Abstract
HIV establishes a latent reservoir early in infection that is resistant to anti-retroviral therapy and has a slow rate of decay. It is thought that the majority of HIV DNA in treated patients is integrated since unintegrated HIV DNA appears to be unstable. Thus, to monitor the HIV latent reservoir, total HIV DNA is commonly measured in PBMC from infected individuals. We investigated how often total approaches integrated HIV DNA in treated patients. To do this, we first assessed how accurate our integration assay is and determined the error in our measurements of total and integrated HIV DNA. We demonstrated an excess of total over integrated HIV DNA was present in a subset of patients, suggesting that measurements of total HIV DNA do not always correlate to the level of integration. Determining the cause of this excess and its frequency may have important implications for understanding HIV latent reservoir maintenance.
Keywords: HIV Integration, Alu-PCR, HIV Reservoir, HIV Latency, HIV Reservoir Maintenance
Introduction
Integration into the host cell genome is a fundamental characteristic of retroviruses. Although some gene expression occurs from unintegrated retroviral DNA, integrated DNA serves as the main template for efficient gene transcription and viral replication (Ansari-Lari, Donehower, and Gibbs, 1995; Cara et al., 1995; Coffin, 1997; Engelman et al., 1995; Englund et al., 1995; Sakai et al., 1993; Stevenson et al., 1990a; Stevenson et al., 1990b; Wiskerchen and Meusing, 1995). This characteristic allows the provirus to be stably expressed by the host machinery and to persist in the infected cell indefinitely. This is particularly important for the survival of HIV in infected individuals. By integrating its genome, HIV can be persistently produced from cells that are resistant to the cytopathic effects of viral replication as occurs in macrophages (Ho, Rota, and Hirsch, 1986; Igarashi et al., 2001; Nicholson et al., 1986; Swingler et al., 2007). Integrated HIV can also remain in a latent state indefinitely, without producing infectious virions, as occurs in resting CD4+ T cells (Bukrinsky et al., 1991; Chun et al., 1995b). This latent form of infection has a very slow apparent rate of decay in infected individuals receiving highly active anti-retroviral therapy (HAART) (Finzi et al., 1999; Han et al., 2007; Stevenson, 2003). Because latent provirus does not produce significant amounts of new particles, it is assumed to be resistant to drug treatment and immune surveillance (Colin and Van Lint, 2009; Fischer et al., 2008). The pool of resting CD4+ T cells harboring latent provirus forms early during initial infection of a host and represents a major obstacle for the eradication of virus from infected patients (Chun et al., 1998).
Given the importance of integration for HIV replication and persistence in the host, measuring this step in the viral life cycle accurately is essential to have a better understanding of HIV biology and pathogenesis. Measuring integration is instrumental for determining how the reservoir of latently infected resting cells forms and how it is maintained. It may also be valuable for monitoring viral replication in combination with other measurements, such as total viral DNA, and for monitoring the effect of therapy on the size of the latent reservoir. A sensitive assay for measuring integration could also help reveal cells that may harbor viral reservoirs but at lower levels than memory CD4+ T cells.
Our group developed a quantitative assay for measuring integration based on Alu-PCR and repetitive sampling (O'Doherty et al., 2002; Yu et al., 2008). The assay is sensitive and the detection limit is defined by the number of replicates performed. We routinely detect integration at levels near 1 in 10,000 in HIV infected samples by performing 42 replicates. This sensitivity makes the assay suitable for the detection of integration in samples both inoculated at very low inoculum in vitro and from infected individuals undergoing combination therapy (Agosto et al., 2007; Yu et al., 2008). This assay incorporates a polyclonal integration standard that parallels the diversity of integration sites found in natural infection to add accuracy to the measurement (Brussel and Sonigo, 2003; Butler, Hansen, and Bushman, 2001; O'Doherty et al., 2002; Yu et al., 2008).
Some studies suggest that total HIV DNA can be used as an estimate of the reservoir in patients on HAART (Chomont et al., 2009; Koelsch et al., 2008). The rationale is that total HIV DNA should approach the level of integrated HIV DNA in patients on HAART since unintegrated HIV DNA has a short half-life in vitro (Koelsch et al., 2008). In order to determine if an excess of total HIV DNA exists in some patients on HAART and to determine if the excess of total DNA is not due to measurement error, we decided to determine the accuracy of our integration assay. In the present study, we demonstrate that our integration estimates are accurate and precise by comparing the level of integration detected by quantitative PCR to the proportion of HIV-positive clones obtained after single-cell cloning of a population of infected cells. We go on to provide evidence that in some patients on HAART, with undetectable plasma viral loads, there is a difference between the levels of total and integrated HIV DNA. Therefore, measuring total HIV DNA is not always a suitable estimate of the levels of integrated HIV DNA and by analogy it is not always a suitable correlate of reservoir size in patients receiving HAART. Based on these findings, we conclude that an accurate measure of integrated HIV DNA is a better indicator of reservoir size than total HIV DNA measurements.
