consistent with a destabilized structure and/or loss of strong interaction mediating solvent occlusion. While wt 2F5 Fab was stabilized by the MPER-lipid environment, this was not the case for the mutants.
It was perhaps surprising to observe changes in CDRL2 when wt 2F5 Fab bound to MPER in liposomes, considering that CDRL2 is at a distance from the MPER core epitope in the crystal structure. Nevertheless, the HX-MS data clearly indicate that CDRL2 is affected by interactions between 2F5 and MPER in liposomes, primarily only in wt 2F5 (Fig. 3b). Two overlapping peptides in the HX-MS experiments (light chain residues 46–53 and 48–54) allowed us to conclude that the region of CDRL2 most involved in this effect is between residues 46–49 because the raw spectra of the shorter peptide (48–54) did not have the bimodal kinetic signature observed in CDRL2 residues 46–53 and CDRH3 100B-100N (as in Fig. 3). The hydrophobic segment (46LLIY49) is not surfaced exposed and hence, not anticipated to interact with membrane or contribute energetically to peptide binding, in contrast to CDRH326. In the crystal structure, residues 46–50 of CDRL2 are directly across from the C-terminal half of the CDRH3, the same region with altered HX upon binding MPER in liposomes (Fig. 2a). One H-bond is observed between CDRL2 D50 and CDRH3 N100L. Therefore, conformational change in CDRH3 may strengthen association of H3 with L2 via additional H-bonds or other contacts upon productive interaction of wt 2F5 with MPER in liposomes, affording protection from deuterium exchange. This interaction is lost in both 2F5m single mutants, F100BS and I100FS.
The 2F5 Fab interaction with MPER in a lipid environment fosters structural change
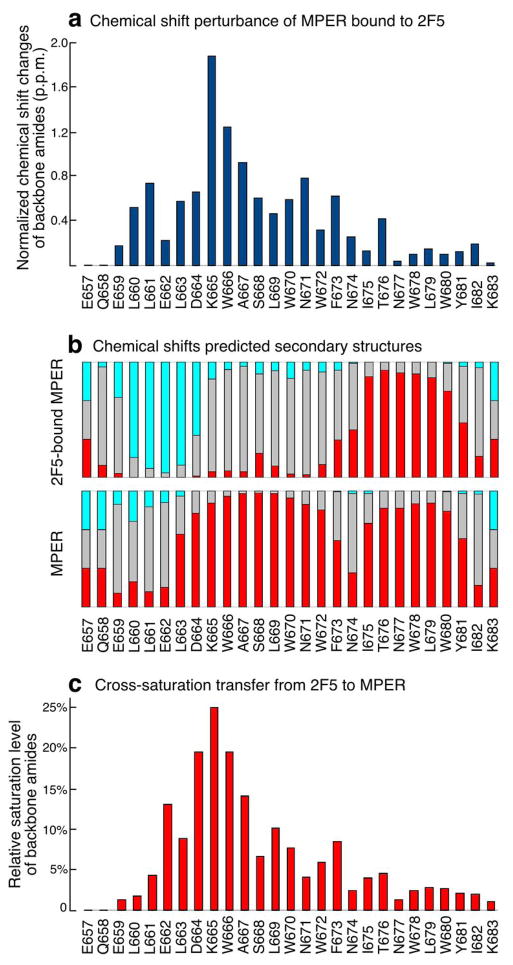
We carried out structural investigation of wt 2F5 interaction with the MPER in DPC micelles using NMR spectroscopy. The fingerprint 2D 15N-TROSY (transverse-relaxation optimized spectroscopy)-HSQC (heteronuclear single quantum coherence) spectrum of 15N-13C-2D triply labeled MPER peptide in the presence of unlabeled 2F5 Fab fragments show large chemical shift changes of the backbone amide peaks from a number of MPER residues (Fig. 4a and Supplementary Fig. 6). MPER residues ranging from L660 to F673 are affected by 2F5 binding, with the most dramatic changes occurring for residues K665, W666, and A667 (numbering according to HxB2). This set of findings indicates that MPER conformational changes subsequent to 2F5 binding extend well beyond the core epitope to include the central hinge region of MPER and possibly T676. On the other hand, residues C-terminal to T676 are less perturbed, suggesting that the conformation of the C-terminal helix of MPER remains largely unchanged35. Secondary structures predicted from the 13C chemical shift values of MPER bound to 2F5 imply dramatic conformational changes to the N-terminal region (Fig. 4b). The N-terminal residues LLELD adopt an extended conformation in contact with the antibody, and the helical conformation of the MPER N-terminal helix including the core epitope plus that of residues C-terminal to the 2F5 binding pocket has been disrupted.
Figure 4. NMR characterization of the HxB2 MPER segment upon 2F5 Fab ligation.
(a) MPER backbone amide chemical shift changes upon binding to 2F5 Fab. The chemical shift values of both amide 15N and 1H are normalized as ((DH2 + (DN/5)2)1/2. (b) Secondary structures predicted from MPER chemical shifts by TALOS+. The red and light blue bars indicate probabilities of helical and beta-strand conformations respectively whereas grey bars represent unstructured regions. (c) Signal reduction observed in 2D-labeled MPER backbone amide peaks as transferred from 1H-saturated 2F5 Fab molecule to the MPER.
