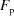A method for growing crystals on cryoloops or micromounts is described, and diffraction patterns of crystals of three proteins grown by both the new method and the conventional drop method are compared. The study investigates the steps for the automation of the crystal growth and manipulation process and describes the design of a tray for the method.
Keywords: protein crystallography, automation, crystal growth, cryoloops, micromounts
Abstract
Protein crystals are usually grown in hanging or sitting drops and generally get transferred to a loop or micromount for cryocooling and data collection. This paper describes a method for growing crystals on cryoloops for easier manipulation of the crystals for data collection. This study also investigates the steps for the automation of this process and describes the design of a new tray for the method. The diffraction patterns and the structures of three proteins grown by both the new method and the conventional hanging-drop method are compared. The new setup is optimized for the automation of the crystal mounting process. Researchers could prepare nanolitre drops under ordinary laboratory conditions by growing the crystals directly in loops or micromounts. As has been pointed out before, higher levels of supersaturation can be obtained in very small volumes, and the new method may help in the exploration of additional crystallization conditions.
1. Introduction
The availability of protein crystals is a prerequisite to the determination of the three-dimensional structure of proteins from X-ray diffraction experiments. The high-throughput production of diffraction-quality crystals remains a major bottleneck and several methods have been suggested to improve the success rate (Page, 2008 ▶). There are several robots that can be used to speed up the crystallization process and reduce human error (Chayen et al., 1990 ▶; Santarsiero et al., 2002 ▶; Hazes & Price, 2005 ▶). These robots also improve screening efficiency by using nanolitre volumes for crystallization experiments (Santarsiero et al., 2002 ▶; Hazes & Price, 2005 ▶; Bodenstaff et al., 2002 ▶).
Protein crystals are usually grown in hanging or sitting drops and generally get transferred to a loop or micromount for cryocooling and data collection. The transfer of crystals from drops is a delicate process that could affect the quality of crystals and may also result in loss of the crystal. However, the method for removing a crystal from the drop has not been fully automated (Dauter, 2006 ▶), so a number of alternatives have been suggested to circumvent this problem. The Universal Micromanipulation Robot can mount crystals from drops (http://www.sqr-1.com/Prod_UMR.html). One approach is to grow crystals in microcapillary tubes (García-Ruiz & Moreno, 1994 ▶; McPherson, 1999 ▶; Kalinin & Thorne, 2005 ▶; Yadav et al., 2005 ▶). This technique was used for the in situ high-resolution (1.8 Å) X-ray data collection of thaumatin crystals (Yadav et al., 2005 ▶). However, to prevent it from drying out, the crystal must remain in the capillary tube, preventing cryocooling and increasing radiation damage (Yadav et al., 2005 ▶). Another recent approach uses an automated procedure to grow crystals in small chips that can be mounted and used for X-ray diffraction experiments (Segelke, 2005 ▶). A semi-automatic protein crystallization system that allows in situ observation of X-ray diffraction from crystals in the drop has also been reported (Watanabe et al., 2002 ▶). McPherson (2000 ▶) has described a procedure for the collection of X-ray data from macromolecular crystals in situ using a prototype device that had sitting drops on small plastic or glass slides. McPherson (2000 ▶) describes several advantages for growing crystals directly on the support. Crystals need never be touched or manipulated, so there is no ‘collateral damage’. Needle crystals, thin-plate crystals, and others of either unfavorable morphology, such as clusters, or small size, otherwise unmountable, can be utilized for data collection. The data from many identical small crystals can be used to yield the data set of a single large crystal (McPherson, 2000 ▶).
In the present paper, we report the results of the crystallization of proteins in cryoloops and micromounts and compare the diffraction patterns and structures of crystals of three proteins grown by the new method and by the conventional hanging-drop method. Directly growing the crystals within the loops/micromounts bypasses the difficulty of retrieving the crystal from a drop and opens up the possibility for automation. We have developed a method to streamline the new crystallization process and describe the design of a tray that could be used for setting up the drops on loops or micromounts.
