Introduction
The nucleus of eukaryotic cells can be sub-divided into three main compartments: chromatin, intranuclear non-membrane bound complexes (which more or less act like cytoplasmic “organelles”), and the nuclear envelope (NE) (Schneider and Grosschedl, 2007; Spector, 2006; Verstraeten et al., 2007). The NE is composed of the outer and inner nuclear membrane (INM), nuclear pore complexes (NPCs) and the nuclear lamina. The latter is a proteinaceous meshwork underlying the INM and is connected to NPCs (Aaronson and Blobel, 1975). There is also evidence for a structural meshwork inside the nucleus (Barboro et al., 2002; Hozak et al., 1995; Vlcek et al., 2001). This internal nucleoskeleton together with the peripheral lamina forms a scaffold which is involved in chromatin organization and the correct spatial and temporal progression of nuclear processes such as DNA replication and transcription (Bridger et al., 2007; Dechat et al., 2008; Dorner et al., 2007). Major components of this scaffold are the nuclear lamins (Shumaker et al., 2003). In this review we discuss recent findings supporting the role of lamins in the organization and regulation of chromatin.
An Overview of the Nuclear Lamins
Nuclear lamins, type V intermediate filament proteins, are divided into A- and B-types based on their sequence homologies (Broers et al., 2006; Goldman et al., 2002; Shumaker et al., 2003). All A-type lamins are encoded by a single gene (LMNA). Their major isoforms are lamins A and C, which are derived by alternative splicing. The two major mammalian B-type lamins, lamins B1 and B2, are encoded by different genes (LMNB1 and LMNB2). At least one lamin isoform is present in every nucleated metazoan cell (Melcer et al., 2007). In mammals, expression of the A- and B-type lamins is developmentally regulated, resulting in cell type-specific complements of lamins (Broers et al., 2006; Dechat et al., 2008; Verstraeten et al., 2007). The B-type lamins are expressed in undifferentiated human and mouse embryonic stem cells and throughout the early stages of mouse development, while A-type lamins are not expressed until day 10 in mouse development (Constantinescu et al., 2006; Rober et al., 1989; Stewart and Burke, 1987).
Lamin monomer structure consists of an α-helical central rod domain with globular N-terminal head and C-terminal tail domains [Figure 1; (Herrmann and Foisner, 2003; Stuurman et al., 1998)]. The central rod domain forms parallel coiled-coil lamin dimers that subsequently assemble into higher order structures. In vitro, these structures tend to take the form of paracrystals rather than typical 10 nm intermediate filaments (Herrmann and Aebi, 2004; Melcer et al., 2007). However, the actual structure of A- and B- type lamin polymers in a living cell remains unknown. Lamins contain a nuclear localization sequence (NLS) within their tail domain close to the C-terminal end of the central rod [Figure 1; (Loewinger and McKeon, 1988)]. Immediately following the NLS, a segment of the tail is folded into a structural motif similar to a type S immunoglobulin fold (Ig-fold) [Figure 1; (Dhe-Paganon et al., 2002; Krimm et al., 2002)]. All lamins, except for lamin C, terminate with a CAAX-box that is involved in numerous post-translational modifications including the farnesylation of the cysteine, removal of the –AAX and carboxymethylation of the cysteine (Rusinol and Sinensky, 2006; Young et al., 2005). These modifications are thought to be important for the efficient targeting of the lamins to the INM (Dechat et al., 2007; Krohne et al., 1989; Rusinol and Sinensky, 2006). While B-type lamins remain farnesylated and carboxymethylated, lamin A is further processed by the zinc metalloproteinase, Zmpste24/FACE1, to remove an additional 15 residues from its C-terminus including the farnesylated and carboxymethylated cysteine (Corrigan et al., 2005).
Figure 1.
Structure of nuclear lamins. Schematic drawing of a lamin polypeptide chain depicting the α-helical central rod domain, the N-terminal globular head domain and the C-terminal globular tail domain. In addition the nuclear localization signal (NLS) and the Ig-fold are indicated.
Lamins provide the nucleus with mechanical stability and nuclear shape and are involved in establishing connections between the nucleoskeleton and the cytoskeleton (Crisp and Burke, 2008; Dahl et al., 2008; Houben et al., 2007; Rowat et al., 2008). Connections between these structural systems are thought to be important for signal transduction (Parnaik, 2008; Stewart et al., 2007). At the onset of mitosis, lamins are disassembled in a phosphorylation dependent manner and subsequently dispersed in the cell (Fields and Thompson, 1995). During the anaphase/telophase transition they start to reassemble around segregating chromosomes (Moir et al., 2000b). Recently, B-type lamins have been shown to be involved in the formation of a matrix-like network essential for the assembly of the mitotic spindle (Tsai et al., 2006). In addition to their structural functions, lamins are involved in several nuclear processes such as DNA replication, transcription, DNA repair and the epigenetic organization of chromatin (Dechat et al., 2008). Over the past 9 years, a large number of mutations in the human LMNA gene have been associated with numerous diseases including autosomal dominant Emery-Dreyfuss muscular dystrophy (AD-EDMD), familial partial lipodystrophy (FPLD), dilated cardiomyopathy, and the progeroid syndromes Hutchinson-Gilford progeria syndrome (HGPS), atypical Werner syndrome, restricted dermopathy, and mandibuloacral dysplasia type A (MADA) (Broers et al., 2006; Capell and Collins, 2006; Worman and Bonne, 2007). These diseases are now collectively termed “laminopathies”.
Nuclear Lamins and Their Association with Chromatin
The organization of interphase chromosomes into specific compartments and territories within the nucleus of eukaryotic cells is essential for proper chromatin function (Dillon, 2008; Fraser and Bickmore, 2007; Kalverda et al., 2008; Lanctot et al., 2007; Misteli, 2007; Trinkle-Mulcahy and Lamond, 2008). There is increasing evidence that nuclear lamins play an important role in this organization. This evidence comes mainly from three experimental approaches: biochemical analyses of the interaction of lamins with chromatin/histones; microscopy; and alteration of chromatin organization and functions due to expression of mutant lamins or lamin deficiencies.