To map the 2F5-bound MPER residues that are in close contact with the Fab, we acquired cross-saturation transfer 15N-TROSY-HSQC spectra36 using the same sample. The aliphatic proton spectral region from the 2F5 Fab was irradiated and the saturation signals transferred to the amide peaks of the MPER through inter-molecular nuclear Overhauser effect (NOE) when residues of the MPER are close in space to the 2F5 surface. In this way, MPER residues nearby the 2F5 Fab are identified. Fig. 4c shows the relative amount of signal reduction as a result of cross saturation transferred from the 2F5 to the Fab-bound MPER residues. The most affected residues are in the core epitope region, but close contacts between the antibody and MPER extend C-terminal to include the MPER central hinge region. The region of intermolecular contact appears to be between E662 to F673, similar in scope with the chemical shift perturbance data. The overall saturation levels are comparable to 4E10-bound MPER, but 2–3 times weaker than Z13e1-bound MPER35 suggesting that 2F5 binding is similar to 4E10, and involves a small but deep epitope binding pocket with a larger paratope contact area. In contrast, Z13e1 ligation is a very rigid type of binding involving many MPER residues. In addition, cross saturation data show that the side-chain amine groups of W670 and W672 are both in close contact with 2F5 (Supplementary Fig. 6b). While W666 in the ELDKWA epitope is expected to make closer contacts with 2F5, its peak is not observed consistent with a rapid relaxation rate in the binding pocket. Overall, our data suggest that there are multiple contacts between 2F5 and MPER outside the ELDKWA core itself, including the region immediately flanking this core and up to the central hinge region (F673-N674).
Differential impact of MPER alanine scanning mutagenesis on binding of wt 2F5 versus 2F5m
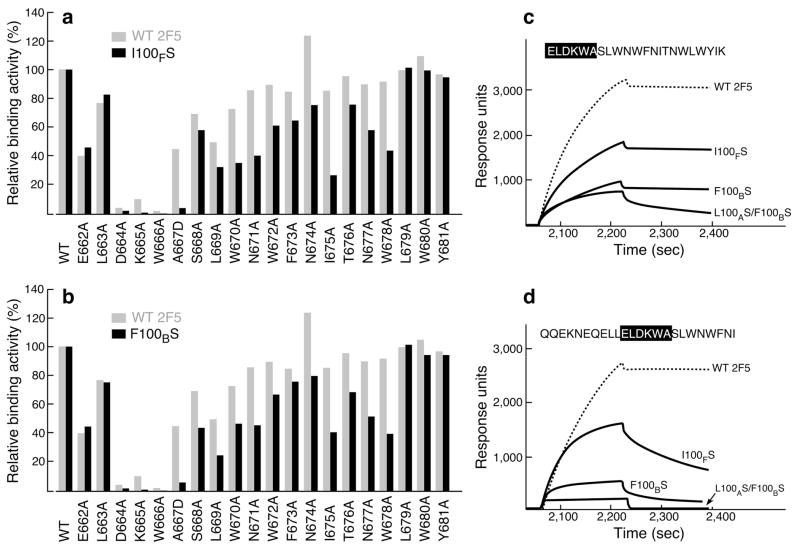
To determine whether broader contacts with MPER residues C-terminal to the 2F5 core epitope were mediated by CDRH3 apex residues, binding of wt 2F5, 2F5m I100FS and 2F5m F100BS was compared through mutational scanning analysis. Single residue alanine substitutions of the MPER showed that the DKW sequence is essential for 2F5 binding to the MPER on the DOPC–DOPG liposome (Fig. 5a), consistent with previous results carried out in solution19,37,38. In contrast to the wt 2F5, the binding of 2F5m I100FS and F100BS was affected by residues in addition to those core DKW residues, extending C-terminal as far as W678 (Fig. 5a and b). An A667D mutation significantly diminished I100FS and F100BS binding (~4–6 % wild type) as did alanine replacement of membrane-embedded L669, W670, W672, F673, I675 and W680 as well as surface exposed residues N671, N674 and N677. Independent peptide analyses using a full-length MPER segment as well as C-terminal synthetic truncation demonstrate that while wt 2F5 binding to the MPER is unaffected by residues C-terminal to the MPER N-helix, truncation of the MPER C-helix diminishes 2F5m I100FS and 2F5m F100BS binding as a result of a faster dissociation rate (Fig. 5c and d). The binding of the 2F5m L100AS F100BS to the C-helix-truncated MPER (Fig. 5d) was abrogated even at the concentration of 100 μg ml−1 tested although binding to full length MPER was detectable. In sum, the results suggest that the membrane interaction of CDRH3 tip may foster transient contact of 2F5 CDRH3 with C-helix residues during initial or intermediate binding states.
Figure 5. Influence of C-terminal MPER residues on 2F5 and 2F5m binding as measured by SPR.
Each bar represents the apparent I100FS (a) and F100BS (b) binding affinities in comparison with those of 2F5 for a series of alanine-substituted MPER peptides versus wt MPER. For A667, a D substitution was introduced. The amount of each MPER variant peptide bound to DOPC-DOPG liposome was normalized to that of wt peptide. 30 μl of wt 2F5 at 20 μg ml−1, 2F5m I100FS at 35 μg ml−1 and 2F5m F100BS at 50 μg ml−1 were injected over peptide–liposome complex at the flow rate of 10 μl min−1. Apparent binding affinities of each antibody for MPER alanine mutants in comparison with the wt peptide were measured by response units taken on the dissociation time point at 3 min following 3 min of association time by BIAcore. (c,d) Binding of 2F5 and mutants to MPER peptides containing or lacking the C-helical MPER region. 20 μg ml−1 of each antibody was injected over peptide-liposome complex except in the case of double mutant L100AS F100BS where 100 μg ml−1 was used.
