Abstract
Peptide-bond isosteres can enable a deep interrogation of the structure and function of a peptide or protein by amplifying or attenuating particular chemical properties. In this minireview, the electronic, structural, and conformational attributes of four such isosteres—thioamides, esters, alkenes, and fluoroalkenes—are examined in detail. In particular, the ability of these isosteres to partake in noncovalent interactions is compared with that of the peptide bond. The consequential perturbations provide a useful tool for chemical biologists to reveal new structure–function relationships, and to endow peptides and proteins with desirable attributes.
Keywords: alkene, ester, fluoroalkene, hydrogen bond, n→π* interaction, thioamide
Introduction
Two centuries have now passed since amino acids were shown to be products of the hydrolysis of proteins.[1] By 1900, most of the 20 canonical amino acids were known. Unclear, however, was the nature of the linkage between these amino acids. In 1902, at the Karlsbad meeting of the Gesellschaft Deutscher Naturforscher und Ärzte, Franz Hofmeister said:[2]
The type of condensation described here through formation of –CO–NH–CH= groups may thus explain both the building up of protein substances in the organism, as well as their breakdown in the intestinal tract and in the tissues. On the basis of these given facts one may therefore consider the proteins as for the most part arising by condensation of α-amino acids, whereby the linkage through the group –CO–NH–CH= has to be regarded as the regularly recurring one.
At the same meeting, Emil Fischer both espoused the Hofmeister theory and introduced the term “peptide”,[3] which became associated with the bonds between constituent amino acids.
Peptide bonds undergo uncatalyzed hydrolysis with a t½ of 400 years,[4] a value that greatly exceeds the lifespan of most organisms. Nonetheless and as noted by Hofmeister,[2] peptide bonds can be much less stable in vivo, due to the action of proteases. This vulnerability, along with a desire to modulate the structure and function of proteins, has motivated chemical biologists to develop surrogates of the peptide bond.
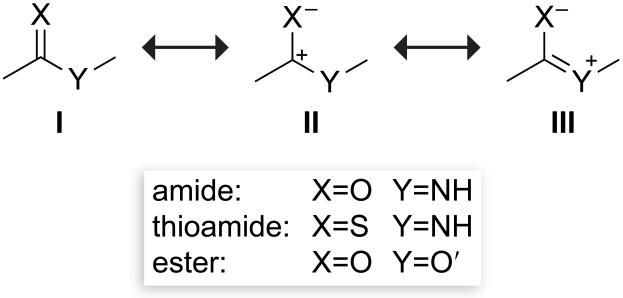
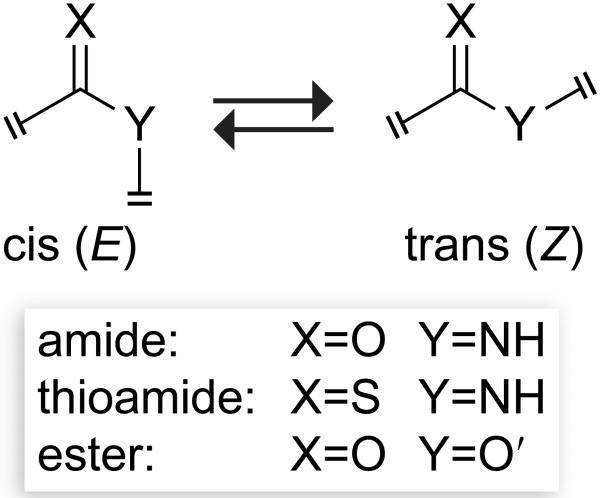
The peptide bond is a resonance hybrid of three structures (Scheme 1 with X=O, Y=NH).[5] The planarity of the peptide bond stems from the delocalization of the nitrogen lone pair into the antibonding orbital (π*) of the carbonyl group, which is reflected in resonance structure III. This delocalization is primarily responsible for the rotational barrier between the cis and trans isomers of the peptide bond, which can limit the rate of protein folding.[6] A survey of peptide-bond surrogates (Scheme 2) reveals that many are neither isosteric nor isoelectronic to the peptide bond. Not surprisingly, replacing peptide bonds with some of these surrogates can lead to a considerable decrease in biological activity.
Scheme 1.
Canonical resonance structures of peptide-bond isosteres.
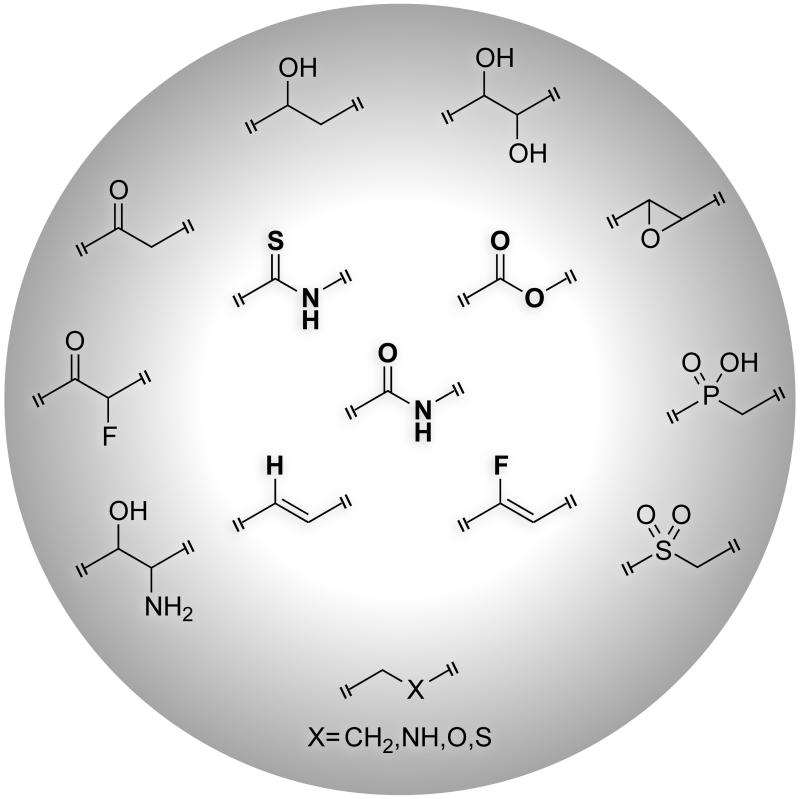
Scheme 2.
Structure of a peptide bond (center) and some surrogates. This minireview is focused on the four isosteres in the inner circle.