Biochemical Analyses
Lamins A/C have been shown to bind to mitotic chromosomes in vitro in a lamin B and membrane independent fashion (Burke, 1990; Glass and Gerace, 1990). In addition, A-type lamins, but not B-type lamins bind to polynucleosomal particles isolated from avian erythrocytes (Yuan et al., 1991). There is evidence that the α-helical central rod domain of lamins alone is responsible for the association of human lamin A/C with mitotic chromosomes (Glass et al., 1993). This association seems to be dependent on the assembly state of the truncated lamin used, as the association is mainly observed under conditions which favor the assembly of the rod domain into higher order structures (Glass et al., 1993). In addition, the C-terminal tail domains of human lamins A, C, B1 and B2 (which do not assemble into higher order structures) also bind to chromatin in vitro (Taniura et al., 1995). For lamin C, the chromatin binding region maps to a 35 amino acid segment between the rod domain and the Ig-fold, suggesting that the NLS and nearby sequences are involved (Taniura et al., 1995). Taken together, these results suggest that there are at least two different chromatin binding sites present in lamin A/C: one in the rod domain, which requires a higher order lamin structure, and one located in the C-terminal tail domain, which is independent of lamin polymerization and requires the NLS. The interaction of lamin A/C with chromatin is most likely mediated by chromosome-associated proteins since trypsinization of chromosomes inhibits their interactions with the rod domain of lamin C (Glass et al., 1993). In further support of this, the lamin C tail binds to a core histone fraction isolated from rat liver, but not to DNA extracted from rat liver chromatin fragments (Taniura et al., 1995).
The C-terminal tail domains of the B-type lamins in Drosophila (lamin Dm0) and C. elegans (LMN-1) also bind to chromosomes (Goldberg et al., 1999; Mattout et al., 2007). The binding of lamin Dm0 to chromosomes is independent of its polymerization state, but requires the NLS and the short region between the NLS and the rod domain. This binding can be displaced with histones H2A and/or H2B, but not with histones H1, H3 or H4 (Goldberg et al., 1999). In agreement with these latter results, lamin Dm0 binds directly to histone H2A (Mattout et al., 2007). This interaction requires the NLS of lamin Dm0 and the N- and C-terminal histone tails. An NLS-dependent interaction with chromosomes and histone H2A is also found for LMN-1 (Mattout et al., 2007).
Besides binding to chromatin via histones, full length lamins may also bind directly to DNA (Rzepecki et al., 1998; Shoeman and Traub, 1990; Stierle et al., 2003). Interestingly, the interaction of lamin A/C with plasmid DNA seems to involve several residues within the Ig-fold and also the NLS (Stierle et al., 2003). Cross-linking experiments using Drosophila Kc cells suggest that interphase but not mitotic lamin Dm0 binds to both DNA and RNA (Rzepecki et al., 1998). The DNA regions thought to be associated with lamins are the scaffold/matrix attachment regions (S/MARs) (Baricheva et al., 1996; Luderus et al., 1992; Luderus et al., 1994; Zhao et al., 1996). These regions are involved in the regulation of transcription, replication and the condensation of chromosomes. They are also thought to play a role in the overall organization of chromatin within the nucleus (Gluch et al., 2008). The binding of lamins to S/MARs appears to require the polymerization of the protein (Zhao et al., 1996).
Microscopy
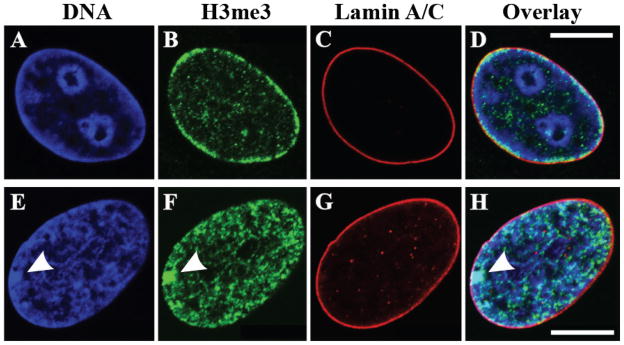
A close association of chromosome ends with the nuclear surface has been recognized since 1887 (Boveri, 1887). Early transmission electron microscopy initially describing the nuclear lamina also reveals its close association with peripheral heterochromatin (Fawcett, 1966; Patrizi and Poger, 1967). Optical sectioning studies of Drosophila salivary gland nuclei show that heterochromatic regions frequently localize at the NE and that the chromocenter is always apposed to the NE (Hochstrasser et al., 1986). A close association of nuclear lamins with peripheral heterochromatin is also evident in combined light and electron microscopy (Belmont et al., 1993; Paddy et al., 1990). In addition, the heterochromatic chromocenter of polytene chromosomes co-localizes with lamin Dm0 at the nuclear periphery of Drosophila salivary gland cells (Baricheva et al., 1996). Confocal immunofluorescence microscopy clearly demonstrates that peripheral nuclear lamins are closely associated with heterochromatin as visualized either by Hoechst staining or by antibodies directed against methylated histones [Figure 2; (Shumaker et al., 2006)]. With respect to the latter, a main fraction of histone H3 trimethylated at lysine 9 (H3K9me3), which is mainly associated with constitutive heterochromatin (Martin and Zhang, 2005; Sarma and Reinberg, 2005), localizes in close association with the inner surface of the nuclear lamina (Figure 2). Furthermore, the inactive X chromosome (Xi), a heterochromatic mass containing histone H3 trimethylated at lysine 27 (H3K27me3) (Plath et al., 2003), is often closely associated with peripheral lamins [Figure 2; (Shumaker et al., 2006)].
Figure 2.
Close association of heterochromatin with the nuclear lamina. Localization of lamins A/C (C, G), histone H3 trimethylated (H3me3) on lysine 9 (H3K9me3) (B), or on lysine 27 (H3K27me3) (F) in human foreskin fibroblasts (A–D) and human dermal fibroblasts from a female donor (E–H). DNA is stained with Hoechst dye (blue; A, E). Note that long stretches of heterochromatin, as revealed by Hoechst staining and by staining for H3K9me3, a histone modification associated mainly with constitutive pericentric heterochromatin, are in close proximity to and partially overlapping with peripheral lamins A/C in human foreskin fibroblasts (A–D). In addition, the Xi, which represents a large heterochromatic mass that can be visualized by staining with Hoechst and H3K27me3 (see arrowheads), is often found associated with lamins at the nuclear lamina (E–H). Scale bars, 10 μM.
The Effects of Mutant Lamins and Altered Lamin Expression on Chromatin Structure and Function
Although all of the studies described above demonstrate that chromatin and lamins are closely associated, at least at the nuclear periphery, they do not provide evidence for a direct involvement of lamins in the organization and function of chromatin. An actual role for lamins in chromatin organization has been revealed by the study of cells derived from LMNA null mice. In LMNA−/− mouse embryonic fibroblasts (MEFs) and cardiomyocytes, a partial loss of peripheral heterochromatin can be observed by electron microscopy (Galiova et al., 2008; Nikolova et al., 2004; Sullivan et al., 1999). This loss is accompanied by the condensation of chromosome territories and by the rearrangement of centromeric heterochromatin (Galiova et al., 2008). Evidence for the role of lamins in chromatin organization has also been derived from studies of Herpes simplex virus infected cells. During lytic infection the cell attempts to silence the viral genome by assembling viral DNA into heterochromatin (Cereghini and Yaniv, 1984). In LMNA−/− MEFs, the viral DNA becomes even more heterochromatic compared to wild type MEFs, leading to reduced viral gene expression, DNA replication, and growth (Silva et al., 2008).