The apex of the 2F5 CDRH3 loop mediates MPER reorientation and epitope extraction
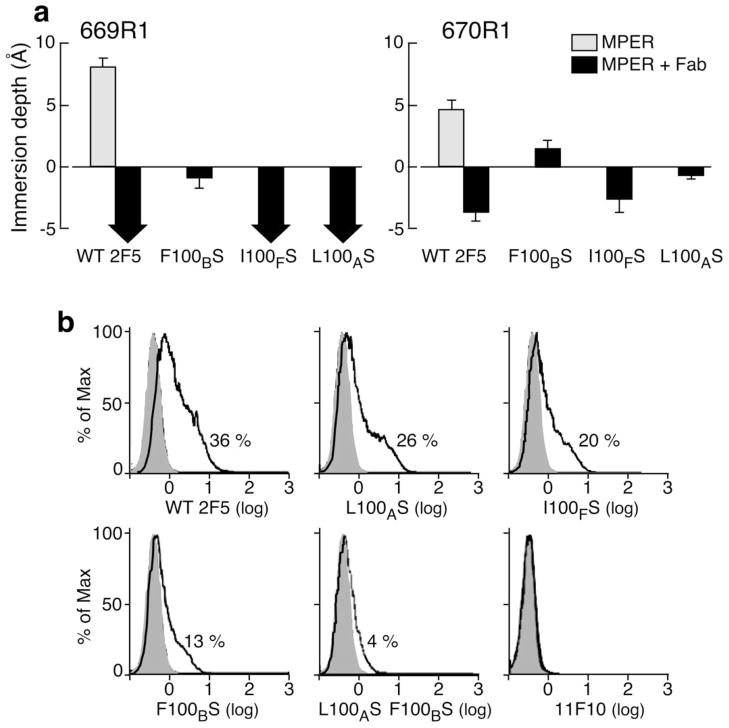
To test the effect of mutations at the tip of CDRH3 loop on MPER reorientation and any correlation with neutralization potency, membrane immersion depths of spin-labeled reference residues L669R1 and W670R135 were measured by EPR for 2F5m L100AS, 2F5m F100BS and 2F5m I100FS. As shown in Fig. 6a, we determined the immersion depth values of spin-labeled L669R1 and W670R1 in the absence and presence of 2F5 Fab. wt 2F5 Fab lifts deeply buried residue L669R1 from the acyl chain region of lipid (depth > 8 Å) out of the membrane surface into the aqueous phase (depth < −5 Å) whereas W670R1 is moved from the lipid acyl chain region into the head group region. In contrast to wt 2F5 Fab, 2F5m F100BS-induced immersion depth changes of L669R1 and W670R1 were attenuated at the head group and acyl chain region, respectively (Fig. 6a). The extraction of L669R1 appears to be unaffected by 2F5m L100AS and 2F5m I100FS, but slightly reduced immersion depth changes were observed with W670R1 upon 2F5m L100AS and 2F5m I100FS binding compared to that of wt 2F5. The results suggest that mutations at different positions of the CDRH3 tip differentially affect the degree of reorientation of the N-helix in the MPER. Of note, comparable EPR mobility spectra changes found for L669R1 and W670R1 (see Supplementary Fig. 7) indicate the presence of similar MPER conformations at the antibody binding interface for the wt and three 2F5m Fabs.
Figure 6. MPER membrane immersion depth changes following 2F5m ligation and their relationship on to trimeric envelope protein binding.
(a) Wild type and mutant 2F5 Fabs-induced membrane immersion depth changes of MPER R1 residues by EPR. Depth values of unligated MPER residues are indicated with gray columns. The values of MPER peptides in complex with wt 2F5 and 2F5m Fabs are shown as black columns. Depth values between −5 Å to 0 Å and larger than 0 Å correspond to lipid headgroup region and acyl chain region, respectively. The precise values of residues exposed to aqueous phase (depth <−5 Å) cannot be determined experimentally and are thus indicated by black arrows. (b) wt 2F5 and 2F5m binding to HIV-1 ADA envelope trimer expressed on 293 T cells. Cells transfected with ADA GP160 or empty vector were incubated with the designated antibody (5 μg ml−1) for 2h at RT followed by phycoerythrin-conjugated goat anti-human secondary antibody. Histogram indicates relative % cell staining by each antibody, respectively, after subtraction of background staining against mock transfected cells. Note the absence of cell staining by 11F10 even at the concentration of 200 μg ml−1.
To test further whether the attenuated extraction of residues in the N-helix of MPER by the 2F5 variants affects antibody interaction with intact GP41 on native membranes, mAb binding to trimeric ADA strain HIV-1 envelope protein GP160 transiently expressed on 293 T cells was analyzed by FACS. As shown in Fig. 6b, 36% of cells stained with wt 2F5, whereas 2F5 mutants binding was reduced: L100AS (26%), I100FS (20%), F100BS (12%) and L100AS F100BS (3.9%). No detectable binding of 11F10 to GP41 was observed even at the concentration of 0.2 mg ml−1, consistent with the result shown in Biacore analysis (Fig. 1b). These results suggest that the reorientation of membrane-embedded residues at the interface of the 2F5 binding pocket mediated by CDRH3 is critical both for binding affinity and the neutralization potency previously shown in Ofek et al.26.