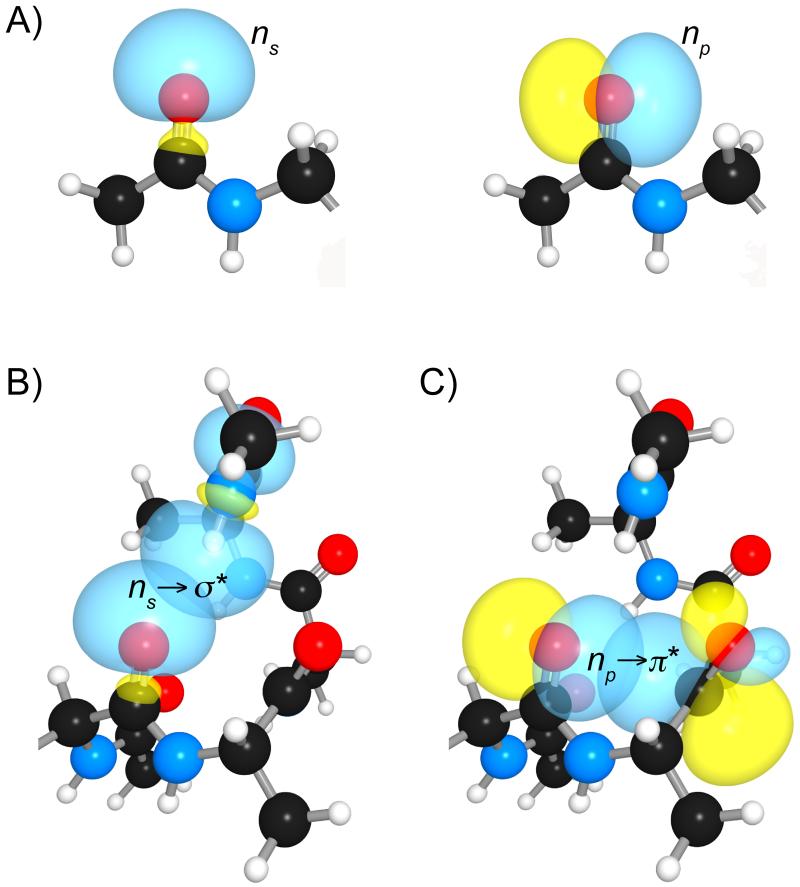
The oxygen lone pairs within a peptide bond can beget noncovalent interactions in peptides and proteins. The orientation and energy of the lone pairs vitally affect these noncovalent interactions. In discord with the conventional picture from VSEPR theory, the oxygen lone pairs are not located in degenerate sp2 hybrid orbitals.[7] Rather, the lower energy lone pair (ns) is considerably s-rich and directed along the C=O bond, whereas the other, higher energy lone pair (np) is p-rich and located orthogonal to the C=O bond (Figure 1A). In many protein secondary structures, the C=O…H-N hydrogen bond between two amide groups involves the delocalization of the s-rich lone pair (ns) into the antibonding orbital (π*) of the N–H bond (Figure 1B).[8] The other, p-rich lone pair (np) delocalizes into the antibonding orbital of the adjacent carbonyl group to give rise to a carbonyl–carbonyl interaction, which we term as an n→π* interaction (Figure 1C).[9] We reasoned that a good peptide-bond mimic should participate in most of these interactions within a polypeptide chain. In this minireview, we explore the electronic and structural properties of four peptide-bond isosteres that resemble closely an amide bond: thioamide, ester, alkene, and fluoroalkene (Scheme 2). Unlike other surrogates, these peptide-bond isosteres are planar and retain two sp2-hybridized atoms in the main chain. We evaluate and compare, in particular, the ability of these four isosteres to partake in noncovalent interactions, and comment on the ensuing consequences.
Figure 1.
(A) Lone pairs of an amide oxygen, and their partial covalent bonds in an α-helix: (B) Hydrogen bond, and (C) n→π* interaction.[9b]
Thioamides
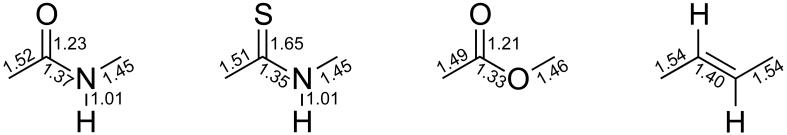
A thioamide is an “isostere” of an amide in the strictest sense, having the same number of atoms and valence electrons arranged in the same manner.[10] (Other chalcogen congeners, such as selenoamides and telluroamides, are unstable, and their incorporation into peptides poses a considerable synthetic challgenge.) The C=S bond in a thioamide is, however, longer than the corresponding C=O bond in an amide; whereas the C–N bond is shorter in a thioamide (Scheme 3). The shorter C–N bond in thioamides arises from the larger contribution from resonance structure III (Scheme 1) in thioamides.[11] On the basis of electronegativity alone, structure III might seem to be more important for amides. Yet, Wiberg and coworkers have argued otherwise.[12] The carbonyl oxygen of an amide is considerably more anionic than is the thiocarbonyl sulfur. Thus, the energetic penalty for its further polarization due to charge transfer from nitrogen is higher for amides. Moreover, the larger size of sulfur better accommodates negative charge than does oxygen.[12] The larger contribution from structure III in thioamides explains two other structural deviations from amides. First, it results in a stiffer out-of-plane bending vibration mode associated with the nitrogen in the thioamides.[12] Second, it increases the rotational barrier across the C-N bond in thioamides over amides by 2–3 kcal/mol.[13] Peptidyl prolyl isomerases, which catalyze the rotation across the C–N bond, are inhibited by substrates containing a thioamide, presumably due to its large rotational barrier.[14]
Scheme 3.
Bond lengths (Å) in peptide-bond isosteres.[17]
Thioamides, like amides, can exist as cis and trans isomers (Scheme 4). Despite the larger rotational barrier, the photoisomerization of a thioamide is more facile than is that of an amide.[15] The photodecomposition during photoisomerization is reduced considerably in thioamides, as the π→π* electronic transition, which is required for photoisomerization, is red-shifted in thioamides (260 nm) relative to amides (200 nm).[16] Photoswitching has been used to increase the population of the cis isomer of prolyl peptide bonds. This photoswitching is fast (t½ < 600 ps) and efficient, and thermal equilibration is slow (t½ > 10 min). Unlike the tertiary thioamide of the prolyl peptide bond,[15a] a secondary thioamide can be switched from the trans to cis isomer, and vice versa, because the ultraviolet–visible absorption spectra of their cis and trans isomers do not overlap significantly.[15b, 15c] For example, photoswitching was used to modulate the enzymatic activity of a semisynthetic ribonuclease S containing a thioamide.[18] The light-induced trans-to-cis isomerization led to a complete loss of enzymatic activity, which was reinstated after thermally induced cis-to-trans isomerization.
Scheme 4.
Cis–trans equilibrium of a peptide bond and isosteres.
Thioamides are poor hydrogen-bond acceptors, but good hydrogen-bond donors. The O=C–N–H…S=C hydrogen bond is weaker than the O=C–N–H…O=C hydrogen bond by ~1.6 kcal/mol, whereas the S=C–N–H…O=C hydrogen bond is stronger than the O=C–N–H…O=C hydrogen bond by ~2 kcal/mol.[19a, 17d, 19b] Interestingly, the proton affinity of sulfur (203 kcal/mol) in a thioamide is greater than that of oxygen (199 kcal/mol) in an amide.[19a] Yet, thioamides are weaker hydrogen-bond acceptors than amides because this enthalpic advantage is negated by the unfavorable entropy that accompanies hydrogen-bond formation in thioamides. Although thioamides are weak hydrogen-bond acceptors, the sulfur in thioamide is a superior donor for an n→π* interaction.[9a] Additionally, the pKa of Cα–H in thioamides is lower than that of amides,[20] suggesting that S=C–Cα–H…O hydrogen bonds are stronger than O=C–Cα–H…O hydrogen bonds.