Results
Accuracy of Alu-PCR Integration Assay
The assay for measuring HIV integration involves two amplification reactions: an end-point PCR amplification and a kinetic PCR amplification (O'Doherty et al., 2002) (Figure 1A). The pre-amplification step, targets integrated HIV specifically by amplifying the region between HIV gag and the nearest Alu repeat (see relative primer locations in Figure 1B). Alu repeats are the most common repetitive elements in the human genome with a frequency of 1 copy every ~3000 base pairs (Jelinek and Schmid, 1982; Mighell, Markham, and Robinson, 1997). Given that HIV integrates at various distances relative to Alu, the distance between integrated HIV and the nearest Alu site determines the limit of detection of the pre-amplification step. The assay described is capable of detecting HIV integration when it occurs within a few thousand base pairs (2–5 kb) of the nearest Alu site (Agosto et al., 2007). Following this pre-amplification step, part of the reaction product is further amplified in a second reaction that is specific for HIV-containing amplicons. This nested-kinetic amplification recognizes the R-U5 region of the HIV long terminal repeat (see relative primer location in Figure 1B). The distribution of cycle thresholds (calculated from the amplification curves) of each reaction (see amplification example in Figure 2A) is used to estimate the number of integration events in the unknown sample by comparing the cycle threshold to a standard curve generated using the integration standard (Agosto et al., 2007). This standard is polyclonal, to reflect the diversity of integration sites that occur in vivo, and contains one integration event per cell. For an integration signal to be considered positive, the average Alu-gag signal must be greater than the gag-only signal (compare Figure 2A to Figure 2B). The gag-only control is included to approximate the signal expected when all the DNA is unintegrated.
Figure 1. Overview of integration assay, lentiviral vector and complementary method to measure integration.
(A) Nested Alu-PCR was used for measuring integration in samples of CEM-SS inoculated in vitro. The assay involves an initial endpoint PCR amplification of the region between gag and the nearest human Alu repeat, followed by a kinetic PCR amplification of the HIV-containing products of the first reaction. (B) The lentiviral vector used for our inoculations is the HIV-based lentiviral vector, VRX494 (Humeau et al., 2004; Lu et al., 2004). The primers used for PCR and their relative priming sites are outlined and described below the figure. (C) A complementary method to measure integration was performed by inoculating CEM-SS cells with VRX494 at 2 different viral dilutions. At 18hr post-inoculation the cells were separated into 2 aliquots: (i) measure integration by nested Alu-PCR and (ii) plated at 0.3 or 1 cell/well in round bottom 96-well plates until colonies grew. Colonies were isolated approximately 2 weeks later and tested for the presence of HIV DNA by late reverse transcription kinetic PCR.
Figure 2. Measuring HIV integration by Alu-PCR.
CEM-SS cells were inoculated with two dilutions of VRX494 1× and 1:5× with or without integrase inhibitor from the beginning of the inoculation. HIV integration was measured by Alu-PCR at 0, 16, 18 and 24hr post-inoculation. (A, B) Sample PCR amplification plots of DNA from cells inoculated at 1× viral inoculum without (A) or with (B) integrase inhibitor at 18hr post-inoculation. Plots show that a signal is positive for integration when the amplification using primers for Alu-gag (gray lines) occurs at an earlier cycle threshold (Ct) than when using only the primer for gag (gag-only – black lines). The Alu-gag signal obtained with an integrase inhibitor gave a similar signal to the gag-only amplification. This confirms that the gag-only signal approximates the signal expected when there is no integration. (C, D) Integration time course in cells inoculated at 1× (C) and 1:5× (D) viral inoculum. Arrows indicate (a) the time point when integrase inhibitor was added to block further integration and (b) the time point when cells were collected to compare the level of integration measured by Alu-PCR and by single-cell cloning. Error bars indicate the standard deviation of the combined measurements of integration from 2 separate experiments. The dashed line indicates the limit of detection of integration. This limit is the average level of amplification obtained from the PCR negative control (gag-only reaction) of the integrase inhibitor inoculation control between 16 and 24hr post-inoculation.
We determined the accuracy of the Alu-PCR integration assay by comparing the level of integration estimated by nested Alu-PCR to the proportion of HIV positive clones after single-cell cloning, both obtained from the same sample of cells inoculated with HIV (Figure 1C). To do this, a CD4+ T cell line (CEM-SS cells) was inoculated at two dilutions of the HIV-based lentiviral vector VRX494 (Figure 1B) and incubated at 37°C. We chose to transduce the CD4+ T cell line with a gene therapy vector to avoid the toxicity of virally encoded genes and to ensure that only one round of infection occurred. We previously titered our virus on the CEM-SS cell line and used viral inoculums that should yield ~1 and ~0.2 proviruses/cell. At 16hr post-inoculation, an integrase inhibitor was added to prevent further integration events. Cells were then incubated for an additional 2hr at 37°C to allow sufficient time for the integrase inhibitor to be internalized by the cells before cloning the cells at limiting dilution. We measured integration 2 hours before, at the time of cloning (18hrs) and 6 hours after cloning to demonstrate that the integrase inhibitor effectively inhibited integration at the time of cloning. In figure 4, we compare the level of integration obtained at 18 hours with the level obtained by cell cloning As a negative control, CEM-SS were inoculated in the presence of integrase inhibitor and were kept exposed to the drug throughout the time course.