Alterations in peripheral heterochromatin caused by a mutation in a nuclear lamina protein were first observed in muscle and cultured skin cells from a patient suffering from X-linked EDMD (Ognibene et al., 1999). This disease is caused by a mutation in the gene encoding the lamin A binding protein, emerin (Holaska and Wilson, 2006). A detachment or loss of heterochromatin from the nuclear periphery and/or a general loss of heterochromatin is also seen in cells derived from patients suffering from diseases caused by mutations in LMNA. These diseases include AD-EDMD (Sabatelli et al., 2001), FPLD (Capanni et al., 2003), MADA (Filesi et al., 2005; Lombardi et al., 2007), and HGPS (Columbaro et al., 2005; Goldman et al., 2004). In HGPS fibroblasts the loss of heterochromatin is accompanied by changes in various epigenetic patterns; specifically, by an overall decrease in both H3K9me3 and H3K27me3, as well as an increase in tri-methylation of histone H4 at lysine 20 (H4K20me3) (Columbaro et al., 2005; Scaffidi and Misteli, 2005; Shumaker et al., 2006). In addition, the association of the Xi with H3K27me3 is lost and co-localization of HP1α with H3K9me3 is dramatically reduced (Shumaker et al., 2006). C2C12 myoblasts expressing a lamin A protein carrying an AD-EDMD mutation A also show a loss of H3K27me3 from the Xi and a dissociation of H3K9me3 from pericentric heterochromatin (Hakelien et al., 2008). These myoblasts fail to hypertrimethylate H3K4 on the myogenin gene promoter leading to impaired differentiation into myotubes. Interestingly, hypertrimethylation of H3K4, which normally leads to gene activation, is also decreased upon over-expression of wild type lamin A in C2C12 myoblasts (Hakelien et al., 2008). Taken together these findings suggest that not only expression of mutant lamins, but also changes in the expression levels of wild type lamins, have an impact on chromatin organization and that defects in the epigenetic regulation of chromatin are a common feature of different types of laminopathies. Interestingly, a decrease in H3K9me3 is also observed in cells derived from healthy elderly individuals (Scaffidi and Misteli, 2006) and these cells express low levels of a mutant lamin A normally associated with HGPS (Cao et al., 2007; McClintock et al., 2007; Scaffidi and Misteli, 2006) suggesting a role for lamins in the normal aging process.
Besides their roles in anchoring heterochromatin to the nuclear periphery and the epigenetic regulation of chromatin, lamins also seem to be involved in chromosome positioning. Evidence for this comes from the finding that chromosome 18 is displaced from the nuclear periphery towards the nuclear interior in cells derived from patients suffering from some types of laminopathy (Charcot-Marie-Tooth type 2 B1, limb girdle muscular dystrophy, MADA, AD-EDMD, and HGPS) (Meaburn et al., 2007) and in MEFs deficient in lamin B1 (Malhas et al., 2007). The change in the localization of chromosomes from the nuclear interior to the nuclear periphery and vice versa might have an impact on the transcriptional regulation of genes located on those chromosomes (see below) and thus contribute to the disease phenotypes.
The Role of Lamins in Gene Regulation
The role of the nuclear lamina and of nuclear lamins in gene expression is highlighted in several recent reviews [see (Bridger et al., 2007; Dechat et al., 2008; Dorner et al., 2007; Heessen and Fornerod, 2007; Shaklai et al., 2007; Verstraeten et al., 2007)]. Therefore we will focus on the most recent studies. In general the nuclear lamina is considered as a transcriptionally inactive region. Initial evidence for the involvement of lamins in transcription comes from a dominant negative lamin mutant which specifically impairs RNA polymerase II (pol II) mediated transcription (Spann et al., 2002). Furthermore, overexpression of lamins A/C in HeLa cells causes a significant decrease in pol II transcription (Kumaran et al., 2002). The downregulation of lamin B1 in HeLa cells by RNAi can also lead to an inhibition of pol II transcription and subsequently also of pol I transcription (Tang et al., 2008). This transcriptional inhibition is accompanied by global nuclear and chromatin changes including alterations in the structural properties of nucleoli and nuclear speckles, the translocation of chromosome territories toward the nuclear periphery, and a loss of general chromosome loop organization in nucleoids. Prolonged downregulation of lamin B1 leads to cell death. However, downregulation of lamin A/C has none of the above described effects on the structural organization of the nucleus or of chromatin (Tang et al., 2008). Defects in the regulation of transcription reflected by changes in gene expression patterns are also seen in lamin B1 deficient MEFs (Malhas et al., 2007) and in various laminopathies (Dechat et al., 2008). Lamin A/C has also been linked to retinoblastoma regulated transcription of E2F target genes (Dechat et al., 2008; Dorner et al., 2007).
The interaction of lamins with DNA in living cells can be mapped by DamID labeling of DNA directly associated with specific lamins (Guelen et al., 2008; Pickersgill et al., 2006). In this approach, the lamin protein fused to the E. coli enzyme DNA adenine methyl transferase (Dam) is expressed and methylated DNA fragments are sequenced to identify DNA sequences bound by the lamin. When applied to lamin Dm0 in Drosophila Kc cells, DNA sequences characterized as transcriptionally inactive and lacking active histone marks are identified (Pickersgill et al., 2006). These DNA sequences are enriched in large intergenic regions that replicate in mid-to late S-phase. In human lung fibroblasts, the lamin B1-associated domains (LADs) of chromosomes are identified by ectopically expressing lamin B1 fused to Dam and hybridizing amplified adenine-methylated DNA to a high-density microarray (Guelen et al., 2008). These LADs are about 1 mega base in size or larger and are interspersed in a distinct pattern with regions of chromosomes associated with very low levels of lamin B1. In general the LADs display a low gene density, an enrichment in heterochromatin as reflected by the presence of pericentromeric regions, H3K27me3 and H3K9me2, and low levels of pol II and H3K4me2 (Guelen et al., 2008). In addition, about three quarters of all gene deserts (exceptionally large gene-free regions) present within the human genome are associated with LADs and genes located within LADs are 5–10 fold less active than genes outside LADs. Taken together these studies suggest that the B-type lamins are associated with a repressive environment with respect to gene expression, even though actively transcribed genes are found within LADs (Guelen et al., 2008).
To investigate the role of the nuclear lamina in gene regulation in more detail three independent studies take advantage of the LacO/LacI system. Briefly the transcriptional activity of either a reporter gene stably integrated into the genome (Kumaran and Spector, 2008; Reddy et al., 2008) or of whole chromosomes (Finlan et al., 2008) is monitored after their relocation from the nuclear interior to the periphery. Tethering to the nuclear lamina is achieved by fusing the LacI to either lamin B1 (Kumaran and Spector, 2008), emerin (Reddy et al., 2008), or the lamina-associated polypeptide (LAP) 2β (Finlan et al., 2008). Data from one of these studies show that the majority of reporter genes can be expressed in the nuclear lamina region when tethered by lamin B1 (Kumaran and Spector, 2008). In support of the idea that these reporter genes are actively transcribed, recruitment of pol II, as well as of the mRNA splicing factor SF2/ASF is observed. These results suggest that the nuclear lamina does not necessarily define a transcriptionally repressive environment.
