Abstract
Cyclooxygenases-1 and -2 (COX-1 and -2) catalyze the committed step in prostaglandin formation. Each isozyme subserves different biological functions. This is, at least in part, a consequence of differences in patterns of COX-1 and COX-2 expression. COX-1 is induced during development, and COX-1 mRNA and COX-1 protein are very stable. These latter properties can explain why COX-1 protein levels usually remain constant in those cells that express this isozyme. COX-2 is usually expressed inducibly in association with cell replication or differentiation. Both COX-2 mRNA and COX-2 protein have short half-lives relative to those of COX-1. Therefore, COX-2 protein is typically present for only a few hr after its synthesis. Here we review and develop the concepts that (a) COX-2 gene transcription can involve at least six different cis-acting promoter elements interacting with trans-acting factors generated by multiple, different signaling pathways, (b) the relative contribution of each cis-acting COX-2 promoter element depends on the cell type, the stimulus and the time following the stimulus and (c) a unique 27 amino acid instability element located just upstream of the C-terminus of COX-2 targets this isoform to the ER-associated degradation system and proteolysis by the cytosolic 26S proteasome.
Keywords: prostaglandin endoperoxide H synthase, aspirin, COX-2, gene regulation, ER-associated protein degradation, nonsteroidal anti-inflammatory drugs
INTRODUCTION
Cyclooxygenases-1 and -2 (COX-1 and -2)1,2 convert arachidonic acid, hydrolyzed from cell membrane phospholipids by a phospholipase A2, to prostaglandin endoperoxide H2 (PGH2), the precursor of the prostanoids--thromboxane A2 and the prostaglandins (PGD2, PGE2, PGF2α and PGI2) (1-4). Nonsteroidal anti-inflammatory drugs (NSAIDs) are commonly used to treat inflammation, pain and fever, and these actions are generally attributed to inhibition of COX-2 (5-7). Prostanoids are lipid mediators that normally act in a paracrine and autocrine manner to coordinate intercellular events stimulated by a circulating hormone (1). Their over-production is associated with pathologies such as tumorigenesis and arthritis whereas atherogenesis is associated with decreased formation of certain prostanoids (8-12).
COX-1 and COX-2 are the products of different genes (1, 13, 14). COX-1 is present in many but not all cell types (15) and when present is usually expressed constitutively. COX-1 gene expression is developmentally controlled and can be upregulated by tumor-promoting phorbol esters or growth factors as seen with primary megakaryocytes and megakaryoblast cell lines (Table 1) (16-20). In contrast to COX-1, COX-2 expression is typically transient. Depending on the cell type COX-2 expression can be rapidly induced by bacterial endotoxin (LPS), cytokines such as IL-1, IL-2, and TNF-α, growth factors, and the tumor promoter phorbol myristate acetate (PMA) (1, 13, 14) (Table 2). It should be noted that some cells in lung (21), brain (22) and kidney (23), pancreatic β-cells (24) , and gastrointestinal carcinomas (11, 25, 26) exhibit constitutive COX-2 expression.
Table 1.
COX-1 inducers
| Cell type | Inducers |
|---|---|
| Fibroblasts | IL-1β (125), TGFβ (126) |
| Vascular endothelial cells | VEGF (127), Shear stress (34), Estrogen (Estradiol-17ß) (36) |
| Tracheal epithelial cells | Phorbol myristate acetate (PMA) (128) |
| Gallbladder | Bradykinin (BK) (35) |
| Neuroblastoma | Retinoic acid (129) |
| Monocytes/Megakaryoblasts | PMA (17, 18, 31), Tobacco carcinogens (130) |
Table 2.
ACOX-2 inducers
| Cell type | Inducers | Signaling pathways |
|---|---|---|
| Fibroblasts | ||
| NIH 3T3 fibroblasts | Serum, PDGF, PMA, v-src (39, 40, 131) | Ras/MEKK-1/JNKK(MKK4)/JNK(39, 40, 131) |
| IL-1β, TNFα, PGE2, PGE3 (42) | Ras/Raf-1/MAPKK/ERK-1/2 (39, 40, 42, 131) | |
| Human foreskin fibroblasts (HFF) | PMA (43), TNFα, IL-1β, LPS (46) | C/EBP (43), AP-1 (43), NF-.B (44, 45) (46) |
| Human intestinal myofibroblasts | IL-1α (47) | PKC, NF-.B, ERK-1/2, JNK, p38 (47) |
| Human synovial fibroblasts | IL-1β (48) | p38, NF-.B, AP-1/ATF (48) |
| Endothelial cells | ||
| Bovine arterial endothelial cells (BAECs) | LPS, PMA (132) | NF-.B, C/EBP (132) |
| Human umbilical vein endothelial cells (HUVEC) | hypoxia (51, 53), VEGF (53) PMA, IL-1ß (53), TNF-a (53), LPS (50), thrombin, cholesterol deprivation (55), IL-1, vanadate (54), platelet derived thromboxane A2 (52) | NF-.B, Sp1 (51), PKC (53) |
| Smooth Muscle cells | ||
| Human pulmonary artery smooth muscle cells (HPASMC) | BK (56), TGFß, IL-1ß, hypoxia (133) | Gs |
| Human airway smooth muscle cells (HASM) | BK, IL-1ß (57), indomethacin, flurbiprofen, NS-398, 15d-PGJ2 (59) | NF-?B, CREB, C/EBP (57)PPAR? (59) |
| Epithelial cells | ||
| Human mammary epithelial cells (HER) | PMA (60) | PKC, ERK-1/2, p38, JNK (60) |
| Human gastric epithelial cells (hGECs) Granulosa Cells | PMA, Helicobacter pylori, (62) Gonadotropin, forskolin, FSH, phorbol didecanoate, LH (65-69) | MEK-1/2, CREB, USF-1/2 (62) PKA (65-69) |
| Bone | ||
| MC3T3-E1 osteoblasts | Bone morphogenic proteins (BMP-2) (71), bFGF, EGF, TGF-α/β, IL-1, (74), thrombin, shear stress (72), TNF-α (73), parathyroid hormone (PTH), PMA (75), BK, epinephrine, prostaglandins, serum (74, 75) | MEKK, JNK, NF-?B (74) C/EBP, AP-1, CREB (72) C/EBP, NF-?B (73), |
| Monocytes/Macrophages | ||
| Human U937 monocytic cells | PMA, LPS, TNF-a, IL-1 (78), MP (79) | C/EBP, NF-?B (78) PI3K/PKC, ERK-1/2, p42/p44 MAPK p38, JNK-1 (79) |
| THP-1 monocytic cells | High glucose (HG) (80, 81) | PKC, p38 MAPK, CREB, NF-?B (80, 81) |
| RAW 264.7 macrophages | Catalase (83), LPS, IL-1, TNFa, peptidoglycan (84), double-stranded RNA (85), | NF-?B, PI3K, ERK, p38, JNK (83) NF-?B, Ras/Raf-1, ERK (84) NF-?B (85) |
In general, COX-2 is completely absent from cells. The absolute levels of COX-2 protein have generally not been measured. While not necessarily a representative example, when murine NIH 3T3 fibroblasts are induced to express COX-2, the maximum protein levels reach those equivalent to about 50% those of COX-1 (i.e. COX-2 represents about one third of the total COX (134)). In the case of 3T3 fibroblasts COX-1 is approximately 1/2000 of total cell protein (unpublished observation).
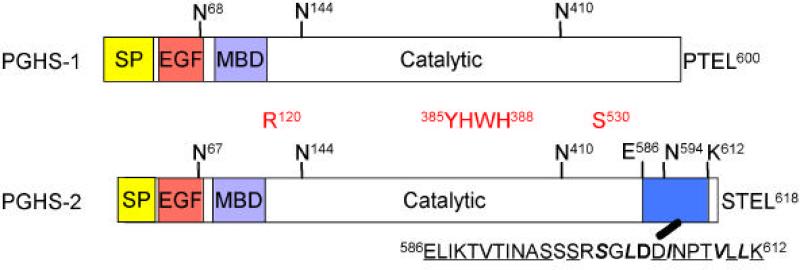
Although the COX-1 and COX-2 proteins are highly homologous, they have some obvious sequence and structural differences including different signal peptides and significant sequence differences in their membrane binding domains (1, 3, 4). Most notably, COX-2 but not COX-1 contains a unique 27 amino acid sequence near its C-terminus (Fig.1). This is an instability element involved in COX-2 protein degradation that is discussed later in this review.
Fig. 1. Comparison of the domain structures of PGHS-1/COX-1 and PGHS-2/COX-2.
Both isoforms have a signal peptide (SP), an epidermal growth factor (EGF)-like domain, a membrane-binding domain (MBD), and a catalytic domain. PGHS-2 has a shorter signal peptide than PGHS-1. The C-terminal PTEL and STEL sequences of PGHS-1 and PGHS-2, respectively, are ER targeting signals. The N-glycosylation sites are also shown; note the additional N-glycosylation site at Asn594 in PGHS-2. R120, Y385 and H388 are important catalysis; S530 is the aspirin acetylation site; the amino acid sequence of the 27 amino acid instability element (27-IE) comprising residues 586-612 is shown in blue. Numbering for PGHS-1/COX-1 begins with the Met at the translation start site. The numbering for PGHS-2/COX-2 parallels the COX-1 numbering in which the start of the mature, processed COX-2 protein has the same number as the start of the mature, processed COX-1.
While relatively little information has been published about the transcriptional regulation of the COX-1 gene or the mechanism of COX-1 or COX-2 protein degradation, the regulation of COX-2 gene expression has been investigated rather extensively. In this review, we focus on recent advances in our understanding of COX-1 and COX-2 gene expression and the degradation of COX-1 and COX-2 proteins.
1. Regulation of COX-1 gene expression
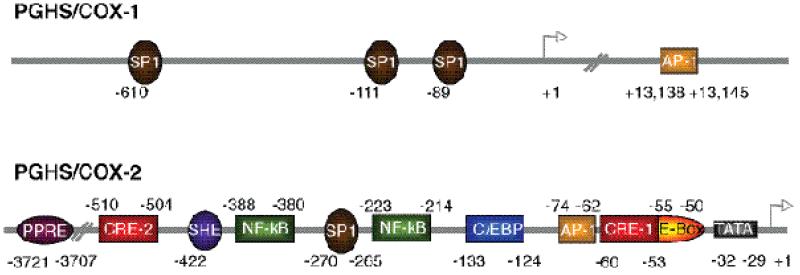
The human COX-1 gene (Ptgs1), located on chromosome 9, is approximately 22 kb in length and contains 11 exons (27, 28). The COX-1 promoter lacks a TATA or CAAT box, has a high GC content, and contains several transcriptional start sites. All of these properties are characteristic of “housekeeping” genes (29, 30). Although COX-1 protein is constitutively expressed in most tissues, COX-1 is upregulated by PMA in some cell types including monocytes (31), human umbilical vein endothelial cells (HUVEC) (32), and primary megakaryocytes and megakaryoblasts (17, 33) (20) as they differentiate during development. COX-1 is also induced by shear stress in HUVEC cells (34), by bradykinin in the gallbladder (35), and by estrogen in endothelial cells (36). Within the 5’ flanking region of the human COX-1 promoter there are three functional Sp1 binding sites at -610, -111, and -89 relative to the ATG start site (Fig. 2). In HUVECs the Sp1 motifs at -610 and -111 contribute to constitutive expression but not to PMA-induced expression of the COX-1 gene (32). The Sp1 binding site at -111 is required for PMA -induced COX-1 transcription in the megakaryoblast cell line MEG-01 (20) and for estradiol induction of COX-1 in ovine endothelial cells (36) where both the Sp1 site at -111 and to an even greater extent the Sp1 site at -89 are required for COX-1 induction. These three Sp1 sites are the only func tional regulatory elements that have been described within 2 kb upstream of the ATG start codon. However, there is an AP-1 site located in intron 8 of the COX-1 gene that is highly conserved across species and that interacts with the -111 SP-1 site of the promoter to regulate PMA-induced expression of COX-1 in MEG-01 cells (20).
Fig. 2. Schematic representation of the functional regulatory elements in the human COX-1 and COX-2 gene promoters1.
Cis-acting elements found within the COX promoters are noted in the shaded boxes and their location relative to the transcriptional start site are noted above or below each element. Note that the COX-1 promoter lacks a TATA box. The correct designations for the COX-1 and COX-2 genes are Ptgs1 and Ptgs2, respectively.