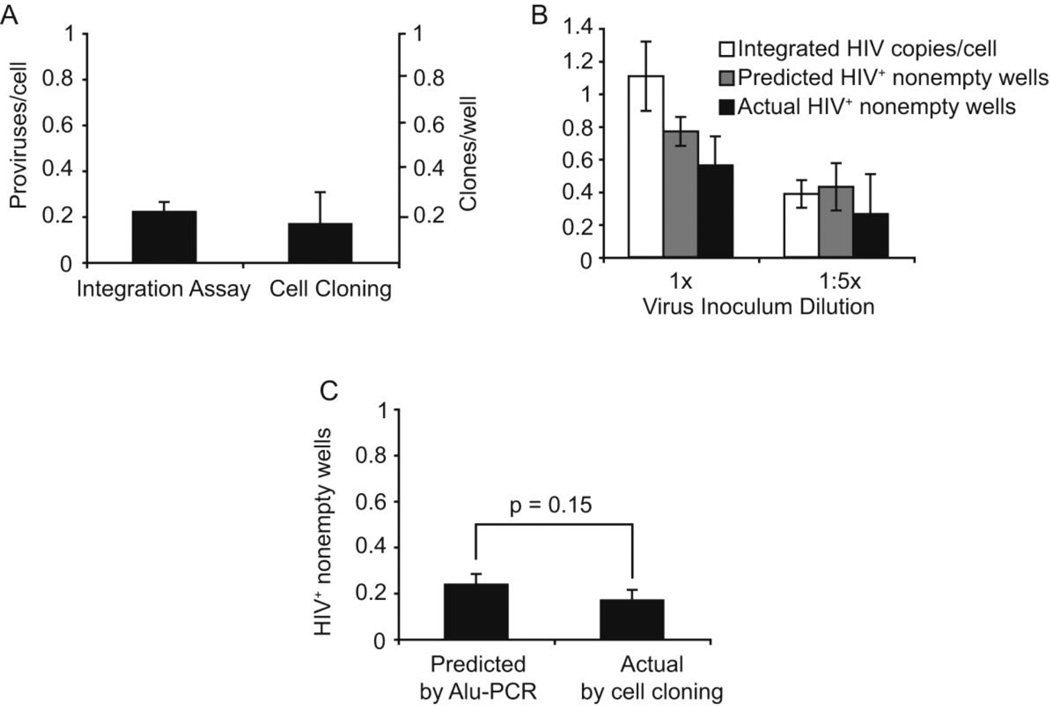
Figure 4. The estimate of integration measured by nested Alu-PCR, closely correlates with the proportion of HIV-positive colonies obtained after single-cell cloning of infected CEM-SS cells.
(A) Comparison of integration measured by Alu-PCR and by cell cloning when the cells were infected at low inoculum and single-cell cloned at 0.3 cells/well. (B) The level of integration detected by Alu-PCR was converted into an estimate of HIV-positive nonempty wells using the Poisson distibution formula (see Supplement). This corrects for multiply-infected cells and wells containing multiple clones at the time of cloning. The standard deviation for the Poisson-based estimate and the actual percent HIV-positive nonempty wells were calculated with the binomial distribution formula. (C) Comparison of predicted vs actual HIV positive nonempty wells. Graph represents the combination of values of (A) and (B) at each virus concentration after values were normalized to the level of late reverse transcription (SST) of each inoculation. p-values were calculated using Student’s T-test.
We show the level of integration at 0, 16, 18, and 24 hr post-inoculation for two viral inoculations (Figure 2C, D). The level of integration appears to decay over time in the cells inoculated at a lower dose. This could be due to slight overgrowth of the uninfected cells after 16hr. This seems reasonable since at higher inoculum, when practically all the cells were infected, we did not see this decay. Alternatively, it may represent error in our assay. The actual level of integration measured by our Alu-based PCR assay for the top dilution at 18hr post-inoculation was 1.1 proviruses/cell and 0.39 provirus/cell for the bottom dilution (Figure 4B) or 0.22 (Figure 4A). At this point, an aliquot of cells was cloned in 96-well plates at on average 1 cell per well (Figure 3 and 4B) or 0.3 cells per well (Figure 4A). Colonies of ~105 cells grew after incubating for 2 weeks. The cloning efficiency was ~60% when we plated at approximately 1 cell per well, which is close to the expected 63% based on the Poisson distribution (Figure 3). Thus, the viral inoculum and the integrase inhibitor had a minimal effect on the cloning efficiency when compared to uninfected or untreated controls (Figure 3). The clones were analyzed for the presence of total HIV DNA (see relative primer position in Figure 1B), until at least 5 colonies were positive for HIV DNA. The majority of the HIV-positive signals were between 0.1 and 1 SST/cell (see distribution of values in Supplemental Figure 1). The range of values below 1 SST/cell can be explained by the possibility that integration may have taken place during the G2 stage of the cell cycle, which has been shown to be the stage at which integration is most efficient (Groschel and Bushman, 2005). At this stage of the cell cycle, a CEMss cell would have ~4 sets of chromosomes and only one set would harbor integrated HIV DNA. Although HIV integration appears to only have a minimal effect on cloning efficiency (Figure 3), it is possible that the daughter cells containing integrated HIV DNA may have different cell division efficiencies, thus affecting the relative proportion of HIV-positive and HIV-negative cells within the colony.
Figure 3. Infection of CEM-SS with VRX494 or treatment with integrase inhibitor, minimally affects cloning efficiency.
Cloning efficiency was determined by calculating the percentage of wells in which cell colonies grew with cloning at ~1 cell/well under the following conditions: viral inoculum with integrase inhibitor added at 16hr, uninfected without integrase inhibitor, uninfected with integrase inhibitor and infected without integrase inhibitor (1× viral dilution). Error bars represent the standard deviation of the combined data of 2 separate experiments.