Different results are obtained in the study using emerin to tether a reporter gene to the NE (Reddy et al., 2008). These analyses show that most of the tethered reporters display no or reduced transcriptional activity at the nuclear lamina. When a mutant emerin that can not tether the reporter to the NE is used, no transcriptional repression is observed (Reddy et al., 2008). These results indicate that transcriptional repression of the reporter gene requires its localization at the NE at least in this system. The seemingly contradictory results from these two studies may be related to the use of different promoters and reporter genes, different tethering proteins or different cell lines. Nevertheless, they indicate that the nuclear lamina contains not only transcriptionally repressive regions, but also regions that permit gene expression. In agreement with this possibility, tethering of whole chromosomes to the nuclear lamina by LAP2β-lacI can lead to the repression of some genes, whereas other genes are unaffected (Finlan et al., 2008).
Lamins in DNA Replication
Lamin B is present at sites of BrdU incorporation and at replication foci containing the proliferating cell nuclear antigen (PCNA) in late S-phase of cultured cells suggesting a role for nuclear lamins in DNA replication (Moir et al., 1994). The functional significance of this finding is supported by additional experiments using the Xenopus egg interphase extract nuclear assembly system. For example, nuclei assembled in a lamin-depleted extract are unable to replicate their DNA (Meier et al., 1991; Newport et al., 1990). In such extracts PCNA is also more extractable, indicating that its association with the nucleoskeleton/chromatin is dependent on lamins (Jenkins et al., 1993). Subsequent studies further show that lamins are involved in the chain elongation phase of replication. When dominant negative lamin mutants are added to interphase extracts containing Xenopus nuclei, they cause the redistribution of endogenous lamins into intranuclear foci and the inhibition of replication (Ellis et al., 1997; Moir et al., 2000a; Spann et al., 1997). The lamin foci formed in this manner also contain the DNA replication elongation factors PCNA and replication factor C (RFC) (Moir et al., 2000a; Spann et al., 1997). However, the distribution of the DNA initiation factors, DNA polymerase α, minichromosome maintenance protein 3 (MCM3) and the origin recognition complex (ORC) protein Orc2 appear unaltered (Moir et al., 2000a). Transferring these lamin-disrupted nuclei to fresh extract leads to a normal lamin distribution and to the reinitiation of the chain elongation phase of replication (Moir et al., 2000a).
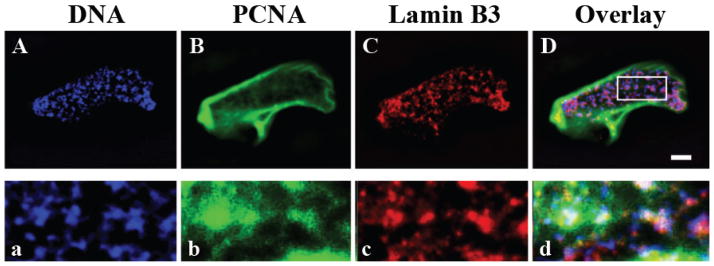
Recent data show that in nuclei assembled in Xenopus egg interphase extracts for 5 to 120 minutes, Xenopus lamin B3 is closely associated with chromatin and PCNA [Figure 3; (Shumaker et al., 2008)]. Furthermore it has been shown that the C-terminal tail domain of lamins can interact directly with PCNA, and that the interaction site is located in the Ig-fold. Both the tail domain and the Ig-fold of Xenopus lamin B3 prevent lamin polymerization as well as the assembly of nuclei in Xenopus egg interphase extracts (Lopez-Soler et al., 2001; Shumaker et al., 2005). When added to the assembly system, the Xenopus lamin B3 tail is closely associated with chromatin and leads to significant decreases in PCNA incorporation into nuclei and in DNA replication (Shumaker et al., 2008). Interestingly, a mutation in the Ig-fold of Xenopus lamin B3, which mimics an LMNA mutation associated with EDMD [R453W; (Bonne et al., 1999)], decreases its binding to PCNA and inhibits DNA replication (Shumaker et al., 2008). These findings suggest that defects in DNA replication caused by the expression of disease causing lamin mutants may contribute to the altered nuclear functions responsible for some types of laminopathies.
Figure 3.
Lamin B3 is closely associated with PCNA and chromatin in in vitro assembled nuclei. Localization of Xenopus lamin B3 (B, b; green) and PCNA (C, c; red) in a nucleus assembled in a Xenopus egg interphase extract for 130 min (A–D). DNA is stained with Hoechst dye (A, a; blue). The area in the box in D is enlarged (3.5×) to show the partial overlap between DNA, lamin B3 and PCNA (a–d). Brightness and contrast are enhanced in b compared to B for better visualization of the internal lamin B3 structures. Scale bar, 5 μM.
Summary
In our model we propose that lamins are major components of a nuclear scaffold which is essential for various nuclear processes such as transcription, DNA replication, chromatin organization and DNA repair (Dechat et al., 2008; Goldman et al., 2002). We further speculate that this lamin based scaffold provides a docking site and organizing center for chromatin and the multicomponent complexes involved in chromatin regulation. Alterations in such a scaffold caused either by changes in lamin expression patterns or by the expression of disease causing mutant lamins can result in the misregulation of nuclear functions leading, for example, to defects in cell cycle progression and differentiation (Dechat et al., 2008). In support of this two recent studies show that adult stem cell differentiation is impaired in HGPS and in premature-aging mice (Espada et al., 2008; Scaffidi and Misteli, 2008).