2. Regulation of COX-2 gene expression
The human COX-2 gene (Ptgs2), located on chromosome 1, is about 8.3 kb long and has 10 exons. Except for the first exon the intron/exon boundaries of the COX-1 and COX-2 genes are the same (29, 37). There are two major transcripts of COX-2--a 4.5 kb full length mRNA and a 2.6 kb polyadenylated variant that lacks the terminal 1.9 kb of the 3’-untranslated region (UTR). The 3’-UTR of the human COX-2 gene contains 23 copies of the ‘ATTTA’ RNA instability element that participates in post-transcriptional regulation of COX-2 expression (38). Sequence analysis of the 5’-flanking region of the human COX-2 gene has identified several potential transcriptional regulatory elements, including a peroxisome proliferator response element (PPRE), two cyclic AMP response elements (CRE), a sterol response element (SRE), two nuclear factor kappa B (NF-κB) sites, an SP1 site, a CAAT enhancer binding protein (C/EBP, or nuclear factor for interleukin-6 expression (NF-IL6)) motif, two AP-2 sites, an E-box, and a TATA box (Fig. 2). The promoter regions of COX-2 genes have sequences of typical immediate early genes (29).
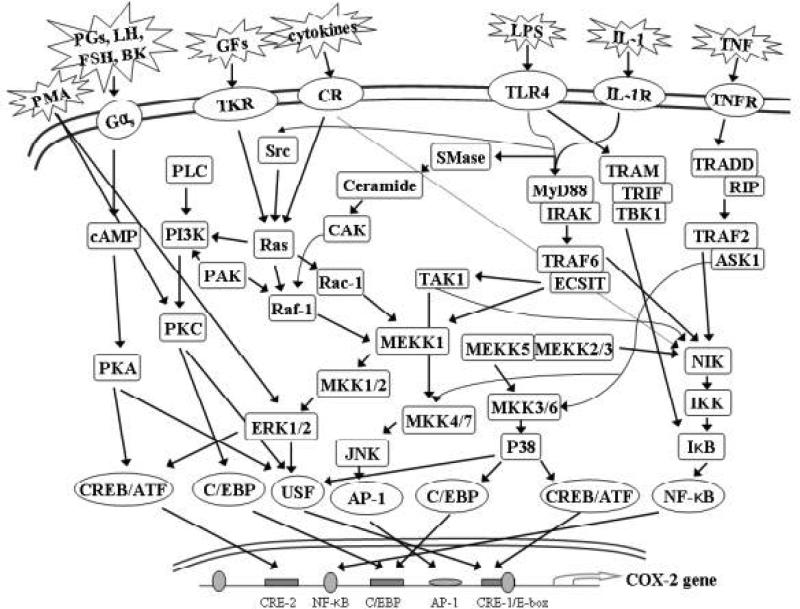
There are some subtle interspecies differences in the sequences of the human, mouse, rat, cow and horse COX-2 genes. For example, the mouse COX-2 promoter has one NF-κB motif and two C/EBP sites instead of the two NF-κB sites and one C/EBP motif found in the human COX-2 promoter. Transcriptional regulation of the COX-2 gene is very complex in that it can involve numerous signaling pathways, and the mechanism varies depending on the specific stimulus and the cell type. Here, we compare COX-2 gene regulation in several different cell types (Table 2) and the regulatory elements (Fig. 2) and signaling pathways (Fig. 3) involved in each system.
Fig. 3. Signaling Pathways involved in PGHS-2/COX-2 gene induction.
The figure illustrates the numerous signaling pathways that can be involved in the transcriptional regulation of PGHS-2/COX-2 expression. As detailed in Table II, only certain of these pathways are operative in individual cell types.
2.1. Transcriptional Regulation of COX-2 in Fibroblasts
COX-2 expression is upregulated in murine NIH 3T3 fibroblast lines in response to fetal calf serum, PDGF, PMA, v-src (39-41), IL-1β, TNFα, PGE2, and PGE3 (Table 2) (42). v-Src oncogene-induced COX-2 expression in NIH 3T3 cells is mediated mainly by CREB and AP-1 transcription factors working through CRE-1. AP-1 activity at the CRE-1 is modulated by the Ras/MEKK-1/JNKK(MKK4)/JNK pathway leading to c-Jun phosphorylation and the Ras/Raf-1/MAPKK/ERK1/2 pathway that activates secondary response genes such as c-fos that form heterodimers with c-Jun (39, 41). In addition to the CRE-1 site, there is an E-box element within the mCOX-2 promoter that is also required for v-src-induced COX-2 expression in NIH 3T3 fibroblasts. Serum, PGE2 and PGE3 also induce COX-2 mRNA through Ras/JNK- and Ras/ERK-mediated signaling pathways (42). Transcriptional activation of COX-2 by serum, PGE2, and PGE3 requires a C/EBP site and the CRE-1 site; however, the NF-κB site and E-box element are not involved. This indicates that COX-2 gene activation is moderated by distinct combinations of cis-regulatory elements and trans-acting factors that are specific for different stimuli.
Human foreskin fibroblasts (HFF) express COX-2 protein after 2 hrs of PMA treatment and the protein level increases over a 4 hr period (43). In contrast, serum-treated NIH 3T3 fibroblasts exhibit more transient COX-2 expression lasting just 2 hrs. COX-2 transcriptional activation in PMA-treated HFF is through the CRE-1 and the C/EBP element. In the basal state, C/EBPd binds to both the C/EBP site and the CRE-1. In response to PMA, C/EBPδ at the C/EBP site is replaced by C/EBPβ, C/EBP β-LAP (activating form) or C/EBPβ-LIP (inhibitory form); binding of C/EBPδ to the CRE-1 site is unchanged even after 4 hr of stimulation. PMA stimulation induces COX-2 gene expression in HFF cells by reducing C/EBPδ protein levels and increasing C/EBPβ phosphorylation which enhances its DNA binding affinity and increases C/EBPβ-LAP binding to the C/EBP element. These changes lead to recruitment of the coactivator p300 to the transcriptional machinery to initiate transcription.
PMA treatment of HFF also induces the binding of the c-Jun/c-fos heterodimer to the CRE-1 site (44). Stimulation by TNFα relies on the inducible binding of the p65/p50 heterodimer to the NF-kB site (44-46). The CRE-1 motif is also bound by CREB2/ATF2 transcription factors; however, their recruitment to the COX-2 promoter does not depend on PMA or TNFα stimulation. In addition to PMA and TNFα, IL-1β and LPS can induce COX-2 expression in HFF (46).
p300 is the predominant transcriptional coactivator in HFF with CBP being barely detectable (46). p300 and PCAF (p300/CBP associated factor) are recruited by transcription factors that are bound to their cognate cis-elements within the COX-2 promoter. p300 and PCAF direct gene expression by acetylating nearby histone tails leading to a “loosened” chromatin structure that is more accessible to transactivators. HDAC-1 (histone deacetylase-1), on the other hand, cleaves acetyl moieties from histone tails and thus negatively regulates COX-2 expression. A balance between acetylation and deacetylation is required to maintain COX-2 expression at physiologically relevant levels (46).
IL-1α induces COX-2 expression in human intestinal myofibroblasts (47). Activation of protein kinase C (PKC), NF-κB, ERK-1/2, and JNK are all required for optimal COX-2 transcriptional activation whereas p38 activation is involved in stabilizing COX-2 mRNA. COX-2 mRNA levels increase after 1 hr of IL-1α treatment peak at 8 hr and then remain the same for the next 16 hr. COX-2 protein expression is observed after 4 hr of IL-1α stimulation and is maximal at 16-24 hr. In contrast, with serum induction, COX-2 mRNA levels decrease to baseline after 4 hr.
In human synovial fibroblasts, IL-1β treatment also results in prolonged expression of COX-2 which is mediated by mRNA stabilization involving p38 MAP kinase (48). IL-1α stimulation induces binding of p65/p50 to the NF-?B motif and of AP-1/ATF transcription factor to the CRE-1 while binding of nuclear proteins to the C/EBP site is not inducible.
2.2. Transcriptional Regulation of COX-2 in Endothelial Cells
LPS and PMA act synergistically to induce COX-2 mRNA in bovine arterial endothelial cells (BAECs) (49). This COX-2 transcriptional activation is mediated by the NF-κB and C/EBP elements and unlike in fibroblasts, the CRE-1 motif appears not to be required. However, the CRE-1 element is required for COX-2 induction when C/EBPδ is activated independently (i.e. by transfection), suggesting that there is an interplay between the CREB and C/EBPδ transcription factors bound to their cognate sites during COX-2 gene activation.
COX-2 expression and prostaglandin synthesis in human umbilical vein endothelial cells (HUVEC) can be stimulated by physical stimuli and by agents, such as, PMA, IL-1β, TNF-α, LPS, vascular endothelial growth factor (VEGF), thrombin, hypoxia, vanadate (an inhibitor of protein-tyrosine phosphatases), cholesterol deprivation and platelet-derived thromboxane A2 (Table 2) (50-55). Hypoxia causes increased binding of p65 to the proximal NF-?B site of the COX-2 promoter in HUVEC without influencing the levels of cytoplasmic p65 or I?Ba , an inhibitory protein that binds cytoplasmic p65/p50. Promoter deletion analysis implicate other regulatory cis-elements and their cognate transcription factors such as C/EBP, AP-1, and the CRE-1/E-box in this response, however their roles have yet to be determined. Hypoxic HUVEC also exhibit nuclear localization of Sp1 while Sp3 protein levels are unaffected, thus elevating the Sp1/Sp3 ratio in the nucleus. Sp1 and Sp3 protein bind to the SP1 motif in the COX-2 promoter and mediate COX-2 transcription (51). High concentrations of cholesterol down regulate COX-2, whereas cholesterol deprivation upregulates COX-2 gene expression in endothelial cells. Cholesterol-dependent COX-2 regulation is mediated by sterol response element binding protein (SREBP) through an SRE located at -422 from the transcription start site (55).
2.3. Transcriptional Regulation of COX-2 in Smooth Muscle Cells
Bradykinin (BK) binds to specific cell surface G protein-coupled receptors and induces COX-2 expression in human pulmonary artery smooth muscle cells (HPASMC) (56). COX-2 mRNA increases 2 fold after 1 hr of BK treatment and returns almost to basal levels within 4 hrs indicating that COX-2 gene activation is transient in this system. BK-induced COX-2 expression is transcriptionally regulated by CREB binding to the CRE-1 motif. COX-2 transcription is also activated by cytosolic phospholipase A2 (cPLA2)-mediated arachidonic acid release and an autocrine loop involving the action of endogenous PGE2 on Gs-linked EP2 and EP4 receptor s (56, 57). BK activates IL-1β mRNA expression in human lung fibroblasts by activating NF-κB, and in HeLa cells BK activates C/EBP during IL-8 gene expression. However, COX-2 gene activation by BK in HPASMC does not involve either the NF-?B or C/EBP motifs. Thus, BK can induce transcription in a gene- and transcription factor-specific manner.
BK and IL-1β activate COX-2 expression in human airway smooth muscle cells (HASM) in a stimulus-specific manner (57). BK induces COX-2 protein expression more quickly (1 hr) than does IL-1β (2 hr). BK induction is also more transient – COX-2 protein disappears within 16 to 24 hrs. With IL-1β-stimulated COX-2 protein can be sustained beyond 24 hrs. Both stimuli involve CREB, but not c-Jun binding to the CRE-1 element. However, IL-1β induced COX-2 transcription requires two other cis-elements, the NF-κB and the C/EBP sites, for maximal COX-2 gene expression. COX-2 transcriptional regulation by BK and IL-1β involving different sites in the COX-2 promoter may due to the different chromatin structure resulting from different patterns of histone modification (57, 58).
While NSAIDs inhibit COX activity, there has been speculation that they induce COX-2 expression in HASM (59). Indomethacin, flurbiprofen, NS-398 (a selective COX-2 inhibitor) and 15d-PGJ2 induce COX-2 expression and enhance IL-1β-induced COX-2 expression through the peroxisome proliferator response element (PPRE) in the COX-2 promoter. NSAID treatment causes nuclear translocation of PPARγ but not NF-κB in HASM cells. PPARγ binds to the PPRE of the COX-2 promoter and can transactivate the gene.