The large size of sulfur, longer C=S bond, and skewed hydrogen-bonding capacity alter the conformational landscape of peptides containing thioamides. Computational studies indicate that the conformational space of the residues both preceding and following the thioamide linkage is reduced considerably.[21] Specifically, the thioamide bond biases the conformation of the subsequent residue towards more negative □ (C’i−1–Ni–Cαi–C’i) and more positive ψ (Ni–Cαi–C’i–Ni+1) dihedral angles.
The effects of thioamide-substitution on the conformation and stability of α- and 310-helices, in particular, have been examined computationally by Burgess and coworkers.[22] α-Helices, 310-helices, and β-sheets have O=C–N–H…O=C hydrogen bonds (2.1 Å) that are shorter than typical O=C–N–H…S=C hydrogen bonds (2.7 Å). It follows that incorporation of thioamides in these canonical structures would engender strong Pauli repulsion between the sulfur lone pairs and the σ-bonding orbital of the N–H bond.[9c] Additionally, the carbonyl dipoles in the adjacent carbonyl groups are in a repulsive orientation in an α-helix.[23] As thioamides have a larger dipole moment than do amides, such repulsions would be more intense. Indeed, Burgess and coworkers reported alteration in the helical parameters to accommodate the longer O=C–N–H…S=C hydrogen bond. Additionally, experimental studies on alanine-based model peptides indicated that a single thioamide substitution can result in helix destabilization by ~1.7 kcal/mol.[24] Interestingly, Miwa and coworkers found that thioamide substitution at slightly “widened” locations of an α-helix can increase stability.[25] These widened locations presumably do not engender strong n)(σ Pauli repulsion between the sulfur lone pair and the σ-bonding orbital of the N–H, but have a stronger hydrogen bond between the thioamide N–H bond and the amide carbonyl acceptor. Additionally, the thioamide likely endows greater conformational stabilization via the n→π* interaction. Miwa and coworkers have also shown that non–hydrogen-bonded turn positions in β-hairpins are compatible with thioamide substitution.[26]
The isosteric replacement of an amide with a thioamide has been exploited to evaluate the contribution of hydrogen bonds to an important protein–ligand interaction.[27] The major histocompatability complex class II protein interacts with its peptidic ligands through a series of hydrogen bonds. One of the hydrogen bonds is between the main-chain oxygen of the peptidic substrate and the side chain of a histidine residue (βHis81). Replacement of that main-chain oxygen with sulfur resulted in a 30-fold increase in the rate of ligand dissociation, in support of the putative role of the hydrogen bond.
A natural thioamide linkage occurs in methyl-coenzyme M reductase, which catalyzes the final step in methane synthesis in a methanogenic archae.[28] An examination of the enzymic crystal structure indicates that the thiocarbonyl group engages in a strong n→π* interaction with the side-chain amide of asparagine α501, orienting that amide to form a hydrogen bond with the sulfhydryl group of coenzyme B. The thioamide sulfur could also act as a one-electron relay during redox catalysis.[28]
Esters
The geometric parameters of an ester are comparable to those of an amide (Scheme 3).[17a] Analogous to amidic resonance, the lone pair of oxygen O’ delocalizes into the antibonding orbital (π*) of the ester carbonyl group.[5b] As with amides and thioamides, this electron delocalization enforces planarity and begets stable cis and trans isomers. The lower rotational barrier in esters (11 kcal/mol) compared with that of amides (20 kcal/mol) can be attributed to the weaker charge transfer from the bridging oxygen (O’) to the carbonyl group.[29] Like amides, the trans conformation of an ester is more stable than its cis conformation. The energy difference between the cis and trans conformations of methyl formate has been estimated to be ΔH° = (4.75 ± 0.19) kcal/mol using matrix-isolation infrared spectroscopy.[30] Delocalization of an O’ lone pair into the antibonding orbital (π*) of the carbonyl group likely contributes to this differential stability.[31]
Amides and esters differ in their ability to form hydrogen bonds, which has important consequences for protein folding and stability.[32] Peptide bonds between all proteinogenic amino acids, except proline, can both donate and accept hydrogen bonds. Esters, however, cannot donate a hydrogen bond and are only weak hydrogen-bond acceptors.[33] This weakness is also reflected in their relatively high volatility and low solvation. For example, replacing the amide bond in the Phe–Phe dipeptide with an ester reduces the free energy of transfer from water to octanol by 0.5 kcal/mol.[34]
An amide-to-ester substitution entails more than diminished hydrogen bonding. In many secondary structures, the distance between the main-chain nitrogen of a hydrogen-bond donor and the main-chain oxygen of a hydrogen-bond acceptor is ~3.0 Å. In ester variants, the corresponding distance increases to 3.1–4.0 Å.[35] This distortion presumably arises because an attractive interaction in amides—the hydrogen bond—is replaced with a repulsive interaction between O’ and the proximal main-chain oxygen. Kelly and coworkers have reported this repulsion to be of ~0.3 kcal/mol in an antiparallel β-sheet structure and ~0.4 kcal/mol in an α-helix.[34, 36]
The effects of amide-to-ester substitution on the stability of common secondary structures have been explored both computationally and experimentally. Cieplak and Surmeili calculated that the instability arising from replacing alanine residues with lactic acid depends on the secondary structure: type I β-turns < 310 helices < α-helices < β-sheets.[37] This instability was attributed to the loss of the hydrogen bond, a decrease in Lewis basicity of the C=O bond, and the aforementioned O’)(O=C repulsion. Ester groups are known to be tolerated in helices, but can cause a change in hydrogen-bonding pattern. In an α- and 310-helix, a hydrogen bond exists between the C=O of residue i and the H–N of residue i + 4 and i + 3, respectively. Replacing amino acid i + 4 in an α-helix with a hydroxyl acid can induce residue i to form a hydrogen bond with residue i + 3 instead.[35d]
