Summary
Herpesviruses, which are major human pathogens, establish life-long persistent infections. Although the α-, β-, and γ-herpesviruses infect different tissues and cause distinct diseases, they each encode a conserved serine/threonine kinase critical for virus replication and spread. The extent of substrate conservation and the key common cell signalling pathways targeted by these kinases are unknown. Using a human protein microarray high-throughput approach we identify shared substrates of the conserved kinases from herpes simplex virus, human cytomegalovirus, Epstein-Barr virus (EBV) and Kaposi's sarcoma associated herpesvirus. DNA damage response (DDR) proteins were statistically enriched and the histone acetyltransferase TIP60, an upstream regulator of the DDR pathway, was required for efficient herpesvirus replication. During EBV replication, TIP60 activation by the BGLF4 kinase triggers EBV-induced DDR and also mediates induction of viral lytic gene expression. Identification of key cellular targets of the conserved herpesvirus kinases will facilitate the development of broadly effective anti-viral strategies.
Introduction
As major human pathogens, herpesviruses establish life-long persistent infections that result in clinical manifestations ranging from mild cold sores, to pneumonitis, birth defects and cancers. Although the α-, β-, and γ-herpesviruses infect different tissues and cause distinct diseases, they confront many of the same challenges in infecting their hosts, reprogramming cell gene expression, sensing and modifying cell cycle state and reactivating the lytic life cycle to produce new virions and spread infection (Arvin et al., 2007). While the α-, β-, and γ- mammalian herpesviruses encode latency and transcriptional regulatory genes that are unique to each sub-family, lytic cycle genes, such as those encoding virion structural components and proteins involved in replication of the viral genomes, are more conserved across the order herpesviridae. Amongst the conserved gene products are the orthologous serine/threonine protein kinases, UL13, UL97, BGLF4 and ORF36, encoded by herpes simplex type 1 (HSV1), human cytomegalovirus (HCMV), Epstein-Barr virus (EBV) and Kaposi's sarcoma associated herpesvirus (KSHV) respectively (Gershburg and Pagano, 2008). These kinases are structurally similar to the cellular kinase cdk2 (Romaker et al., 2006) and are recognized to phosphorylate a number of cyclin dependent kinase cellular targets including pRb (Hume et al., 2008), condensin (Lee et al., 2007), stathmin (Chen et al., 2010), lamin A/C (Hamirally et al., 2009; Lee et al., 2008; Meng et al., 2010), elongation factor 1 delta (Kato et al., 2001; Kawaguchi and Kato, 2003; Kawaguchi et al., 2003), MCM4 (Kudoh et al., 2006) and p27/KIP1 (Iwahori et al., 2009) as well as viral targets including KSHV bZIP (RAP) (Izumiya et al., 2007), EBV EBNA1 and virion proteins (Zhu et al., 2009) and HCMV UL69 (Rechter et al., 2009). Deletion of the protein kinases or inhibition of their activity has been shown to impair virus replication of HCMV and EBV in cultured cells (Gershburg et al., 2007; Prichard et al., 1999; Wolf et al., 2001) and to reduce the titer of HSV1 and murine gamma herpesvirus 68 (γ-HV68) in infected mice (Shibaki et al., 2001; Tarakanova et al., 2007). Herpesvirus replication takes place against a background of cell cycle arrest overlaid with a pseudo S phase environment whereby virus replication becomes dissociated from cellular DNA replication but selectively utilizes machinery normally activated during S-phase (Kudoh et al., 2005; Li and Hayward, 2011). The mimicry of cyclin dependent kinase activity by the conserved herpesvirus protein kinases contributes to the creation of the pseudo S-phase replication environment. This includes breakdown of the nuclear membrane, which is required for egress of virus capsids from the nucleus, and is dependent in infected cells on the viral protein kinase phosphorylation of lamin A/C (Hamirally et al., 2009; Lee et al., 2008; Meng et al., 2010).
Herpesvirus infection and lytic replication trigger the cellular DNA damage response. The induced DNA damage response is blunted during the establishment of latent herpesvirus infection, in EBV by the latency protein EBNA3C (Nikitin et al., 2010) and in HSV1 by the ICP0 protein (Lilley et al., 2010a) and this attenuation of the response is necessary for effective establishment of viral latency. Conversely, aspects of the DNA damage pathway are selectively incorporated into the herpesvirus lytic replication program (Gaspar and Shenk, 2006; Kudoh et al., 2005; Lilley et al., 2005; Shin et al., 2006) and are necessary for efficient viral replication. In particular, early events such as activation of the DNA damage response kinase ATM (Ataxia telangiectasia mutated protein) and phosphorylation of the ATM target H2AX are detected in cells undergoing lytic herpesvirus replication. The γ-HV68 protein kinase (orf36) and the EBV protein kinase BGLF4 have been shown to phosphorylate and activate ATM and H2AX (Tarakanova et al., 2007).
The nucleoside analog drugs acyclovir and ganciclovir that are used to treat herpesvirus infections require a mono-phosphorylation step that occurs in herpesvirus infected cells but not in uninfected cells and conserved protein kinases can mediate this phosphorylation (Gershburg et al., 2004; Meng et al., 2010; Moore et al., 2001; Sullivan et al., 1992). The multiple contributions of the conserved protein kinases to herpesvirus replication and spread also makes these kinases potential anti-viral drug targets although to date only one inhibitor of protein kinase enzymatic activity, maribavir, has entered clinical trials (Prichard, 2009).
The herpesvirus protein kinases have a broader substrate recognition than cellular cdks (Baek et al., 2002a; Cano-Monreal et al., 2008; Romaker et al., 2006; Zhu et al., 2009) and neither the full range of their substrates nor the degree to which the substrates of individual conserved protein kinases overlap is known. Comprehensive knowledge of common host targets would provide valuable insight into key host factors that facilitate herpesvirus replication and identify signalling pathways whose targeting in combination could enhance the effectiveness of anti-viral treatments. Using a human protein microarray screen, we have identified more than 100 shared substrates of the α, β and γ herpesvirus conserved kinases. Bioinformatic analyses of these shared substrates revealed a statistical enrichment of proteins involved in the DNA damage response. Follow-up experimentation highlighted the key contribution to herpesvirus replication of protein kinase mediated phosphorylation of the histone acetlytransferase TIP60, a regulator of the DNA damage response and of chromatin remodeling.
Results
Common host substrate identification for conserved herpesvirus protein kinases
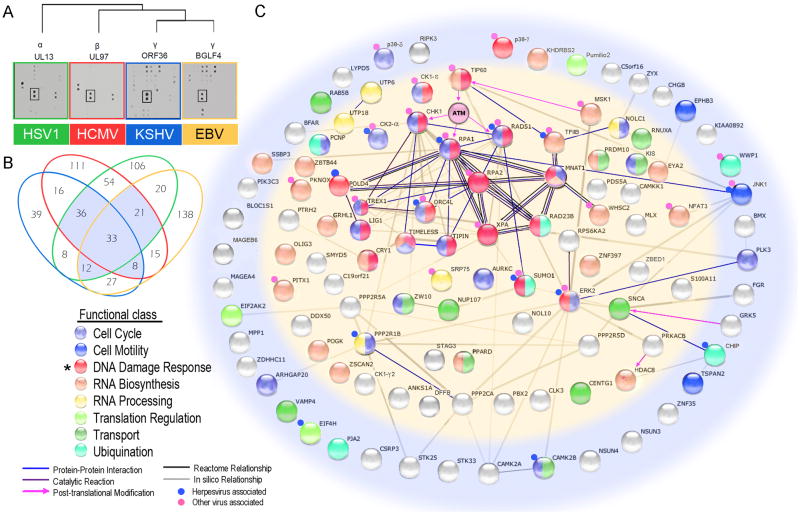
To identify common substrates for the herpesvirus conserved protein kinases, we performed assays on a human protein microarray composed of 4,191 unique human proteins using the UL13, UL97, BGLF4 and ORF36 orthologous kinases encoded by the α-, β-, and γ-viruses HSV1, HCMV, and EBV and KSHV, respectively (Figure 1A). Using normalized amounts of purified viral kinases, as determined by autophosphorylation reactions, we identified 273, 178, 290, and 294 substrates of BGLF4, ORF36, UL97, and UL13, respectively, at a cut-off value of standard deviation (SD) ≥3 (Figure 1A, Figure S1 & Table S1). Of the 643 non- redundant substrates collectively identified by the four kinases, 110 are shared by at least three kinases (Figure 1B). Gene ontology (GO) analysis of these 110 common substrates revealed involvement in eight major functional classes while statistical analysis indicated that the DDR was significantly enriched (p=0.004; hypergeometric test) (Figure 1B & 1C). In addition, proteins in this DDR category are also enriched for known association with viral infections (p=0.016; Table S2).