2.4. Transcriptional Regulation of COX-2 in Epithelial Cells
In contrast to what is reported for NSAID-treated smooth muscle cells, PPARγ ligands such as ciglitazone and 15d-PGJ2 inhibit PMA-mediated induction of COX-2 in human mammary epithelial cells (184B5/HER) (60). PMA-stimulated COX-2 induction in these cells is mediated by the CRE-1. PMA treatment leads to c-Jun, c-Fos, and ATF-2 binding to the CRE-1 site; this can be prevented by PPARγ ligands by inhibition of AP -1 activity. PMA treatment activates PKC, ERK-1/2, p38, and JNK signaling pathways in human mammary epithelial cells. Activated ERK-1/2 induces c-fos expression, p38 phosphorylates ATF-2 and ATF-2 dimerizes with c-Jun and induces more c-Jun expression. Activated JNK both induces c-Jun expression and phosphorylates c-Jun, allowing it to activate COX-2 gene expression. PMA-induced COX-2 transcription in this system also requires a functional CBP/p300 coactivator complex having HAT activity. Retinoic acid (RA) and other nuclear receptor ligands, and carnosol, a phenolic antioxidant isolated from rosemary oil, suppress PMA-mediated COX-2 transcription. RA is thought to downregulate COX-2 expression by a receptor-dependent mechanism whereby the ligand-bound receptor complex sequesters the CBP/p300 that is available for AP-1 mediated induction of COX-2. Carnosol reduces binding of AP-1 to the CRE-1 element by inhibiting PKC, ERK1/2, p38, and JNK signal transduction pathway (61).
Helicobacter pylori and PMA stimulate transient COX-2 transcription (1.5 hr to 4.5 hr after stimulation) in human gastric epithelial cells (hGECs) through the CRE-1/E-box, but not via the C/EBP, AP-1 or NF-κB sites (62). Helicobacter pylori activates the MEK-1/2 kinase cascade, which in turn activates CREB and USF-1/2 transcription factors. Unlike in PMA-stimulated HER cells, in hGECs CREB protein but not c-Jun binds to the CRE-1 site. USF-1/2 binds to the E-box and activates COX-2 gene transcription. Helicobacter pylori treatment does not require any cis-regulatory element other than the CRE-1/E-box element. Whether there are interactions between CREB and USF-1/2 transcription factors bound to this site remains to be determined.
2.5. Transcriptional Regulation of COX-2 in Granulosa Cells
Prostaglandins play important roles in several aspects of female reproduction. For example, follicular prostaglandin levels increase dramatically prior to ovulation (63) and NSAIDs such as indomethacin block ovulation (63). COX-2 deficient female mice are infertile because of problems with ovulation, fertilization, implantation and decidualization (64).
COX-2 is induced by gonadotropins in granulosa cells prior to ovulation, and also by forskolin, follicle-stimulating hormone (FSH), luteinizing hormone (LH), and phorbol didecanoate all acting through the protein kinase A (PKA) signaling pathway (65-69). The duration of COX-2 induction in granulosa cells varies from 2-4 hr in rats to 18-30 hr in cows and mares. Bovine and rat COX-2 promoter analyses revealed that the proximal 150-200 bp upstream of the transcriptional start site is sufficient to confer inducible promoter activity. This region of the COX-2 promoter contains the C/EBP, CRE-1 and E-box elements, and mutational analyses have suggested that only the E-box element is required for promoter activity in rat granulosa cells but that both the C/EBP and E-box elements are required in bovine granulosa cells. Antibody supershift EMSAs have established that USF-1 and USF-2 bind to the E-box element of both the bovine and the rat COX-2 promoters and that gonadotropin treatment has no effect on the binding of protein complexes to the E-box. In contrast, C/EBPβ binds to the bovine C/EBP site in the absence of stimulation and gonadotropin treatment decreases C/EBPß binding. However, in rat granulosa cells, there is no C/EBPβ binding to the C/EBP site prior to treatment but C/EBPβ binding rapidly increases upon treatment with gonadotropin (68, 69). This difference may explain the molecular basis for the variation in the duration of COX-2 expression among species.
Recent studies have indicated that phosphorylation of members of the upstream signaling factor (USF)-transcription factors is involved in the regulation of COX-2 promoter activity in granulosa cells (70). PKA-mediated USF phosphorylation increases USF binding to the E-box promoter element thereby enhancing COX-2 promoter activity. In contrast, activation of the PKC pathway with PMA had no stimulatory effect on the COX-2 promoter. Thus, activation of the cAMP/PKA pathway by luteinizing hormone and consequent phosphorylation of USF proteins that bind to the E-box appears to be essential for induction of COX-2 in granulosa cells.
2.6. Transcriptional Regulation of COX-2 in Bone
COX-2 plays a significant role in bone resorption and formation. The regulation of COX-2 expression has been extensively studied in MC3T3-E1 osteoblasts. MC3T3-E1 cells established from newborn mouse calvaria, have the capacity to differentiate into osteoblasts and osteocytes and to form calcified bone tissue in vitro. COX-2 can be up-regulated by many stimuli in MC3T3-E1 cells including BMP-2, bFGF, EGF, TGF-α/β, IL-1, TNF-α, PTH, thrombin, bradykinin, forskolin, epinephrine, and prostaglandins (PGI2, PGE2, PGF2α) or their stable analogues (Table 2) (29, 71-75).
TNF-α treated MC3T3-E1 cells exhibit a triphasic change in COX-2 mRNA expression. There is an initial increase that reaches a maximum 2 hr after TNF-α stimulation. This is followed by a decrease at 3 hr, and then a second increase in COX-2 mRNA at 6-12 hr. The second increase is due to PGE2 produced by the COX-2 formed in the first phase (73). Binding of p65/p50 to the NF-κB site and C/EBPβ to the C/EBP-1 site at 1 hr are necessary for TNF-α induced COX-2 expression in MC3T3-E1 osteoblasts. However, different transcription factors and cis-elements may be involved during the first and second increases in COX-2 transcription.
bFGF, PDGF, PGE2 or a combination of TNF-α and IL-1β stimulation causes after 4 hr the activation of c-Jun, MEKK and JNK signaling pathways in MC3T3-E1 cells (74). Promoter analysis identified a second C/EBP motif in the murine COX-2 promoter that is required together with the C/EBP-1 site for optimal promoter activity. C/EBPβ and C/EBPδ transactivate murine COX-2 gene expression via C/EBP sites. The CRE-1 element is necessary for COX-2 transcriptional activity while neither the NF-κB site nor the E-box are not required for bFGF, PDGF, PGE2, or TNFα+IL-1β induced COX-2 expression in MC3T3-E1 osteoblasts.
When MC3T3-E1 cells are treated with PMA or serum for 3 hr, the -371/+70 region of the COX-2 promoter, which does not include the CRE-2 or the NF-?B sites, is sufficient to activate COX-2 transcription (75). PMA or serum treatments cause inducible binding of c-Jun and c-fos to the AP-1 site and constitutive binding of nuclear protein to the CRE-1 site. This suggests that there is a cooperative interaction between transcription factors bound to the AP-1 site and the CRE-1 in trans-activating COX-2 transcription. The CRE-1 is quantitatively more important for the serum response and the AP-1 site is more important for the PMA response (75).
Mechanical loading is crucial for maintaining bone mass and integrity. This generates extracellular matrix deformation and fluid flow. Mechanical stimuli such as loading and fluid shear stress cause the production of signaling factors. In bone forming cells, prostaglandins are formed that modulate the overall process of bone metabolism. Fluid sheer stress induces COX-2 expression through a process involving the activation of a cytoskeleton-associated Ca2+ channel, phospholipase C, PKA, PKC and phospholipase A2 (76, 77). COX-2 mRNA levels increase after 1 hr of fluid sheer stress and show a sustained increase for up to 9 hr of treatment. Sustained COX-2 gene activation likely occurs via the PKA pathway. The CRE-2, NF-κB, C/EBP-2 and E-box appear not to play roles in fluid sheer stress-mediated COX-2 transcription in MC3T3-E1 cells. Instead, the process involves the C/EBP-1 site bound inducibly by C/EBPß, the AP-1 element bound inducibly by c-Jun/AP-1 transcription factors, and the CRE-1 constitutively bound by CREB. This is similar to the mechanisms by which PMA and serum induce COX-2 gene activation.
2.7. Transcriptional Regulation of COX-2 in Monocytes/Macrophages
Monocytes/macrophages are crucial to the development of the immune response because of their ability to present antigens and secrete mediators of inflammation including cytokines and prostaglandins. These products regulate cell proliferation, differentiation, and, in general, the acquisition of immune functions. Regulation of prostaglandin synthesis in monocytes is important in immune responses
Human U937 monocytic cells undergo morphologic and functional changes and differentiate to macrophage-like cells when treated with PMA. U937 cells do not express COX-2 mRNA or protein in the undifferentiated state, but during differentiation, low levels of COX-2 are expressed. COX-2 is further induced by inflammatory stimuli such as LPS, TNF-α, or IL-1 (Table 2) (78). LPS-induced COX-2 expression in U937 cells involves the CRE-1, the C/EBP site and the downstream NF-κB elements.
Platelet microparticles (MP), formed by platelet activation, also activate COX-2 gene expression and lead to prostaglandin production in U937 cells (79). MP activates PI-3-kinase resulting in the transient activation of several PKC isoforms (PKC-β/δ/ς/?), ERK-1/2, p42/p44 MAPK, p38, and the sustained activation of JNK-1 as well as activation of c-Jun and Elk-1 transcription factors. Curiously, MP-induced COX-2 expression does not involve the CRE-1.
Prostaglandins play a role in complications of diabetes such as hyperglycemia, accelerated atherosclerotic and inflammatory disease, oxidant stress and the formation of advanced glycation end products (AGEs) (80-82). High glucose (HG) or AGE treatment of THP-1 monocytic cells, which are similar to U937 monocytes, leads to a significant increase in COX-2 mRNA and protein. The increase in COX-2 mRNA is predominantly due to transcriptional upregulation (81). AGEs act via the RAGE (receptor for AGE) and both AGEs and S100b, a specific ligand for RAGE, activate multiple signaling pathways including those involving p38, MEK/ERK, oxidant stress, PKC, and NF-κB; however, the JNK pathway is not activated in THP-1 cells in response to AGEs (82). Interestingly, AGEs and S100b induce COX-2 via the distal NF-κB site (-455/-428) while HG induces COX-2 transcription via the proximal NF-κB site (-232/-205) (80).
HG treatment of THP-1 monocytes activates PKC and the p38 MAPK pathway but not the ERK or JAK-STAT pathways. HG-induced COX-2 expression requires the CRE-1 element and activation of the CREB transcription factor as well as NF-?B activation (80). The association of CBP/p300, p/CAP (p300/CBP associated protein) and the p65 NF-?B transcription factor with the COX-2 promoter were evaluated by chromatin immunoprecipitation assays (ChIPs) with HG treated THP-1 cells (81). CBP, p/CAP, and p65 are recruited to the COX-2 promoter sequentially. p65 and CBP association occurs after 16 hr of HG treatment, peaks after 24 hr, and decreases at 48 to 72 hr. p/CAP and p50 are recruited to the COX-2 promoter in parallel, appear as early as 16 hr after HG stimulation, increase over time, and remain elevated at 72 hr. The transcriptional repressor, HDAC-1 is associated with the COX-2 promoter under basal conditions. After HG treatment, the association decreases with time as the binding of CBP increases. This suggests that recruitment of activated transcription factors such as p65 to the COX-2 promoter after HG stimulation in THP-1 monocytes is enabled by dissociation of HDAC-1 from the promoter.
The murine macrophage cell line RAW 264.7 has been used extensively as a model for examining macrophage activation and the inflammatory response. Various inflammatory mediators and cytokines, catalase, peptidoglycan (a cell wall component of gram-positive bacteria), double-stranded RNA, viral infection, and LPS stimulate COX-2 induction and prostaglandin formation in RAW 264.7 macrophages (29, 83-85).