Acknowledgments
We wish to thank the NIA, NCI and the Ellison Medical Research Foundation for supporting our studies of the nuclear lamins.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aaronson RP, Blobel G. Isolation of nuclear pore complexes in association with a lamina. Proc Natl Acad Sci U S A. 1975;72:1007–11. doi: 10.1073/pnas.72.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboro P, D'Arrigo C, Diaspro A, Mormino M, Alberti I, Parodi S, Patrone E, Balbi C. Unraveling the organization of the internal nuclear matrix: RNA-dependent anchoring of NuMA to a lamin scaffold. Exp Cell Res. 2002;279:202–18. doi: 10.1006/excr.2002.5605. [DOI] [PubMed] [Google Scholar]
- Baricheva EA, Berrios M, Bogachev SS, Borisevich IV, Lapik ER, Sharakhov IV, Stuurman N, Fisher PA. DNA from Drosophila melanogaster beta-heterochromatin binds specifically to nuclear lamins in vitro and the nuclear envelope in situ. Gene. 1996;171:171–6. doi: 10.1016/0378-1119(96)00002-9. [DOI] [PubMed] [Google Scholar]
- Belmont AS, Zhai Y, Thilenius A. Lamin B distribution and association with peripheral chromatin revealed by optical sectioning and electron microscopy tomography. J Cell Biol. 1993;123:1671–85. doi: 10.1083/jcb.123.6.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonne G, Di Barletta MR, Varnous S, Becane HM, Hammouda EH, Merlini L, Muntoni F, Greenberg CR, Gary F, Urtizberea JA, Duboc D, Fardeau M, Toniolo D, Schwartz K. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat Genet. 1999;21:285–8. doi: 10.1038/6799. [DOI] [PubMed] [Google Scholar]
- Boveri T. Zellen-Studien. 1887. Die Bildung der Richtungskörper bei Ascaris megalocephala und Ascaris lumbricoides. [Google Scholar]
- Bridger JM, Foeger N, Kill IR, Herrmann H. The nuclear lamina. Both a structural framework and a platform for genome organization. Febs J. 2007;274:1354–61. doi: 10.1111/j.1742-4658.2007.05694.x. [DOI] [PubMed] [Google Scholar]
- Broers JL, Ramaekers FC, Bonne G, Yaou RB, Hutchison CJ. Nuclear lamins: laminopathies and their role in premature ageing. Physiol Rev. 2006;86:967–1008. doi: 10.1152/physrev.00047.2005. [DOI] [PubMed] [Google Scholar]
- Burke B. On the cell-free association of lamins A and C with metaphase chromosomes. Exp Cell Res. 1990;186:169–76. doi: 10.1016/0014-4827(90)90223-w. [DOI] [PubMed] [Google Scholar]
- Cao K, Capell BC, Erdos MR, Djabali K, Collins FS. A lamin A protein isoform overexpressed in Hutchinson-Gilford progeria syndrome interferes with mitosis in progeria and normal cells. Proc Natl Acad Sci U S A. 2007;104:4949–54. doi: 10.1073/pnas.0611640104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capanni C, Cenni V, Mattioli E, Sabatelli P, Ognibene A, Columbaro M, Parnaik VK, Wehnert M, Maraldi NM, Squarzoni S, Lattanzi G. Failure of lamin A/C to functionally assemble in R482L mutated familial partial lipodystrophy fibroblasts: altered intermolecular interaction with emerin and implications for gene transcription. Exp Cell Res. 2003;291:122–34. doi: 10.1016/s0014-4827(03)00395-1. [DOI] [PubMed] [Google Scholar]
- Capell BC, Collins FS. Human laminopathies: nuclei gone genetically awry. Nat Rev Genet. 2006;7:940–52. doi: 10.1038/nrg1906. [DOI] [PubMed] [Google Scholar]
- Cereghini S, Yaniv M. Assembly of transfected DNA into chromatin: structural changes in the origin-promoter-enhancer region upon replication. EMBO J. 1984;3:1243–53. doi: 10.1002/j.1460-2075.1984.tb01959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Columbaro M, Capanni C, Mattioli E, Novelli G, Parnaik VK, Squarzoni S, Maraldi NM, Lattanzi G. Rescue of heterochromatin organization in Hutchinson-Gilford progeria by drug treatment. Cell Mol Life Sci. 2005;62:2669–78. doi: 10.1007/s00018-005-5318-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinescu D, Gray HL, Sammak PJ, Schatten GP, Csoka AB. Lamin A/C expression is a marker of mouse and human embryonic stem cell differentiation. Stem Cells. 2006;24:177–85. doi: 10.1634/stemcells.2004-0159. [DOI] [PubMed] [Google Scholar]
- Corrigan DP, Kuszczak D, Rusinol AE, Thewke DP, Hrycyna CA, Michaelis S, Sinensky MS. Prelamin A endoproteolytic processing in vitro by recombinant Zmpste24. Biochem J. 2005;387:129–38. doi: 10.1042/BJ20041359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp M, Burke B. The nuclear envelope as an integrator of nuclear and cytoplasmic architecture. FEBS Lett. 2008;582:2023–32. doi: 10.1016/j.febslet.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Dahl KN, Ribeiro AJ, Lammerding J. Nuclear shape, mechanics, and mechanotransduction. Circ Res. 2008;102:1307–18. doi: 10.1161/CIRCRESAHA.108.173989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Shimi T, Adam SA, Rusinol AE, Andres DA, Spielmann HP, Sinensky MS, Goldman RD. Alterations in mitosis and cell cycle progression caused by a mutant lamin A known to accelerate human aging. Proc Natl Acad Sci U S A. 2007;104:4955–60. doi: 10.1073/pnas.0700854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, Goldman RD. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008;22:832–53. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhe-Paganon S, Werner ED, Chi YI, Shoelson SE. Structure of the globular tail of nuclear lamin. J Biol Chem. 2002;277:17381–4. doi: 10.1074/jbc.C200038200. [DOI] [PubMed] [Google Scholar]
- Dillon N. The impact of gene location in the nucleus on transcriptional regulation. Dev Cell. 2008;15:182–6. doi: 10.1016/j.devcel.2008.07.013. [DOI] [PubMed] [Google Scholar]
- Dorner D, Gotzmann J, Foisner R. Nucleoplasmic lamins and their interaction partners, LAP2alpha, Rb, and BAF, in transcriptional regulation. Febs J. 2007;274:1362–73. doi: 10.1111/j.1742-4658.2007.05695.x. [DOI] [PubMed] [Google Scholar]
- Ellis DJ, Jenkins H, Whitfield WG, Hutchison CJ. GST-lamin fusion proteins act as dominant negative mutants in Xenopus egg extract and reveal the function of the lamina in DNA replication. J Cell Sci. 1997;110:2507–18. doi: 10.1242/jcs.110.20.2507. [DOI] [PubMed] [Google Scholar]
- Espada J, Varela I, Flores I, Ugalde AP, Cadinanos J, Pendas AM, Stewart CL, Tryggvason K, Blasco MA, Freije JM, Lopez-Otin C. Nuclear envelope defects cause stem cell dysfunction in premature-aging mice. J Cell Biol. 2008;181:27–35. doi: 10.1083/jcb.200801096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett DW. On the occurrence of a fibrous lamina on the inner aspect of the nuclear envelope in certain cells of vertebrates. Am J Anat. 1966;119:129–45. doi: 10.1002/aja.1001190108. [DOI] [PubMed] [Google Scholar]