Figure 1. Identification of common host protein substrates for α-, β- and γ-human herpesvirus protein kinases: Enrichment of proteins in the DNA damage pathway.
(A) Autoradiograph showing representative sections of typical protein array phosphorylation assays performed using the four viral kinases. All substrates are printed in duplicate. The rectangle highlights a common substrate, Pumilio2.
(B) Venn diagram illustrating the overlap in substrate specificity of the herpesvirus protein kinases. Of 643 total substrates, the highlighted 110 host proteins were phosphorylated by at least three kinases. See also Tables S1 & Figure S1
(C) Interaction network for the 110 shared host proteins. Proteins are color-coded by their functional classes. An asterisk indicates the enriched functional class of 19 proteins involved in the DDR. Proteins in the inner oval (light yellow) are nuclear proteins while the proteins in the outer ring are in other cellular compartments. Edges between the proteins represent known or predicted connections, such as protein-protein interactions, catalytic reactions, and enzyme-substrate relationships, obtained from the database STRING (http://string-db.org/). Note that ATM was not present on the human protein array. See also Tables S1 & S2.
An effective means for a virus to exploit the host is to target a master regulator that controls multiple downstream signalling pathways. To identify such a master regulator, we applied orthogonal analysis to the shared substrates by incorporating different types of data (e.g., protein-protein and enzyme-substrate interactions), and found a highly connected cluster of 12 proteins, all involved in the DDR (Figure 1C). Intriguingly, several proteins are either the direct targets (e.g., CHK1, RPA1, and RAD51) or downstream effectors of Ataxia telangiectasia mutated (ATM) kinase, which plays a crucial role in the DDR (Harper and Elledge, 2007). ATM was not present on our protein microarrays. However, recent studies have shown that the activation of ATM's kinase activity in response to DNA damage is dependent upon TIP60 (Sun et al., 2005), one of the substrates that was common to the herpesvirus conserved protein kinases (Figure 1C). Because TIP60 plays an important role in both the DDR and transcription regulation through chromatin remodeling (Sapountzi et al., 2006; Squatrito et al., 2006), it is a candidate master regulator of the herpesvirus life cycle. Therefore, we focused on TIP60 and its role in herpesvirus replication.
EBV BGLF4 regulates lytic replication through the phosphorylation and activation of TIP60
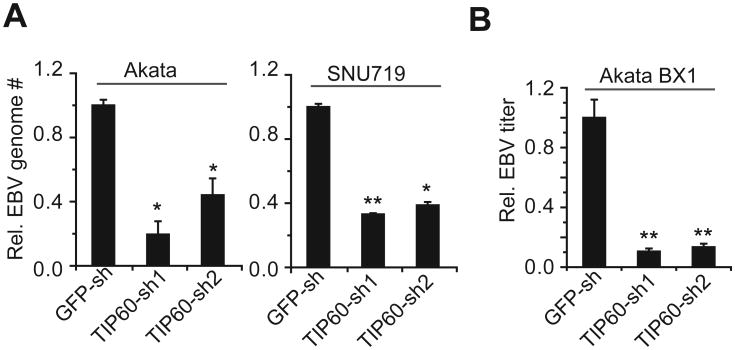
Choosing EBV as the primary model, we first tested whether TIP60 expression affected viral DNA replication. Both Akata (EBV+) B cells and SNU719 (EBV+) gastric carcinoma cells were transformed with individual shRNA lentiviral constructs to knockdown TIP60 expression. As a surrogate for viral DNA replication, we measured the EBV genome copy following lytic induction of EBV by IgG cross-linking and bortezomib treatment, respectively. Knockdown of TIP60 was incomplete (Figure S2) but none-the-less reduced the number of EBV genomes by 60 to 80% on both cell backgrounds and with both lytic induction treatments (Figure 2A). Measurement of extracellular infectious virus found an approximately 90% reduction upon TIP60 silencing (Figure 2B). Since the observed decrease was shown with two different shRNAs, the phenotype is unlikely to be due to off-target shRNA effects. These results indicate TIP60 is a physiologically relevant substrate in the EBV life cycle.
Figure 2. TIP60 is required for efficient EBV lytic replication.
(A) TIP60 silencing impairs lytic DNA replication. Relative viral genome copy numbers measured by qPCR in lytically induced Akata (EBV+) and SNU719 (EBV+) cells carrying control shRNA (GFP-sh) or TIP60 shRNAs (TIP60-sh1, TIP60-sh2). The experiments were performed in triplicate with ± SD shown. *p<0.02, **p<0.01. See also Figure S2A.
(B) TIP60 silencing reduces infectious virus production. Relative EBV titer produced by lytically induced Akata BX1(EBV+) cells carrying control shRNA (GFP-sh) or TIP60 shRNAs (TIP60-sh1, TIP60-sh2) was measured using Raji cell infection assay. The experiments were carried out in triplicate with ± SD shown. **p<0.01. See also Figure S2B.
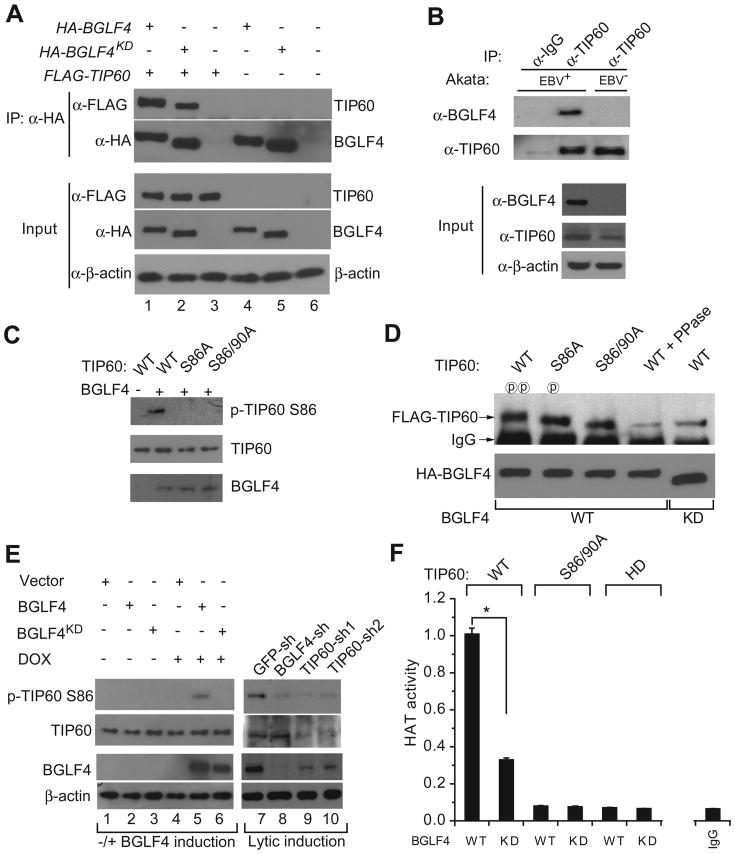
To demonstrate that TIP60 is a target of the EBV kinase BGLF4 in cells, we first showed that TIP60 interacted with both wild-type (WT) BGLF4 and a kinase-dead mutant (BGLF4KD) in transfected cells using reciprocal co-immunoprecipitations (co-IPs) (Figure 3A & Figure S3A). Note that while the loss of BGLF4 kinase activity did not affect its interaction with TIP60, there was a change in TIP60 mobility with BGLF4KD, indicating that BGLF4 plays a role in TIP60 phosphorylation. BGLF4-TIP60 interaction during EBV infection was validated using EBV-positive Akata (EBV+) cells induced into the lytic cycle by treatment with IgG to cross-link the B cell receptor and antibodies against endogenously expressed TIP60 (Figure 3B). Autologous EBV negative Akata 4E3 cells (EBV-) served as a negative control. Having shown that BGLF4 directly phosphorylated TIP60 in vitro (Figure S3B), we sought to determine which sites on TIP60 were phosphorylated. In a previous study, phosphorylation at Ser86 and Ser90 of TIP60 was shown to enhance its HAT activity in vitro using histones as substrates and in addition, GSK3β and CDK1/cyclin B were found to in vitro phosphorylate Ser86 and Ser90, respectively (Charvet et al., 2011; Lemercier et al., 2003). Because BGLF4 and CDK1/cyclin B have overlapping substrate recognition (Hume et al., 2008; Zhu et al., 2009), we created TIP60 constructs carrying single- or double-mutations at Ser86 and Ser90. To show that BGLF4 directly phosphorylates TIP60 at Ser86, an in vitro phosphorylation assay was performed. We found that the TIP60 pSer86 specific antibody detected phosphorylation of WT TIP60 but not phosphorylation of the S86A or S86/90A mutants (Figure 3C). Further, immunoblot analysis of TIP60 co-precipitated from WT TIP60, S86A or S86/90A transfected cell extracts by anti-BGLF4 antibody revealed that the S86A and S90A mutations each affected TIP60 mobility with the effects of the double-mutation being additive (Figure 3D). Phosphatase treatment, increased the mobility of WT TIP60 co-precipitated with WT BGLF4 to equal that of WT TIP60 co-precipitated with BGLF4KD and also that of the S86/90A double-mutant. This indicates that TIP60 Ser86/90 are major sites of phosphorylation by BGLF4. To further confirm Ser86 phosphorylation of TIP60 in vivo, we monitored Ser86 phosphorylation of endogenous TIP60 upon induction of WT BGLF4 or BGLF4KD and found that the Ser86 phosphorylation was dependent on presence of BGLF4 kinase activity (Figure 3E, left panel). These results were further supported in lytically induced Akata (EBV+) cells, where Ser86 phosphorylation of TIP60 was almost completely abolished upon BGLF4 knockdown (Figure 3E; right panel).