2. Experimental methods
2.1. Tray preparation
Normal hanging-drop trays (Nextal plates) were used to test the feasibility of crystal growth directly in loops and micromounts. Mounted cryoloops of 10 and 20 µm thickness, ranging in loop sizes from 0.05 to 0.5 mm diameter, were purchased from Hampton Research. MicroMounts, MicroGrippers and MicroMeshes were purchased from MiTeGen (Stum et al., 2004 ▶). The lengths of the pins were shortened to fit in the 18 mm copper magnetic base, and the loops or the MiTeGen products (MicroMounts, MicroGrippers and MicroMeshes) were then fastened using Duco cement (Hampton Research) to the inside of the hanging-drop wells, with one to four loops in each well. The loops were oriented such that the loop face would be parallel to the bottom of the well, for easy crystal viewing (Fig. 1 ▶ and supplementary material,1 Video 1). The loops were handled with tweezers to prevent them from touching the skin and becoming contaminated with grease or oil.
Figure 1.
Photographs showing the loops mounted in a Nextal tray. (a) Plate with mounted loops. (b) Delivery of a film onto the loop using a pipette. (c) View of a well showing the mounted loops and the hanging drop on the cover.
2.2. Protein sample preparation
The reservoir was filled with 500 µl of the mother solution, and a drop containing 1.2 µl of reservoir and 1.2 µl of protein solution was placed on the coverslip and mixed well. Then a 0.6 µl portion of the drop was pipetted and held as a drop at the tip of the pipette. The drop was brushed against the loops in the well while being viewed through a microscope (supplementary material, Video 1). Approximately 125 films were drawn from a 1 µl drop (from lysozyme well solution) using 0.2 mm Hampton loops indicating a volume of ∼8 nl per loop. The precise amount of the solution depends on the surface tension and on the orientation of the loop. The 0.2 mm MicroMounts hold around 23 nl per mount (stable films require filling the wicking aperture with the solution).
We have used the above procedure to prepare around 400 films (Hampton loops and the three MiTeGen mounts). We used ∼275 Hampton loops and 25 MiTeGen MicroMounts for our initial experiments. We tested MiTeGen MicroGrippers and MicroMeshes after the initial trials, and these products improved the stability of the film. The method was then improved by incorporating magnets for loop manipulation (Fig. 2 ▶ and supplementary material, Video 2). In this case, the loops attached to the magnets were drawn through a drop containing protein and reservoir solution. This setup uses handle magnets to move the loops in and out of the drops (supplementary material, Video 2). This design increases the reproducibility of the films on the loops and has a greater potential to speed up or automate the transfer of loops to the goniometer. It is envisaged that through using this system it may be possible to place the films in the loops at different time intervals in a closed system.
Figure 2.

Crystallization well design with magnets. The base of the pin is glued to a magnet. This assembly is held in place by the handle magnet on the outside of the well. The drops on the loops are formed by slowly moving the pin into the hanging drop or sitting drop using the handle magnet. The sitting drops were made by placing the drops on a glass coverslip mounted on a post. The magnets are mounted into Teflon pieces (not shown) for ease of handling and smooth control from outside the tray.
Ten different proteins [lysozyme (Sigma), thaumatin (Sigma), RNase (Sigma), two mutants of the Thermotoga maritima protein TM0449 (JCSG), T. maritima proteins TM0574 (JCSG), TM0559 (JCSG) and TM1389 (JCSG), an intein protein (New England Biolabs), and phosphoadenylyl sulfate reductase (Dr David Stout)] were tested (Fig. 3 ▶). These proteins were chosen to cover diverse crystallization conditions, from high salt to very high poly(ethylene glycol) (PEG). The crystals of lysozyme, thaumatin, RNase and T. maritima protein (a mutant of TM0449) were tested for diffraction (Table 1 ▶).
Figure 3.

Photographs showing the crystallization results. Images (a) and (b) show lysozyme crystals grown in a loop and a micromount. Images (c) and (d) show one of the TM0449 mutants grown in a loop and a micromount. (e) Plate-like crystals of TM1389. (f) Crystals of TM0574. (g) Crystals of the intein protein. (h) Crystals of TM0559. (i) Crystals of RNase. (j) Crystals of thaumatin. The TM1389, TM0574, intein and TM0559 proteins were tested during the early stages using Hampton loops only.