Ester substitution has also been used to study the role of hydrogen bonding on the conformational stability of several proteins. For example, we have used amide-to-ester substitution to evaluate the strength of the repetitive interstrand hydrogen bonds in the collagen triple helix.[38] The strands of collagen have a repeating sequence: Xaa–Yaa–Gly, where Xaa and Yaa are often proline or proline derivatives. The central axis of a collagen triple helix has a ladder of interstrand hydrogen bonds formed by the C=O of each Xaa residue and the H–N of each glycine. Amide-to-ester substitution revealed that each of these hydrogen bonds contributes ~2.0 kcal/mol to triple-helix stability. Likewise, Kelly and coworkers have studied the effects of amide-to-ester substitution in a Pin 1 WW domain, which folds into a twisted three-stranded β-sheet structure.[34] Deletion of a hydrogen bond in the hydrophobic core of this structure decreases stability by ~0.8 kcal/mol. Additionally, Koksch and coworkers have examined the effects of amide-to-ester substitution on the stability of coiled-coil proteins.[39] They found that the contribution of hydrogen bonding to stability depends on the local environment. Specifically, the contributions of solvent-exposed hydrogen bonds are minimal, but those of buried hydrogen bonds are substantial.[40] Finally, we note that crystal structures have shown that amide-to-ester substitutions lead to a slight distortion of the main-chain geometry.[41]
Other attributes of esters are also relevant in this context. They are vulnerable to hydrolysis. (Esters are, however, 102-fold more resistant than thioesters to amine and thiolate nucleophiles.[42]) In addition, their torsional flexibility engenders amorphicity. These detriments, along with their weaker intermolecular associations, make esters less attractive linkages within compact proteins.[43]
Alkenes
Alkene isosteres are another genre of peptide-bond surrogates. In an alkene isostere, a C=C bond replaces the C–N bond (Scheme 2). On geometric grounds alone, an alkene isostere mimics closely the salient features of a peptide bond. For example, the distance between Ciα and Ci+1α in both amides and their alkene isostere is ~3.8 Å,[17b, 17c] and other bond lengths and angles are likewise comparable (Scheme 3). Nevertheless, the electronic and electrostatic properties of the amide and alkene isostere differ dramatically. The dipole moment of an alkene isostere (0.1 D) is much lower than that of an amide bond (3.6 D).[44] Thus, dipolar interactions involving the alkene isostere will be much weaker. Additionally, its lack of a heteroatom precludes important noncovalent interactions. For example, the alkene isostere can be neither a strong hydrogen-bond donor nor a hydrogen-bond acceptor. Hence, the alkene isostere is hydrophobic, which is reflected in its desolvation energies. Kelly and coworkers determined the water-to-octanol transfer free energies to be −1.4 kcal/mol for a Phe–Phe dipeptidic system and −2.4 kcal/mol for its alkene isostere.[34] The alkenic C–H bond can act as only a weak hydrogen-bond donor, and reduces the strength of a Cα–H…O hydrogen bonds. We note as well that a C–H…O hydrogen bond differs from a N–H…O hydrogen bonds not only in strength, but also in nature.[46] For example, a hydrogen-bonded amide N–H bond shows bond lengthening and a red shift in infrared stretching frequency, whereas a hydrogen-bonded C–H bond exhibits bond shortening and a blue shift in infrared stretching frequency.[46]
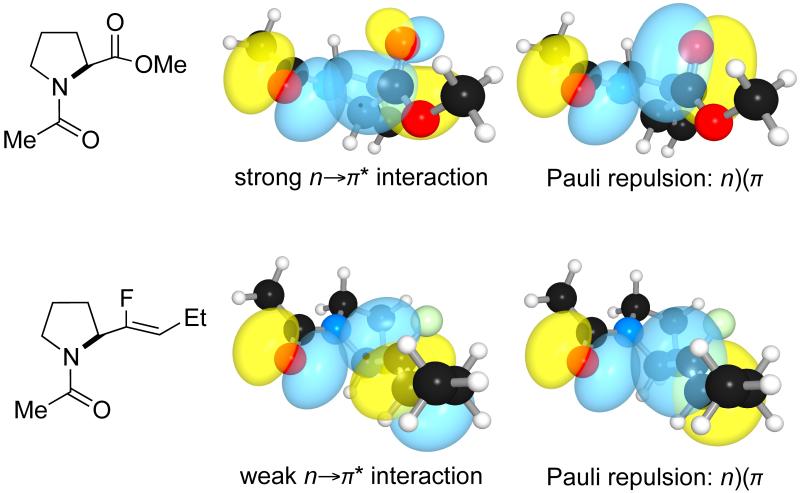
An alkene isostere cannot be a donor for an n→π* interaction and is only a weak acceptor.[9c] At least two factors make this isostere a weak acceport for an n→π* interaction. A strong n→π* interaction requires a small energy gap between the lone pair (n) of the donor and the π* orbital of the acceptor. The energy gap between a carbonyl lone pair (n) and the π* orbital of an alkene is ~10-fold greater than the analogous energy difference when the π* orbital of an amide carbonyl group is the acceptor. Moreover, the orientation of the π* orbital of the amide carbonyl and that of the π* orbital of the alkene differ substantially relative to the main-chain atoms—the π* orbital of the carbonyl is located along the C=O bond, whereas the π* orbital of the alkene is along the C=C bond. This altered orientation reduces considerably the overlap of the donor lone pair (n) and the acceptor π* orbital.
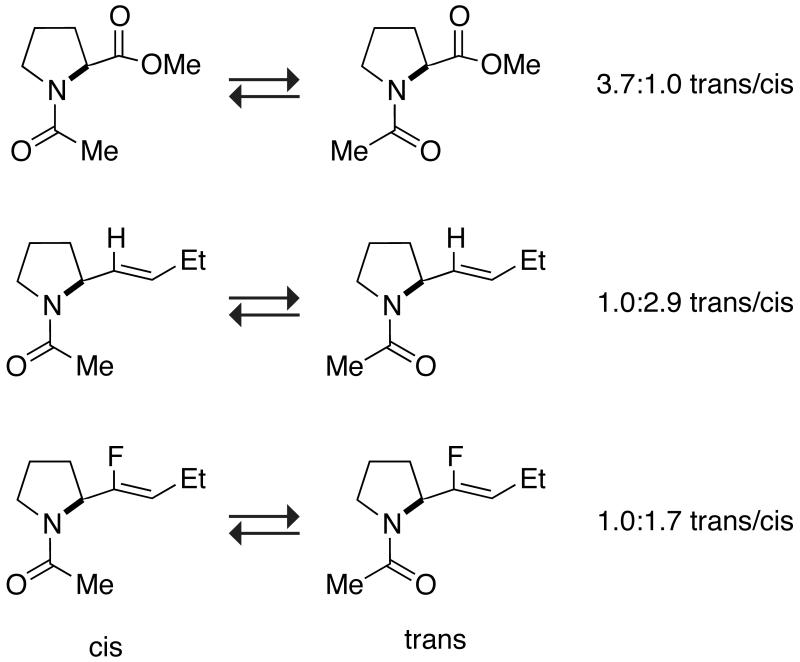
Pauli repulsion between the lone pair of a donor carbonyl group (n) and a π-orbital exists in both an amide and its alkene isostere. Only in an amide, however, is this destabilizing interaction balanced by a stabilizing n→π* interaction. We demonstrated this dichotomy by using a Xaa–Pro dipeptidic system, wherein the equilibrium constant (Ktrans/cis) between the cis and trans isomers reports on the donor–acceptor interaction because the stabilizing n→π* interaction and the destabilizing n)(π Pauli repulsion can operate only in the trans conformation of this system (Figure 2).[47] We found that the trans isomer is more stable than the cis in the ester, but the cis isomer is more stable than the trans in the alkene isostere (Scheme 5). Miller and coworkers have also observed the destabilizing effect of n)(π Pauli repulsion in a β-turn peptidic model system (Scheme 6).[45] This β-turn is stabilized by a hydrogen bond, allylic strain across the Pro–D-Val amide bond, a dipole-dipole interaction, and an n→π* interaction. Replacing the Pro–D-Val amide bond with an alkene isostere disrupts the structure of this particular β-turn by reducing the population of the trans conformation across the Xaa–Pro bond from 100 to 78%.[45]
Figure 2.
n→π* interaction and n)(π Pauli repulsion in the trans isomer of Xaa–Pro isosteres.[9a]
Scheme 5.