Figure 3. BGLF4 interacts with, phosphorylates and activates TIP60.
(A) Both EBV BGLF4 and kinase dead BGLF4 interact with TIP60. Western blot analysis of transfected 293T cell extracts showing co-precipitation of TIP60 with BGLF4. BGLF4KD, BGLF4 kinase dead mutant. Input, 2% whole cell lysate used for IP. See also Figure S3A.
(B) Interaction between endogenous TIP60 and EBV BGLF4. Lytically induced Akata (EBV+) and Akata 4E3 (EBV-) cell extracts were immunoprecipitated with control IgG or anti-TIP60 antibodies and the precipitated proteins were immunoblotted with the indicated antibodies. Input, 1% whole cell lysate used for IP.
(C) BGLF4 phosphorylates TIP60 at S86 in vitro. Western blot analysis after in vitro phosphorylation reactions with indicated combinations of BGLF4 and wild type or mutant TIP60.
(D) TIP60 Ser86 (S86) and Ser90 (S90) are substrates for BGLF4. Immunoblot comparing the mobility of BGLF4- and kinase dead BGLF4-coprecipitated wild type and mutant FLAG-TIP60 with and without phosphatase treatment. 293T cells were transfected as indicated and then treated with 20 μM roscovitine for 12 hrs before harvest. See also Figure S3B.
(E) BGLF4 induces TIP60 S86 phosphorylation in vivo. Western blot analysis of cell extracts from Akata (EB V+) cell carrying empty vector, BGLF4 or BGLF4KD with or without doxycycline (DOX, 20 ng/ml) treatment, and cell extracts from lytically induced Akata (EBV+) cells carrying control GFP, BGLF4 or TIP60 shRNAs.
(F) BGLF4 increases TIP60 HAT activity. Relative HAT activity of wild type TIP60 (WT), phosphorylation deficient TIP60 (S86/90A) and HAT-dead TIP60 (HD) immunoprecipitated from 293T cells transfected with wild type BGLF4 (WT) or BGLF4KD (KD) constructs. The experiments were carried out in triplicate with ± SD shown. *p<0.01. Immunoprecipitated TIP60 loading controls are shown in Figure S3C.
To investigate whether the histone acetyltransferase (HAT) activity of TIP60 is affected by BGLF4 phosphorylation, we compared the HAT activity of WT and S86/90A TIP60 co-expressed with either WT BGLF4 or BGLF4KD. TIP60 was immunoprecipitated and its HAT activity was measured in vitro (Figure 3F & Figure S3C). TIP60's HAT activity in the presence of WT BGLF4 was 3-fold greater than that seen with BGLF4KD, indicating that BGLF4's phosphorylation of TIP60 substantially enhances its HAT activity. This result was further supported by the observation that the S86/90A double-mutation reduced TIP60's HAT activity to that of a HAT deficient TIP60 mutant regardless of the presence or absence of WT BGLF4 or BGLF4KD (Figure 3F & Figure S3C). Taken together, the data establish that BGLF4 interacts with and phosphorylates TIP60 to increase TIP60's HAT activity.
BGLF4 induces the DNA damage response and chromatin remodeling through TIP60
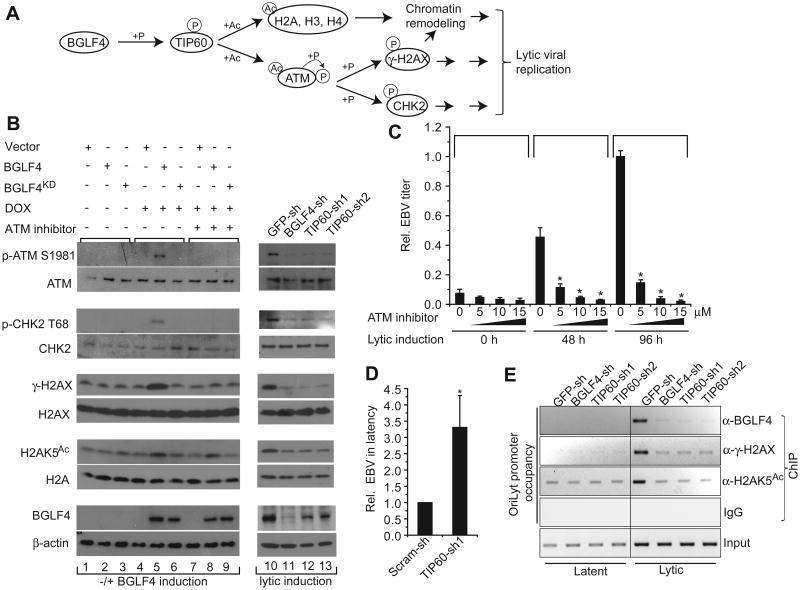
TIP60 mediates chromatin remodeling and TIP60 acetylation of ATM activates ATM autophosphorylation and ATM transphosphorylation of downstream targets such as those illustrated in Figure 4A. Although the DDR and chromatin remodeling have been implicated in herpesvirus replication, the molecular mechanisms are poorly understood (Lilley et al., 2010b). Therefore, we examined whether BGLF4 regulates the DDR and chromatin remodeling via TIP60. As shown in Figure 4B, the presence of a series of DNA damage markers, including pSer1981 of ATM, pThr68 of CHK2, and pSer139 of histone H2AX (γ-H2AX) is dependent on induction of WT BGLF4, but not BGLF4KD, in Akata (EBV+) cells, and inhibition of ATM abolishes these effects. In addition, Lys5 acetylation of histone H2A (H2AK5Ac), a known target of TIP60, is substantially enhanced upon BGLF4 induction regardless of ATM inhibition (Figure 4B, lanes 5 and 8). Moreover, in a time course of lytic induction, in Akata (EBV+) cells, BGLF4 appearance coincides with TIP60 phosphorylation, ATM activation and γ-H2AX generation (Figure S4A). Consistent with this result, when endogenous BGLF4 is knocked down after lytic induction, ATM Ser1981 autophosphorylation is reduced to the same level as that seen in TIP60-knockdown cells, suggesting that the BGLF4-induced DDR depends on TIP60. Moreover, in the same context, the phosphorylation of ATM's downstream effectors CHK2 (pThr68) and H2AX (pSer139 or y-H2AX), and the acetylation of TIP60's direct target H2AK5Ac, are also BGLF4-dependent (Figure 4B; right panel).
Figure 4. BGLF4 induces the DNA damage response through TIP60.
(A) Schematic illustration of BGLF4's potential function in the DDR and chromatin remodeling through TIP60 phosphorylation. P, phosphorylation; Ac, acetylation.
(B) BGLF4 induces histone acetylation and ATM activation via TIP60 phosphorylation. Western blot analysis of cell extracts from Akata (EBV+) cells carrying empty vector, BGLF4 or BGLF4KD with or without doxycycline (DOX, 20 ng/ml) or ATM inhibitor (KU55933, 15μM) treatment as indicated, and cell extracts from lytically induced Akata (EBV+) cells carrying control GFP, BGLF4, or TIP60 shRNAs. See also Figure S4A. The data are representative of at least two independent biological replicates.
(C) ATM inhibitor inhibits EBV lytic replication. Relative EBV titer of lytically induced Akata BX1(EBV+) cells in the absence or presence of ATM inhibitor (KU55933) as indicated was measured using Raji cell infection assay. The experiments were carried out in triplicate with ± SD shown. *p<0.01. See also Figure S4B.