A model of 2F5 Fab binding to the MPER
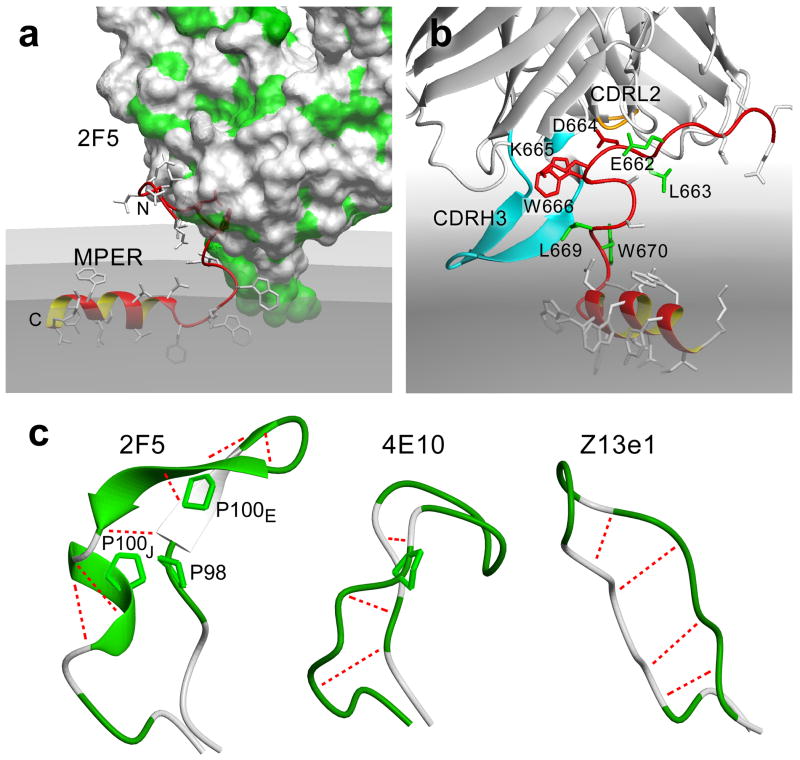
Based on these NMR, EPR, HX-MS and functional results, we have modeled the full-length MPER in complex with the wt 2F5 Fab on a liposome surface. Details of the modeling are provided in Methods. As shown in Fig. 7a, the N-terminal segment of the MPER has been lifted vertically out from the membrane while contacts between the hydrophobic CDRH3 loop and MPER juxta-hinge residues likely contribute to complex stabilization. We propose that the CDRH3 loop acts like a scoop to extract the 2F5 core epitope residues into its binding pocket. As viewed from a 90° rotation of the MPER-2F5 Fab complex model shown in Fig. 7b, extraction causes D664 K665 W666 to fit into the deep 2F5 binding pocket, while E662 L663 and L669 W670 line the entrance and exit, respectively. The hydrophobic tip of the 2F5 CDRH3 loop is positioned in the membrane to be compatible with the HX-MS data. Notably, the 2F5 CDRH3 loop is rigidified by a cross-strand hydrogen bond network and the presence of three proline residues to facilitate extraction. Fig. 7c offers a comparison of crystallographically defined CDRH3 loops of BNAbs 2F5, 4E10 and Z13e1 mAbs. The proline-containing scoop-like structures of CDRH3 loops of the extracting mAbs, 2F5 and 4E10, contrast to the flat, paddle-like CDRH3 structure of the non-extracting Z13e1 mAb35.
Figure 7. MPER in complex with 2F5 Fab on viral membrane surface.
(a) Docking model showing 2F5 lifting up the N-terminal segment of the MPER. The MPER backbone ribbon is colored in red. The 2F5 Fab is represented by Van der Waals surface with hydrophobic patches colored in green. The membrane is colored grey with the solvent-lipid headgroup interface, phosphate-containing plane and lipid headgroup-aliphatic-chain interface in progressively darker shades. (b) Alternative view of the MPER-2F5 docking model showing the scoop-like action of 2F5 CDRH3 (light blue). Key 2F5 epitope residues D664 K665 W666 are shown in red, residues critical for initial binding (E662 L663) and residues used for EPR immersion depth probes (L669 W670) are shown in green. 2F5 CDRL2 loop closely coupled to CDRH3 loop is colored in orange towards the back of the model. (c) Comparison of X-ray crystallographic structures of CDRH3 loops of 2F5 (PDB:1TJI)19, 4E10 (PDB:1TZG)19 and Z13e1 (PDB:3FNO)54 neutralizing antibodies. The hydrophobic residues are colored in green, (including proline side-chains in stick representations), and the hydrogen bonds are shown as dotted blue lines.
DISCUSSION
A multi-step process of 2F5 binding to lipid-embedded MPER
2F5 binding to the MPER proceeds through several distinct steps. Given the weak interaction of 2F5 with lipid membranes yielding undetectable alterations by the HX-MS experiments (see Supplementary Fig. 5), we hypothesize that 2F5 initially interacts with the solvent exposed MPER residues E662, L663 and possibly D664, lifting up MPER residues N-terminal to D664. As a result, the 2F5 CDRH3 loop comes into contact with the MPER at the peptide–lipid interface on the W672 side. The hydrophobic residues in the CDRH3 loop then inserts deeper into the membrane to stabilize a beta-sheet hairpin structure, and together with the proline containing base of the loop, form a rigidified scoop. We suggest that this scoop lifts up the underside of the N-terminal helical segment of the MPER. That action involves a concerted movement in which the CDRH3 loop sweeps across the N-terminal front of the MPER, contacting D664, K665 and W666, and extracts the latter two residues from the membrane into the 2F5 binding pocket, resulting in a tight complex. The tip of the CDRH3 loop with F100B anchored into the membrane is crucial to stabilize the 2F5-MPER complex in this process. The single population of 2F5 CDRH3 and CDRL2 peptides protected from deuterium exchange in the presence of MPER-embedded in liposome is likely an HX-MS correlate of this process. 2F5m F100BS, and to a lesser extent other 2F5m L100AS and I100FS, are less effective in excavating the buried core epitope residues, thereby resulting in partial extraction with prolonged extraction kinetics as evidenced by slower on-rate constants (Fig.1d). Those 2F5 mutations prevent the CDRH3 regions from interacting strongly with the MPER in lipid resulting in the much more rapid appearance of highly deuterated species in HX-MS (Figs. 2–3). This incomplete extraction may explain the observation that 2F5m 100FS and F100BS are sensitive to MPER alanine mutations in the central hinge and the first turn of the C-terminal helix (Fig. 5). Although there may be no contacts with these MPER residues in the final 2F5-MPER complex, during the intermediate stage of extraction, structural changes linked to the MPER alanine mutants and/or loss of direct side chain contacts may interfere with the extraction process mediated by these 2F5 CDRH3 mutants.