Cis–trans equilibrium of an Xaa–Pro peptide bond and isosteres.[9c]
Scheme 6.
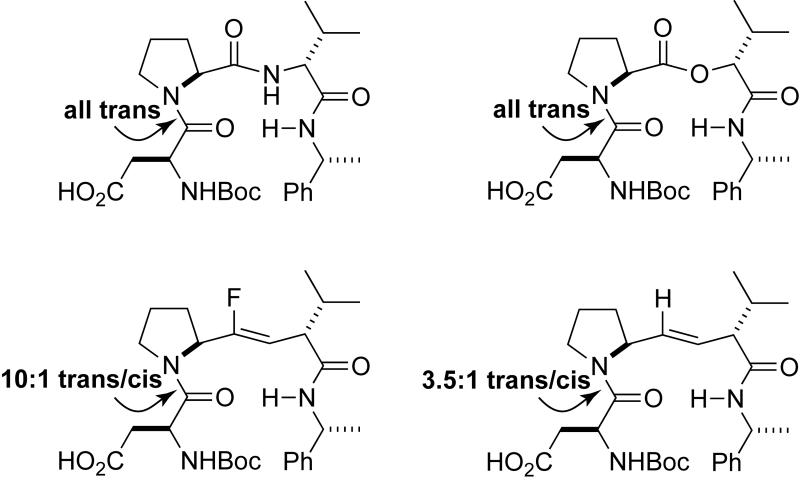
Cis–trans equilibrium of a peptide bond and isosteres in a β-turn.[45]
Other forces can contribute to the conformation of alkene isosteres. For example, A1,3 and A1,2 strain restricts the conformational space in tetra- and trisubstituted alkene isosteres, and has been used by Gellman, Wipf, and their coworkers to create β-turn mimics.[48a, 44, 48b, 48c] Interestingly, the disubstituted structures have a more open bend, clearly implicating steric effects in limiting accessible □,ψ conformational space. Analogously, Hoffman and coworkers have employed gauche pentane interference in the design of β-turn mimics.[49] Some of these β-turn mimics have been shown to be strong nucleators of β-sheet formation in a Pin 1 WW domain.[50]
Despite its success in mimicking a peptide bond in a β-turn context, alkene isosteres have proven to be poor mimics in other systems. The mimicry has been studied most extensively in the collagen triple helix.[38, 51] In unfolded collagen strands, prolyl peptide bonds exist as both cis and trans isomers, whereas they are only in the trans conformation in a triple helix. Accordingly, substitution of a prolyl peptide bond with its E-alkene isostere should increase triple-helix stability by preorganizing the prolyl peptide bond in the requisite trans conformation. Yet, replacing any of the three peptide bonds in the triple repeat with an E-alkene decreases triple-helix stability.[38, 51] This observation is consistent with the loss of a favorable n→π* interaction and the gain of unfavorable n)(π Pauli repulsion (Figure 2).[9c] In studies unrelated to collagen, Oishi and coworkers reported that the substitution of a peptide bond with an alkene isostere leads to severe disruption of α-helical structure,[52] which can be attributed to the loss of hydrogen bonds and n→π* interactions and the gain of disruptive n)(π Pauli repulsion.
The insurmountable rotational barrier between the cis and the trans isomers of an alkene isostere has been exploited by Etzkorn and coworkers.[53] Enzymes can have a higher affinity for the cis or trans isomers of their peptidic substrates. For example, Etzkorn and coworkers synthesized alkene isosteres that mimic cis and trans prolyl peptide bonds and demonstrated that Pin 1 prefers to bind the cis isostere. Both alkene and fluoroalkene isosteres have been used to study the activity of peptidyl prolyl isomerases.[54]
The inability of the alkene isosteres to form hydrogen bonds has been employed by Kelly and coworkers to probe the requirements of Alzheimer’s amyloidogenesis.[34, 55] It has been argued that the main-chain hydrogen bonds are important for the formation of the cross–β-sheet formed by the Aβ peptide in amyloid fibrils. Accordingly, deleting the main-chain hydrogen bond by replacing a peptide bond with an alkene isostere could deter fibril formation. Indeed, Aβ peptides containing the alkene isostere do not form fibrillar aggregates.
Fluoroalkenes
In a fluoroalkene isostere, one of the olefinic hydrogens of the alkene isostere is replaced with fluorine (Scheme 2). The highly electronegative fluorine mimics the carbonyl oxygen of the amide and endows a fluoroalkene with a sizeable dipole moment (1.4 D).[44] Despite this enhancement, a fluoroalkene isostere is still unable to mimic many essential features of a peptide bond. For example, the fluoroalkene isostere is neither a good hydrogen-bond donor nor a good hydrogen-bond acceptor, which makes it considerably hydrophobic. In addition, the fluoroalkene isostere remains a poor acceptor for an n→π* interaction, and prefers a cis prolyl peptide bond in the aforementioned Xaa–Pro dipeptidic system (Scheme 5).[9c]
Like an alkene isostere, a fluoroalkene disrupts β-turn structures by reducing the population of the trans conformation across the Xaa–Pro bond (Scheme 6).[45] Again, the loss of the stabilizing n→π* interaction and gain of a destabilizing n)(π repulsion is detrimental to β-turn structure. Fluoroalkene isosteres, like alkene isosteres, also disrupt α-helical conformation, presumably due to their inability to participate in hydrogen bonds and n→π* interactions, and also because of their inherent n)(π Pauli repulsion (Figure 2).[52]
The Z- and E-fluoroalkene isosteres have beend used to mimic the trans and cis isomers of peptide bonds. For example, in a peptide nucleic acid (PNA), a tertiary amide links a peptide-like main chain to side-chain nucleobases. This amide has two isomers. Leumann and coworkers replaced this amide with fluoroalkene and alkene isosteres, and found that both the E- and Z-olefinic peptide nucleic acids (OPAs) bound to DNA with the same affinity.[56] This surprising result indicated that both cis and trans isomers can form a duplex with DNA. In addition, the OPAs, unlike PNAs, do not form a triplex and bind preferentially in the parallel mode to DNA. Nonetheless, the duplexes of OPAs and fluoroalkenic peptide nucleic acid (F-OPAs) with DNA are less stable than the corresponding PNA:DNA duplex. Accordingly, an aspect of the tertiary amide is important for PNA:DNA duplex stability. In addition, the OPAs show a higher tendency to self-associate, presumably due to their enhanced hydrophobicity. A close examination of the crystal structures of PNA:PNA and PNA:DNA duplexes reveals that the carbonyl group of the tertiary amide engages in a lone pair-π interaction[57] with the aromatic ring of the nucleobases. Such a stabilizing interaction would be less significant for the fluoroalkene isostere and absent from the alkene isostere.