(D) TIP60 knockdown increases the efficiency of EBV latency establishment. EBV BX1 virus was used to infect Raji cells carrying control scramble shRNA (Scram-sh) or TIP60 shRNA (TIP60-sh1). Phorbol-12-myristate-13-acetate (TPA) (20 ng/ml) and sodium butyrate (3 mM) were added six days post infection and the number of the GFP-positive Raji cells was calculated to determine the efficiency of latency establishment. *p<0.01. See also Figure S4C.
(E) Lytic induction results in the recruitment of BGLF4 and γ-H2AX to the EBV lytic replication origin (OriLyt) promoter and enrichment of histone acetylation (H2A Lys5 acetylation, H2AK5Ac) on this promoter. ChIP-PCR analysis performed on Akata (EBV+) cells carrying indicated shRNAs showing BGLF4, γ-H2AX and H2AK5Ac enrichment at the OriLyt promoter after lytic induction.
To test whether BGFL4/TIP60 dependent activation of the DDR via ATM plays a role in EBV lytic replication, we measured extracellular virus produced by Akata BX1 (EBV+) cells in the latent state (0 hr) or after lytic induction (48 and 96 hrs) in the absence or presence of an ATM inhibitor. We found that EBV lytic replication was suppressed in a dose-dependent manner by ATM inhibition (Figure 4C and S4B), demonstrating the critical role of ATM in EBV lytic replication.
A recent study showed that inhibition of the DDR kinases ATM and Chk2 markedly increases the efficiency of EBV latency establishment in B cells (Nikitin et al., 2010). Since TIP60 acts upstream of ATM and CHK2, we asked whether inhibition of TIP60 also increases EBV latency establishment. Using GFP-tagged virus produced by AkataBX1 cells and a Raji B cell infection assay, we found that latency establishment was increased in Raji cells carrying TIP60 shRNA compared to cells carrying control shRNA (Figure 4D & Figure S4C).
BGLF4 induces the expression of key lytic viral genes through TIP60
To further illustrate the integration of BGLF4 and the DDR into EBV DNA replication, we demonstrated that BGLF4 was recruited to the EBV lytic replication origin (OriLyt) upon lytic induction and its presence induced the recruitment of γ-H2AX and the accumulation of H2AK5Ac at the same locus (Figure 4E). Since TIP60 is known to acetylate histones and regulate gene expression (Avvakumov and Cote, 2007; Baek et al., 2002b; Ikura et al., 2000), we reasoned that the accumulation of H2AK5Ac at this promoter induced by TIP60 could also contribute to viral gene expression. We therefore investigated whether the OriLyt (BHLF1) promoter or other promoters are targeted by TIP60 during lytic induction.
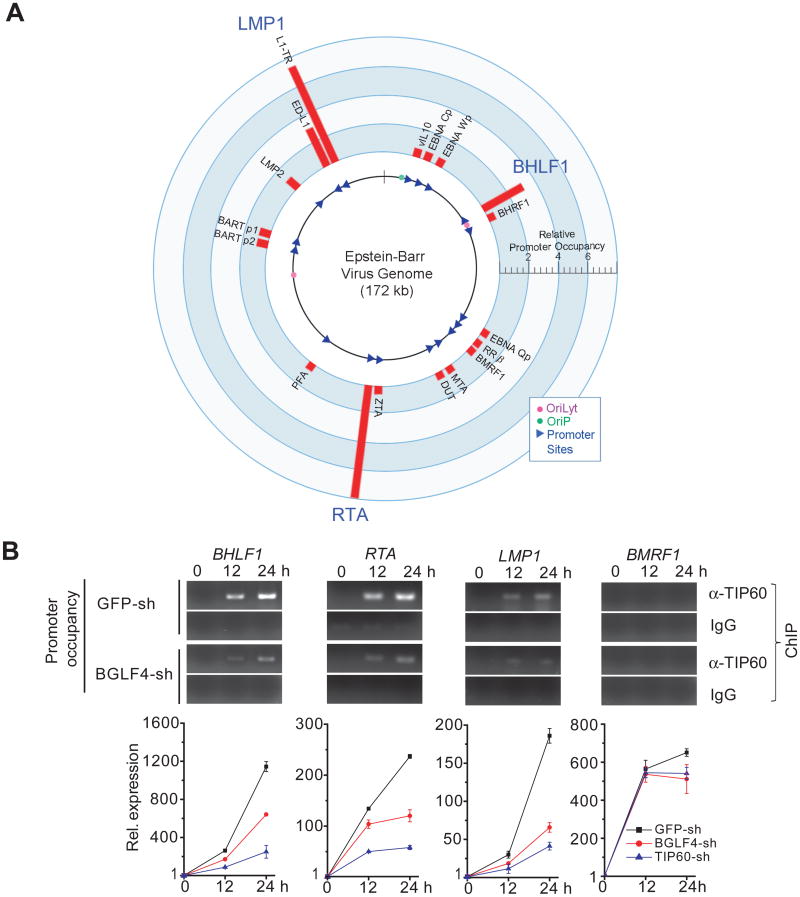
We performed chromatin immunoprecipitation (ChIP) assays coupled with real-time PCR to quantitatively survey 18 well-annotated EBV promoter regions, including the OriLyt (BHLF1) promoter, for TIP60 occupancy. The selected promoters are distributed across the EBV genome (de Jesus et al., 2003) and control 22 EBV genes (Figure 5A). Using antibody against endogenous TIP60 in lytically induced Akata (EBV+) cells, we found that TIP60 associated with the BHLF1 (OriLyt) and RTA promoters and both promoters (ED-L1 and L1-TR) that regulate LMP1 (Figure 5A), while no significant enrichment of TIP60 was observed on the other tested promoters. These results indicate that TIP60 associates with specific EBV promoters. We next examined the dynamics of this relationship to compare TIP60 occupancy of the BHLF1 (OriLyt), RTA, and LMP1 promoters during latency and post lytic induction. TIP60 association was not detected during latent infection of Akata (EBV+) cells but TIP60 was recruited to all three promoters at 12 hr post-induction and remained associated at 24 hr (Figure 5B, upper panels). In contrast, the BMRF1 lytic promoter was not occupied by TIP60 during the course of lytic induction. To determine BGLF4's role in this process, we used shRNA lentiviral constructs to knockdown BGLF4 expression in Akata (EBV+) B cells and then examined TIP60's recruitment to the BHLF1, RTA and LMP1 promoters during the course of EBV lytic induction (Figure 5B, middle panel). Quantitative measurement by qPCR showed that TIP60's occupancy on the three promoters was reduced by at least 50% between 12 to 24 hr post-induction (Figure S5A). Thus, BGLF4 enhances TIP60's recruitment to these three viral promoters.
Figure 5. Regulation of EBV lytic gene expression through TIP60.
(A) TIP60 recruitment to EBV promoters. The EBV genome annotated with the 18 tested promoters (triangles) and origins of DNA replication (dots). Red bars: Relative TIP60 occupancy normalized to the IgG control.
(B) Impact of TIP60 recruitment and BGLF4 activity on EBV lytic gene expression. Upper panel: ChIP-PCR analysis performed on Akata (EBV+) cells showing TIP60 enrichment at the BHLF1, RTA and LMP1 (L1-TR) promoters after lytic induction but not the BMRF1 lytic promoter. Middle panel: Recruitment of TIP60 to the BHLF1, RTA and LMP1 (L1-TR) promoters was reduced in BGLF4-sh Akata (EBV+) cells. Lower panel: RT-qPCR analysis of mRNA levels for the corresponding genes and a non-enriched gene (BMRF1) in Akata (EBV+) cells carrying the indicated shRNAs. The experiments were carried out in triplicate with ± SD shown. See also Figure S5.
Importantly, the three EBV genes targeted by TIP60 play key roles in viral replication. RTA is one of the two key transcriptional activators that drive early and late lytic EBV gene expression (Zalani et al., 1996). The BHLF1 (OriLyt) promoter is an essential component of the viral lytic origin of replication (Schepers et al., 1993). LMP1 is a latency gene but its expression is up-regulated in the lytic cycle where LMP1 provides key functions for cell survival and virus release (Ahsan et al., 2005; Dirmeier et al., 2005; Uchida et al., 1999). To correlate TIP60 recruitment and BGLF4 function with the efficiency of expression of these EBV genes, we generated Akata (EBV+) cells that expressed BGLF4 shRNA (BGLF4-sh), TIP60 shRNA (TIP60-sh), or control GFP shRNA (GFP-sh). In the control GFP-sh Akata cells, as expected, these 3 genes, and BMRF1, were highly upregulated at 12- and 24-hr post induction (Figure 5B, lower panels). However, in BGLF4-sh and TIP60-sh cells (Figure 5B, lower panels; Figure S5B), the expression level of BHLF1, RTA, and LMP1 was significantly reduced at both time points, whereas BMRF1 expression was minimally affected. TIP60 expression was not altered by BGLF4-sh (Figure S5C). Interestingly, TIP60 knockdown had a greater negative impact than BGLF4 knockdown (Figure 5B, lower panel). To summarize, the results reveal that EBV exploits TIP60 via BGLF4 phosphorylation to drive lytic viral gene expression. RTA induced transcription of BGLF4 leads to reinforced RTA transcription and consequently to enhanced expression of the RTA regulated lytic viral replication program (Wang et al., 2010).