Table 1. Diffraction results for crystals grown by the loop/micromount method.† .
| Protein (number of crystals) | Loop/micromount | Beamline | Resolution range () |
|---|---|---|---|
| Lysozyme (5) | Loop | SSRL BL9-2 | 0.941.27 |
| TM0449 mutant (1) | Loop | ALS 8.3.1 | 1.60 |
| Lysozyme (3) | Loop | ALS 8.3.1 | 1.061.30 |
| TM0449 mutant (5) | Loop | ALS 5.0.3 | 1.451.85 |
| Lysozyme (5) | Loop | ALS 5.0.3 | 1.151.35 |
| TM0449 mutant (4) | MicroMount | ALS 5.0.1 | 2.003.00 |
| Lysozyme (2) | Loop | ALS 5.0.1 | 1.361.80 |
| Lysozyme (2) | MicroMount | SSRL 7-1 | 1.401.60 |
| Thaumtin (4) | MicroMounts and MicroMesh | SSRL 11-1 | 1.401.70 |
| RNase (3) | Loops and MicroMesh | SSRL 11-1 | 1.451.60 |
| TM0449 mutant (3) | Loops and MicroMesh | SSRL 11-1 | 2.32.5 |
The diffraction limit was checked by visually inspecting the diffraction images.
Thaumatin crystals were grown from a well solution containing 0.9 M sodium/potassium tartrate, 0.1 M 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, pH 7.3) and 15% glycerol (Nanao et al., 2005 ▶). The well solution used for RNase contained 1.7 M ammonium sulfate, 2 M NaCl and 0.1 M sodium acetate (Nanao et al., 2005 ▶), and the mutant of TM0449 was crystallized from a well solution containing 49% PEG 200 and 0.1 M HEPES (pH 7.6).
Complete data sets for crystals of thaumatin, RNase and the TM0449 mutant were collected. As a control, diffraction data for these proteins from crystals grown in a normal hanging-drop experiment, within the same well as the crystals used for loops/mounts, were collected.
2.3. Cryoprotection
The proteins selected for the tests had crystallization conditions ranging from high salt (1.5 M NaCl) to high PEG (up to 60% PEG 200). Typically the pins were removed from the side of the well using tweezers (supplementary material, Video 1). Those crystals grown in the high-PEG conditions were suitable for flash cooling without the need of a cryoprotectant, and after being removed from the well they were quickly transferred to a copper base containing glue and stored in liquid nitrogen. We tested Duco cement, Super Glue and 5 Minute Epoxy as the glue, and the copper bases with 5 Minute Epoxy showed the best stability after cooling at 100 K. (Note: The reusable bases from MiTeGen should simplify this process.) For the remainder, the loop containing the crystal was plunged into the cryosolution and flash cooled using another loop.
2.4. Data collection and refinement
X-ray diffraction data were collected using ADSC Q315R or MAR325 CCD detectors, at SSRL (Stanford Synchrotron Radiation Laboratory, California, USA) beamlines 7-1 or 11-1 controlled through the Blu-Ice (McPhillips et al., 2002 ▶) interface. Photographs of the crystals grown in the support are shown in Fig. 4 ▶.
Figure 4.
Crystals grown in loops used for the structural study. The left panels show pictures of the crystals in the tray. The right panels show the same crystals mounted on the beamline for data collection.
The data sets for the crystals grown directly in the loops were solved using molecular replacement and the protein models derived from this were used as models to solve and refine the data sets collected from crystals grown in the hanging-drop experiment [Protein Data Bank (PDB; Berman et al., 2000 ▶) entry used: 1ly0 (thaumatin; Charron et al., 2002 ▶), 3dh5 (RNase; Kurpiewska et al., 2009 ▶), 1o26 (TM0449 mutant; Mathews et al., 2003 ▶)]. The data collection and refinement statistics are summarized in Table 2 ▶.
Table 2. Crystallographic parameters, data collection and refinement statistics.