Functional consequences of 2F5m cannot be explained by loss of “non-specific” hydrophobic membrane interactions as a singular prerequisite for subsequent MPER binding suggested previously28. Instead, 2F5 extraction of epitope residues on the MPER segment requires, in our view, three key steps: 1) an initial MPER capturing paratope adjacent to the core binding pocket (i.e. imparting specificity); 2) a semi-rigid kinematic CDRH3 loop with flexible joints that can interact simultaneously with residues C-terminal to the binding pocket and lipid membrane necessary to extract core epitopes shielded by head group or acyl chains (i.e. offering hydrophobic interaction energetics for lifting the MPER); and 3) a tight paratope binding pocket for stable complex formation with the fully exposed core epitope (i.e. yielding a high affinity interaction).
A potential proline-based molecular switching mechanism for the 2F5 CDRH3 extraction scoop
From a survey of 516 antibody structures in the PDB database, only about 5% possess rigid and long (≥14 residues) CDRH3 loops (26 of 516); 2% among non-HIV mAbs (10 of 459) compared with 28% of HIV-related mAbs (16 of 57). The CDRH3 loop of 2F5 is remarkable in its length (22 residues) and function noted above. The energy gained from favorable interactions of the CDRH3 tip with the membrane and MPER N-terminal segment can be efficiently transmitted through the rigid segments of CDRH3 loop to facilitate extraction. Notably, the 2F5 CDRH3 loop contains three proline residues, a distinctive feature among known natural antibody structures in the PDB aside from the tyrosine sulfated 412D antibody directed against HIV-1 GP12039. In the case of 2F5, glycine residues N-terminal to two of the three prolines (P98 and P100J) may afford segmental flexibility in structure around these maneuverable “joints”. We envision that the cis-trans isomerization of these proline residues can act like a molecular switch40,41 to mediate dynamic robotic movement of the largely rigid CDRH3 loop for extraction (Fig. 7b and 7c), locking it into a trans-proline state. This notion is consistent with the conformational change of the 2F5 CDRH3 loop observed by HX-MS upon 2F5 binding to membrane-bound MPER. Moreover, that transition may place the CDRH3 tip hydrophobic residues on one surface for membrane interaction in an energetically favorable manner26 that can provide the energy source and trigger for this switching mechanism.
Implication for immunogen design
CDRH3 is the major determinant of antibody diversity and a key contributor to antigen affinity and specificity42. The selection bias of 2F5 progenitor B cells for long CDRH3 loop may be driven by the context in which the MPER antigen is presented, being largely sterically occluded at the base of the glycosylated GP160 ectodomain in proximity to viral membrane43. A correlation between CDRH3 length and the nature of the antigen being recognized offers a precedent for such selection44,45. Although some amino acids in the putative 2F5 germline antibody participated in certain core epitope residue interaction, the germline-like antibody most closely corresponding to 2F5 lacks soluble GP140 binding46,47. Somatic hypermutation may have occurred during prolonged antigen exposure, facilitating generation of a higher affinity binder capable of neutralizing HIV-148. Crystal structures of other germline and affinity-matured antibodies have revealed structural plasticity of CDRH3 with an evolution of antibody affinity and specificity49. Hence, the high affinity antigen binding site of 2F5 may have evolved into an optimized binding site through conformational adaptation of the CDRH3 loop in proximity to the membrane. Likely, one or more hydrophobic residue in the CDRH3 including proline coevolved for the extraction process and, as a result, enhanced antibody binding affinity by extending the 2F5 progenitor’s binding site contacts to buried residues. The weaker affinity of 2F5 CDRH3 tip variants and importance of additional MPER C-terminal residue contacts for their binding may be reminiscent of the evolving 2F5 progenitor during maturation.
A 2F5-like antibody with high affinity is achieved through conformational adaptation of the long CDRH3 loop in a membrane environment via extensive contact with MPER residues plus membrane constituents. In turn, the MPER geometry on the virus along with antibody approach angle relative to the viral surface must be considered in immunogen design. Given the rarity of naturally elicited anti-MPER BNAbs in infected patients50, focusing the immune response on the MPER requires epitope-specific immunogens to avoid the immunodominance of other regions in gp41 and gp120. That said, display of the buried core epitope alone in an immunogen scaffold cannot generate a BNAb which must first recognize the membrane-exposed MPER elements prior to core extraction. Antigen-driven B cell selection of neutralizing antibody may thus require a successive, complementary immunization strategy.
Antibodies as tools to modify membrane-embedded structures
The multi-step process beginning with initial antibody encounter with surface-embedded MPER, extraction and final ligation resulting in structural rearrangement of the target is not unique to 2F5. How 4E10 CDRH3 function differs from that of 2F5 will be of interest to determine. The distinction in size, shape and number of prolines in the 2F5 vs. 4E10 CDRH3 scoops implies that significant differences will be found in the extraction process of these BNAbs and immunogens needed for their elicitation. Moreover, our findings have implications for exploiting antibodies to regulate key cellular receptor functions through induction of membrane-dependent reorganizations in macromolecular structure. Examples of the latter might include T cell receptors whose mechanosensor function is dependent on physical force transmitted upon ectodomain ligation51, growth receptors such as the epidermal growth factor receptor (EGFR) family members dependent upon asymmetrical dimerization of kinase domain52, and plexins, receptors for semaphorins inducing dimerization to control axonal guidance in the nervous system and cellular migration more generally53. Development of antibodies that can either interdict or mimic ligand-dependent events through membrane-directed structural rearrangements could create a new class of molecular robotic tools to regulate cellular processes. The HIV-1 GP41 example shows that membrane-active antibodies have potent biologic effects capable of being studied in explicit molecular terms.