Conserved role for TIP60 in herpesvirus replication
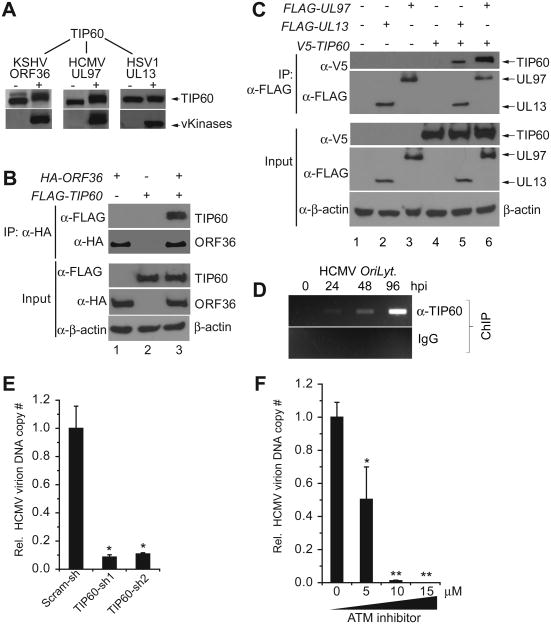
Finally, we tested whether the interplay between the viral kinases and TIP60 is conserved in the other herpesviruses. Using the same approaches described above, we showed that KSHV ORF36, HCMV UL97 and, to a lesser extent, HSV1 UL13 phosphorylated and increased the mobility of TIP60 in co-transfected Hela cells (Figure 6A) and interacted with TIP60 in transfected 293T cells (Figures 6B, 6C, S6A & S6B). In addition, we tested for recruitment of TIP60 at the HCMV lytic replication origin (OriLyt) and found that, similar to EBV, TIP60 was recruited to HCMV oriLyt at 24, 48 and 96 hours post infection (hpi) (Figure 6D). Furthermore, knockdown of TIP60 in HCMV-infected cells significantly reduced production of extracellular HCMV virion DNA (Figure 6E & Figure S6C). HCMV lytic replication was also significantly suppressed by an ATM inhibitor in a dose-dependent manner (Figures 6F and S6D) suggesting that the mechanism of inhibition parallels that shown for EBV. These results demonstrate that the viral kinase-TIP60 partnership is conserved and represents a common virus-host interaction.
Figure 6. Conserved role for TIP60 in herpesvirus replication.
(A) Western blot showing that KSHV ORF36 and HCMV UL97increase the mobility of TIP60 in transfected Hela cells. (B) and (C) TIP60 co-precipitates with (B) KSHV ORF36, (C) HCMV UL97 and HSV1 UL13 using co-transfected 293T cells. Reciprocal immunoprecipitations are presented in Figure S6A & S6B. Input, 2% whole cell lysate used for IP.
(D) TIP60 is recruited to HCMV lytic replication origin (OriLyt) at 24, 48 and 96 hours post-infection (hpi).
(E) TIP60 is required for efficient HCMV replication. Relative supernatant virion DNA from HCMV-infected HF cells (96 hpi) carrying control scramble shRNA (Scram-sh) or TIP60 shRNA (TIP60-sh) was determined with qPCR. The experiments were carried out in triplicate with ± SD shown. *p<0.01. See also Figure S6C
(F) ATM inhibitor inhibits HCMV replication. Relative supernatant virion DNA from HCMV-infected HF cells (96 hpi) in the absence or presence ATM inhibitor (KU55933) was determined with qPCR. The experiments were carried out in triplicate with ± SD shown. *p<0.01, **p< 0.005. See also Figure S6D.
Discussion
High-throughput technology is emerging as a powerful tool for discovery of factors involved in pathogen-host interactions (Brass et al., 2009; Calderwood et al., 2007; Karlas et al., 2010; Konig et al., 2010; Shapira et al., 2009). Here, we took a protein microarray approach to identify enzyme-substrate interactions for four conserved human herpesvirus kinases, with the hypothesis that the common substrates would reveal host pathways that are critical for replication across the herpesvirus family. By analyzing over 100 shared host substrates we identified the DDR pathway as a central target of the conserved herpesvirus kinases. Mechanistic studies showed that in the absence of external DNA damage cues, the EBV kinase phosphorylated and activated an upstream master regulator of the DDR, the histone acetyltransferase TIP60. TIP60 was additionally integrated to the virus lytic program by recruitment to the viral chromatin where TIP60 activated specific EBV genes critical for viral replication.
TIP60 was originally identified as a partner of the human immunodeficiency virus type 1 (HIV-1) transactivator Tat (Kamine et al., 1996) and is targeted by several viruses. Human T cell lymphotropic virus type 1 (HTLV-1) p30II enhances Myc transforming activity through stabilizing Myc-TIP60 transcriptional interactions (Awasthi et al., 2005). TIP60 interaction with viral TAT, E6 and UL27 proteins encoded by HIV-1, human papillomavirus (HPV) and HCMV, respectively, induces TIP60 degradation (Col et al., 2005; Jha et al., 2010; Reitsma et al., 2011), which is believed to enable establishment of viral latency and enhance virus-induced oncogenesis. In the case of HCMV, viruses deleted or mutated for the UL97 protein kinase escape through secondary mutations in the UL27 protein that degrades TIP60 (Chou, 2009; Reitsma et al., 2011). A recent study by Nikitin et al. found that the DDR induced upon EBV infection is a robust host anti-viral defense and EBV employs counter-measures to overcome the growth inhibitory effects of the host DDR in order to establish latency (Nikitin et al., 2010). These authors found that treatment of B cells with an ATM inhibitor increased latency establishment. We find that TIP60 inhibition with shRNA also increases latency establishment implying that TIP60 is an upstream mediator of the DNA damage response induced upon EBV infection. Interestingly, BGLF4 is present in the EBV tegument (Asai et al., 2006) and is consequently introduced into cells upon EBV infection. BGLF4 would therefore be available to initiate a transient activation of TIP60 and the DDR and the BGLF4/TIP60 partnership may therefore be an important factor in inducing a cellular environment that is hostile to latency establishment.
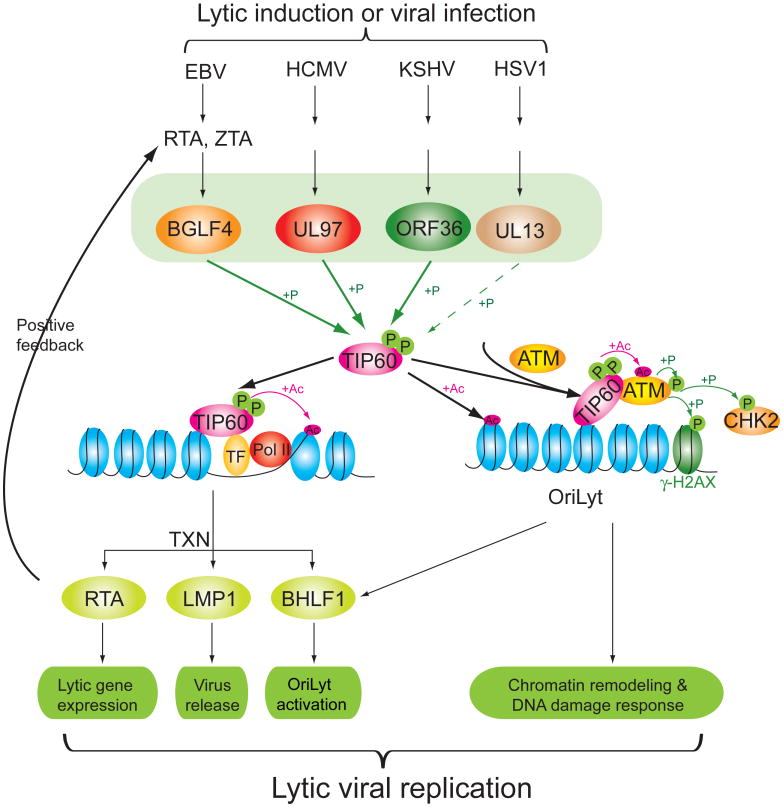
In counterpoint, we demonstrate that TIP60 plays a positive role in the lytic replication of herpesviruses: TIP60 shRNA significantly reduces virus production from β and γ herpesvirus infected cells. In the case of EBV, TIP60 HAT activity is enhanced via phosphorylation by the EBV-encoded protein kinase BGLF4 at the same sites phosphorylated by CDC2 and GSK3β (Charvet et al., 2011; Lemercier et al., 2003) and this interaction is sufficient to trigger the DDR. The DDR plays an important role in the lytic viral life cycle. EBV lytic replication elicits a DDR by triggering ATM autophosphorylation and cativation. Activated ATM phosphorylates its downstream targets, such as H2AX, p53, CHK2 and RPA2, and phosphorylated ATM, RPA2 and Mre11/Rad50/Nbs1 (MRN) complexes are recruited to replication compartments in nuclei during EBV lytic replication (Kudoh et al., 2005; Kudoh et al., 2009). However, the mechanism of virus-triggered ATM activation has been elusive. Although γ-HV68 kinase orf36 and EBV BGLF4 have been found to directly phosphorylate H2AX, this phosphorylation was significantly reduced in ATM deficient cells (Tarakanova et al., 2007) and, as shown in Figure 4B in the present study, in cells treated with an ATM inhibitor. As summarized in Figure 7, our experiments mechanistically link the viral kinases to ATM and its downstream targets CHK2 and H2AX via TIP60.