We then sought to determine if the two methods of quantitation would yield similar results. We found when we plated cells at 0.3 cells per well (single cell cloning conditions) and with a low proviral level (0.22 proviruses per cell by our Alu-PCR based assay), both methods gave similar results (Figure 4A). We obtained 0.22 proviruses per cell by our integration assay and 0.17 (17%) of clones were positive for HIV by single cell cloning. We also cloned the cells at higher levels of integration (1.1 proviruses per cell and 0.39 proviruses per cell) and at a higher concentration of cells (on average 1 cell/well). We found that 56% of the nonempty wells were positive for HIV at 1.1 proviruses per cell and ~23% were positive at 0.39 proviruses per cell by counting the number of nonempty wells that were positive for HIV. At these higher inoculums, we needed to perform two corrections before we could compare the two methods: we needed to first compensate for multiply infected cells and then to compensate for the increased probability of multiple clones per well. We did not need to make these corrections, when we cloned the cells at 0.3 cells/well with 0.22 proviruses per cell since the chance of multiply infected cells and multiple clones per well was low. In Supplement 2, we provide a detailed description of how we converted the proviruses per cell as estimated by Alu-PCR to the predicted percentage of nonempty wells that were positive for HIV. Briefly, we converted the proviral number to the expected number of clones that would be infected which was then converted the to the number of nonempty wells expected to be positive for HIV (Fig. 4B). Finally, we normalized the number of nonempty wells that were positive for HIV to the level of infection by dividing by the number of reverse transcripts present at the time of cloning. We averaged these measurements and performed a Student’s t test and found the measurement by both methods were not significantly different (Fig. 4C).
An excess of total DNA is present in a subset of patients receiving HAART
Because unintegrated HIV DNA has a short half-life in vitro, it stands to reason that the level of total HIV DNA could approach the level of integrated HIV DNA after prolonged HAART. In other words, the ratio of total over integrated should approach 1. Studies support that this indeed occurs at least in some patients on HAART (Chomont et al., 2009; Koelsch et al., 2008). We wanted to determine how often total HIV DNA and integrated HIV DNA are equivalent in patients after at least one year of suppressive HAART therapy. To do this, we needed to account for measurement errors of total HIV DNA and integrated HIV DNA since some elevated ratios could be due to measurement error. To determine the error in our measurements, we used our integration standard, which was designed to have a ratio of 1 total HIV DNA per integrated DNA since all the HIV is integrated in our standard (Agosto et al., 2007). Thus, any variation from this value would be due to errors. To mimic the conditions in a patient sample, we diluted our integration standard so that there was 1 integration standard cell in 10,000 uninfected cells. Total DNA and integrated HIV DNA were measured 7 times. The ratio of total DNA over integrated DNA obtained was 0.93 +/− 0.40 (Table 1). Because the levels of total DNA were measured independently from the levels of integrated HIV DNA, it is not completely appropriate to match any individual measurement of total HIV DNA with any individual measurement of integrated DNA. For this reason, we also calculated the average total and the average integrated HIV DNA and then calculated the ratio of total over integrated. We also calculated the standard deviation using a formula for propagating error in quotients (Lindberg, 2010). Using this method, we obtained a ratio of total over integrated of 0.8 +/− 0.5. Finally, we also combined every permutation of total over integrated measurements from our 7 replicates and found that the ratios ranged from 0.3 to 1.9 with a 96% confidence interval of 0.4 to 1.9. Thus, all three methods gave similar confidence intervals. We decided conservatively, that a ratio greater than 2.1 (p< 0.01, using the standard deviation from our most conservative estimate in Table 1) would be indicative of a true excess.
Table 1. Calculating the measurement error of total over integrated HIV DNA, to determine the ratio that indicates real excesses of unintegrated HIV DNA.
In this experiment, a master stock of our integration standard (IS) diluted in PBMC DNA was used to mimic a patient sample (1 copy of IS in 10,000 PBMC). DNA corresponding to 2×106 cells, 1 × 106 and 0.2 × 106 was assayed for total DNA (SST/cell) 7 times with 10 replicates per assay. DNA corresponding to 7.5×103 cells was assayed for integrated DNA 7 times with 42 replicates per assay. The assays were then paired in the order that they were performed. These pairings were then averaged and the standard deviation was calculated. Using these numbers, we calculated that a significant excess occurs when the ratio is greater than 2.0 (using 2.58 standard deviations, for a p=0.01). We also averaged the measurements for total HIV DNA and divided that number by the average number of proviruses and then calculated a standard deviation. Using this method, we calculated that a significant excess occurred when the ratio was greater than 2.1 (i.e. 2.58 standard deviations above the mean, p=0.01).
| Experiment number |
HIV copies per 10,000 cells (n=10) |
Provirus copies per 10,000 cells (n=42) |
Ratio SST/Provirus |
||
|---|---|---|---|---|---|
| 1 | 1.01 | 0.68 | 1.49 | ||
| 2 | 1.27 | 0.86 | 1.48 | ||
| 3 | 1.28 | 2.45 | 0.52 | ||
| 4 | 0.95 | 1.46 | 0.65 | ||
| 5 | 0.92 | 1.31 | 0.70 | ||
| 6 | 0.83 | 0.86 | 0.97 | ||
| 7 | 0.90 | 1.27 | 0.71 | ||
| Average | 1.02 | 1.27 | |||
| Average Ratioa | 0.81 | Average Ratiob | 0.93 | ||
| Standard Deviation | 0.5 | Standard Deviation | 0.40 | ||
| p=0.01 cutoff | 2.10 | p=0.01 cutoff | 1.96 | ||
Average of HIV copies per 10,000 cells for 7 experiments divided by average of provirus copies per 10,000 cells for 7 experiments
Average of individual ratios for each of the 7 experiments
We evaluated if the estimate of total HIV DNA correlates with the level of integrated DNA in PBMC from closely monitored individuals receiving suppressive HAART. The patients have been on HAART for at least a year on an efavirenz-based regimen along with 2–3 nucleoside analogs (Table 2). The patients sustained undetectable viral loads (<75 RNA copies/ml) for at least a year and the viral load was monitored every 3 months prior to PBMC sample collection and analysis. We then measured integration by Alu-PCR in samples from patients on HAART. We found a significant excess of total HIV DNA in 3 out of 7 patients (Figure 5), suggesting that the total HIV DNA levels do not always correlate with the level of HIV integration in patients on HAART. Our results also suggests that excesses of total over integrated DNA may occur commonly in treated patients which may have implications for understanding latency and reservoir maintenance in the presence of effective therapy.