LPS forms a complex with LPS-binding protein (LBP) and this complex interacts with a monocyte differentiation antigen CD14 (86-88). The binding of the LPS/LBP complex to CD14 and the toll-like receptor 4 (TLR4) induces receptor dimerization. Members of the toll-like receptor family recognize conserved microbial structures such as LPS. LPS activates multiple signaling pathways that are critical for induction of the immune response causing the release of TNF-α, IL-1β, and prostaglandins from macrophage and monocyte cells (89). Upon receptor dimerization, the intracellular Toll/IL-1R (TIR) domain of TLR4 associates with two pairs of adaptor proteins: (a) myeloid differentiation factor (MyD88) plus in IL-1R associated kinase (IRAK) and (b) Toll/IL-1R domain-containing adaptor inducing IFN-β (TRIF; TICAM-1) plus TRIF-related adaptor molecule (TRAM). This facilitates the assembly of the toll receptor complex (90, 91). MyD88/IRAK recruits TNF receptor associated kinase-6 (TRAF-6), and the evolutionarily conserved signaling intermediate in toll (ECSIT) (87, 88), leading to activation of transforming growth factor β-activated kinase (TAK-1), which is a MAPKKK that initiates JNK, ERK, and p38 MAP kinase cascades. TRAF-6 can also activate the IκBα kinase complex (IKK) through NF-κB inducing kinase (NIK), leading to NF-κB activation (88). In RAW 264.7 macrophages, activated MAP kinases and NIK up-regulate COX-2 expression as well as expression of other pro-inflammatory cytokines (89). Another signaling pathway, which is MyD88 independent, and involves TRIF/TRAM adaptor proteins leads to delayed NF-κB activation (90, 91).
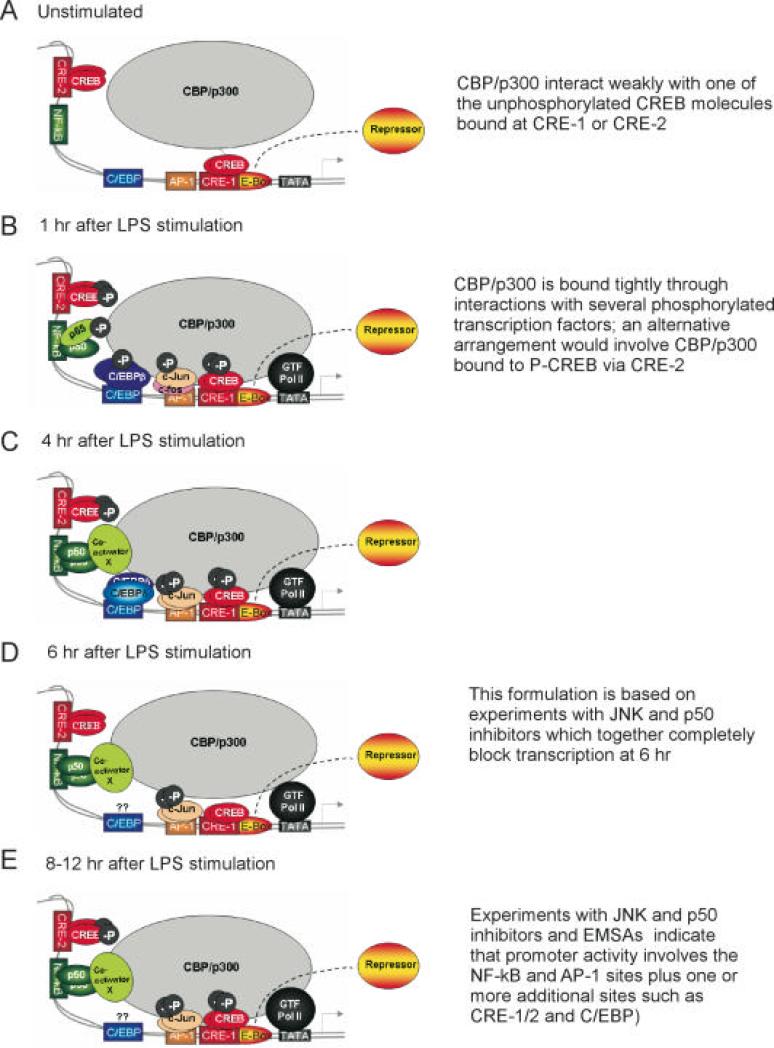
It has been shown that COX-2 induction in LPS-stimulated RAW 264.7 cells consists of an early phase of rapid induction of COX-2 mRNA expression after 1 hr of LPS-treatment followed by a phase of sustained mRNA expression (92). It was suggested that these different phases of mRNA expression required different sets of transcriptional activators. Consistent with this idea, the early phase of COX-2 expression was shown to be independent of de novo protein synthesis, whereas in the second phase, synthesis of C/EBPδ was required. Furthermore, a C/EBP β homodimer was bound to a C/EBP element in the initial phase while a C/CBP β·δ heterodimer bound to the C/EBP element during the second phase (92, 93). It has also been demonstrated that CREB and NF-κB are important in LPS-induced COX-2 transcription in RAW 264.7 cells. Taken together, these studies suggested that the NF-κB and C/EBP sites and the CRE-1 are important for regulating COX-2 transcription in LPS-stimulated macrophages and that COX-2 transcription in this system consists of several phases that lead to persistent gene activation (Fig. 4).
Fig. 4. Model depicting the actions of transcription factors at the COX-2 promoter at different times after initiating LPS-induced COX-2 expression in RAW 264.7 macrophages.
As reported previously (94) , during the first 4 hr of LPS treatment (Fig. 4B), we have proposed that CREB bound at CRE-1 and CRE-2 becomes phosphorylated enhancing the recruitment of CBP and p300 coactivators. The 65/p50 heterodimer is the major species bound to the NF-κB site and p65 is presumably phosphorylated, and the C/EBPβ homodimer binds to the C/EBP element. In the proximal region of the COX-2 promoter a heterodimer of newly phosphorylated c-Jun/c-fos becomes bound to the AP-1 site. Phosphorylated p65, C/EBPβ, and phosphorylated c-Jun have all been shown to interact with CBP/p300. This suggests that the various activated transcription factors acting in concert may be recruiting CBP/p300 and general transcription factors to the COX-2 promoter leading to active transcription during the first 4 hr; as depicted in Fig. 4C, at about 4 hr a C/EBPδ/C/EBPβ heterodimer replaces a C/EBPβ homodimer at the C/EBP site (92). COX-2 transcription slows during the middle phase (4-8 hr); this phase is dominated by the AP-1 and NF-κB sites and involves phosphorylated c-Jun homodimers bound to the AP-1 site and p50 homodimers bound to the NF-κB site (94). The rate of COX-2 transcription increases again after about 8 hr of LPS treatment, and the transcription rate remains elevated at 12 hr (Fig. 4E). During this late stage (8-12 hr), the concentration of phosphorylated c-Jun increases leading to a relatively higher level of phosphorylated c-Jun homodimer bound to the AP-1 site; binding of p50/p50 to the NF-κB site also occurs at levels comparable to those seen during the middle phase of induction (94). The amount of CREB bound to the CRE-2 and CRE-1 elements remains the same before and throughout the LPS treatment, however CREB may be phosphorylated during the late phase of LPS stimulation. Binding of C/EBP transcription factors to the C/EBP site may also contribute to enhanced transcription during the late phase. Again, all these transcriptional activators are likely to be facilitating transcription by binding to CBP/p300. It is clear that repression is mediated via the E-Box, but the mechanism and proteins involved have not been determined (94). From Ref. (94) with permission.
Using nuclear run-on assays, RT-PCR and northern blot analyses we have shown recently that COX-2 gene transcription is rapidly increased and sustained during the 12 hr after LPS stimulation in RAW 264.7 cells (94). These findings indicate that COX-2 is mainly regulated at the transcriptional level in this system. In addition to a previously identified CRE-1, we identified a second functional cAMP response element (CRE-2) within the COX-2 promoter. The CRE-2 is constitutively bound by CREB. On the other hand, the p65/p50 heterodimer inducibly binds to the NF-κB site after 1 hr of LPS treatment but after 4-12 hr is replaced by p50 homodimer (94). Thus, it is possible that the p65/p50 heterodimer together with CREB is required to initiate COX-2 gene transcription, and the p50 homodimer together with CREB is required to sustain the activation by interacting with a different set of transcriptional coactivators. It will be important to determine if there is a cooperative interplay between the factors bound to the CRE-2 and the NF-κB site in the COX-2 gene promoter.
We also found that an AP -1 element is required for optimal induction of COX-2 in LPS-stimulated RAW 264.7 cells (94). Consistent with what has been observed in PMA-treated MC3T3 cells, the CRE-1 site is constitutively bound by CREB/ATF transcription factor family members and the AP-1 element is inducibly bound by phosphorylated c-Jun and c-fos in the initial phase and by phosphorylated c-Jun dimers at the late phase of COX-2 transcription in LPS-treated RAW 264.7 macrophages (94). It is clear that COX-2 gene transcription is activated through multiple redundant mechanisms by LPS in macrophages. Fig. 4 provides a model of the events occurring at different times at the COX-2 promoter after treating RAW264.7 cells with LPS.
3. COX-1 and COX-2 protein degradation
There are large differences between the t1/2 values for the degradation of COX-1 and COX-2 proteins (26, 95-99). In several different cell types, co-expressing the enzymes, COX-1 degradation is slow or undetectable while the t1/2 for COX-2 protein degradation varies from 2-7 hr. These studies suggest that COX-2 degradation is specifically programmed to limit the amount of COX-2. There is anecdotal evidence that prolonged, overexpression of COX-2 is not well tolerated by certain types of cultured cells (R. Langenbach, personal communication), and COX-2 overexpression in vivo is associated with pathologies such as colon cancer (100-104).
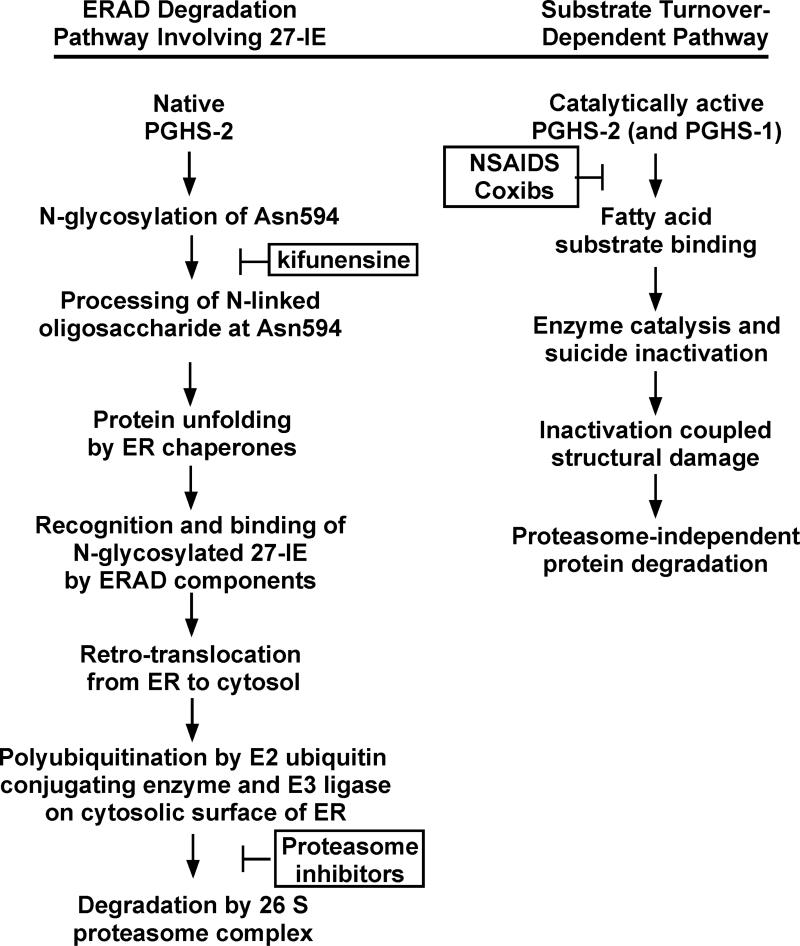
COX-2 is embedded in the luminal membrane of the ER. Its degradation appears to involve processing of the N-linked oligosaccharide at Asn594, unfolding presumably by ER chaperones such as BiP and calnexin, retrograde transport to the cytoplasm via the ER-associated degradation (ERAD) system (105-112), and finally ubiquitination and proteolysis by the 26S proteasome (26, 98, 99, 113) (Fig. 5). There is no literature on the mechanism of COX-1 degradation; however, both COX-1 and COX-2 are localized to the luminal surfaces of the ER and associated membranes of the nuclear envelope (114-119) and share many structural properties.