Figure 7. Model for conserved herpesvirus kinases in regulating vrial replication through TIP60.
The contribution of TIP60 activation by the conserved herpesvirus kinases to lytic replication as illustrated mechanistically for EBV-infected cells. TXN, transcription; P, phosphorylation; Ac, acetylation; TF, transcription factor; Pol II, RNA polymerase II; OriLyt, lytic replication origin.
We also demonstrate that TIP60 plays a positive role in transcriptional regulation of key lytic viral genes (Figure 7). BGLF4 has been implicated in facilitating viral egress from the nucleus by phosphorylating lamins (Lee et al., 2008). Interestingly, we find that TIP60 is recruited to the LMP1 promoters after lytic induction and is needed for achieving normal levels of lytic LMP1 transcription. LMP1 downstream signalling is important for nuclear egress of virions (Ahsan et al., 2005) and our data suggest that TIP60 mediated activation of LMP1 expression represents another mechanism by which BGLF4 promotes this aspect of infectious EBV production. TIP60's negative role in establishment of latency and positive role in lytic viral replication place TIP60 at the decision point between viral latency establishment and productive lytic replication (Figure 2 & Figure 4D).
This work illustrates the value of high throughput, unbiased approaches for the discovery of conserved viral targets. There are few drugs available to treat herpesvirus infections and viral escape mutants develop upon extensive use of this limited repertoire. The herpesvirus protein kinases are attractive anti-viral drug targets. However, developing broadly effectively drugs requires knowledge of their common cellular substrates. The information provided by our common substrate identification will assist in the design of assays for new broadly effective anti-herpesvirus therapeutics.
Experimental Procedures
Kinase assay
Phosphorylation of proteins on human protein arrays by herpesvirus protein kinases was assayed as previously described (Ptacek et al., 2005; Zhu et al., 2009). The list of the 4,191 unique proteins on this array can be found in Table S2 of Hu et al (Hu et al., 2009). Detailed information was described in the Supplemental Experimental Procedures.
Immunoprecipitation and ChIP assays
Cells were transfected using Lipofectamine 2000 (Invitrogen) or calcium phosphate and the amount of DNA in each sample was equalized using vector DNA. Transfected cells were harvested 48 hrs post-transfection using RIPA lysis buffer [50 mM Tris-HCl (pH7.4), 150 mM NaCl, 1% (v/v) NP40, 1 % (w/v) deoxycholate 0.1% (w/v) SDS, 1 mM EDTA] containing protease inhibitors and phosphatase cocktail I and II (Sigma) (Li et al., 2007). In figure 3D, cells were treated with 20 μM roscovitine for 12 hrs before harvest to minimize the contribution of CDC2. Immunoprecipitation and ChIP were carried out as described previously (Zhu et al., 2009). For phosphatase treatment, the immunoprecipitated complex was re-suspended in 1× NEBuffer and incubated with 10 units of calf intestinal phosphatase (New England Biolabs) at 37 °C for 1 h. The complex was then eluted with Laemmli sample buffer and subsequently analyzed by SDS-PAGE and immunoblotting.
Histone acetyltransferase (HAT) assay
TIP60 HAT activity was assayed using Flag-TIP60, Flag-TIP60S86/90A and HAT dead Flag-TIP60 immunoprecipitated from 293T cells cotransfected with, HA-BGLF4 or HA-BGLF4 kinase dead mutant. Cells were treated with 20 μM roscovitine for 12 hrs before harvest and TIP60 HAT activity was assayed using the HAT Assay Kit (Millipore) modified according to Sun et al. (Sun et al., 2005).
Virus infection
For HCMV infection, HF cells were seeded into 24 well plates one day before infection. The cells were washed with PBS and HCMV-luciferase virus (MOI = 1) was added to each well and incubated for 1.5 hrs in 200 μl serum-free DMEM. Free viruses were removed with washing and cells were incubated in medium containing 4% fetal bovine serum for 96 hrs. To induce the EBV lytic cycle, Akata (EBV+) cells were treated with 50 μg/ml of goat anti-human IgG (MP Biomedicals) for 24 hrs and SNU719 (EBV+) cells were treated for 24 hrs with 20 nM of bortezomib (Fu et al., 2008).
Statistical Analysis
Statistical analyses employed a two-tailed Student's t test. A p-value of ≤ 0.05 was considered statistically significant. The data are representative of at least two independent experiments, and values are given as the mean of replicate experiments ± standard deviation (SD).
Supplementary Material
Highlights.
Host substrates identified for the conserved herpesvirus protein kinases
Common targeting of the DNA damage response (DDR) pathway by the conserved kinases
TIP60 activation mediates EBV kinase BGLF4-induced DDR and viral gene expression
TIP60 knockdown impairs β and γ herpesvirus replication.
Acknowledgments
J.Z. was supported by American Heart Association Predoctoral Fellowship 0715295U. This work is supported in part by the NIH (R01 CA30356 and R37 CA42245 to SDH, R21 CA138163 to SDH and HZ, RR020839 and R01 GM076102 to HZ, R01 EY017589 to JQ). We thank Jef Boeke and Seth Blackshaw for critical comments and suggestions and Ravit Arav-Boger and Ran He for assistance with HCMV infection. We also thank Lindsey Hutt-Fletcher for providing recombinant EBV-BX1 virus. Author Contributions: J.Z., R. L., S.D.H., and H.Z. conceived the project. J.Z., R.L., S.D.H., and H.Z. designed experiments. S.T., R.A., and G.S.H. had input into experimental design. R.L. and J.Z. performed most of the experiments. Z.X., J.L., and J.Q. performed informatics and statistical analyses. S.H. and C.W. generated the human protein arrays. G.L. and M.R.C. generated reagents. R.L., J.L., J.Z., P.D., and G.S.H. designed and performed HCMV infection assays. H.Z., R.L., J.Z. and S.D.H. wrote the manuscript.