Table2.
Patient data. Provides the range of CD4 counts during the year the patients viral load and CD4 counts were monitored every 3 months. The nadir CD4 for patients is prior to treatment and the drug regimen.
| Patient ID Number | CD4 Counta | Count Collectionb | Nadir CD4 c | Viral Loadd | Drug Regimene |
|---|---|---|---|---|---|
| 1 | 657 – 780 | 689 | 156 | <75 | EFV, ABC, ZDV, 3TC |
| 2 | 208 – 435 | 435 | 172 | <75 | EFV, 3TC, D4T |
| 3 | 386 – 651 | 611 | N/A | <75 | EFV, ABC, 3TC |
| 4 | 197 – 317 | 315 | 34 | <75 | EFV, ABC, ZDV, 3TC |
| 5 | 538 – 725 | 725 | 38 | <75 | EFV, D4T, DDI |
| 6 | 269 – 385 | 321 | N/A | <75 | EFV, ZDV, 3TC |
| 7 | 399 – 527 | 399 | N/A | <75 | EFV, ABC, 3TC, TDF |
CD4 cell count (cells/ml) was monitored every 3 months for the length of the study (12 months).
CD4 cell count (cells/ml) at the time of sample collection for PCR analysis (12 months).
Nadir count (cells/ml) detected at least 7 months prior to starting in the study.
Viral load (RNA copies/ml) was measured every 3 months for the length of the study. Viral load was never detected above the limit of detection (75 copies/ml).
Efavirenz (EFV), Abacavir (ABC), Zidovudine (ZDV), Lamivudine (3TC), Stavudine (d4T), Didanosine (DDI), Tenofovir (TDF).
Figure 5. An excess of total DNA exists in a subset of patients receiving HAART.
Total (SST) and integrated DNA were measured in PBMC samples from patients on HAART. The numbers above each black bar represents the ratio of total DNA over integrated DNA. The boxed numbers indicate measurements that show a true excess of unintegrated HIV DNA. These ratios are greater than 2.58 standard deviations above the integration standard ratio for a p > 0.01. We measured 15,000 genomes per well to measure integration (7,500 cell equivalents). We measured 0.2– 2 × 106 cell equivalents for total, depending on sample availability. Error bars represent the standard deviation of 3 measurements of total DNA and 42 measurements of integrated DNA.
Discussion
Many studies have measured integration in patient samples providing useful information about the pathogenesis of HIV (Chun et al., 1997; Chun et al., 1995a; Ibanez et al., 1999; Izopet et al., 2002; Ostrowski et al., 1999). We recently also described an assay that was suitable for clinical samples (Liszewski, Yu, and O'Doherty, 2009; Yu et al., 2008). In the present study, we set out to determine the accuracy and precision (error) of our integration assay and applied it to samples from HIV infected patients on HAART to determine how frequently an excess of unintegrated HIV DNA can be detected. While our prior work demonstrated that our integration assay can discriminate between integrated and unintegrated HIV DNA (Yu et al., 2008), here we demonstrate that our Alu-PCR integration measurement is accurate by comparing it to the the proportion of HIV DNA-positive clones after single cell cloning. This indicates that our Alu-PCR provides reliable measurements of integration from samples obtained from HIV-infected patients or from cells inoculated in vitro. Using this assay, we demonstrate that in a subset of patients on suppressive HAART there is an excess of unintegrated HIV DNA.
Many exciting approaches for stimulating out reservoirs are being investigated (Coiras et al., 2009; Colin and Van Lint, 2009; Dahl and Palmer, 2009). As eradicating HIV reservoirs becomes a goal and as treatments that target reservoirs become available, it will become even more important to accurately measure the reservoir. HIV integration levels may be a good surrogate marker for reservoirs and therefore measuring integration accurately could be used to monitor treatments that target reservoirs. As therapies are in development, ideally we would want sensitive assays able to detect small changes in reservoir size; an assay with a small error would be able to detect smaller changes. We found in some patients on HAART the level of total HIV approached the level of integrated HIV DNA consistent with previous work (Chomont et al., 2009; Koelsch et al., 2008). However, we also found patients where total was in excess of integrated. Given that excesses do occur in some patients, we suggest measuring total HIV DNA in patients receiving HAART is not always representative of viral reservoirs.