METHODS
Deuterium labeling and mass analysis
To prepare for labeling, all protein solutions were brought to a concentration of 15 μM. For experiments with antibodies alone, 2 μL of 150 μM protein (2F5, 2F5 F100BS or 2F5 I100FS) was diluted with 18 μL of 20 mM Tris, 150 mM NaCl, pH 7.4. For experiments with antibodies bound to MPER, 2 μL of 150 μM protein was diluted with 18 μL of 130 μM MPER in 11% DMSO (v/v). For experiments with antibodies bound to both MPER and liposomes, 2 μL of 150 μM protein was diluted with 18 μL of 23 mM DOPC-DOPG liposomes containing 130 μM MPER in 150 mM KCl, 10 mM Hepes pH 7.4. To initiate deuterium labeling, 2 μL of each 15 μM protein solution was diluted 25-fold with 48 μL labeling buffer (20 mM Tris, 150 mM NaCl, 99% D2O, pH 7.4) at room temperature. At selected time points ranging from 15 seconds to 4 hours, the labeling reaction was quenched by adding 50 μL ice-cold quench buffer (150 mM KH2PO4, 150 mM K2HPO4, 4M Guanidine HCl, 1M TCEP, pH 2.5, 100% H2O) and frozen immediately on dry ice an conserved overnight at −80°C.
Mass analysis was performed as described 32. Totally frozen quenched samples (100 μL) were rapidly thawed on ice and injected in custom-designed nanoACQUITY UPLC system (Waters, Milford, MA) for hydrogen-deuterium exchange55. In this system, online pepsin digestion was performed at 15.0°C with a 2.1mm × 50mm stainless column packed with pepsin immobilized on POROS-20AL beads (PerSeptive Biosystems), as described in 56. A peptide trap (Waters VanGuard pre-column ACQUITY UPLC BEH C18 1.7 μm beads, 2.1 × 5 mm) was used to trap the digested peptides. A second, identical trap was placed in front of the analytical column (Waters ACQUITY UPLC BEH C18 1.7 μm beads, 1.0 × 100 mm) as a second guard to block lipids from entering the analytical column and mass spectrometer. A 5% to 40% gradient of acetonitrile over 9 minutes at a flow rate of 40 μL min−1 was used to separate peptides. Both chromatographic mobile phases contained 0.05% (v/v) formic acid giving a pH of 2.6. Mass analysis was performed with a Waters SYNAPT G1 mass spectrometer equipped with standard ESI source and lockmass correction. Glu-Fibrinopeptide was used to maintain the calibration at 3–5 ppm throughout analysis. The mass spectrometer settings were: ESI+ mode; capillary 3500 V; cone 45 V; desolvation and source temperatures were 175°C and 80°C, respectively; nitrogen desolvation gas flow 500 L hr−1; mass acquisition range 50–1700 m/z; scan rate 0.23 scans S−1; instrument always collecting data in MSE mode. Peptic peptides were identified using MSE and IdentityE software within Waters ProteinLynx Global Server 2.4 (PLGS). The deuterium exchange levels were determined by subtracting the centroid mass of undeuterated peptide from the centroid mass of deuterated peptides using the program HX-Express57. Throughout the manuscript, data that are plotted on the same graph were obtained using identical experimental conditions, thus negating the need for back-exchange correction for comparison purposes 31. Caution must be used comparing data not on the same graph because in order to compare such data on an absolute deuterium basis, a back-exchange correction which takes into account experimental variation from day-to-day must be applied. As we wished to determine the relative difference between protein states, no such back-exchange correction was made. At least two total replicate analyses, starting from protein production and labeling, were obtained for each experiment. The error of each data point in deuterium uptake graphs was no greater than 0.25 Da, as determined by replicate analysis, analysis of peptide-protein standards and prior work with this experimental system 58.
Nuclear magnetic resonance
A 15N-13C-2D-triply labeled MPER 27mer (EQELLELDKWASLWNWFNITNWLWYIK) was expressed and purified as previously described 18. 2F5 mAb was digested into Fab fragments before being mixed with the triply labeled MPER. The sample is in 10 mM NaPO4, pH 6.8 and contains 120 mM d38-DPC detergent and 10% D2O for field locking. MPER concentration is 0.66mM and in excess of Fab (0.47mM). The backbone 15N and 13C chemical shifts including Cb were assigned using TROSY-type triple resonance experiments as described previously 18 on a Bruker-800MHz spectrometers at 35°C. The cross-saturation transfer experiment 59 uses 100ms saturating WURST2 decoupling pulse train alternating between on-resonance (aliphatic region) and off-resonance (reference) FIDs, and was acquired on a Bruker-600MHz spectrometers at 35°C. NMR data were processed and analyzed using the software nmrPipe and CARA. Secondary structure and backbone dihedral angle predictions were obtained using the software TALOS+. Modeling of 2F5-MPER complex is performed using XPLOR-NIH.
Exploratory modeling of 2F5-MPER complex was performed using XPLOR-NIH 10. The MPER N-terminal residues up to A667 are fixed together with the 2F5-Fab according to the crystallographic structure (PDB: 1TJI), and the C-terminal residues starting from F673 are rigidified using pseudo-distance constraints converted from inter-atomic distances obtained from the NMR structure (PDB: 2PV6). The intervening residues are modeled through simulated annealing starting from the backbone conformation assumed from TALOS+ chemical shift predictions. EPR membrane immersion depths were translated into pseudo-distance constraints from CB atoms of each residue to the center of a fictitious liposome with a radius of 500Å. Hypothetical contact between I100F in the 2F5 CDRH3 loop and W672 of MPER has been introduced based on our NMR NOE data. Structural validation using PDB server indicates 8 local geometry deviations but no close contact clashes. Coordinates of this complex model are available upon request.
Additional methods
Preparation of liposomes, SPR measurements, EPR, Fab generation, transfection and FACS analysis methods are detailed in Supplemental Methods.