Fig. 5.
Pathways for PGHS-2/COX-2 protein degradation.
The molecular basis for the differences in the stabilities of COX-1 and COX-2 is due in part to the presence of a 27 amino acid instability element (27-IE) that is located near the C-terminus of COX-2 but absent from COX-1 (Fig. 1) (99) ; the 27-IE has no sequence or structural parallels in GenBank or the Protein Data Bank. We have recently found that a functional N-glycosylation site present at Asn594 (120, 121) is involved along with at least some other segments of the 27-IE with entry into the ERAD pathway (99). Presumably the oligosaccharide at Asn594 undergoes modification involving addition and cleavage of glucose and mannose residues generating part of the tag for entry into the ERAD pathway (107-109, 122).
NSAIDs and COX-2 inhibitors retard COX-2 protein degradation in 3T3 cells and in HEK293 cells expressing COX-2 heterologously, and COX-2 degradation can be promoted by substrate turnover in a manner that is independent of the 27-IE of the enzyme (unpublished observations). Thus, there appears to be a second pathway for COX-2 degradation. Substrate turnover by COXs leads to their suicide inactivation and attendant denaturation (123, 124). This second pathway could be initiated by suicide inactivation of COX-2. It is possible that this could also occur with COX-1.
CONCLUDING COMMENTS
Regulation of COX gene expression is a complex process that varies in different cell types and even between the same cell types in different species. COX-1 and COX-2 genes are activated by a wide variety of stimuli acting through numerous signaling pathways and the relative contribution of each depends upon the stimulus, the cell type, and the time of stimulation. These factors and conditions determine which transcription factors are associated with the COX gene response elements. Although we now have a general understanding of the factors involved in COX-2 gene regulation, the interplay between the various regulatory transcription factors remains to be elucidated. At least some of the broad features of COX-2 protein degradation are now known. It will be important to characterize the pathways of COX-2 protein degradation involving both the 27-IE and substrate-dependent suicide inactivation.
ACKNOWLEDGEMENTS
The portion of the work reviewed in this paper that was performed in the laboratory of the corresponding author was supported by NIH Grant GM68848.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The abbreviations used are: COX, cyclooxygenase; PGH2, prostaglandin endoperoxide H2; NSAIDs, nonsteroidal anti-inflammatory drugs; LPS, lipopolysaccharide; CRE, cAMP response element; SRE, sterol response element; NF-κB, nuclear factor-kappa B; AP-1, Activator Protein-1; IL-1, interleukin-1; TNF; tumor necrosis factor; PMA, phorbol myristate acetate; HUVEC, human umbilical vein endothelial cells; PPRE, peroxisome proliferator response element; TKR, Tyrosine Kinase receptor; CR, Cytokine Receptor; TLR4, Toll Like Receptor 4; IL-1R, Interleukin-1 Receptor; TNFR, Tumor Necrosis Factor Receptor; PKA, cAMP dependent Protein Kinase; CREB, cAMP Response Element Binding Protein; ATF, Activating Transcription Factor; PLC, Phospholipase C; PI3K, Phosphatidyl-inositol-3-kinase; PKC, Calcium dependent Protein Kinase; PAK, p21 Associated Kinase; ERK, Extracellular signal-Regulated Kinase; MKK, Mitogen Activated Protein Kinase Kinase, also called MEK; MKKK, Mitogen Activated Protein Kinase Kinase Kinase, also called MEKK; JNK, c-Jun N-term Kinase; C/EBP, CAAT Enhancer Binding Protein; AP-1, Activator Protein-1; NF-?B, Nuclear Factor-kappa B; USF, upstream signaling factors; CAK, Ceramide Activated Kinase; TAK-1, Transforming growth factor-beta Activated Kinase-1; MyD88, Myocyte Differentiation Factor; IRAK, Interleukin-1 Receptor Associated Kinase; TRAF, Tumor Necrosis Factor Receptor Associated Factor; ECSIT, Evolutionary Conserved Signaling Intermediate in Toll; TRADD, Tumor Necrosis Factor Receptor Associated Death Domain; RIP, TNF Receptor Interacting Protein; ASK1, Apoptosis Signal-regulating Kinase; NIK, Nuclear Factor-kappa B Inducing Kinase; IKK, Inhibitor of κB Kinase; I?B, Inhibitor of κB; TRIF, TIR (Toll/IL-1 Receptor) domain-containing adaptor-inducing IFN-ß; TRAM, TRIF-related adaptor molecule; TBK1, TANK-Binding Kinase 1.
The generic name COX for the enzyme that is more accurately termed PGHS has been used in most of the manuscript. PGHS is a more accurate term because the enzyme has both a peroxidase (POX) and a COX activity.
REFERENCES
- 1.Smith W, DeWitt D, Garavito R. Cyclooxygenases: structural, cellular, and molecular biology. Annu Rev Biochem. 2000;69:145–182. doi: 10.1146/annurev.biochem.69.1.145. [DOI] [PubMed] [Google Scholar]
- 2.Smith WL, Murphy RC. The eicosanoids: cyclooxygenase, lipoxygenase, and epoxygenase pathways. In: Vance DE, Vance JE, editors. Biochemistry of Lipids, Lipoproteins and Membranes. 4th ed. Elsevier; Amsterdam: 2002. pp. 341–372. [Google Scholar]
- 3.van der Donk W, Tsai A, Kulmacz R. The cyclooxygenase reaction mechanism. Biochemistry. 2002;41:15451–15458. doi: 10.1021/bi026938h. [DOI] [PubMed] [Google Scholar]
- 4.Rouzer C, Marnett L. Mechanism of free radical oxygenation of polyunsaturated fatty acids by cyclooxygenases. Chem Rev. 2003;103:2239–2304. doi: 10.1021/cr000068x. [DOI] [PubMed] [Google Scholar]
- 5.Masferrer JL, Zweifel BS, Colburn SM, Ornberg RL, Salvemini D, Isakson P, Seibert K. The Role of Cyclooxygenase-2 in Inflammation. Am J Ther. 1995;2:607–610. doi: 10.1097/00045391-199509000-00005. [DOI] [PubMed] [Google Scholar]
- 6.Seibert K, Masferrer J, Zhang Y, Gregory S, Olson G, Hauser S, Leahy K, Perkins W, Isakson P. Mediation of inflammation by cyclooxygenase-2. Agents Actions Suppl. 1995;46:41–50. doi: 10.1007/978-3-0348-7276-8_5. [DOI] [PubMed] [Google Scholar]
- 7.Cao C, Matsumura K, Ozaki M, Watanabe Y. Lipopolysaccharide Injected into the Cerebral Ventricle Evokes Fever through Induction of Cyclooxygenase-2 in Brain Endothelial Cells. J. Neurosci. 1999;19:716–725. doi: 10.1523/JNEUROSCI.19-02-00716.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narumiya S, FitzGerald GA. Genetic and pharmacological analysis of prostanoid receptor function. Journal of Clinical Investigation. 2001;108:25–30. doi: 10.1172/JCI13455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubois RN, Abramson SB, Crofford L, Gupta RA, Simon LS, Van-De-Putte LB, Lipsky PE. Cyclooxygenase in biology and disease. FASEB J. 1998;12:1063–1073. [PubMed] [Google Scholar]
- 10.Marnett LJ, DuBois RN. COX-2: a target for colon cancer prevention. Annu Rev Pharmacol Toxicol. 2002;42:55–80. doi: 10.1146/annurev.pharmtox.42.082301.164620. [DOI] [PubMed] [Google Scholar]
- 11.Carlson ML, Wilson ET, Prescott SM. Regulation of COX-2 transcription in a colon cancer cell line by Pontin52/TIP49a. Mol Cancer. 2003;2:42. doi: 10.1186/1476-4598-2-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grosser T, Fries S, FitzGerald GA. Biological basis for the cardiovascular consequences of COX-2 inhibition: therapeutic challenges and opportunities. J. Clin. Invest. 2006;116:4–15. doi: 10.1172/JCI27291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herschman HR. Prostaglandin synthase 2. Biochim. Biophys. Acta. 1996;1299:125–140. doi: 10.1016/0005-2760(95)00194-8. [DOI] [PubMed] [Google Scholar]
- 14.Tanabe T, Tohnai N. Cyclooxygenase isozymes and their gene structures and expression. Prostaglandins Other Lipid Mediat. 2002;68-69:95–114. doi: 10.1016/s0090-6980(02)00024-2. [DOI] [PubMed] [Google Scholar]
- 15.Smith WL. Localization of enzymes responsible for prostaglandin formation. In: Willis AL, editor. Handbook of Eicosanoids: Prostaglandins and Related Lipids. CRC Press; Boca Raton, FL: 1987. pp. 175–191. [Google Scholar]
- 16.Murakami M, Bingham CO, 3rd., Matsumoto R, Austen KF, Arm JP. IgE-dependent activation of cytokine-primed mouse cultured mast cells induces a delayed phase of prostaglandin D2 generation via prostaglandin endoperoxide synthase-2. J. Immunol. 1995;155:4445–4453. [PubMed] [Google Scholar]
- 17.Matijevic-Aleksic N, Sanduja SK, Wang LH, Wu KK. Differential expression of thromboxane A synthase and prostaglandin H synthase in megakaryocytic cell line. Biochim. Biophys. Acta. 1995;1269:167–175. doi: 10.1016/0167-4889(95)00116-a. [DOI] [PubMed] [Google Scholar]
- 18.Ueda N, Yamashita R, Yamamoto S, Ishimura K. Induction of cyclooxygenase-1 in a human megakaryoblastic cell line (CMK) differentiated by phorbol ester. Biochim. Biophys. Acta. 1997;1344:103–110. doi: 10.1016/s0005-2760(96)00131-2. [DOI] [PubMed] [Google Scholar]
- 19.Mroske C, Plant MH, Franks DJ, Laneuville O. Characterization of prostaglandin endoperoxide H synthase-1 enzyme expression during differentiation of the megakaryocytic cell line MEG-01. Exp Hematol. 2000;28:411–421. doi: 10.1016/s0301-472x(00)00125-9. [DOI] [PubMed] [Google Scholar]
- 20.Delong CJ, Smith WL. An intronic enhancer regulates cyclooxygenase-1 gene expression. Biochem Biophys Res Commun. 2005;338:53–61. doi: 10.1016/j.bbrc.2005.07.184. [DOI] [PubMed] [Google Scholar]
- 21.Asano K, Lilly CM, Drazen JM. Prostaglandin G/H synthase-2 is the constitutive and dominant isoform in cultured human lung epithelial cells. Am J Physiol Lung Cell Mol Physiol. 1996;271:L126–131. doi: 10.1152/ajplung.1996.271.1.L126. [DOI] [PubMed] [Google Scholar]
- 22.Kaufmann WE, Worley PF, Taylor CV, Bremer M, Isakson PC. Cyclooxygenase-2 expression during rat neocortical development and in Rett syndrome. Brain. Dev. 1997;19:25–34. doi: 10.1016/s0387-7604(96)00047-2. [DOI] [PubMed] [Google Scholar]
- 23.Harris RC, Breyer MD. Physiological regulation of cyclooxygenase-2 in the kidney. Am J Physiol Renal Physiol. 2001;281:F1–11. doi: 10.1152/ajprenal.2001.281.1.F1. [DOI] [PubMed] [Google Scholar]
- 24.Robertson RP. Dominance of cyclooxygenase-2 in the regulation of pancreatic islet prostaglandin synthesis. Diabetes. 1998;47:1379–1383. doi: 10.2337/diabetes.47.9.1379. [DOI] [PubMed] [Google Scholar]