Crystals grown in loops.
| RNase | Thaumatin | TM0449 mutant | |
|---|---|---|---|
| Crystallographic parameters† | |||
| Space group | P3221 | P41212 | P212121 |
| Unit-cell dimensions (, ) | 64.2, 64.2, 63.9 | 57.9, 57.9, 150.4 | 54.5, 116.8, 141.0 |
| 90.0, 90.0, 120.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | |
| Data collection statistics | |||
| Resolution limits () | 29.01.50 | 29.01.50 | 35.62.3 |
| No. of observed reflections | 149216 | 197807 | 200943 |
| No. of unique reflections | 24805 | 41945 | 40897 |
| Completeness | |||
| Overall/outer shell | 99.9/99.9 | 99.8/100 | 100/100 |
| Rsym ‡ (%) | |||
| Overall/outer shell, I/ | 8.1/86.7, 16.0 | 7.1/57.6, 16.1 | 10.2/79.0, 11.1 |
| Refinement statistics | |||
| Resolution limits | 29.01.50 | 29.01.50 | 30.02.3 |
| Number of reflections/% | 23536/100.0 | 39810/99.8 | 38732/99.8 |
| Reflections used for R free | 1269 | 2135 | 2046 |
| R factor § (%) | 17.6 | 15.4 | 19.8 |
| R free (%) | 19.3 | 17.7 | 24.9 |
| Model contents/average B (2) | |||
| Protein atoms | 951/12.4 | 1545/9.8 | 7135/32.2 |
| Ligand atoms | 0 | 0 | 292/52.1 |
| Ions/buffer atoms | 9/28.8 | 20/17.5 | 0 |
| Water molecules | 140/19.7 | 219/21.3 | 39/28.5 |
| r.m.s. deviations | |||
| Bond length () | 0.018 | 0.018 | 0.016 |
| Bond angle () | 1.77 | 1.69 | 1.84 |
Crystals grown in drops.
| RNase | Thaumatin | TM0449 mutant | |
|---|---|---|---|
| Crystallographic parameters† | |||
| Space group | P3221 | P41212 | P212121 |
| Unit-cell dimensions (, ) | 64.2, 64.2, 63.7 | 57.9, 57.9, 150.24 | 54.6, 116.7, 141.2 |
| 90.0, 90.0, 120.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | |
| Data collection statistics | |||
| Resolution limits () | 29.01.50 | 29.01.50 | 40.02.3 |
| No. of observed reflections | 145316 | 195309 | 184294 |
| No. of unique reflections | 24750 | 41768 | 40698 |
| Completeness | |||
| Overall/outer shell | 99.8/99.7 | 99.4/99.9 | 99.5/98.7 |
| R sym † (%) | |||
| Overall/outer shell, I/ | 4.6/44.6, 23.6 | 4.0/20.2, 26.3 | 8.9/52.0, 12.8 |
| Refinement statistics | |||
| Resolution limits | 29.01.50 | 29.01.50 | 30.02.3 |
| Number of reflections/% | 23475/99.8 | 39637/99.6 | 38582/99.4 |
| Reflections used for R free | 1249 | 2083 | 2037 |
| R factor ‡ (%) | 17.6 | 14.6 | 19.6 |
| R free (%) | 19.3 | 16.3 | 24.8 |
| Model contents/average B (2) | |||
| Protein atoms | 951/12.3 | 1545/8.3 | 7135/25.8 |
| Ligand atoms | 0 | 0 | 292/41.6 |
| Ions/buffer atoms | 9/27.9 | 20/15.1 | 0 |
| Water molecules | 140/20.2 | 223/18.9 | 46/22.2 |
| r.m.s. deviations | |||
| Bond length () | 0.018 | 0.016 | 0.019 |
| Bond angle () | 1.70 | 1.60 | 1.95 |
The coordinates for the structures are deposited in the PDB [codes 3mzr (RNase, grown in loop), 3mzq (RNase, grown from drop), 3n02 (thaumatin, grown in loop), 3n03 (thaumatin, grown from drop), 3n0b (TM0449 mutant, grown in loop) and 3n0c (TM0449 mutant, grown from drops)].
3. Results and discussion
3.1. Success rate and crystal quality
We prepared 400 films on the loops or micromounts for the crystallization trials. Around 71% of the loops retained the liquid and 52% of those that retained the liquid produced crystals. The success rate was improved by using MiTeGen MicroMeshes or MicroGrippers. Loop manipulation using tweezers (to avoid oil or grease from hands) as well as washing the loops with dilute hydrochloric acid helped in producing a stable film for the higher-salt conditions.