Footnotes
Supplementary Information: Supplemental materials contain Tables S1 and S2, 6 supplemental figures with legends, and supplementary experimental procedures and references.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahsan N, Kanda T, Nagashima K, Takada K. Epstein-Barr virus transforming protein LMP1 plays a critical role in virus production. J Virol. 2005;79:4415–4424. doi: 10.1128/JVI.79.7.4415-4424.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K. Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. Cambridge: Cambridge University Press; 2007. [PubMed] [Google Scholar]
- Asai R, Kato A, Kato K, Kanamori-Koyama M, Sugimoto K, Sairenji T, Nishiyama Y, Kawaguchi Y. Epstein-Barr virus protein kinase BGLF4 is a virion tegument protein that dissociates from virions in a phosphorylation-dependent process and phosphorylates the viral immediate-early protein BZLF1. J Virol. 2006;80:5125–5134. doi: 10.1128/JVI.02674-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avvakumov N, Cote J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene. 2007;26:5395–5407. doi: 10.1038/sj.onc.1210608. [DOI] [PubMed] [Google Scholar]
- Awasthi S, Sharma A, Wong K, Zhang J, Matlock EF, Rogers L, Motloch P, Takemoto S, Taguchi H, Cole MD, et al. A human T-cell lymphotropic virus type 1 enhancer of Myc transforming potential stabilizes Myc-TIP60 transcriptional interactions. Mol Cell Biol. 2005;25:6178–6198. doi: 10.1128/MCB.25.14.6178-6198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek MC, Krosky PM, He Z, Coen DM. Specific phosphorylation of exogenous protein and peptide substrates by the human cytomegalovirus UL97 protein kinase. Importance of the P+5 position. J Biol Chem. 2002a;277:29593–29599. doi: 10.1074/jbc.M202312200. [DOI] [PubMed] [Google Scholar]
- Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and beta-amyloid precursor protein. Cell. 2002b;110:55–67. doi: 10.1016/s0092-8674(02)00809-7. [DOI] [PubMed] [Google Scholar]
- Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood MA, Venkatesan K, Xing L, Chase MR, Vazquez A, Holthaus AM, Ewence AE, Li N, Hirozane-Kishikawa T, Hill DE, et al. Epstein-Barr virus and virus human protein interaction maps. Proc Natl Acad Sci U S A. 2007;104:7606–7611. doi: 10.1073/pnas.0702332104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-Monreal GL, Tavis JE, Morrison LA. Substrate specificity of the herpes simplex virus type 2 UL13 protein kinase. Virology. 2008;374:1–10. doi: 10.1016/j.virol.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvet C, Wissler M, Brauns-Schubert P, Wang SJ, Tang Y, Sigloch FC, Mellert H, Brandenburg M, Lindner SE, Breit B, et al. Phosphorylation of Tip60 by GSK-3 Determines the Induction of PUMA and Apoptosis by p53. Mol Cell. 2011;42:584–596. doi: 10.1016/j.molcel.2011.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PW, Lin SJ, Tsai SC, Lin JH, Chen MR, Wang JT, Lee CP, Tsai CH. Regulation of microtubule dynamics through phosphorylation on stathmin by Epstein-Barr virus kinase BGLF4. J Biol Chem. 2010;285:10053–10063. doi: 10.1074/jbc.M109.044420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S. Diverse cytomegalovirus UL27 mutations adapt to loss of viral UL97 kinase activity under maribavir. Antimicrob Agents Chemother. 2009;53:81–85. doi: 10.1128/AAC.01177-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Col E, Caron C, Chable-Bessia C, Legube G, Gazzeri S, Komatsu Y, Yoshida M, Benkirane M, Trouche D, Khochbin S. HIV-1 Tat targets Tip60 to impair the apoptotic cell response to genotoxic stresses. Embo J. 2005;24:2634–2645. doi: 10.1038/sj.emboj.7600734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jesus O, Smith PR, Spender LC, Elgueta Karstegl C, Niller HH, Huang D, Farrell PJ. Updated Epstein-Barr virus (EBV) DNA sequence and analysis of a promoter for the BART (CST, BARF0) RNAs of EBV. J Gen Virol. 2003;84:1443–1450. doi: 10.1099/vir.0.19054-0. [DOI] [PubMed] [Google Scholar]
- Dirmeier U, Hoffmann R, Kilger E, Schultheiss U, Briseno C, Gires O, Kieser A, Eick D, Sugden B, Hammerschmidt W. Latent membrane protein 1 of Epstein-Barr virus coordinately regulates proliferation with control of apoptosis. Oncogene. 2005;24:1711–1717. doi: 10.1038/sj.onc.1208367. [DOI] [PubMed] [Google Scholar]
- Fu DX, Tanhehco Y, Chen J, Foss CA, Fox JJ, Chong JM, Hobbs RF, Fukayama M, Sgouros G, Kowalski J, et al. Bortezomib-induced enzyme-targeted radiation therapy in herpesvirus-associated tumors. Nat Med. 2008;14:1118–1122. doi: 10.1038/nm.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar M, Shenk T. Human cytomegalovirus inhibits a DNA damage response by mislocalizing checkpoint proteins. Proc Natl Acad Sci U S A. 2006;103:2821–2826. doi: 10.1073/pnas.0511148103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershburg E, Marschall M, Hong K, Pagano JS. Expression and localization of the Epstein-Barr virus-encoded protein kinase. J Virol. 2004;78:12140–12146. doi: 10.1128/JVI.78.22.12140-12146.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershburg E, Pagano JS. Conserved herpesvirus protein kinases. Biochim Biophys Acta. 2008;1784:203–212. doi: 10.1016/j.bbapap.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershburg E, Raffa S, Torrisi MR, Pagano JS. Epstein-Barr virus-encoded protein kinase (BGLF4) is involved in production of infectious virus. J Virol. 2007;81:5407–5412. doi: 10.1128/JVI.02398-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamirally S, Kamil JP, Ndassa-Colday YM, Lin AJ, Jahng WJ, Baek MC, Noton S, Silva LA, Simpson-Holley M, Knipe DM, et al. Viral mimicry of Cdc2/cyclin-dependent kinase 1 mediates disruption of nuclear lamina during human cytomegalovirus nuclear egress. PLoS Pathog. 2009;5:e1000275. doi: 10.1371/journal.ppat.1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Hu S, Xie Z, Onishi A, Yu X, Jiang L, Lin J, Rho HS, Woodard C, Wang H, Jeong JS, et al. Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell. 2009;139:610–622. doi: 10.1016/j.cell.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume AJ, Finkel JS, Kamil JP, Coen DM, Culbertson MR, Kalejta RF. Phosphorylation of retinoblastoma protein by viral protein with cyclin-dependent kinase function. Science. 2008;320:797–799. doi: 10.1126/science.1152095. [DOI] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- Iwahori S, Murata T, Kudoh A, Sato Y, Nakayama S, Isomura H, Kanda T, Tsurumi T. Phosphorylation of p27Kip1 by Epstein-Barr virus protein kinase induces its degradation through SCFSkp2 ubiquitin ligase actions during viral lytic replication. J Biol Chem. 2009;284:18923–18931. doi: 10.1074/jbc.M109.015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumiya Y, Izumiya C, Van Geelen A, Wang DH, Lam KS, Luciw PA, Kung HJ. Kaposi's sarcoma-associated herpesvirus-encoded protein kinase and its interaction with K-bZIP. J Virol. 2007;81:1072–1082. doi: 10.1128/JVI.01473-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S, Vande Pol S, Banerjee NS, Dutta AB, Chow LT, Dutta A. Destabilization of TIP60 by human papillomavirus E6 results in attenuation of TIP60-dependent transcriptional regulation and apoptotic pathway. Mol Cell. 2010;38:700–711. doi: 10.1016/j.molcel.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamine J, Elangovan B, Subramanian T, Coleman D, Chinnadurai G. Identification of a cellular protein that specifically interacts with the essential cysteine region of the HIV-1 Tat transactivator. Virology. 1996;216:357–366. doi: 10.1006/viro.1996.0071. [DOI] [PubMed] [Google Scholar]
- Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, et al. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 2010;463:818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- Kato K, Kawaguchi Y, Tanaka M, Igarashi M, Yokoyama A, Matsuda G, Kanamori M, Nakajima K, Nishimura Y, Shimojima M, et al. Epstein-Barr virus-encoded protein kinase BGLF4 mediates hyperphosphorylation of cellular elongation factor 1delta (EF-1delta): EF-1delta is universally modified by conserved protein kinases of herpesviruses in mammalian cells. J Gen Virol. 2001;82:1457–1463. doi: 10.1099/0022-1317-82-6-1457. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Kato K. Protein kinases conserved in herpesviruses potentially share a function mimicking the cellular protein kinase cdc2. Rev Med Virol. 2003;13:331–340. doi: 10.1002/rmv.402. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Kato K, Tanaka M, Kanamori M, Nishiyama Y, Yamanashi Y. Conserved protein kinases encoded by herpesviruses and cellular protein kinase cdc2 target the same phosphorylation site in eukaryotic elongation factor 1delta. J Virol. 2003;77:2359–2368. doi: 10.1128/JVI.77.4.2359-2368.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig R, Stertz S, Zhou Y, Inoue A, Hoffmann HH, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y, et al. Human host factors required for influenza virus replication. Nature. 2010;463:813–817. doi: 10.1038/nature08699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudoh A, Daikoku T, Ishimi Y, Kawaguchi Y, Shirata N, Iwahori S, Isomura H, Tsurumi T. Phosphorylation of MCM4 at sites inactivating DNA helicase activity of the MCM4-MCM6-MCM7 complex during Epstein-Barr virus productive replication. J Virol. 2006;80:10064–10072. doi: 10.1128/JVI.00678-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudoh A, Fujita M, Zhang L, Shirata N, Daikoku T, Sugaya Y, Isomura H, Nishiyama Y, Tsurumi T. Epstein-Barr virus lytic replication elicits ATM checkpoint signal transduction while providing an S-phase-like cellular environment. J Biol Chem. 2005;280:8156–8163. doi: 10.1074/jbc.M411405200. [DOI] [PubMed] [Google Scholar]