- Fields AP, Thompson LJ. The regulation of mitotic nuclear envelope breakdown: a role for multiple lamin kinases. Prog Cell Cycle Res. 1995;1:271–86. doi: 10.1007/978-1-4615-1809-9_22. [DOI] [PubMed] [Google Scholar]
- Filesi I, Gullotta F, Lattanzi G, D'Apice MR, Capanni C, Nardone AM, Columbaro M, Scarano G, Mattioli E, Sabatelli P, Maraldi NM, Biocca S, Novelli G. Alterations of nuclear envelope and chromatin organization in mandibuloacral dysplasia, a rare form of laminopathy. Physiol Genomics. 2005 doi: 10.1152/physiolgenomics.00060.2005. [DOI] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–7. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- Galiova G, Bartova E, Raska I, Krejci J, Kozubek S. Chromatin changes induced by lamin A/C deficiency and the histone deacetylase inhibitor trichostatin A. Eur J Cell Biol. 2008;87:291–303. doi: 10.1016/j.ejcb.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Glass CA, Glass JR, Taniura H, Hasel KW, Blevitt JM, Gerace L. The alpha-helical rod domain of human lamins A and C contains a chromatin binding site. Embo J. 1993;12:4413–24. doi: 10.1002/j.1460-2075.1993.tb06126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass JR, Gerace L. Lamins A and C bind and assemble at the surface of mitotic chromosomes. J Cell Biol. 1990;111:1047–57. doi: 10.1083/jcb.111.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluch A, Vidakovic M, Bode J. Scaffold/matrix attachment regions (S/MARs): relevance for disease and therapy. Handb Exp Pharmacol. 2008:67–103. doi: 10.1007/978-3-540-72843-6_4. [DOI] [PubMed] [Google Scholar]
- Goldberg M, Harel A, Brandeis M, Rechsteiner T, Richmond TJ, Weiss AM, Gruenbaum Y. The tail domain of lamin Dm0 binds histones H2A and H2B. Proc Natl Acad Sci U S A. 1999;96:2852–7. doi: 10.1073/pnas.96.6.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman RD, Gruenbaum Y, Moir RD, Shumaker DK, Spann TP. Nuclear lamins: building blocks of nuclear architecture. Genes Dev. 2002;16:533–47. doi: 10.1101/gad.960502. [DOI] [PubMed] [Google Scholar]
- Goldman RD, Shumaker DK, Erdos MR, Eriksson M, Goldman AE, Gordon LB, Gruenbaum Y, Khuon S, Mendez M, Varga R, Collins FS. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A. 2004;101:8963–8. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, van Steensel B. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–51. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Hakelien AM, Delbarre E, Gaustad KG, Buendia B, Collas P. Expression of the myodystrophic R453W mutation of lamin A in C2C12 myoblasts causes promoter-specific and global epigenetic defects. Exp Cell Res. 2008;314:1869–80. doi: 10.1016/j.yexcr.2008.02.018. [DOI] [PubMed] [Google Scholar]
- Heessen S, Fornerod M. The inner nuclear envelope as a transcription factor resting place. EMBO Rep. 2007;8:914–9. doi: 10.1038/sj.embor.7401075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann H, Foisner R. Intermediate filaments: novel assembly models and exciting new functions for nuclear lamins. Cell Mol Life Sci. 2003;60:1607–12. doi: 10.1007/s00018-003-3004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann H, Aebi U. Intermediate filaments: molecular structure, assembly mechanism, and integration into functionally distinct intracellular Scaffolds. Annu Rev Biochem. 2004;73:749–89. doi: 10.1146/annurev.biochem.73.011303.073823. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M, Mathog D, Gruenbaum Y, Saumweber H, Sedat JW. Spatial organization of chromosomes in the salivary gland nuclei of Drosophila melanogaster. J Cell Biol. 1986;102:112–23. doi: 10.1083/jcb.102.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holaska JM, Wilson KL. Multiple roles for emerin: implications for Emery-Dreifuss muscular dystrophy. Anat Rec A Discov Mol Cell Evol Biol. 2006;288:676–80. doi: 10.1002/ar.a.20334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben F, Ramaekers FC, Snoeckx LH, Broers JL. Role of nuclear lamina-cytoskeleton interactions in the maintenance of cellular strength. Biochim Biophys Acta. 2007;1773:675–86. doi: 10.1016/j.bbamcr.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Hozak P, Sasseville AM, Raymond Y, Cook PR. Lamin proteins form an internal nucleoskeleton as well as a peripheral lamina in human cells. J Cell Sci. 1995;108 ( Pt 2):635–44. doi: 10.1242/jcs.108.2.635. [DOI] [PubMed] [Google Scholar]
- Jenkins H, Holman T, Lyon C, Lane B, Stick R, Hutchison C. Nuclei that lack a lamina accumulate karyophilic proteins and assemble a nuclear matrix. J Cell Sci. 1993;106:275–85. doi: 10.1242/jcs.106.1.275. [DOI] [PubMed] [Google Scholar]
- Kalverda B, Roling MD, Fornerod M. Chromatin organization in relation to the nuclear periphery. FEBS Lett. 2008;582:2017–22. doi: 10.1016/j.febslet.2008.04.015. [DOI] [PubMed] [Google Scholar]
- Krimm I, Ostlund C, Gilquin B, Couprie J, Hossenlopp P, Mornon JP, Bonne G, Courvalin JC, Worman HJ, Zinn-Justin S. The Ig-like structure of the C-terminal domain of lamin A/C, mutated in muscular dystrophies, cardiomyopathy, and partial lipodystrophy. Structure (Camb) 2002;10:811–23. doi: 10.1016/s0969-2126(02)00777-3. [DOI] [PubMed] [Google Scholar]
- Krohne G, Waizenegger I, Hoger TH. The conserved carboxy-terminal cysteine of nuclear lamins is essential for lamin association with the nuclear envelope. J Cell Biol. 1989;109:2003–11. doi: 10.1083/jcb.109.5.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran RI, Muralikrishna B, Parnaik VK. Lamin A/C speckles mediate spatial organization of splicing factor compartments and RNA polymerase II transcription. J Cell Biol. 2002;159:783–93. doi: 10.1083/jcb.200204149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol. 2008;180:51–65. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104–15. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- Loewinger L, McKeon F. Mutations in the nuclear lamin proteins resulting in their aberrant assembly in the cytoplasm. Embo J. 1988;7:2301–9. doi: 10.1002/j.1460-2075.1988.tb03073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi F, Gullotta F, Columbaro M, Filareto A, D'Adamo M, Vielle A, Guglielmi V, Nardone AM, Azzolini V, Grosso E, Lattanzi G, D'Apice MR, Masala S, Maraldi NM, Sbraccia P, Novelli G. Compound Heterozygosity for Mutations in LMNA in a Patient with a Myopathic and Lipodystrophic Mandibuloacral Dysplasia Type A Phenotype. J Clin Endocrinol Metab. 2007 doi: 10.1210/jc.2007-0116. [DOI] [PubMed] [Google Scholar]