The success of the crystallization in loops was monitored by comparing the drop on the coverslip with the corresponding loops. A comparison of the crystals grown in loops and grown by the hanging-drop method for lysozyme and a TM0449 mutant is shown in Fig. 5 ▶. The well solution for lysozyme for this trial ranges from 1.2 to 0.8 M NaCl and 50 mM sodium acetate (pH 4.5). The TM0449 mutant well solution varies from 55 to 40% PEG 200 and 0.1 M HEPES (pH 7.5). The appearance of crystals in the films and the absence of crystals in the corresponding drops highlight the effect of nanolitre drop volume on the crystallization (Bodenstaff et al., 2002 ▶).
Figure 5.
Comparison of crystals in loops and the corresponding hanging drops. The images in the left and right panels show crystallization results for lysozyme and one of the TM0449 mutants, respectively (the precipitant concentration decreases as we go down the figure). The pictures in each panel shows the hanging drops on the left side and the corresponding loops on the right side. The loop rods appear as dark blurred lines directly under many of the drops.
Generally the crystals grown in the loops were smaller and fewer in number. Lowering the precipitant concentration seemed to help with the crystallization of some of the protein samples. The best procedure consists of using two loops per well, a small amount of glue and 20 µm-thick loops with a loop diameter of 0.2 to 0.5 mm. The 10 µm-thick loops and loops larger than 0.5 mm in loop diameter dehydrated too frequently. The stability of the film and the sizes of the crystals improved by using the MicroMeshes and MicroGrippers. The results of a comparison study of the lysozyme crystallization with Hampton loops, MicroMounts, MicroMeshes and MicroGrippers are given in Table 3 ▶.
Table 3. Comparison of results (films retaining solution/films with good crystals) of films from Hampton loops (0.2mm) and MiTeGen MicroMounts (0.2mm), MicroGrippers (0.3mm) and MicroMesh (0.4 0.05 mesh).† .
| Crystallization condition | Hampton loops‡ | MicroMount | MicroGripper§ | MicroMesh§ |
|---|---|---|---|---|
| 1.2M NaCl, 50mM sodium acetate (pH 4.5); protein solution at 25mgml1 in water | 10/5 | 13/9 | 15/13 | 15/14 |
Fifteen films were set up for each method. The films were checked on the third day.
Six of the films with Hampton loops had needle-shaped microcrystals.
The crystals grown on the MicroGripper and MicroMesh were larger.
Crystallization in loops/micromounts using the new method (Fig. 2 ▶) helped to retain the film and gave improved results. This improved setup, whereby the films on the loops/micromounts were prepared in a closed system, showed greater reproducibility in the stability of the film.
3.2. Data quality and model comparison
We tested the diffraction of 37 crystals grown by the loop method (17 lysozyme crystals, and 13 crystals of a TM0449 mutant, four of thaumatin and three of RNase). The diffraction results are given in Table 1 ▶. In general, the diffraction quality of the crystals grown in the loop/MiTeGen mounts was lower than the diffraction reported for the same protein grown by conventional methods. However, it may be noted that the best resolution we observed for the TM0449 mutant crystals grown by the hanging-drop method is 1.75 Å.
We also compared the diffraction of crystals grown by the loop method with the diffraction of crystals from the drops of the corresponding wells, and the diffraction quality generally depended on the crystal size. The diffraction patterns for some of the crystals grown by the loop method showed elongation of the spots. This was observed for the lysozyme crystals grown from high-salt conditions, which had to be cryocooled using ethylene glycol as the cryoprotectant. However, the lysozyme crystals grown from the 30% PEG 5000 monomethyl ether conditions did not show this from either the loop-grown or the hanging-drop-grown crystals.
We selected crystals of thaumatin, RNase and the TM0449 mutant for complete data collection. Data sets were collected for crystals grown in the loops/MiTeGen mounts and by the conventional hanging-drop method.