Wipf and coworkers have argued that a trifluoromethyl group is a better electrostatic mimic of the carbonyl oxygen than is the fluorine or hydrogen of the fluoroalkene and alkene isosteres. This argument is supported by the dipole moment of the trifluoromethylalkene isostere (2.3 D) being closer to that of the amide (3.6 D). Yet, incorporating a trifluoromethyl group in the cyclodecapeptide gramicidin led to considerable structural perturbation as revealed by circular dichroism spectroscopy and X-ray crystallography.[58] Apparently, the trifluoromethyl group was not well accommodated in the middle of the β-hairpin. (A-Values indicate that a trifluoromethyl group is at least as large as a isopropyl group; Taft-type steric parameters indicate it to be as large as an isobutyl group.[59]) Interestingly, the minimum inhibitory concentration (MIC) value of the trifluoromethylalkene isostere was equivalent to that of gramicidin, as the structural feature necessary for the biological activity—the extension of the ornithine side chains in an orthogonal fashion from the periphery o f t h e β-sheet—is preserved in both structures. Hence, trifluoromethylalkene isosteres could be useful as peptide-bond surrogates at locations that are not involved directly in molecular recognition.
Outlook
We have reviewed the electronic and structural properties of four peptide-bond isosteres, along with their ability to participate in noncovalent interactions. We find that employing these isosteres enhances some attributes of the amide bond, but diminishes others (Table 1). These isosteres enable the structural and biological consequences of the peptide bond to be probed as thoroughly as possible, within the constraints imposed by the periodic table. Our conclusions derive mainly from work with peptides. Although much effort has been directed to the incorporation of amino acids with nonnatural side chains into proteins, less work has been done on replacing peptide bonds in proteins. We encourage those efforts, which could provide much insight.
Table 1.
Qualitative comparison of peptide-bond isosteres.
| Hydrogen bond | n→π* interaction | |||||
|---|---|---|---|---|---|---|
| Isostere | Dipole moment |
cis-trans rigidity |
donor | acceptor | donor | acceptor |
| amide | 0 | 0 | 0 | 0 | 0 | 0 |
| thioamide | + | + | + | − | + | − |
| ester | − | − | − | − | + | |
| alkene | − − | + + | − − | − | ||
| fluoroalkene | − | + + | − | − | − | − |
Acknowledgements
Work in our laboratory on peptide-bond isosteres is supported by grants R01 GM044783 and R01 CA073808 (NIH).
References
- [1].Vickery HB, Schmidt CLA. Chem. Rev. 1931;9:169–318. [Google Scholar]
- [2].a) Hofmeister F. Ergebnisse der Physiologie. 1902;1:759–802. [Google Scholar]; b) Teich M, Needham DM. A Documentary History of Biochemistry. Fairleigh Dickinson University Press; Rutherford, NJ: 1992. pp. 1770–1940. [Google Scholar]
- [3].Fischer E. Untersuchungen über Aminosäuren, Polypeptide und Proteïne (1899–1906) Verlag von Julius Springer; Berlin: 1906. [Google Scholar]
- [4].Radzicka A, Wolfenden R. J. Am. Chem. Soc. 1996;118:6105–6109. [Google Scholar]
- [5].a) Pauling L. The Nature of the Chemical Bond. 3rd ed. Cornell University Press; Ithaca, NY: 1960. [Google Scholar]; b) Wiberg KB, Laidig KE. J. Am. Chem. Soc. 1987;109:5935–5943. [Google Scholar]
- [6].Brandts JF, Halvorson HR, Brennan M. Biochemistry. 1975;14:4853–4963. doi: 10.1021/bi00693a026. [DOI] [PubMed] [Google Scholar]
- [7].a) Gray HB. In: Electrons and Chemical Bonding. Benjamin WA, editor. Inc.; New York: 1965. [Google Scholar]; b) Raber DJ, Raber NK, Chandrasekhar J, Scheleyer P. v. R. Inorg. Chem. 1984;23:4076–4080. [Google Scholar]; c) Laing M. J. Chem. Educ. 1987;64:124–128. [Google Scholar]
- [8].a) Isaacs ED, Shukla A, Platzman PM, Barbiellini DR, Tulk CA. Phys. Rev. Lett. 1999;82:600–603. [Google Scholar]; b) Weinhold F, Landis CR. Valency and Bonding: A Natural Bond Orbital Donor–Acceptor Perspective. Cambridge University Press; Cambridge, UK: 2005. [Google Scholar]; c) Weinhold F. Advances in Protein Chemistry: Peptide Solvation and H-Bonds. Vol. 72. Elsevier Academic Press; San Diego, CA: 2006. pp. 121–155. [DOI] [PubMed] [Google Scholar]
- [9].a) Choudhary A, Gandla D, Krow GR, Raines RT. J. Am. Chem. Soc. 2009;131:7244–7246. doi: 10.1021/ja901188y. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Bartlett GA, Choudhary A, Raines RT, Woolfson DN. Nat. Chem. Biol. 2010;6:615–620. doi: 10.1038/nchembio.406. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Jakobsche CE, Choudhary A, Miller SJ, Raines RT. J. Am. Chem. Soc. 2010;132:6651–6653. doi: 10.1021/ja100931y. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Choudhary A, Raines RT. Protein Sci. 2011;20:1077–1081. doi: 10.1002/pro.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Langmuir I. J. Am. Chem. Soc. 1919;41:1543–1559. [Google Scholar]
- [11].Glendening ED, Hrabal JA., II J. Am. Chem. Soc. 1997;119:12940–12946. [Google Scholar]
- [12].Wiberg KB, Rablen PR. J. Am. Chem. Soc. 1995;117:2201–2209. [Google Scholar]
- [13].a) Wiberg KB. Acc. Chem. Res. 1999;32:922–929. [Google Scholar]; b) Wiberg KB, Rush DJ. J. Am. Chem. Soc. 2001;123:2038–2046. doi: 10.1021/ja003586y. [DOI] [PubMed] [Google Scholar]
- [14].Schutkowski M, Wollner S, Fischer G. Biochemistry. 1995;34:13016–13026. doi: 10.1021/bi00040a012. [DOI] [PubMed] [Google Scholar]