Measuring total HIV DNA in PBMCs from patients is one of the common methods for monitoring reservoir size. However, as shown here, there is variability in the level of total DNA relative to the level of integrated DNA. Another method employed for estimating the size of the reservoir is the infectious units per million-assay (IUPM) (Siliciano and Siliciano, 2005). This method involves serial dilution of stimulated PBMC from an infected individual followed by culturing the diluted cells in the presence of activated CD4+ T blasts. The IUPM assay, while powerful, is costly, laborious, with inherently wide errors, and thus, not ideal for monitoring the reservoir size in HIV infected individuals routinely. Therefore, we propose that measuring integration may be a more feasible method for routinely monitoring the effect of treatment on reservoir size. This method could also be employed for determining if novel drugs that target the reservoir directly, such as histone deacetylase inhibitors, can decrease the level of integration over time as an indirect measure of reservoir size (Archin et al., 2009; Lehrman et al., 2005). However, because the number of replication competent proviruses has been estimated to be far less than the quantity of integrated HIV DNA, it is possible that the reservoir size would not correlate with the number of integration events. Future work will be required to determine if measuring integration correlates with reservoir size as measured by the Infectious Unit Per Million (IUPM) assay. Nonetheless, we reason measuring total and integrated HIV DNA may still be useful since excesses of unintegrated HIV DNA may be physiologically relevant. It is currently unclear what an excess of unintegrated HIV DNA indicates or what processes lead to the excess. Because total DNA has a short half-life in vitro (Koelsch et al., 2008), this excess of total DNA over integrated DNA may represent new rounds of infection, as previously proposed (Koelsch et al., 2008). This would suggest that some low level of viral replication might be taking place in some patients despite suppressive therapy. This low level of ongoing viral replication could contribute to the replenishment of the latent reservoir in at least a subset of patients (Buzon et al.; Ramratnam et al., 2000; Ramratnam et al., 2004). Any level of viral replication would have to be quite low, because several studies indicate that drug resistance does not frequently develop on HAART (Hermankova et al., 2001; Persaud et al., 2004) and viral evolution cannot be detected in many patients on HAART (Kieffer et al., 2004; Nettles et al., 2004; Nottet et al., 2009; Shen and Siliciano, 2008; Tobin et al., 2005). Alternative explanations, such as undetected viral blips or undetected low level viremia and 2-LTR circles that have accumulated over time prior to the initiation of therapy, could also account for this excess of total HIV DNA. Some studies suggest these 2-LTR forms are more stable forms at least in vitro (Butler, Johnson, and Bushman, 2002; Pierson et al., 2002) and thus could potentially accumulate in vivo, though there is also evidence that these forms may be less stable in vivo than in vitro (Sharkey et al., 2005). More work would be required to determine the true nature of this excess of total HIV DNA, under what circumstance does it occur and how often does it occur. If this excess represents new rounds of replication, monitoring the level of both integrated and total HIV DNA using assays similar to the ones that we described could be useful as a cost effective, sensitive and reliable method for detecting ongoing replication in patients receiving HAART.
Materials and Methods
Cell line and virus
The CD4+ T lymphoblastoid cell line CEM-SS, a subclonal cell line derived from the CEM cell line (Foley et al., 1965; Nara and Fischinger, 1988), was maintained at 1−5×105 cells/ml. The culture medium used to maintain the cells was RPMI 1640 (Invitrogen) supplemented with 10% heat inactivated FBS (Invitrogen) and 100ug/ml of penicillin-streptomycin (Mediatech Inc.). The lentiviral vector VRX494, was pseudotyped with VSV-G and was donated by VIRxSYS (Humeau et al., 2004; Lu et al., 2004).
Cell inoculations
The cells were inoculated with VRX494 in 48-well flat-bottom plates at 5×105 cells/ml by spinoculation (1,200×g, 25°C, 2hr) (O'Doherty, Swiggard, and Malim, 2000). The viral inoculum was diluted to give a relative activity of 1.1, 0.39 or 0.22 proviruses/cell. After inoculation, the cells were washed twice to remove unbound virus and were resuspended in culture medium. The cells were then incubated at 37°C and aliquots of cells were collected at 0, 16, 18 and 24hr post-inoculation for measuring the level of integration. The integrase inhibitor raltegravir (obtained from AIDS Research and Reference Reagents Program, NIAID, catalog #11680 and donated by Merck & Co., Inc.) or L870,812 (donated by Merck & Co., Inc.) was added to the cells at 16hr post-inoculation to prevent any further integration events. Raltegravir added at 1µM (one cloning experiment) or L870,812 (one cloning experiment) added at 0.8µM inhibit integration at >90%. At 18hr post-inoculation, cells were collected for both measuring integration and for single-cell cloning.
Measuring total and integrated HIV DNA
Total HIV DNA represents the level of reverse transcripts that have completed the second strand transfer step (SST). Total HIV DNA was measured by kinetic PCR in 20 µl reaction volumes, following the conditions and protocol previously described (Swiggard et al., 2005). The following primers were used: HIV-U5 Forward primer 5' GCCTCAATAAAGCTTGCCTTGAGTG-3' and HIV-gag reverse primer 5' CAGCAAGCCAGATCCTGCG-3'. The kinetic PCR probe used is 5'FAM-CCAGAGTCACACAACAGACG-TAMRA 3'. HIV integration was measured following the conditions and protocol previously described (Agosto et al., 2009), but using different primers which are described in (O'Doherty et al., 2002). For the patient samples a slightly different protocol was used as described (Liszewski et al., 2009). Cell numbers were estimated by measuring the number of β-globin copies per DNA sample by kinetic PCR (O'Doherty, Swiggard, and Malim, 2000). In our previous study (Agosto et al., 2007), we reported that 25 µl at 2 µg/ml was equivalent to 15,000 genomes, which is equivalent to 7,500 diploid cells. In our prior paper, we occasionally substituted the word “cells” incorrectly when we intended the word “genomes” since there are two genomes per diploid cell. The kinetic PCR instrument used is the ABI 7500 Fast Real-Time PCR System.