The data sets of the crystals grown by the hanging-drop method showed consistent improvement in the data quality (Table 2 ▶). This is partly due to the larger crystal size, which is also reflected in the higher I/σ values for the data sets of the crystals grown by the hanging-drop method. The electron density maps for the structures from the loops/MiTeGen mounts and the hanging-drop method showed no noticeable differences.
3.3. New crystallization tray
The preliminary design of a new crystallization tray is shown in Fig. 6 ▶. The new tray is designed to enable automation of the crystallization protocol described here. The aim is to automate the loading of the film, dislodging of the loop, and placement of the loop on a beamline or a storage device. The holder magnets could be moved manually or using a robot to place the film on the loops/MiTeGen mounts. The basic steps are also outlined in Fig. 2 ▶, and Video 2 in the supplementary materials shows the placing of the films and manual transfer of the pin to the goniometer.
Figure 6.

Design of the crystallization tray. The tray designed for automatic placement of the drops into loops. The loops can be moved into and out of the drops (figure shows hanging drops; it can also be used for sitting drops on a coverslip; see supplementary material, Video 2) by a robot or manually using the holder magnets. The loops with crystals could be transferred to the goniometer or a storage device.
4. Conclusions
Large-scale structural genomics programs rely heavily on automated methods for crystallization, crystal visualization and handling. In the area of crystallization, the main effort is focused on the automation of screening procedures to identify important leads. Most of the steps, including mounting the crystals on the sample goniometer, data collection and data processing, have been fully automated (Abola et al., 2000 ▶; Cohen et al., 2002 ▶; Dauter, 2006 ▶). The method of removing a crystal from a drop has not been fully automated before, and this presents a major challenge (Dauter, 2006 ▶). Although a number of new techniques have been proposed to overcome this problem (McPherson, 2000 ▶; Yadav et al., 2005 ▶), each of them presents new challenges.
We believe that crystallization in loops will help to automate the task of crystal selection and, if optimized, could eliminate the need for manual crystal manipulation during mounting. Since crystals would be already grown in loops, robots will be able to pick them directly from the newly designed trays and mount them into a storage device or onto a goniometer. The approach described here using magnets and the new tray design or a similar device should help with the automation of the mounting process. The crystallization of proteins directly in loops and MiTeGen mounts can be easily reproduced in a typical laboratory, allowing researchers to test the effect of nanolitre drop volumes on crystallization or crystal quality.
Supplementary Material
. DOI: 10.1107/S0021889810040409/ea5129sup1.pdf
Legends for the supplementary movies
. DOI: 10.1107/S0021889810040409/ea5129sup2.wmv
Video 1: Preparation of the Nextal tray
. DOI: 10.1107/S0021889810040409/ea5129sup3.wmv
Video 2: Incorporating magnets for loop manipulation
Acknowledgments
We thank Drs G. Card, P. Dunten and C. Smith for critical reading of the manuscript and Professor A. McPherson for valuable suggestions. We thank Dr T. Doukov and L. Aguila for help with the video. We thank Professor D. Stout (Scripps Research Institute, La Jolla, CA, USA) for providing a sample of phosphoadenylyl sulfate reductase, Dr F. Perler (New England BioLabs, Beverly, MA, USA) for the intein protein and the Joint Center for Structural Genomics (JCSG) for the T. maritima proteins. We also thank members of the Structural Molecular Biology group at SSRL and the members of the JCSG at SSRL for helpful discussions and support. This work was supported by an SSRL summer student internship to MAB and JHD. Portions of this research were carried out at the SSRL and Advanced Light Source (ALS). SSRL is a national user facility operated by Stanford University on behalf of the US Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. The ALS is supported by the Director, Office of Science, Office of Basic Energy Sciences, Materials Sciences Division, of the US Department of Energy under contract No. DE-AC03-76SF00098 at Lawrence Berkeley National Laboratory.
Footnotes
Supplementary material, including the videos discussed in this paper, is available from the IUCr electronic archives (Reference: EA5129). Services for accessing this material are described at the back of the journal.