- 25.Ristimaki A, Honkanen N, Jankala H, Sipponen P, Harkonen M. Expression of cyclooxygenase-2 in human gastric carcinoma. Cancer Res. 1997;57:1276–1280. [PubMed] [Google Scholar]
- 26.Shao J, Sheng H, Inoue H, Morrow JD, DuBois RN. Regulation of Constitutive Cyclooxygenase-2 Expression in Colon Carcinoma Cells. J. Biol. Chem. 2000;275:33951–33956. doi: 10.1074/jbc.M002324200. [DOI] [PubMed] [Google Scholar]
- 27.Yokoyama C, Tanabe T. Cloning of the human gene encoding prostaglandin endoperoxide synthase and primary structure of the enzyme. Biochem. Biophys. Res. Commun. 1989;165:888–894. doi: 10.1016/s0006-291x(89)80049-x. [DOI] [PubMed] [Google Scholar]
- 28.Kraemer SA, Meade SA, DeWitt DL. Prostaglandin endoperoxide synthase gene structure: identification of the transcriptional start site and 5'-flanking regulatory sequences. Arch. Biochem. Biophys. 1992;293:391–400. doi: 10.1016/0003-9861(92)90411-o. [DOI] [PubMed] [Google Scholar]
- 29.Tanabe T, Tohnai N. Cyclooxygenase isozymes and their gene structures and expression. Prostaglandins Other Lipid Mediat. 2002;68-69:95–114. doi: 10.1016/s0090-6980(02)00024-2. [DOI] [PubMed] [Google Scholar]
- 30.Wang L, Hajubeigi A, Xu X, Loose-Mitchell D, Wu KK. Characterization of the promoter of human prostaglandin H synthase-1 gene. Biochem. Biophys. Res. Commun. 1993;190:406–411. doi: 10.1006/bbrc.1993.1062. [DOI] [PubMed] [Google Scholar]
- 31.Smith CJ, Morrow JD, Roberts LJ, Marnett LJ. Differentiation of monocytoid THP-1 cells with phorbol ester induces expression of prostaglandin endoperoxide synthase-1 (Cox-1). Biochem. Biophys. Res. Commun. 1993;192:787–793. doi: 10.1006/bbrc.1993.1483. [DOI] [PubMed] [Google Scholar]
- 32.Xu XM, Tang JL, Chen X, Wang LH, Wu KK. Involvement of two Sp1 elements in basal endothelial prostaglandin H synthase-1 promoter activity. J. Biol. Chem. 1997;272:6943–6950. doi: 10.1074/jbc.272.11.6943. [DOI] [PubMed] [Google Scholar]
- 33.Ueda N, Yamashita R, Yamamoto S, Ishimura K. Induction of cyclooxygenase-1 in a human megakaryoblastic cell line (CMK) differentiated by phorbol ester. Biochim Biophys Acta. 1997;1344:103–110. doi: 10.1016/s0005-2760(96)00131-2. [DOI] [PubMed] [Google Scholar]
- 34.Okahara K, Sun B, Kambayashi J. Upregulation of prostacyclin synthesis-related gene expression by shear stress in vascular endothelial cells. Arterioscler Thromb Vasc Biol. 1998;18:1922–1926. doi: 10.1161/01.atv.18.12.1922. [DOI] [PubMed] [Google Scholar]
- 35.Bogar LJ, Bartula LL, Parkman HP, Myers SI. Enhanced bradykinin-stimulated prostaglandin release in the acutely inflamed guinea pig gallbladder is due to new synthesis of cyclooxygenase 1 and prostacyclin synthase. J Surg Res. 1999;84:71–76. doi: 10.1006/jsre.1999.5612. [DOI] [PubMed] [Google Scholar]
- 36.Gibson LL, Hahner L, Osborne-Lawrence S, German Z, Wu KK, Chambliss KL, Shaul PW. Molecular basis of estrogen-induced cyclooxygenase type 1 upregulation in endothelial cells. Circ Res. 2005;96:518–525. doi: 10.1161/01.RES.0000158967.96231.88. [DOI] [PubMed] [Google Scholar]
- 37.Smith WL, DeWitt DL. Prostaglandin endoperoxide H synthases-1 and -2. In: Dixon FJ, editor. Advances in Immunology. Academic Press; San Diego, CA: 1996. pp. 167–215. [DOI] [PubMed] [Google Scholar]
- 38.Sawaoka H, Dixon DA, Oates JA, Boutaud O. Tristetraprolin Binds to the 3'-Untranslated Region of Cyclooxygenase-2 mRNA. A polyadenylation variant in a cancer cell line lacks the binding site. J. Biol. Chem. 2003;278:13928–13935. doi: 10.1074/jbc.M300016200. [DOI] [PubMed] [Google Scholar]
- 39.Xie W, Fletcher BS, Andersen RD, Herschman HR. v-src induction of the TIS10/PGS2 prostaglandin synthase gene is mediated by an ATF/CRE transcription response element. Mol. Cell. Biol. 1994;14:6531–6539. doi: 10.1128/mcb.14.10.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie W, Herschman HR. Transcriptional regulation of prostaglandin synthase 2 gene expression by platelet-derived growth factor and serum. J. Biol. Chem. 1996;271:31742–31748. doi: 10.1074/jbc.271.49.31742. [DOI] [PubMed] [Google Scholar]
- 41.Xie W, Herschman HR. v-src induces prostaglandin synthase 2 gene expression by activation of the c-Jun N-terminal kinase and the c-Jun transcription factor. J. Biol. Chem. 1995;270:27622–27628. doi: 10.1074/jbc.270.46.27622. [DOI] [PubMed] [Google Scholar]
- 42.Bagga D, Wang L, Farias-Eisner R, Glaspy JA, Reddy ST. Differential effects of prostaglandin derived from omega -6 and omega -3 polyunsaturated fatty acids on COX-2 expression and IL-6 secretion. PNAS. 2003;100:1751–1756. doi: 10.1073/pnas.0334211100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu Y, Saunders MA, Yeh H, Deng WG, Wu KK. Dynamic regulation of cyclooxygenase-2 promoter activity by isoforms of CCAAT/enhancer-binding proteins. J Biol Chem. 2002;277:6923–6928. doi: 10.1074/jbc.M108075200. [DOI] [PubMed] [Google Scholar]
- 44.Deng WG, Zhu Y, Montero A, Wu KK. Quantitative analysis of binding of transcription factor complex to biotinylated DNA probe by a streptavidin-agarose pulldown assay. Anal Biochem. 2003;323:12–18. doi: 10.1016/j.ab.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 45.Deng WG, Saunders MA, Gilroy DW, He XZ, Yeh H, Zhu Y, Shtivelband MI, Ruan KH, Wu KK. Purification and characterization of a cyclooxygenase-2 and angiogenesis suppressing factor produced by human fibroblasts. FASEB J. 2002;16:1286–1288. doi: 10.1096/fj.01-0844fje. [DOI] [PubMed] [Google Scholar]
- 46.Deng WG, Zhu Y, Wu KK. Role of p300 and PCAF in regulating cyclooxygenase-2 promoter activation by inflammatory mediators. Blood. 2004;103:2135–2142. doi: 10.1182/blood-2003-09-3131. [DOI] [PubMed] [Google Scholar]
- 47.Mifflin RC, Saada JI, Di Mari JF, Adegboyega PA, Valentich JD, Powell DW. Regulation of COX-2 expression in human intestinal myofibroblasts: mechanisms of IL-1-mediated induction. Am J Physiol Cell Physiol. 2002;282:C824–834. doi: 10.1152/ajpcell.00388.2001. [DOI] [PubMed] [Google Scholar]
- 48.Faour WH, He Y, He QW, de Ladurantaye M, Quintero M, Mancini A, Di Battista JA. Prostaglandin E(2) regulates the level and stability of cyclooxygenase-2 mRNA through activation of p38 mitogen-activated protein kinase in interleukin-1 beta-treated human synovial fibroblasts. J Biol Chem. 2001;276:31720–31731. doi: 10.1074/jbc.M104036200. [DOI] [PubMed] [Google Scholar]
- 49.Inoue H, Yokoyama C, Hara S, Tone Y, Tanabe T. Transcriptional regulation of human prostaglandin-endoperoxide synthase-2 gene by lipopolysaccharide and phorbol ester in vascular endothelial cells. Involvement of both nuclear factor for interleukin-6 expression site and cAMP response element. J. Biol. Chem. 1995;270:24965–24971. doi: 10.1074/jbc.270.42.24965. [DOI] [PubMed] [Google Scholar]
- 50.Jones DA, Carlton DP, McIntyre TM, Zimmerman GA, Prescott SM. Molecular cloning of human prostaglandin endoperoxide synthase type II and demonstration of expression in response to cytokines. J. Biol. Chem. 1993;268:9049–9054. [PubMed] [Google Scholar]
- 51.Xu Q, Ji YS, Schmedtje JF., Jr. Sp1 increases expression of cyclooxygenase-2 in hypoxic vascular endothelium. Implications for the mechanisms of aortic aneurysm and heart failure. J Biol Chem. 2000;275:24583–24589. doi: 10.1074/jbc.M003894200. [DOI] [PubMed] [Google Scholar]
- 52.Caughey GE, Cleland LG, Gamble JR, James MJ. Up-regulation of Endothelial Cyclooxygenase-2 and Prostanoid Synthesis by Platelets. Role of Thromboxane A2. J. Biol. Chem. 2001;276:37839–37845. doi: 10.1074/jbc.M010606200. [DOI] [PubMed] [Google Scholar]
- 53.Wu G, Mannam AP, Wu J, Kirbis S, Shie JL, Chen C, Laham RJ, Sellke FW, Li J. Hypoxia induces myocyte-dependent COX-2 regulation in endothelial cells: role of VEGF. Am J Physiol Heart Circ Physiol. 2003;285:H2420–2429. doi: 10.1152/ajpheart.00187.2003. [DOI] [PubMed] [Google Scholar]
- 54.Hirai K, Takayama H, Tomo K, Okuma M. Protein-tyrosine-kinase-dependent expression of cyclo-oxygenase-1 and -2 mRNAs in human endothelial cells. Biochem J. 1997;322(Pt 2):373–377. doi: 10.1042/bj3220373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith LH, Petrie MS, Morrow JD, Oates JA, Vaughan DE. The sterol response element binding protein regulates cyclooxygenase-2 gene expression in endothelial cells. J Lipid Res. 2005;46:862–871. doi: 10.1194/jlr.M500021-JLR200. [DOI] [PubMed] [Google Scholar]
- 56.Bradbury DA, Newton R, Zhu YM, El-Haroun H, Corbett L, Knox AJ. Cyclooxygenase-2 induction by bradykinin in human pulmonary artery smooth muscle cells is mediated by the cyclic AMP response element through a novel autocrine loop involving endogenous prostaglandin E2, E-prostanoid 2 (EP2), and EP4 receptors. J Biol Chem. 2003;278:49954–49964. doi: 10.1074/jbc.M307964200. [DOI] [PubMed] [Google Scholar]
- 57.Nie M, Pang L, Inoue H, Knox AJ. Transcriptional regulation of cyclooxygenase 2 by bradykinin and interleukin-1beta in human airway smooth muscle cells: involvement of different promoter elements, transcription factors, and histone h4 acetylation. Mol Cell Biol. 2003;23:9233–9244. doi: 10.1128/MCB.23.24.9233-9244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pang L, Knox AJ. Bradykinin stimulates IL-8 production in cultured human airway smooth muscle cells: role of cyclooxygenase products. J Immunol. 1998;161:2509–2515. [PubMed] [Google Scholar]
- 59.Pang L, Nie M, Corbett L, Knox AJ. Cyclooxygenase-2 expression by nonsteroidal anti-inflammatory drugs in human airway smooth muscle cells: role of peroxisome proliferator-activated receptors. J Immunol. 2003;170:1043–1051. doi: 10.4049/jimmunol.170.2.1043. [DOI] [PubMed] [Google Scholar]
- 60.Subbaramaiah K, Lin DT, Hart JC, Dannenberg AJ. Peroxisome roliferator-activated receptor gamma ligands suppress the transcriptional activation of cyclooxygenase-2. Evidence for involvement of activator protein-1 and CREB-binding protein/p300. J. Biol. Chem. 2001;276:12440–12448. doi: 10.1074/jbc.M007237200. [DOI] [PubMed] [Google Scholar]