- [15].a) Frank R, Jakob M, Thunecke F, Fischer G, Schutkowski M. Angew. Chem. Int. Ed. 2000;39:1120–1122. [PubMed] [Google Scholar]; b) Zhao J, Wildemann D, Jakob M, Vargas C, Schiene-Fischer C. Chem. Commun. 2003:2810–2811. doi: 10.1039/b309927j. [DOI] [PubMed] [Google Scholar]; c) Zhao J, Micheau J-C, Vargas C, Schiene-Fischer C. Chem. Eur. J. 2004;10:6093–6101. doi: 10.1002/chem.200400400. [DOI] [PubMed] [Google Scholar]; d) Helbing J, Bregy H, Bredenbeck J, Pfister R, Hamm P, Huber R, Wachtveitl J, Vico LD, Olivucci M. J. Am. Chem. Soc. 2004;2004:8823–8834. doi: 10.1021/ja049227a. [DOI] [PubMed] [Google Scholar]; e) Satzger H, Root C, Gilch P, Zinth W, Wildemann D, Fischer G. J. Phys. Chem. B. 2005;109:4770–4775. doi: 10.1021/jp045151t. [DOI] [PubMed] [Google Scholar]; f) Bregy H, Heimgartner H, Helbing J. J. Phys. Chem. B. 2009;113:1756–1762. doi: 10.1021/jp8089402. [DOI] [PubMed] [Google Scholar]
- [16].Harada I, Tasumi M. Chem. Phys. Lett. 1980;70:279–282. [Google Scholar]
- [17].a) Ramakrishnan C, Mitra J. Proc. Indian Acad. Sci. Sect. A. 1978;87:13–21. [Google Scholar]; b) Hann MM, Sammes PG. Chem. Commun. 1980:234–235. [Google Scholar]; c) Hann MM, Sammes PG, Kennewell PD, Taylor JB. J. Chem. Soc. Perkin Trans. I. 1982:307–314. [Google Scholar]; d) Aleman C. J. Phys. Chem. A. 2001;105:6717–6723. [Google Scholar]
- [18].Wildemann D, Schiene-Fischer C, Aumuller T, Bachmann A, Kiefhaber T, Lucke C, Fischer G. J. Am. Chem. Soc. 2007;129:4910–4918. doi: 10.1021/ja069048o. [DOI] [PubMed] [Google Scholar]
- [19].a) Min BK, Lee H-J, Choi YS, Park J, Yoon C-J, Yu J-A. J. Mol. Struc. 1998;471:283–288. [Google Scholar]; b) Lee H-J, Choi Y-S, Lee K-B, Park J, Yoon C-J. J. Phys. Chem. A. 2002;106:7010–7017. [Google Scholar]
- [20].Bordwell FG. Acc. Chem. Res. 1988;21:456–463. [Google Scholar]
- [21].a) Artis DR, Lipton MA. J. Am. Chem. Soc. 1998;120:12200–12206. [Google Scholar]; b) Tran TT, Burgess AW, Treutlein H, Zeng J. J. Mol. Graph. Mod. 2001;20:245–256. doi: 10.1016/s1093-3263(01)00118-8. [DOI] [PubMed] [Google Scholar]; c) Tran TT, Treutlein H, Burgess AW. J. Comput. Chem. 2001;22:1010–1025. [Google Scholar]; d) Tran TT, Treutlein H, Burgess AW. J. Comput. Chem. 2001;22:1026–1037. [Google Scholar]
- [22].Tran TT, Zeng J, Treutlein H, Burgess AW. J. Am. Chem. Soc. 2002;124:5222–5230. doi: 10.1021/ja011916o. [DOI] [PubMed] [Google Scholar]
- [23].a) Perczel A, Angyan JG, Kajtar M, Viviani W, Rivail JL, Marcoccia JF, Csizmadia IG. J. Am. Chem. Soc. 1991;113:6256–6265. [Google Scholar]; b) Yang A-S, Honig B. J. Mol. Biol. 1995;252:351–365. doi: 10.1006/jmbi.1995.0502. [DOI] [PubMed] [Google Scholar]
- [24].Reiner A, Wildemann D, Fischer G, Kiefhaber T. J. Am. Chem. Soc. 2008;130:8079–8084. doi: 10.1021/ja8015044. [DOI] [PubMed] [Google Scholar]
- [25].Miwa JH, Pallivathucal L, Gowda S, Lee KE. Org. Lett. 2002;4:4655–4657. doi: 10.1021/ol027056d. [DOI] [PubMed] [Google Scholar]
- [26].Miwa JH, Patel AK, Vivatrat N, Popek SM, Meyer AM. Org. Lett. 2001;3:3373–3375. doi: 10.1021/ol0166092. [DOI] [PubMed] [Google Scholar]
- [27].McFarland BJ, Katz JF, Sant AJ, Beesom C. J. Mol. Biol. 2005;350:170–183. doi: 10.1016/j.jmb.2005.04.069. [DOI] [PubMed] [Google Scholar]
- [28].Grabarse W, Mahlert F, Shima S, Thauer RK, Ermler U. J. Mol. Biol. 2000;303:329–344. doi: 10.1006/jmbi.2000.4136. [DOI] [PubMed] [Google Scholar]
- [29].a) Kallies B, Mitzner R. J. Chem. Soc. Perkin Trans. 1996;2:1397–1401. [Google Scholar]; b) Kallies B, Mitzner R. J. Chem. Soc. Perkin Trans. 1996;2:1403–1408. [Google Scholar]
- [30].Blom CE, Gunthard HH. Chem. Phys. Lett. 1981;84:267–271. [Google Scholar]
- [31].a) Deslongchamps P. Heterocycles. 1977;7:1271–1317. [Google Scholar]; b) Beaulieu N, Deslongchamps P. Can. J. Chem. 1980;58:164–167. [Google Scholar]; c) Deslongchamps P, Beaulieu N, Chênevert R, Dickinson RA. Can. J. Chem. 1980;58:1051–1058. [Google Scholar]; d) Kirby AJ. The Anomeric Effect and Related Stereoelectronic Effects at Oxygen. Springer; New York: 1983. [Google Scholar]
- [32].a) Koh JT, Cornish VW, Schultz PG. Biochemistry. 1997;36:11314–11322. doi: 10.1021/bi9707685. [DOI] [PubMed] [Google Scholar]; b) Powers ET, Deechongkit S, Kelly JW. Adv. Protein Chem. 2005;72:39–78. doi: 10.1016/S0065-3233(05)72002-7. [DOI] [PubMed] [Google Scholar]
- [33].Arnett EM, Mitchell EJ, Murty TSSR. J. Am. Chem. Soc. 1974;96:3875–3891. [Google Scholar]
- [34].Fu Y, Gao J, Bieschke J, Dendle MA, Kelly JW. J. Am. Chem. Soc. 2006;128:15948–15949. doi: 10.1021/ja065303t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].a) Ohyama T, Oku H, Hiroki A, Maekawa Y, Yoshida M, Katakai R. Biopolymers. 2000;54:375–378. doi: 10.1002/1097-0282(200011)54:6<375::AID-BIP10>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]; b) Karle IL, Das C, Balaram P. Biopolymers. 2001;59:279–289. doi: 10.1002/1097-0282(20011005)59:4<276::AID-BIP1024>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]; c) Ohyama T, Oku H, Yoshida M, Katakai R. Biopolymers. 2001;58:636–642. doi: 10.1002/1097-0282(200106)58:7<636::AID-BIP1036>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]; d) Aravinda S, Shamala N, Das C, Balaram P. Biopolymers. 2002;64:255–267. doi: 10.1002/bip.10192. [DOI] [PubMed] [Google Scholar]; e) Oku H, Ohyama T, Hiroki A, Yamada K, Fukuyama K, Kawaguchi H, Katakai R. Biopolymers. 2004;75:242–254. doi: 10.1002/bip.20117. [DOI] [PubMed] [Google Scholar]