- Lopez-Soler RI, Moir RD, Spann TP, Stick R, Goldman RD. A role for nuclear lamins in nuclear envelope assembly. J Cell Biol. 2001;154:61–70. doi: 10.1083/jcb.200101025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luderus ME, de Graaf A, Mattia E, den Blaauwen JL, Grande MA, de Jong L, van Driel R. Binding of matrix attachment regions to lamin B1. Cell. 1992;70:949–59. doi: 10.1016/0092-8674(92)90245-8. [DOI] [PubMed] [Google Scholar]
- Luderus ME, den Blaauwen JL, de Smit OJ, Compton DA, van Driel R. Binding of matrix attachment regions to lamin polymers involves single-stranded regions and the minor groove. Mol Cell Biol. 1994;14:6297–305. doi: 10.1128/mcb.14.9.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhas A, Lee CF, Sanders R, Saunders NJ, Vaux DJ. Defects in lamin B1 expression or processing affect interphase chromosome position and gene expression. J Cell Biol. 2007;176:593–603. doi: 10.1083/jcb.200607054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–49. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Mattout A, Goldberg M, Tzur Y, Margalit A, Gruenbaum Y. Specific and conserved sequences in D. melanogaster and C. elegans lamins and histone H2A mediate the attachment of lamins to chromosomes. J Cell Sci. 2007;120:77–85. doi: 10.1242/jcs.03325. [DOI] [PubMed] [Google Scholar]
- McClintock D, Ratner D, Lokuge M, Owens DM, Gordon LB, Collins FS, Djabali K. The Mutant Form of Lamin A that Causes Hutchinson-Gilford Progeria Is a Biomarker of Cellular Aging in Human Skin. PLoS ONE. 2007;2:e1269. doi: 10.1371/journal.pone.0001269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn KJ, Cabuy E, Bonne G, Levy N, Morris GE, Novelli G, Kill IR, Bridger JM. Primary laminopathy fibroblasts display altered genome organization and apoptosis. Aging Cell. 2007;6:139–53. doi: 10.1111/j.1474-9726.2007.00270.x. [DOI] [PubMed] [Google Scholar]
- Meier J, Campbell KH, Ford CC, Stick R, Hutchison CJ. The role of lamin LIII in nuclear assembly and DNA replication, in cell- free extracts of Xenopus eggs. J Cell Sci. 1991;98:271–9. doi: 10.1242/jcs.98.3.271. [DOI] [PubMed] [Google Scholar]
- Melcer S, Gruenbaum Y, Krohne G. Invertebrate lamins. Exp Cell Res. 2007;313:2157–66. doi: 10.1016/j.yexcr.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Moir RD, Montag-Lowy M, Goldman RD. Dynamic properties of nuclear lamins: lamin B is associated with sites of DNA replication. J Cell Biol. 1994;125:1201–12. doi: 10.1083/jcb.125.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir RD, Spann TP, Herrmann H, Goldman RD. Disruption of nuclear lamin organization blocks the elongation phase of DNA replication. J Cell Biol. 2000a;149:1179–92. doi: 10.1083/jcb.149.6.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir RD, Spann TP, Lopez-Soler RI, Yoon M, Goldman AE, Khuon S, Goldman RD. Review: the dynamics of the nuclear lamins during the cell cycle-- relationship between structure and function. J Struct Biol. 2000b;129:324–34. doi: 10.1006/jsbi.2000.4251. [DOI] [PubMed] [Google Scholar]
- Newport JW, Wilson KL, Dunphy WG. A lamin-independent pathway for nuclear envelope assembly. J Cell Biol. 1990;111:2247–59. doi: 10.1083/jcb.111.6.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolova V, Leimena C, McMahon AC, Tan JC, Chandar S, Jogia D, Kesteven SH, Michalicek J, Otway R, Verheyen F, Rainer S, Stewart CL, Martin D, Feneley MP, Fatkin D. Defects in nuclear structure and function promote dilated cardiomyopathy in lamin A/C-deficient mice. J Clin Invest. 2004;113:357–69. doi: 10.1172/JCI19448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ognibene A, Sabatelli P, Petrini S, Squarzoni S, Riccio M, Santi S, Villanova M, Palmeri S, Merlini L, Maraldi NM. Nuclear changes in a case of X-linked Emery-Dreifuss muscular dystrophy. Muscle Nerve. 1999;22:864–9. doi: 10.1002/(sici)1097-4598(199907)22:7<864::aid-mus8>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Paddy MR, Belmont AS, Saumweber H, Agard DA, Sedat JW. Interphase nuclear envelope lamins form a discontinuous network that interacts with only a fraction of the chromatin in the nuclear periphery. Cell. 1990;62:89–106. doi: 10.1016/0092-8674(90)90243-8. [DOI] [PubMed] [Google Scholar]
- Parnaik VK. Role of nuclear lamins in nuclear organization, cellular signaling, and inherited diseases. Int Rev Cell Mol Biol. 2008;266:157–206. doi: 10.1016/S1937-6448(07)66004-3. [DOI] [PubMed] [Google Scholar]
- Patrizi G, Poger M. The ultrastructure of the nuclear periphery. The zonula nucleum limitans. J Ultrastruct Res. 1967;17:127–36. doi: 10.1016/s0022-5320(67)80025-x. [DOI] [PubMed] [Google Scholar]
- Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet. 2006;38:1005–14. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–5. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–7. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- Rober RA, Weber K, Osborn M. Differential timing of nuclear lamin A/C expression in the various organs of the mouse embryo and the young animal: a developmental study. Development. 1989;105:365–78. doi: 10.1242/dev.105.2.365. [DOI] [PubMed] [Google Scholar]
- Rowat AC, Lammerding J, Herrmann H, Aebi U. Towards an integrated understanding of the structure and mechanics of the cell nucleus. Bioessays. 2008;30:226–36. doi: 10.1002/bies.20720. [DOI] [PubMed] [Google Scholar]
- Rusinol AE, Sinensky MS. Farnesylated lamins, progeroid syndromes and farnesyl transferase inhibitors. J Cell Sci. 2006;119:3265–72. doi: 10.1242/jcs.03156. [DOI] [PubMed] [Google Scholar]
- Rzepecki R, Bogachev SS, Kokoza E, Stuurman N, Fisher PA. In vivo association of lamins with nucleic acids in Drosophila melanogaster. J Cell Sci. 1998;111 ( Pt 1):121–9. doi: 10.1242/jcs.111.1.121. [DOI] [PubMed] [Google Scholar]
- Sabatelli P, Lattanzi G, Ognibene A, Columbaro M, Capanni C, Merlini L, Maraldi NM, Squarzoni S. Nuclear alterations in autosomal-dominant Emery-Dreifuss muscular dystrophy. Muscle Nerve. 2001;24:826–9. doi: 10.1002/mus.1076. [DOI] [PubMed] [Google Scholar]