References
- Abola, E., Kuhn, P., Earnest, T. & Stevens, R. C. (2000). Nat. Struct. Biol. 7, 973–977. [DOI] [PubMed]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed]
- Bodenstaff, E. R., Hoedemaeker, F. J., Kuil, M. E., de Vrind, H. P. M. & Abrahams, J. P. (2002). Acta Cryst. D58, 1901–1906. [DOI] [PubMed]
- Charron, C., Kadri, A., Robert, M.-C., Giegé, R. & Lorber, B. (2002). Acta Cryst. D58, 2060–2065. [DOI] [PubMed]
- Chayen, N. E., Shaw Stewart, P. D., Maeder, D. L. & Blow, D. M. (1990). J. Appl. Cryst. 23, 297–302.
- Cohen, A. E., Ellis, P. J., Miller, M. D., Deacon, A. M. & Phizackerley, R. P. (2002). J. Appl. Cryst. 35, 720–726. [DOI] [PMC free article] [PubMed]
- Dauter, Z. (2006). Acta Cryst. D62, 1–11. [DOI] [PubMed]
- García-Ruiz, J. M. & Moreno, A. (1994). Acta Cryst. D50, 484–490. [DOI] [PubMed]
- Hazes, B. & Price, L. (2005). Acta Cryst. D61, 1165–1171. [DOI] [PubMed]
- Kabsch, W. (2010a). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010b). Acta Cryst. D66, 133–144. [DOI] [PMC free article] [PubMed]
- Kalinin, Y. & Thorne, R. (2005). Acta Cryst. D61, 1528–1532. [DOI] [PubMed]
- Kurpiewska, K., Font, J., Ribo, M., Vilanova, M. & Lewinski, K. (2009). Proteins, 77, 658–669. [DOI] [PubMed]
- Leslie, A. G. W. (1999). Acta Cryst. D55, 1696–1702. [DOI] [PubMed]
- Mathews, I. I., Deacon, A. M., Canaves, J. M., McMullan, D., Lesley, S. A., Agarwalla, S. & Kuhn, P. (2003). Structure, 11, 677–690. [DOI] [PubMed]
- McPherson, A. (1999). Crystallization of Biological Macromolecules, pp. 193–194. Cold Spring Harbor Laboratory Press.
- McPherson, A. (2000). J. Appl. Cryst. 33, 397–400.
- McPhillips, T. M., McPhillips, S. E., Chiu, H.-J., Cohen, A. E., Deacon, A. M., Ellis, P. J., Garman, E., Gonzalez, A., Sauter, N. K., Phizackerley, R. P., Soltis, S. M. & Kuhn, P. (2002). J. Synchrotron Rad. 9, 401–406. [DOI] [PubMed]
- Nanao, M. H., Sheldrick, G. M. & Ravelli, R. B. G. (2005). Acta Cryst. D61, 1227–1237. [DOI] [PubMed]
- Page, R. (2008). Methods Mol. 426, 345–362. [DOI] [PubMed]
- Santarsiero, B. D., Yegian, D. T., Lee, C. C., Spraggon, G., Gu, J., Scheibe, D., Uber, D. C., Cornell, E. W., Nordmeyer, R. A., Kolbe, W. F., Jin, J., Jones, A. L., Jaklevic, J. M., Schultz, P. G. & Stevens, R. C. (2002). J. Appl. Cryst. 35, 278–281.
- Segelke, B. (2005). Expert Rev. Proteomics, 2, 165–172. [DOI] [PubMed]
- Stum, Z., Kmetko, J., O’Neill, K., Gillilan, R., Bartnick, A. & Thorne, R. E. (2004). Synchroton Rad. News, 17, 31–32.
- Watanabe, N., Murai, H. & Tanaka, I. (2002). Acta Cryst. D58, 1527–1530. [DOI] [PubMed]
- Yadav, M. K., Gerdts, C. J., Sanishvili, R., Smith, W. W., Roach, L. S., Ismagilov, R. F., Kuhn, P. & Stevens, R. C. (2005). J. Appl. Cryst. 38, 900–905. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
. DOI: 10.1107/S0021889810040409/ea5129sup1.pdf
Legends for the supplementary movies
. DOI: 10.1107/S0021889810040409/ea5129sup2.wmv
Video 1: Preparation of the Nextal tray
. DOI: 10.1107/S0021889810040409/ea5129sup3.wmv
Video 2: Incorporating magnets for loop manipulation