- 61.Subbaramaiah K, Cole PA, Dannenberg AJ. Retinoids and carnosol suppress cyclooxygenase-2 transcription by CREB-binding protein/p300-dependent and -independent mechanisms. Cancer Res. 2002;62:2522–2530. [PubMed] [Google Scholar]
- 62.Juttner S, Cramer T, Wessler S, Walduck A, Gao F, Schmitz F, Wunder C, Weber M, Fischer SM, Schmidt WE, Wiedenmann B, Meyer TF, Naumann M, Hocker M. Helicobacter pylori stimulates host cyclooxygenase-2 gene transcription: critical importance of MEK/ERK-dependent activation of USF1/-2 and CREB transcription factors. Cell Microbiol. 2003;5:821–834. doi: 10.1046/j.1462-5822.2003.00324.x. [DOI] [PubMed] [Google Scholar]
- 63.Sirois J, Sayasith K, Brown KA, Stock AE, Bouchard N, Dore M. Cyclooxygenase-2 and its role in ovulation: a 2004 account. Hum Reprod Update. 2004;10:373–385. doi: 10.1093/humupd/dmh032. [DOI] [PubMed] [Google Scholar]
- 64.Scherle PA, Ma W. -g., Lim H, Dey SK, Trzaskos JM. Regulation of Cyclooxygenase-2 Induction in the Mouse Uterus During Decidualization. An Event of Early Pregnancy. J. Biol. Chem. 2000;275:37086–33206. doi: 10.1074/jbc.M006168200. [DOI] [PubMed] [Google Scholar]
- 65.Tsai SJ, Wiltbank MC, Bodensteiner KJ. Distinct mechanisms regulate induction of messenger ribonucleic acid for prostaglandin (PG) G/H synthase-2, PGE (EP3) receptor, and PGF2 alpha receptor in bovine preovulatory follicles. Endrocrinology. 1996;137:3348–3355. doi: 10.1210/endo.137.8.8754761. [DOI] [PubMed] [Google Scholar]
- 66.Wong WYL, DeWitt DL, Smith WL, Richards JS. Rapid induction of prostaglandin endoperoxide synthase in rat preovulatory follicles by luteinizing hormone and cAMP is blocked by inhibitors of transcription and translation. Mol. Endocrinol. 1989;3:1714–1723. doi: 10.1210/mend-3-11-1714. [DOI] [PubMed] [Google Scholar]
- 67.Sirois J. Induction of prostaglandin endoperoxide synthase-2 by human chorionic gonadotropin in bovine preovulatory follicles in vivo. Endocrinology. 1994;135:841–848. doi: 10.1210/endo.135.3.8070377. [DOI] [PubMed] [Google Scholar]
- 68.Liu J, Antaya M, Boerboom D, Lussier JG, Silversides DW, Sirois J. The Delayed Activation of the Prostaglandin G/H Synthase-2 Promoter in Bovine Granulosa Cells Is Associated with Down-regulation of Truncated Upstream Stimulatory Factor-2. J. Biol. Chem. 1999;274:35037–35045. doi: 10.1074/jbc.274.49.35037. [DOI] [PubMed] [Google Scholar]
- 69.Sirois J, Richards JS. Transcriptional regulation of the rat prostaglandin endoperoxide synthase 2 gene in granulosa cells. J. Biol. Chem. 1993;268:21931–21938. [PubMed] [Google Scholar]
- 70.Sayasith K, Lussier JG, Sirois J. Role of upstream stimulatory factor phosphorylation in the regulation of the prostaglandin G/H synthase-2 promoter in granulosa cells. J. Biol. Chem. 2005:M413434200. doi: 10.1074/jbc.M413434200. [DOI] [PubMed] [Google Scholar]
- 71.Chikazu D, Li X, Kawaguchi H, Sakuma Y, Voznesensky OS, Adams DJ, Xu M, Hoshio K, Katavic V, Herschman HR, Raisz LG, Pilbeam CC. Bone morphogenetic protein 2 induces cyclo-oxygenase 2 in osteoblasts via a Cbfal binding site: role in effects of bone morphogenetic protein 2 in vitro and in vivo. J Bone Miner Res. 2002;17:1430–1440. doi: 10.1359/jbmr.2002.17.8.1430. [DOI] [PubMed] [Google Scholar]
- 72.Ogasawara A, Arakawa T, Kaneda T, Takuma T, Sato T, Kaneko H, Kumegawa M, Hakeda Y. Fluid shear stress-induced cyclooxygenase-2 expression is mediated by C/EBP beta , cAMP-response element-binding protein, and AP-1 in osteoblastic MC3T3-E1 cells. J. Biol. Chem. 2001;276:7048–7054. doi: 10.1074/jbc.M008070200. [DOI] [PubMed] [Google Scholar]
- 73.Yamamoto K, Arakawa T, Ueda N, Yamamoto S. Transcriptional roles of nuclear factor kappa B and nuclear factor -interleukin-6 in the tumor necrosis factor alpha-dependent induction of cyclooxygenase-2 in MC3T3-E1 cells. J. Biol. Chem. 1995;270:31315–31320. doi: 10.1074/jbc.270.52.31315. [DOI] [PubMed] [Google Scholar]
- 74.Wadleigh DJ, Herschman HR. Transcriptional regulation of the cyclooxygenase-2 gene by diverse ligands in murine osteoblasts. Biochem Biophys Res Commun. 1999;264:865–870. doi: 10.1006/bbrc.1999.1606. [DOI] [PubMed] [Google Scholar]
- 75.Okada Y, Voznesensky O, Herschman H, Harrison J, Pilbeam C. Identification of multiple cis-acting elements mediating the induction of prostaglandin G/H synthase-2 by phorbol ester in murine osteoblastic cells. J Cell Biochem. 2000;78:197–209. [PubMed] [Google Scholar]
- 76.Ajubi NE, Klein-Nulend J, Alblas MJ, Burger EH, Nijweide PJ. Signal transduction pathways involved in fluid flow-induced PGE2 production by cultured osteocytes. Am J Physiol. 1999;276:E171–178. doi: 10.1152/ajpendo.1999.276.1.E171. [DOI] [PubMed] [Google Scholar]
- 77.Wadhwa S, Choudhary S, Voznesensky M, Epstein M, Raisz L, Pilbeam C. Fluid flow induces COX-2 expression in MC3T3-E1 osteoblasts via a PKA signaling pathway. Biochem Biophys Res Commun. 2002;297:46–51. doi: 10.1016/s0006-291x(02)02124-1. [DOI] [PubMed] [Google Scholar]
- 78.Inoue H, Nanayama T, Hara S, Yokoyama C, Tanabe T. The cyclic AMP response element plays an essential role in the expression of the human prostaglandin-endoperoxide synthase 2 gene in differentiated U937 monocytic cells. FEBS Letters. 1994;350:51–54. doi: 10.1016/0014-5793(94)00731-4. [DOI] [PubMed] [Google Scholar]
- 79.Barry OP, Kazanietz MG, Pratico D, FitzGerald GA. Arachidonic acid in platelet microparticles up-regulates cyclooxygenase-2-dependent prostaglandin formation via a protein kinase C/mitogen-activated protein kinase-dependent pathway. J. Biol. Chem. 1999;274:7545–7556. doi: 10.1074/jbc.274.11.7545. [DOI] [PubMed] [Google Scholar]
- 80.Shanmugam N, Gaw Gonzalo IT, Natarajan R. Molecular mechanisms of high glucose-induced cyclooxygenase-2 expression in monocytes. Diabetes. 2004;53:795–802. doi: 10.2337/diabetes.53.3.795. [DOI] [PubMed] [Google Scholar]
- 81.Miao F, Gonzalo IG, Lanting L, Natarajan R. In vivo chromatin remodeling events leading to inflammatory gene transcription under diabetic conditions. J Biol Chem. 2004;279:18091–18097. doi: 10.1074/jbc.M311786200. [DOI] [PubMed] [Google Scholar]
- 82.Shanmugam N, Kim YS, Lanting L, Natarajan R. Regulation of cyclooxygenase-2 expression in monocytes by ligation of the receptor for advanced glycation end products. J Biol Chem. 2003;278:34834–34844. doi: 10.1074/jbc.M302828200. [DOI] [PubMed] [Google Scholar]
- 83.Jang BC, Kim DH, Park JW, Kwon TK, Kim SP, Song DK, Park JG, Bae JH, Mun KC, Baek WK, Suh MH, Hla T, Suh SI. Induction of cyclooxygenase-2 in macrophages by catalase: role of NF-kappaB and PI3K signaling pathways. Biochem Biophys Res Commun. 2004;316:398–406. doi: 10.1016/j.bbrc.2004.02.060. [DOI] [PubMed] [Google Scholar]
- 84.Chen B-C, Chang Y-S, Kang J-C, Hsu M-J, Sheu J-R, Chen T-L, Teng C-M, Lin C-H. Peptidoglycan Induces Nuclear Factor-{kappa}B Activation and Cyclooxygenase-2 Expression via Ras, Raf-1, and ERK in RAW 264.7 Macrophages. J. Biol. Chem. 2004;279:20889–20897. doi: 10.1074/jbc.M311279200. [DOI] [PubMed] [Google Scholar]
- 85.Steer SA, Moran JM, Maggi LB, Jr., Buller RML, Perlman H, Corbett JA. Regulation of Cyclooxygenase-2 Expression by Macrophages in Response to Double-Stranded RNA and Viral Infection. J. Immunol. 2003;170:1070–1076. doi: 10.4049/jimmunol.170.2.1070. [DOI] [PubMed] [Google Scholar]
- 86.Guha M, Mackman N. LPS induction of gene expression in human monocytes. Cell Signal. 2001;13:85–94. doi: 10.1016/s0898-6568(00)00149-2. [DOI] [PubMed] [Google Scholar]
- 87.Takeda K, Akira S. TLR signaling pathways. Semin Immunol. 2004;16:3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 88.Barton GM, Medzhitov R. Toll-like receptor signaling pathways. Science. 2003;300:1524–1525. doi: 10.1126/science.1085536. [DOI] [PubMed] [Google Scholar]
- 89.Hwang D. Modulation of the expression of cyclooxygenase-2 by fatty acids mediated through Toll-like receptor 4 derived signaling pathways. FASEB J. 2001;15:2556–2564. doi: 10.1096/fj.01-0432com. [DOI] [PubMed] [Google Scholar]
- 90.Yamamoto M, Sato S, Hemmi H, Uematsu S, Hoshino K, Kaisho T, Takeuchi O, Takeda K, Akira S. TRAM is specifically involved in the Toll-like receptor 4-mediated MyD88-independent signaling pathway. Nat Immunol. 2003;4:1144–1150. doi: 10.1038/ni986. [DOI] [PubMed] [Google Scholar]
- 91.Jiang Z, Georgel P, Du X, Shamel L, Sovath S, Mudd S, Huber M, Kalis C, Keck S, Galanos C, Freudenberg M, Beutler B. CD14 is required for MyD88-independent LPS signaling. Nat Immunol. 2005;6:565–570. doi: 10.1038/ni1207. [DOI] [PubMed] [Google Scholar]
- 92.Caivano M, Gorgoni B, Cohen P, Poli V. The Induction of Cyclooxygenase-2 mRNA in Macrophages Is Biphasic and Requires both CCAAT Enhancer-binding protein beta (C/EBPbeta ) and C/EBPdelta Transcription Factors. J. Biol. Chem. 2001;276:48693–48701. doi: 10.1074/jbc.M108282200. [DOI] [PubMed] [Google Scholar]
- 93.Gorgoni B, Caivano M, Arizmendi C, Poli V. The transcription factor C/EBPbeta is essential for inducible expression of the cox-2 gene in macrophages but not in fibroblasts. J. Biol. Chem. 2001;276:40769–48701. doi: 10.1074/jbc.M106865200. [DOI] [PubMed] [Google Scholar]
- 94.Kang Y-J, Wingerd BA, Arakawa T, Smith WL. Cyclooxygenase-2 gene transcription in a macrophage model of inflammation. J. Immunol. 2006 doi: 10.4049/jimmunol.177.11.8111. in press. [DOI] [PubMed] [Google Scholar]
- 95.Rich G, Yoder E, Moore S. Regulation of prostaglandin H synthase-2 expression in cerebromicrovascular smooth muscle by serum and epidermal growth factor. J Cell Physiol. 1998;176:495–505. doi: 10.1002/(SICI)1097-4652(199809)176:3<495::AID-JCP6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 96.DeWitt DL, Meade EA. Serum and glucocorticoid regulation of gene transcription and expression of the prostaglandin H synthase-1 and prostaglandin H synthase-2 isozymes. Arch. Biochem. Biophys. 1993;306:94–102. doi: 10.1006/abbi.1993.1485. [DOI] [PubMed] [Google Scholar]