Supplementary Material
Acknowledgments
This work was supported by NIH grants RO1AI84785 and U19AI91693 to ELR and GW and a grant from the Gates Foundation, The Collaboration for AIDS Vaccine Discovery (CAVD) Program to ELR, GW and JRE. JRE was also supported by NIH RO1-GM086507 and funding through a cooperative research agreement with the Waters Corporation. KDR was supported by The Danish Council for Independent Research in Natural Sciences (FNU grant 09-063876).
JRE would like to thank Prof. T.E. Wales for expert technical assistance.
Footnotes
AUTHOR CONTRIBUTIONS
Z.J.S. designed, conducted NMR experiments, analyzed the data and wrote the paper; GW directed NMR effort and did analysis of NMR structure; A.F.F. contributed to the analysis of CDRH3 loop; J.H.W contributed to the analysis of complex structure; X.S. and K.D.R. designed and carried out HX-MS experiments and contributed to the data analysis; J.R.E. contributed to design, data analysis of HX-MS and wrote the paper; L.S. performed EPR experiments; S.M. prepared 2F5 and its variant Fabs and labeling of the MPER segment; Y.C. carried out cell transfection and FACS analysis; G. O., Y. Y. and P.D.K. provided 2F5 and mutant antibodies; M.K. designed and carried out Biacore experiments, analyzed the data and wrote the paper; E.L.R directed the project and wrote the paper.
References
- 1.McElrath MJ, Haynes BF. Induction of immunity to human immunodeficiency virus type-1 by vaccination. Immunity. 2010;33:542–554. doi: 10.1016/j.immuni.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han Y, Wind-Rotolo M, Yang HC, Siliciano JD, Siliciano RF. Experimental approaches to the study of HIV-1 latency. Nat Rev Microbiol. 2007;5:95–106. doi: 10.1038/nrmicro1580. [DOI] [PubMed] [Google Scholar]
- 3.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 4.Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 5.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci U S A. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwong PD, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 8.Wei X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 9.Simek MD, et al. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol. 2009;83:7337–7348. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cardoso RM, et al. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity. 2005;22:163–173. doi: 10.1016/j.immuni.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 11.Muster T, et al. Cross-neutralizing activity against divergent human immunodeficiency virus type 1 isolates induced by the gp41 sequence ELDKWAS. J Virol. 1994;68:4031–4034. doi: 10.1128/jvi.68.6.4031-4034.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson JD, et al. An affinity-enhanced neutralizing antibody against the membrane-proximal external region of human immunodeficiency virus type 1 gp41 recognizes an epitope between those of 2F5 and 4E10. J Virol. 2007;81:4033–4043. doi: 10.1128/JVI.02588-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trkola A, et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker LM, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou T, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou T, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salzwedel K, West JT, Hunter E. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J Virol. 1999;73:2469–2480. doi: 10.1128/jvi.73.3.2469-2480.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun ZY, et al. HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity. 2008;28:52–63. doi: 10.1016/j.immuni.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 19.Ofek G, et al. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J Virol. 2004;78:10724–10737. doi: 10.1128/JVI.78.19.10724-10737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Julien JP, Bryson S, Nieva JL, Pai EF. Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site. J Mol Biol. 2008;384:377–392. doi: 10.1016/j.jmb.2008.09.024. [DOI] [PubMed] [Google Scholar]
- 21.Alam SM, et al. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J Immunol. 2007;178:4424–4435. doi: 10.4049/jimmunol.178.7.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanchez-Martinez S, et al. Specific phospholipid recognition by human immunodeficiency virus type-1 neutralizing anti-gp41 2F5 antibody. FEBS Lett. 2006;580:2395–2399. doi: 10.1016/j.febslet.2006.03.067. [DOI] [PubMed] [Google Scholar]
- 23.Scherer EM, Leaman DP, Zwick MB, McMichael AJ, Burton DR. Aromatic residues at the edge of the antibody combining site facilitate viral glycoprotein recognition through membrane interactions. Proc Natl Acad Sci U S A. 2010;107:1529–1534. doi: 10.1073/pnas.0909680107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Julien JP, et al. Ablation of the complementarity-determining region H3 apex of the anti-HIV-1 broadly neutralizing antibody 2F5 abrogates neutralizing capacity without affecting core epitope binding. J Virol. 2010;84:4136–4147. doi: 10.1128/JVI.02357-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu H, et al. Interactions between lipids and human anti-HIV antibody 4E10 can be reduced without ablating neutralizing activity. J Virol. 2010;84:1076–1088. doi: 10.1128/JVI.02113-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ofek G, et al. Relationship between antibody 2F5 neutralization of HIV-1 and hydrophobicity of its heavy chain third complementarity-determining region. J Virol. 2010;84:2955–2962. doi: 10.1128/JVI.02257-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zwick MB, et al. The long third complementarity-determining region of the heavy chain is important in the activity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5. J Virol. 2004;78:3155–3161. doi: 10.1128/JVI.78.6.3155-3161.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alam SM, et al. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc Natl Acad Sci U S A. 2009;106:20234–20239. doi: 10.1073/pnas.0908713106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guenaga J, et al. Heterologous epitope-scaffold prime:boosting immuno-focuses B cell responses to the HIV-1 gp41 2F5 neutralization determinant. PLoS One. 2011;6:e16074. doi: 10.1371/journal.pone.0016074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ofek G, et al. Elicitation of structure-specific antibodies by epitope scaffolds. Proc Natl Acad Sci U S A. 2010;107:17880–17887. doi: 10.1073/pnas.1004728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 32.Wales TE, Fadgen KE, Gerhardt GC, Engen JR. High-speed and high-resolution UPLC separation at zero degrees celsius. Anal Chem. 2008;80:6815–6820. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miranker A, Robinson CV, Radford SE, Aplin RT, Dobson CM. Detection of transient protein folding populations by mass spectrometry. Science. 1993;262:896–900. doi: 10.1126/science.8235611. [DOI] [PubMed] [Google Scholar]