- Sarma K, Reinberg D. Histone variants meet their match. Nat Rev Mol Cell Biol. 2005;6:139–49. doi: 10.1038/nrm1567. [DOI] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat Med. 2005 doi: 10.1038/nm1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312:1059–63. [Google Scholar]
- Scaffidi P, Misteli T. Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nat Cell Biol. 2008;10:452–9. doi: 10.1038/ncb1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Grosschedl R. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev. 2007;21:3027–43. doi: 10.1101/gad.1604607. [DOI] [PubMed] [Google Scholar]
- Shaklai S, Amariglio N, Rechavi G, Simon AJ. Gene silencing at the nuclear periphery. Febs J. 2007;274:1383–92. doi: 10.1111/j.1742-4658.2007.05697.x. [DOI] [PubMed] [Google Scholar]
- Shoeman RL, Traub P. The in vitro DNA-binding properties of purified nuclear lamin proteins and vimentin. J Biol Chem. 1990;265:9055–61. [PubMed] [Google Scholar]
- Shumaker DK, Kuczmarski ER, Goldman RD. The nucleoskeleton: lamins and actin are major players in essential nuclear functions. Curr Opin Cell Biol. 2003;15:358–66. doi: 10.1016/s0955-0674(03)00050-4. [DOI] [PubMed] [Google Scholar]
- Shumaker DK, Lopez-Soler RI, Adam SA, Herrmann H, Moir RD, Spann TP, Goldman RD. Functions and dysfunctions of the nuclear lamin Ig-fold domain in nuclear assembly, growth, and Emery-Dreifuss muscular dystrophy. Proc Natl Acad Sci U S A. 2005;102:15494–9. doi: 10.1073/pnas.0507612102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker DK, Dechat T, Kohlmaier A, Adam SA, Bozovsky MR, Erdos MR, Eriksson M, Goldman AE, Khuon S, Collins FS, Jenuwein T, Goldman RD. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci U S A. 2006;103:8703–8. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker DK, Solimando L, Sengupta K, Shimi T, Adam SA, Grunwald A, Strelkov SV, Aebi U, Cardoso MC, Goldman RD. The highly conserved nuclear lamin Ig-fold binds to PCNA: its role in DNA replication. J Cell Biol. 2008;181:269–80. doi: 10.1083/jcb.200708155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva L, Cliffe A, Chang L, Knipe DM. Role for A-type lamins in herpesviral DNA targeting and heterochromatin modulation. PLoS Pathog. 2008;4:e1000071. doi: 10.1371/journal.ppat.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann TP, Moir RD, Goldman AE, Stick R, Goldman RD. Disruption of nuclear lamin organization alters the distribution of replication factors and inhibits DNA synthesis. J Cell Biol. 1997;136:1201–12. doi: 10.1083/jcb.136.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann TP, Goldman AE, Wang C, Huang S, Goldman RD. Alteration of nuclear lamin organization inhibits RNA polymerase II-dependent transcription. J Cell Biol. 2002;156:603–8. doi: 10.1083/jcb.200112047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL. SnapShot: Cellular bodies. Cell. 2006;127:1071. doi: 10.1016/j.cell.2006.11.026. [DOI] [PubMed] [Google Scholar]
- Stewart C, Burke B. Teratocarcinoma stem cells and early mouse embryos contain only a single major lamin polypeptide closely resembling lamin B. Cell. 1987;51:383–92. doi: 10.1016/0092-8674(87)90634-9. [DOI] [PubMed] [Google Scholar]
- Stewart CL, Roux KJ, Burke B. Blurring the boundary: the nuclear envelope extends its reach. Science. 2007;318:1408–12. doi: 10.1126/science.1142034. [DOI] [PubMed] [Google Scholar]
- Stierle V, Couprie J, Ostlund C, Krimm I, Zinn-Justin S, Hossenlopp P, Worman HJ, Courvalin JC, Duband-Goulet I. The carboxyl-terminal region common to lamins A and C contains a DNA binding domain. Biochemistry. 2003;42:4819–28. doi: 10.1021/bi020704g. [DOI] [PubMed] [Google Scholar]
- Stuurman N, Heins S, Aebi U. Nuclear lamins: their structure, assembly, and interactions. J Struct Biol. 1998;122:42–66. doi: 10.1006/jsbi.1998.3987. [DOI] [PubMed] [Google Scholar]
- Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–20. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CW, Maya-Mendoza A, Martin C, Zeng K, Chen S, Feret D, Wilson SA, Jackson DA. The integrity of a lamin-B1-dependent nucleoskeleton is a fundamental determinant of RNA synthesis in human cells. J Cell Sci. 2008;121:1014–24. doi: 10.1242/jcs.020982. [DOI] [PubMed] [Google Scholar]
- Taniura H, Glass C, Gerace L. A chromatin binding site in the tail domain of nuclear lamins that interacts with core histones. J Cell Biol. 1995;131:33–44. doi: 10.1083/jcb.131.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L, Lamond AI. Nuclear functions in space and time: gene expression in a dynamic, constrained environment. FEBS Lett. 2008;582:1960–70. doi: 10.1016/j.febslet.2008.04.029. [DOI] [PubMed] [Google Scholar]
- Tsai MY, Wang S, Heidinger JM, Shumaker DK, Adam SA, Goldman RD, Zheng Y. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science. 2006;311:1887–93. doi: 10.1126/science.1122771. [DOI] [PubMed] [Google Scholar]
- Verstraeten VL, Broers JL, Ramaekers FC, van Steensel MA. The nuclear envelope, a key structure in cellular integrity and gene expression. Curr Med Chem. 2007;14:1231–48. doi: 10.2174/092986707780598032. [DOI] [PubMed] [Google Scholar]
- Vlcek S, Dechat T, Foisner R. Nuclear envelope and nuclear matrix: interactions and dynamics. Cell Mol Life Sci. 2001;58:1758–65. doi: 10.1007/PL00000815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ, Bonne G. “Laminopathies”: a wide spectrum of human diseases. Exp Cell Res. 2007;313:2121–33. doi: 10.1016/j.yexcr.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SG, Fong LG, Michaelis S. Prelamin A, Zmpste24, misshapen cell nuclei, and progeria--new evidence suggesting that protein farnesylation could be important for disease pathogenesis. J Lipid Res. 2005;46:2531–58. doi: 10.1194/jlr.R500011-JLR200. [DOI] [PubMed] [Google Scholar]
- Yuan J, Simos G, Blobel G, Georgatos SD. Binding of lamin A to polynucleosomes. J Biol Chem. 1991;266:9211–5. [PubMed] [Google Scholar]
- Zhao K, Harel A, Stuurman N, Guedalia D, Gruenbaum Y. Binding of matrix attachment regions to nuclear lamin is mediated by the rod domain and depends on the lamin polymerization state. FEBS Lett. 1996;380:161–4. doi: 10.1016/0014-5793(96)00034-8. [DOI] [PubMed] [Google Scholar]