- 97.Perkins D, Kniss D. Rapid and transient induction of cyclo-oxygenase 2 by epidermal growth factor in human amnion-derived WISH cells. Biochem J. 1997;321(Pt 3):677–681. doi: 10.1042/bj3210677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Figueiredo-Pereira ME, Li Z, Jansen M, Rockwell P. N-Acetylcysteine and Celecoxib Lessen Cadmium Cytotoxicity Which Is Associated with Cyclooxygenase-2 Up-regulation in Mouse Neuronal Cells. J. Biol. Chem. 2002;277:25283–25289. doi: 10.1074/jbc.M109145200. [DOI] [PubMed] [Google Scholar]
- 99.Mbonye UR, Wada M, Rieke CJ, Tang H-Y, DeWitt DL, Smith WL. The 19 amino acid cassette of cyclooxygenase-2 mediates entry of the protein into the ER-associated degradation system. J. Biol. Chem. 2006:M608281200. doi: 10.1074/jbc.M608281200. [DOI] [PubMed] [Google Scholar]
- 100.Kargman SL, O'Neill GP, Vicker PJ, Evans JF, Mancini JA, Jothy S. Expression of Prostaglandin G/H synthase-1 and -2 protein in human colon cancer. Cancer Res. 1995;55:2556–2559. [PubMed] [Google Scholar]
- 101.Shao J, Sheng H, Aramandla R, Pereira MA, Lubet RA, Hawk E, Grogan L, Kirsch IR, Washington MK, Beauchamp RD, DuBois RN. Coordinate regulation of cyclooxygenase-2 and TGF-beta1 in replication error-positive colon cancer and azoxymethane-induced rat colonic tumors. Carcinogenesis. 1999;20:185–191. doi: 10.1093/carcin/20.2.185. [DOI] [PubMed] [Google Scholar]
- 102.Nagasaka T, Sasamoto H, Notohara K, Cullings HM, Takeda M, Kimura K, Kambara T, MacPhee DG, Young J, Leggett BA, Jass JR, Tanaka N, Matsubara N. Colorectal Cancer With Mutation in BRAF, KRAS, and Wild-Type With Respect to Both Oncogenes Showing Different Patterns of DNA Methylation. J. Clin. Oncol. 2004;22:4584–4594. doi: 10.1200/JCO.2004.02.154. [DOI] [PubMed] [Google Scholar]
- 103.Dixon DA, Tolley ND, King PH, Nabors LB, McIntyre TM, Zimmerman GA, Prescott SM. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J. Clin. Invest. 2001;108:1657–1665. doi: 10.1172/JCI12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang D, Mann J, Dubois R. The role of prostaglandins and other eicosanoids in the gastrointestinal tract. Gastroenterology. 2005;128:1445–1461. doi: 10.1053/j.gastro.2004.09.080. [DOI] [PubMed] [Google Scholar]
- 105.DeMartino GN, Slaughter CA. The Proteasome, a Novel Protease Regulated by Multiple Mechanisms. J. Biol. Chem. 1999;274:22123–22126. doi: 10.1074/jbc.274.32.22123. [DOI] [PubMed] [Google Scholar]
- 106.Gardner RG, Shearer AG, Hampton RY. In Vivo Action of the HRD Ubiquitin Ligase Complex: Mechanisms of Endoplasmic Reticulum Quality Control and Sterol Regulation. Mol. Cell. Biol. 2001;21:4276–4291. doi: 10.1128/MCB.21.13.4276-4291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tsai B, Ye Y, Rapoport T. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat Rev Mol Cell Biol. 2002;3:246–255. doi: 10.1038/nrm780. [DOI] [PubMed] [Google Scholar]
- 108.Kostova Z, Wolf DH. New EMBO member's review: For whom the bell tolls: protein quality control of the endoplasmic reticulum and the ubiquitin-proteasome connection. EMBO J. 2003;22:2309–2317. doi: 10.1093/emboj/cdg227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.McCracken A, Brodsky J. Evolving questions and paradigm shifts in endoplasmic-reticulum-associated degradation (ERAD). Bioessays. 2003;25:868–877. doi: 10.1002/bies.10320. [DOI] [PubMed] [Google Scholar]
- 110.Khanna R, Lee EJ, Papazian DM. Transient calnexin interaction confers long-term stability on folded K+ channel protein in the ER. J. Cell Sci. 2004;117:2897–2908. doi: 10.1242/jcs.01141. [DOI] [PubMed] [Google Scholar]
- 111.Shearer AG, Hampton RY. Structural Control of Endoplasmic Reticulum-associated Degradation: Effect of Chemical Chaperones on 3-Hydroxy-3-Methylglutaryl-CoA Reductase. J. Biol. Chem. 2004;279:188–196. doi: 10.1074/jbc.M307734200. [DOI] [PubMed] [Google Scholar]
- 112.Ismail N, Ng DT. Have you HRD? Understanding ERAD is DOAble! Cell. 2006;126:237–239. doi: 10.1016/j.cell.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 113.Rockwell P, Yuan H, Magnusson R, Figueiredo-Pereir M. Proteasome inhibition in neuronal cells induces a proinflammatory response manifested by upregulation of cyclooxygenase-2, its accumulation as ubiquitin conjugates, and production of the prostaglandin PGE(2). Arch Biochem Biophys. 2000;374:325–333. doi: 10.1006/abbi.1999.1646. [DOI] [PubMed] [Google Scholar]
- 114.Picot D, Loll PJ, Garavito M. The X-ray crystal structure of the membrane protein prostaglandin H2 synthase-1. Nature. 1994;367:243–249. doi: 10.1038/367243a0. [DOI] [PubMed] [Google Scholar]
- 115.Kurumbail RG, Stevens AM, Gierse JK, McDonald JJ, Stegeman RA, Pak JY, Gildehaus D, Miyashiro JM, Penning TD, Seibert K, Isakson PC, Stallings WC. Structural basis for selective inhibition of cyclooxygenase-2 by anti-inflammatory agents. Nature. 1996;384:644–648. doi: 10.1038/384644a0. [DOI] [PubMed] [Google Scholar]
- 116.Otto JC, Smith WL. The orientation of prostaglandin endoperoxide synthases-1 and -2 in the endoplasmic reticulum. J. Biol. Chem. 1994;269:19868–19875. [PubMed] [Google Scholar]
- 117.Spencer AG, Woods JW, Arakawa T, Singer II, Smith WL. Subcellular localization of prostaglandin endoperoxide H synthases-1 and -2 by immunoelectron microscopy. J. Biol. Chem. 1998;273:9886–9893. doi: 10.1074/jbc.273.16.9886. [DOI] [PubMed] [Google Scholar]
- 118.Spencer AG, Thuresson EA, Otto JC, Song I, Smith T, DeWitt DL, Garavito RM, Smith WL. The membrane binding domains of prostaglandin endoperoxide H synthase-1 and -2: Peptide mapping and mutational analysis. J. Biol. Chem. 1999;274:32936–32942. doi: 10.1074/jbc.274.46.32936. [DOI] [PubMed] [Google Scholar]
- 119.MirAfzali Z, Leipprandt JR, McCracken JL, DeWitt DL. Topography of the prostaglandin endperoxide H2 Synthase-2 in membranes. J. Biol. Chem. 2006:M605206200. doi: 10.1074/jbc.M605206200. [DOI] [PubMed] [Google Scholar]
- 120.Otto JC, DeWitt DL, Smith WL. N-glycosylation of prostaglandin endoperoxide synthases-1 and -2 and their orientations in the endoplasmic reticulum. J. Biol. Chem. 1993;268:18234–18242. [PubMed] [Google Scholar]
- 121.Percival M, Bastien L, Griffin P, Kargman S, Ouellet M, O'Neill G. Investigation of human cyclooxygenase-2 glycosylation heterogeneity and protein expression in insect and mammalian cell expression systems. Protein Expr Purif. 1997;9:388–398. doi: 10.1006/prep.1996.0685. [DOI] [PubMed] [Google Scholar]
- 122.Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 123.Song I, Ball TM, Smith WL. Different suicide inactivation processes for the peroxidase and cyclooxygenase activities of prostaglandin endoperoxide H synthase-1. Biochem. Biophys. Res. Commun. 2001;289:869–875. doi: 10.1006/bbrc.2001.6071. [DOI] [PubMed] [Google Scholar]
- 124.Mevkh A, Miroshnikov K, Igumnova N, Varfolomeev S. Prostaglandin H synthase. Inactivation of the enzyme in the course of catalysis is accompanied by fast and dramatic changes in protein structure. FEBS Lett. 1993;321:205–208. doi: 10.1016/0014-5793(93)80109-8. [DOI] [PubMed] [Google Scholar]
- 125.Kirtikara K, Morham SG, Raghow R, Laulederkind SJ, Kanekura T, Goorha S, Ballou LR. Compensatory prostaglandin E2 biosynthesis in cyclooxygenase 1 or 2 null cells. J. Exp. Med. 1998;187:517–523. doi: 10.1084/jem.187.4.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Diaz A, Chepenik KP, Korn JH, Reginato AM, Jimenez SA. Differential regulation of cyclooxygenases 1 and 2 by interleukin-1 beta, tumor necrosis factor-alpha, and transforming growth factor-beta 1 in human lung fibroblasts. Exp Cell Res. 1998;241:222–229. doi: 10.1006/excr.1998.4050. [DOI] [PubMed] [Google Scholar]
- 127.Bryant CE, Appleton I, Mitchell JA. Vascular endothelial growth factor upregulates constitutive cyclooxygenase 1 in primary bovine and human endothelial cells. Life Sci. 1998;62:2195–2201. doi: 10.1016/s0024-3205(98)00197-0. [DOI] [PubMed] [Google Scholar]
- 128.Kitzler J, Hill E, Hardman R, Reddy N, Philpot R, Eling TE. Analysis and quantitation of splicing variants of the TPA-inducible PGHS-1 mRNA in rat tracheal epithelial cells. Arch. Biochem. Biophys. 1995;316:856–863. doi: 10.1006/abbi.1995.1115. [DOI] [PubMed] [Google Scholar]
- 129.Schneider N, Lanz S, Ramer R, Schaefer D, Goppelt-Struebe M. Up-regulation of cyclooxygenase-1 in neuroblastoma cell lines by retinoic acid and corticosteroids. J Neurochem. 2001;77:416–424. doi: 10.1046/j.1471-4159.2001.00264.x. [DOI] [PubMed] [Google Scholar]
- 130.Rioux N, Castonguay A. The induction of cyclooxygenase-1 by a tobacco carcinogen in U937 human macrophages is correlated to the activation of NF-{kappa}B. Carcinogenesis. 2000;21:1745–1751. doi: 10.1093/carcin/21.9.1745. [DOI] [PubMed] [Google Scholar]
- 131.Xie W, Herschman HR. v-src induces prostaglandin synthase 2 gene expression by activation of the c-Jun N-terminal kinase and the c-Jun transcription factor. J Biol Chem. 1995;270:27622–27628. doi: 10.1074/jbc.270.46.27622. [DOI] [PubMed] [Google Scholar]
- 132.Inoue H, Yokoyama C, Hara S, Tone Y, Tanabe T. Transcriptional regulation of human prostaglandin-endoperoxide synthase-2 gene by lipopolysaccharide and phorbol ester in vascular endothelial cells. Involvement of both nuclear factor for interleukin-6 expression site and cAMP response element. J Biol Chem. 1995;270:24965–24971. doi: 10.1074/jbc.270.42.24965. [DOI] [PubMed] [Google Scholar]
- 133.Bradbury DA, Newton R, Zhu YM, Stocks J, Corbett L, Holland ED, Pang LH, Knox AJ. Effect of bradykinin, TGF-beta1, IL-1beta, and hypoxia on COX-2 expression in pulmonary artery smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2002;283:L717–725. doi: 10.1152/ajplung.00070.2002. [DOI] [PubMed] [Google Scholar]
- 134.Bhattacharyya DK, Lecomte M, Dunn J, Morgans DJ, Smith WL. Selective inhibition of prostaglandin endoperoxide synthase-1 (cyclooxygenase-1) by valerylsalicylic acid. Arch. Biochem. Biophys. 1995;317:19–24. doi: 10.1006/abbi.1995.1130. [DOI] [PubMed] [Google Scholar]