- 34.Weis DD, Wales TE, Engen JR, Hotchko M, Ten Eyck LF. Identification and characterization of EX1 kinetics in H/D exchange mass spectrometry by peak width analysis. J Am Soc Mass Spectrom. 2006;17:1498–1509. doi: 10.1016/j.jasms.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 35.Song L, et al. Broadly neutralizing anti-HIV-1 antibodies disrupt a hinge-related function of gp41 at the membrane interface. Proc Natl Acad Sci U S A. 2009;106:9057–9062. doi: 10.1073/pnas.0901474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi H, Nakanishi T, Kami K, Arata Y, Shimada I. A novel NMR method for determining the interfaces of large protein-protein complexes. Nat Struct Biol. 2000;7:220–223. doi: 10.1038/73331. [DOI] [PubMed] [Google Scholar]
- 37.Bryson S, Julien JP, Hynes RC, Pai EF. Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications. J Virol. 2009;83:11862–11875. doi: 10.1128/JVI.01604-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian Y, et al. Structure-affinity relationships in the gp41 ELDKWA epitope for the HIV-1 neutralizing monoclonal antibody 2F5: effects of side-chain and backbone modifications and conformational constraints. J Pept Res. 2002;59:264–276. doi: 10.1034/j.1399-3011.2002.02988.x. [DOI] [PubMed] [Google Scholar]
- 39.Huang CC, et al. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science. 2007;317:1930–1934. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andreotti AH. Native state proline isomerization: an intrinsic molecular switch. Biochemistry. 2003;42:9515–9524. doi: 10.1021/bi0350710. [DOI] [PubMed] [Google Scholar]
- 41.Lummis SCR, et al. Cis-trans isomerization at a proline opens the pore of a neurotransmitter-gated ion channel. Nature. 2005;438:248–252. doi: 10.1038/nature04130. [DOI] [PubMed] [Google Scholar]
- 42.Padlan EA. Anatomy of the antibody molecule. Mol Immunol. 1994;31:169–217. doi: 10.1016/0161-5890(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 43.Klein JS, et al. Examination of the contributions of size and avidity to the neutralization mechanisms of the anti-HIV antibodies b12 and 4E10. Proc Natl Acad Sci U S A. 2009;106:7385–7390. doi: 10.1073/pnas.0811427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Collis AV, Brouwer AP, Martin AC. Analysis of the antigen combining site: correlations between length and sequence composition of the hypervariable loops and the nature of the antigen. J Mol Biol. 2003;325:337–354. doi: 10.1016/s0022-2836(02)01222-6. [DOI] [PubMed] [Google Scholar]
- 45.Johnson G, Wu TT. Preferred CDRH3 lengths for antibodies with defined specificities. Int Immunol. 1998;10:1801–1805. doi: 10.1093/intimm/10.12.1801. [DOI] [PubMed] [Google Scholar]
- 46.Kunert R, Ruker F, Katinger H. Molecular characterization of five neutralizing anti-HIV type 1 antibodies: identification of nonconventional D segments in the human monoclonal antibodies 2G12 and 2F5. AIDS Res Hum Retroviruses. 1998;14:1115–1128. doi: 10.1089/aid.1998.14.1115. [DOI] [PubMed] [Google Scholar]
- 47.Xiao X, et al. Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens. Biochem Biophys Res Commun. 2009;390:404–409. doi: 10.1016/j.bbrc.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao X, Chen W, Feng Y, Dimitrov DS. Maturation pathways of cross-reactive HIV-1 neutralizing antibodies. Viruses. 2009;1:802–817. doi: 10.3390/v1030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yin J, Beuscher AEt, Andryski SE, Stevens RC, Schultz PG. Structural plasticity and the evolution of antibody affinity and specificity. J Mol Biol. 2003;330:651–656. doi: 10.1016/s0022-2836(03)00631-4. [DOI] [PubMed] [Google Scholar]
- 50.Mikell I, et al. Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS Pathog. 2011;7:e1001251. doi: 10.1371/journal.ppat.1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim ST, et al. The alphabeta T cell receptor is an anisotropic mechanosensor. J Biol Chem. 2009;284:31028–31037. doi: 10.1074/jbc.M109.052712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmitz KR, Ferguson KM. Interaction of antibodies with ErbB receptor extracellular regions. Exp Cell Res. 2009;315:659–670. doi: 10.1016/j.yexcr.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Janssen BJ, et al. Structural basis of semaphorin-plexin signalling. Nature. 2010;467:1118–1122. doi: 10.1038/nature09468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pejchal R, et al. A conformational switch in human immunodeficiency virus gp41 revealed by the structures of overlapping epitopes recognized by neutralizing antibodies. J Virol. 2009;83:8451–8462. doi: 10.1128/JVI.00685-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wales TE, Fadgen KE, Gerhardt GC, Engen JR. High-speed and high-resolution UPLC separation at zero degrees Celsius. Anal Chem. 2008;80:6815–6820. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang L, Pan H, Smith DL. Hydrogen exchange-mass spectrometry: optimization of digestion conditions. Mol Cell Proteomics. 2002;1:132–138. doi: 10.1074/mcp.m100009-mcp200. [DOI] [PubMed] [Google Scholar]
- 57.Weis DD, Engen JR, Kass IJ. Semi-automated data processing of hydrogen exchange mass spectra using HX-Express. J Am Soc Mass Spectrom. 2006;17:1700–1703. doi: 10.1016/j.jasms.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 58.Houde D, Berkowitz SA, Engen JR. The utility of hydrogen/deuterium exchange mass spectrometry in biopharmaceutical comparability studies. J Pharm Sci. 2010 doi: 10.1002/jps.22432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shimada I. NMR techniques for identifying the interface of a larger protein-protein complex: cross-saturation and transferred cross-saturation experiments. Methods Enzymol. 2005;394:483–506. doi: 10.1016/S0076-6879(05)94020-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.