- [36].Gao J, Kelly JW. Protein Sci. 2008;17:1096–1101. doi: 10.1110/ps.083439708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cieplak AS, Surmeli NB. J. Org. Chem. 2004;69:3250–3261. doi: 10.1021/jo0358372. [DOI] [PubMed] [Google Scholar]
- [38].Jenkins CL, Vasbinder MM, Miller SJ, Raines RT. Org. Lett. 2005;7:2619–2622. doi: 10.1021/ol050780m. [DOI] [PubMed] [Google Scholar]
- [39].Scheike JA, Baldauf C, Spengler J, Albericio F, Pisabarro MT, Koksch B. Angew. Chem. Int. Ed. 2007;46:7766–7769. doi: 10.1002/anie.200702218. [DOI] [PubMed] [Google Scholar]
- [40].Gao J, Bosco DA, Powers ET, Kelly JW. Nat. Struct. Mol. Biol. 2009;16:684–690. doi: 10.1038/nsmb.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].a) Bateman KS, Huang K, Anderson S, Lu W, Qasim MA, Laskowski M, James MNG. J. Mol. Biol. 2001;305:839–849. doi: 10.1006/jmbi.2000.4343. [DOI] [PubMed] [Google Scholar]; b) Valiyaveetil FI, Sekedat M, MacKinnon R, Muir TW. J. Am. Chem. Soc. 2006;128:11591–11599. doi: 10.1021/ja0631955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].a) Yang W, Drueckhammer DG. J. Am. Chem. Soc. 2001;123:11004–11009. doi: 10.1021/ja010726a. [DOI] [PubMed] [Google Scholar]; b) McGrath NA, Raines RT. Acc. Chem. Res. 2011;44:752–761. doi: 10.1021/ar200081s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Weber AL, Miller SL. J. Mol. Evol. 1981;17:273–284. doi: 10.1007/BF01795749. [DOI] [PubMed] [Google Scholar]
- [44].Wipf P, Henninger TC, Geib SJ. J. Org. Chem. 1998;63:6088–6089. doi: 10.1021/jo981057v. [DOI] [PubMed] [Google Scholar]
- [45].Jakobsche CE, Peris G, Miller SJ. Angew. Chem. Int. Ed. 2008;47:6707–6711. doi: 10.1002/anie.200802223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Scheiner S, Grabowski SJ, Kar T. J. Phys. Chem. A. 2001;105:10607–10612. [Google Scholar]
- [47].a) Bretscher LE, Jenkins CL, Taylor KM, DeRider ML, Raines RT. J. Am. Chem. Soc. 2001;123:777–778. doi: 10.1021/ja005542v. [DOI] [PubMed] [Google Scholar]; b) DeRider ML, Wilkens SJ, Waddell MJ, Bretscher LE, Weinhold F, Raines RT, Markley JL. J. Am. Chem. Soc. 2002;124:2497–2505. doi: 10.1021/ja0166904. [DOI] [PubMed] [Google Scholar]; c) Hinderaker MP, Raines RT. Protein Sci. 2003;12:1188–1194. doi: 10.1110/ps.0241903. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Jenkins CL, Lin G, Duo J, Rapolu D, Guzei IA, Raines RT, Krow GR. J. Org. Chem. 2004;69:8565–8573. doi: 10.1021/jo049242y. [DOI] [PubMed] [Google Scholar]; e) Hodges JA, Raines RT. Org. Lett. 2006;8:4695–4697. doi: 10.1021/ol061569t. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Choudhary A, Fry CG, Raines RT. ARKIVOC. 2010:251–262. [PMC free article] [PubMed] [Google Scholar]; g) Choudhary A, Pua KH, Raines RT. Amino Acids. 2011;41:181–186. doi: 10.1007/s00726-010-0504-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].a) Gardner RR, Liang G-B, Gellman SH. J. Am. Chem. Soc. 1995;117:3280–3281. [Google Scholar]; b) Gardner RR, Liang G-B, Gellman SH. J. Am. Chem. Soc. 1999;121:1806–1816. [Google Scholar]; c) Wipf P, Xiao J, Geib SJ. Adv. Syn. Catal. 2005;347:1605–1613. [Google Scholar]
- [49].a) Schopfer U, Stahl M, Brandl T, Hoffmann RW. Angew. Chem. Int. Ed. 1997;36:1745–1747. [Google Scholar]; b) Hoffmann RW, Schopfer U, Muller G, Brandl T. Helv. Chim. Acta. 2002;85:4424–4441. [Google Scholar]
- [50].Fuller AA, Du D, Liu F, Davoren JE, Bhabha G, Kroon G, Case DA, Dyson HJ, Powers ET, Wipf P, Gruebele M, Kelly JW. Proc. Natl. Acad. Sci. U.S.A. 2009;106:11067–11072. doi: 10.1073/pnas.0813012106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].a) Dai N, Wang XJ, Etzkorn FA. J. Am. Chem. Soc. 2008;130:5396–5397. doi: 10.1021/ja711021m. [DOI] [PubMed] [Google Scholar]; b) Dai N, Etzkorn FA. J. Am. Chem. Soc. 2009;131:13728–13732. doi: 10.1021/ja904177k. [DOI] [PubMed] [Google Scholar]
- [52].Oishi S, Kamitani H, Kodera Y, Watanabe K, Kobayashi K, Narumi T, Tomita K, Ohno H, Naito T, Kodama E, Matsuoka M, Fujii N. Org. Biomol. Chem. 2009;7:2872–2877. doi: 10.1039/b907983a. [DOI] [PubMed] [Google Scholar]
- [53].a) Hart SA, Etzkorn FA. J. Org. Chem. 1999;64:2998–2999. doi: 10.1021/jo990409a. [DOI] [PubMed] [Google Scholar]; b) Wang XJ, Xu B, Mulllins AB, Neiler FK, Etzkorn FA. J. Am. Chem. Soc. 2004;126:15533–15542. doi: 10.1021/ja046396m. [DOI] [PubMed] [Google Scholar]
- [54].Wang XJ, Etzkorn FA. Biopolymers. 2006;84:125–146. doi: 10.1002/bip.20240. [DOI] [PubMed] [Google Scholar]
- [55].Bieschke J, Siegel SJ, Fu Y, Kelly JW. Biochemistry. 2008;47:50–59. doi: 10.1021/bi701757v. [DOI] [PubMed] [Google Scholar]
- [56].a) Schultz R, Cantin M, Roberts C, Greiner B, Uhlmann E, Leumann C. Angew. Chem. Int. Ed. 2000;39:1250–1253. doi: 10.1002/(sici)1521-3773(20000403)39:7<1250::aid-anie1250>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]; b) Hollenstein M, Leumann C. J. Org. Chem. 2004;70:3205–3217. doi: 10.1021/jo047753e. [DOI] [PubMed] [Google Scholar]
- [57].a) Toth G, Kover KE, Murphy RF, Lovas S. J. Phys. Chem. B. 2004;108:9287–9296. [Google Scholar]; b) Gung BW, Zou Y, Xu Z, Amicangelo JC, Irwin DG, Ma S, Zhou H-C. J. Org. Chem. 2008;73:689–693. doi: 10.1021/jo702170j. [DOI] [PubMed] [Google Scholar]
- [58].Xiao J, Weisblum B, Wipf P. J. Am. Chem. Soc. 2005;127:5742–5743. doi: 10.1021/ja051002s. [DOI] [PubMed] [Google Scholar]
- [59].Kitazume T, Yamazaki T. Introduction to Experimental Methods in Organic Fluorine Chemistry. Kodansha Ltd.; Tokyo: 1998. [Google Scholar]