- Kudoh A, Iwahori S, Sato Y, Nakayama S, Isomura H, Murata T, Tsurumi T. Homologous recombinational repair factors are recruited and loaded onto the viral DNA genome in Epstein-Barr virus replication compartments. J Virol. 2009;83:6641–6651. doi: 10.1128/JVI.00049-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CP, Chen JY, Wang JT, Kimura K, Takemoto A, Lu CC, Chen MR. Epstein-Barr virus BGLF4 kinase induces premature chromosome condensation through activation of condensin and topoisomerase II. J Virol. 2007;81:5166–5180. doi: 10.1128/JVI.00120-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CP, Huang YH, Lin SF, Chang Y, Chang YH, Takada K, Chen MR. Epstein-Barr virus BGLF4 kinase induces disassembly of the nuclear lamina to facilitate virion production. J Virol. 2008;82:11913–11926. doi: 10.1128/JVI.01100-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemercier C, Legube G, Caron C, Louwagie M, Garin J, Trouche D, Khochbin S. Tip60 acetyltransferase activity is controlled by phosphorylation. J Biol Chem. 2003;278:4713–4718. doi: 10.1074/jbc.M211811200. [DOI] [PubMed] [Google Scholar]
- Li R, Hayward SD. The Ying-Yang of the virus-host interaction: control of the DNA damage response. Future Microbiology. 2011;6:379–383. doi: 10.2217/fmb.11.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li RF, Shang Y, Liu D, Ren ZS, Chang Z, Sui SF. Differential ubiquitination of Smad1 mediated by CHIP: implications in the regulation of the bone morphogenetic protein signaling pathway. J Mol Biol. 2007;374:777–790. doi: 10.1016/j.jmb.2007.09.082. [DOI] [PubMed] [Google Scholar]
- Lilley CE, Carson CT, Muotri AR, Gage FH, Weitzman MD. DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc Natl Acad Sci U S A. 2005;102:5844–5849. doi: 10.1073/pnas.0501916102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley CE, Chaurushiya MS, Boutell C, Landry S, Suh J, Panier S, Everett RD, Stewart GS, Durocher D, Weitzman MD. A viral E3 ligase targets RNF8 and RNF168 to control histone ubiquitination and DNA damage responses. Embo J. 2010a;29:943–955. doi: 10.1038/emboj.2009.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley CE, Chaurushiya MS, Weitzman MD. Chromatin at the intersection of viral infection and DNA damage. Biochim Biophys Acta. 2010b;1799:319–327. doi: 10.1016/j.bbagrm.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q, Hagemeier SR, Fingeroth JD, Gershburg E, Pagano JS, Kenney SC. The Epstein-Barr virus (EBV)-encoded protein kinase, EBV-PK, but not the thymidine kinase (EBV-TK), is required for ganciclovir and acyclovir inhibition of lytic viral production. J Virol. 2010;84:4534–4542. doi: 10.1128/JVI.02487-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SM, Cannon JS, Tanhehco YC, Hamzeh FM, Ambinder RF. Induction of Epstein-Barr virus kinases to sensitize tumor cells to nucleoside analogues. Antimicrob Agents Chemother. 2001;45:2082–2091. doi: 10.1128/AAC.45.7.2082-2091.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitin PA, Yan CM, Forte E, Bocedi A, Tourigny JP, White RE, Allday MJ, Patel A, Dave SS, Kim W, et al. An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein-Barr virus transformation of primary human B cells. Cell Host Microbe. 2010;8:510–522. doi: 10.1016/j.chom.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prichard MN. Function of human cytomegalovirus UL97 kinase in viral infection and its inhibition by maribavir. Rev Med Virol. 2009;19:215–229. doi: 10.1002/rmv.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prichard MN, Gao N, Jairath S, Mulamba G, Krosky P, Coen DM, Parker BO, Pari GS. A recombinant human cytomegalovirus with a large deletion in UL97 has a severe replication deficiency. J Virol. 1999;73:5663–5670. doi: 10.1128/jvi.73.7.5663-5670.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- Rechter S, Scott GM, Eickhoff J, Zielke K, Auerochs S, Muller R, Stamminger T, Rawlinson WD, Marschall M. Cyclin-dependent Kinases Phosphorylate the Cytomegalovirus RNA Export Protein pUL69 and Modulate Its Nuclear Localization and Activity. J Biol Chem. 2009;284:8605–8613. doi: 10.1074/jbc.M805693200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitsma JM, Savaryn JP, Faust K, Sato H, Halligan BD, Terhune SS. Antiviral Inhibition Targeting the HCMV Kinase pUL97 Requires pUL27-Dependent Degradation of Tip60 Acetyltransferase and Cell-Cycle Arrest. Cell Host Microbe. 2011;9:103–114. doi: 10.1016/j.chom.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Romaker D, Schregel V, Maurer K, Auerochs S, Marzi A, Sticht H, Marschall M. Analysis of the structure-activity relationship of four herpesviral UL97 subfamily protein kinases reveals partial but not full functional conservation. J Med Chem. 2006;49:7044–7053. doi: 10.1021/jm060696s. [DOI] [PubMed] [Google Scholar]
- Sapountzi V, Logan IR, Robson CN. Cellular functions of TIP60. Int J Biochem Cell Biol. 2006;38:1496–1509. doi: 10.1016/j.biocel.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Schepers A, Pich D, Mankertz J, Hammerschmidt W. cis-acting elements in the lytic origin of DNA replication of Epstein-Barr virus. J Virol. 1993;67:4237–4245. doi: 10.1128/jvi.67.7.4237-4245.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira SD, Gat-Viks I, Shum BO, Dricot A, de Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE, et al. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139:1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibaki T, Suzutani T, Yoshida I, Ogasawara M, Azuma M. Participation of type I interferon in the decreased virulence of the UL13 gene-deleted mutant of herpes simplex virus type 1. J Interferon Cytokine Res. 2001;21:279–285. doi: 10.1089/107999001300177466. [DOI] [PubMed] [Google Scholar]
- Shin YC, Nakamura H, Liang X, Feng P, Chang H, Kowalik TF, Jung JU. Inhibition of the ATM/p53 signal transduction pathway by Kaposi's sarcoma-associated herpesvirus interferon regulatory factor 1. J Virol. 2006;80:2257–2266. doi: 10.1128/JVI.80.5.2257-2266.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squatrito M, Gorrini C, Amati B. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 2006;16:433–442. doi: 10.1016/j.tcb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Sullivan V, Talarico CL, Stanat SC, Davis M, Coen DM, Biron KK. A protein kinase homologue controls phosphorylation of ganciclovir in human cytomegalovirus-infected cells. Nature. 1992;358:162–164. doi: 10.1038/358162a0. [DOI] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarakanova VL, Leung-Pineda V, Hwang S, Yang CW, Matatall K, Basson M, Sun R, Piwnica-Worms H, Sleckman BP, Virgin HWt. Gamma-herpesvirus kinase actively initiates a DNA damage response by inducing phosphorylation of H2AX to foster viral replication. Cell Host Microbe. 2007;1:275–286. doi: 10.1016/j.chom.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida J, Yasui T, Takaoka-Shichijo Y, Muraoka M, Kulwichit W, Raab-Traub N, Kikutani H. Mimicry of CD40 signals by Epstein-Barr virus LMP1 in B lymphocyte responses. Science. 1999;286:300–303. doi: 10.1126/science.286.5438.300. [DOI] [PubMed] [Google Scholar]
- Wang JT, Chuang YC, Chen KL, Lu CC, Doong SL, Cheng HH, Chen YL, Liu TY, Chang Y, Han CH, et al. Characterization of Epstein-Barr virus BGLF4 kinase expression control at the transcriptional and translational levels. J Gen Virol. 2010;91:2186–2196. doi: 10.1099/vir.0.019729-0. [DOI] [PubMed] [Google Scholar]
- Wolf DG, Courcelle CT, Prichard MN, Mocarski ES. Distinct and separate roles for herpesvirus-conserved UL97 kinase in cytomegalovirus DNA synthesis and encapsidation. Proc Natl Acad Sci U S A. 2001;98:1895–1900. doi: 10.1073/pnas.98.4.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalani S, Holley-Guthrie E, Kenney S. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc Natl Acad Sci U S A. 1996;93:9194–9199. doi: 10.1073/pnas.93.17.9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Liao G, Shan L, Zhang J, Chen MR, Hayward GS, Hayward SD, Desai P, Zhu H. Protein array identification of substrates of the Epstein-Barr virus protein kinase BGLF4. J Virol. 2009;83:5219–5231. doi: 10.1128/JVI.02378-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.