Single-cell cloning
At 18hr post-inoculation, an aliquot of cells was diluted to 10 cells/ml or 3 cells/ml. 100µl of this diluted cell aliquot was plated in 96-well round bottom plates to obtain on average 1 or 0.3 cells per well. Colonies were allowed to grow for ~2 weeks in the presence of integrase inhibitor, reaching a density of ~104 – 105 cells/well. Cells from individual wells were collected and DNA was purified using the QIAamp DNA Micro Kit (Qiagen). The DNA samples were then assayed for HIV late reverse transcripts. Colonies were assayed for HIV late reverse transcripts until at least 5 colonies were positive. A signal was considered positive for HIV DNA if the level was above the detection limit of the total DNA assay (0.0001SST/cell), when DNA from 104 cells were assayed in duplicate.
Data analysis
Microsoft Excel was used for data analysis and the statistics software MyStat 12 was used to prepare the box plot shown in Figure 4A.
Supplementary Material
Acknowledgements
We gratefully acknowledge Daria Hazuda for her intellectual contributions and the CFAR at Penn for reagents and facilities. We thank Michele Di Mascio for helpful discussions on measurement of error in our system. We thank Robert Gross and Jennifer Chapman for providing patient samples. We thank Laurent Humeau and Nikolay Kirokhov for providing us with VRX494 pseudotyped with VSV-G. This work was supported by Merck, PKC Pharmaceuticals and NIH grants 7-K08-AI-073102-02, 5K02AI078766-03, R21 AI081215-01 A1, and T32AI07324.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agosto LM, Yu JJ, Dai J, Kaletsky R, Monie D, O'Doherty U. HIV-1 integrates into resting CD4+ T cells even at low inoculums as demonstrated with an improved assay for HIV-1 integration. Virology. 2007;368(1):60–72. doi: 10.1016/j.virol.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agosto LM, Yu JJ, Liszewski MK, Baytop C, Korokhov N, Humeau LM, O'Doherty U. The CXCR4-tropic human immunodeficiency virus envelope promotes more efficient gene delivery to resting CD4+ T cells than the vesicular stomatitis virus glycoprotein G envelope. J Virol. 2009;83(16):8153–8162. doi: 10.1128/JVI.00220-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari-Lari MA, Donehower LA, Gibbs RA. Analysis of human immunodeficiency virus type 1 integrase mutants. Virology. 1995;211(1):332–335. doi: 10.1006/viro.1995.1412. [DOI] [PubMed] [Google Scholar]
- Archin NM, Keedy KS, Espeseth A, Dang H, Hazuda DJ, Margolis DM. Expression of latent human immunodeficiency type 1 is induced by novel and selective histone deacetylase inhibitors. AIDS. 2009;23(14):1799–1806. doi: 10.1097/QAD.0b013e32832ec1dc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brussel A, Sonigo P. Analysis of early human immunodeficiency virus type 1 DNA synthesis by use of a new sensitive assay for quantifying integrated provirus. J Virol. 2003;77(18):10119–10124. doi: 10.1128/JVI.77.18.10119-10124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukrinsky MI, Stanwick TL, Dempsey MP, Stevenson M. Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection. Science. 1991;254:423–427. doi: 10.1126/science.1925601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler SL, Hansen MST, Bushman FD. A quantitative assay for HIV DNA integration in vivo. Nature Medicine. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- Butler SL, Johnson EP, Bushman FD. Human Immunodeficiency Virus cDNA Metabolism: Notable Stability of Two-Long Terminal Repeat Circles. Journal of Virology. 2002;76(8):3739–3747. doi: 10.1128/JVI.76.8.3739-3747.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzon MJ, Massanella M, Llibre JM, Esteve A, Dahl V, Puertas MC, Gatell JM, Domingo P, Paredes R, Sharkey M, Palmer S, Stevenson M, Clotet B, Blanco J, Martinez-Picado J. HIV-1 replication and immune dynamics are affected by raltegravir intensification of HAART-suppressed subjects. Nat Med. doi: 10.1038/nm.2111. [DOI] [PubMed] [Google Scholar]
- Cara A, Guarnaccia F, Reitz MS, Gallo RC, Lori F. Self-limiting, cell type-dependent replication of an integrase-defective human immunodeficiency virus type 1 in human primary macrophages but not T lymphocytes. Virology. 1995;208(1):242–248. doi: 10.1006/viro.1995.1148. [DOI] [PubMed] [Google Scholar]
- Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy JP, Haddad EK, Sekaly RP. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15(8):893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun T-W, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo Y-H, Brookmeyer R, Zeiger MA, Barditch-Crovo P, Siliciano RF. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- Chun T-W, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nature Medicine. 1995a;1:1284–1290. doi: 10.1038/nm1295-1284. [DOI] [PubMed] [Google Scholar]
- Chun TW, Engel D, Berrey MM, Shea T, Corey L, Fauci AS. Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proceedings of the National Academy of Sciences USA. 1998;95:8869–8873. doi: 10.1073/pnas.95.15.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med. 1995b;1(12):1284–1290. doi: 10.1038/nm1295-1284. [DOI] [PubMed] [Google Scholar]
- Coffin JM. In: Retroviruses. Coffin JM, Hughes SH, Varmus HE, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- Coiras M, Lopez-Huertas MR, Perez-Olmeda M, Alcami J. Understanding HIV-1 latency provides clues for the eradication of long-term reservoirs. Nat Rev Microbiol. 2009;7(11):798–812. doi: 10.1038/nrmicro2223. [DOI] [PubMed] [Google Scholar]
- Colin L, Van Lint C. Molecular control of HIV-1 postintegration latency: implications for the development of new therapeutic strategies. Retrovirology. 2009;6:111. doi: 10.1186/1742-4690-6-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl V, Palmer S. Establishment of drug-resistant HIV-1 in latent reservoirs. J Infect Dis. 2009;199(9):1258–1260. doi: 10.1086/597760. [DOI] [PubMed] [Google Scholar]
- Engelman A, Englund G, Orenstein JM, Martin MA, Craigie R. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. Journal of Virology. 1995;69:2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund G, Theodore TS, Freed EO, Engelman A, Martin MA. Integration is required for productive infection of monocyte-derived macrophages by Human Immunodeficiency Virus type 1. Journal of Virology. 1995;69:3216–3219. doi: 10.1128/jvi.69.5.3216-3219.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange SJ, Gallant J, Siliciano RF. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nature Medicine. 1999;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- Fischer M, Joos B, Niederost B, Kaiser P, Hafner R, von Wyl V, Ackermann M, Weber R, Gunthard HF. Biphasic decay kinetics suggest progressive slowing in turnover of latently HIV-1 infected cells during antiretroviral therapy. Retrovirology. 2008;5:107. doi: 10.1186/1742-4690-5-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley GE, Handler AH, Lynch PM, Wolman SR, Stolberg CS, Eagle H. Loss of neoplastic properties in vitro. II. Observations on KB sublines. Cancer Research. 1965;25:1254–1261. [PubMed] [Google Scholar]
- Groschel B, Bushman F. Cell cycle arrest in G2/M promotes early steps of infection by human immunodeficiency virus. J Virol. 2005;79(9):5695–5704. doi: 10.1128/JVI.79.9.5695-5704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Wind-Rotolo M, Yang HC, Siliciano JD, Siliciano RF. Experimental approaches to the study of HIV-1 latency. Nat Rev Microbiol. 2007;5(2):95–106. doi: 10.1038/nrmicro1580. [DOI] [PubMed] [Google Scholar]
- Hermankova M, Ray SC, Ruff C, Powell-Davis M, Ingersoll R, D'Aquila RT, Quinn TC, Siliciano JD, Siliciano RF, Persaud D. HIV-1 drug resistance profiles in children and adults with viral load of <50 copies/ml receiving combination therapy. JAMA. 2001;286(2):196–207. doi: 10.1001/jama.286.2.196. [DOI] [PubMed] [Google Scholar]
- Ho DD, Rota TR, Hirsch MS. Infection of monocyte/macrophages by human T lymphotropic virus type III. The Journal of Clinical Investigation. 1986;77:1712–1715. doi: 10.1172/JCI112491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humeau LM, Binder GK, Lu X, Slepushkin V, Merling R, Echeagaray P, Pereira M, Slepushkina T, Barnett S, Dropulic LK, Carroll R, Levine BL, June CH, Dropulic B. Efficient lentiviral vector-mediated control of HIV-1 replication in CD4 lymphocytes from diverse HIV+ infected patients grouped according to CD4 count and viral load. Mol Ther. 2004;9(6):902–913. doi: 10.1016/j.ymthe.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Ibanez A, Puig T, Elias J, Clotet B, Ruiz L, Martinez MA. Quantification of integrated and total HIV-1 DNA after long-term highly active antiretroviral therapy in HIV-1-infected patients. Aids. 1999;13(9):1045–1049. doi: 10.1097/00002030-199906180-00007. [DOI] [PubMed] [Google Scholar]
- Igarashi T, Brown CR, Endo Y, Buckler-White A, Plishka R, Bischofberger N, Hirsch V, Martin MA. Macrophage are the principal reservoir and sustain high virus loads in rhesus macaques after the depletion of CD4+ T cells by a highly pathogenic simian immunodeficiency virus/HIV type 1 chimera (SHIV): Implications for HIV-1 infections of humans. Proc Natl Acad Sci U S A. 2001;98(2):658–663. doi: 10.1073/pnas.021551798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izopet J, Cazabat M, Pasquier C, Sandres-Saune K, Bonnet E, Marchou B, Massip P, Puel J. Evolution of total and integrated HIV-1 DNA and change in DNA sequences in patients with sustained plasma virus suppression. Virology. 2002;302(2):393–404. doi: 10.1006/viro.2002.1621. [DOI] [PubMed] [Google Scholar]
- Jelinek WR, Schmid CW. Repetitive sequences in eukaryotic DNA and their expression. Annual Review of Biochemistry. 1982;51:813–844. doi: 10.1146/annurev.bi.51.070182.004121. [DOI] [PubMed] [Google Scholar]
- Kieffer TL, Finucane MM, Nettles RE, Quinn TC, Broman KW, Ray SC, Persaud D, Siliciano RF. Genotypic analysis of HIV-1 drug resistance at the limit of detection: virus production without evolution in treated adults with undetectable HIV loads. J Infect Dis. 2004;189(8):1452–1465. doi: 10.1086/382488. [DOI] [PubMed] [Google Scholar]
- Koelsch KK, Liu L, Haubrich R, May S, Havlir D, Gunthard HF, Ignacio CC, Campos-Soto P, Little SJ, Shafer R, Robbins GK, D'Aquila RT, Kawano Y, Young K, Dao P, Spina CA, Richman DD, Wong JK. Dynamics of total, linear nonintegrated, and integrated HIV-1 DNA in vivo and in vitro. J Infect Dis. 2008;197(3):411–419. doi: 10.1086/525283. [DOI] [PubMed] [Google Scholar]
- Lehrman G, Hogue IB, Palmer S, Jennings C, Spina CA, Wiegand A, Landay AL, Coombs RW, Richman DD, Mellors JW, Coffin JM, Bosch RJ, Margolis DM. Depletion of latent HIV-1 infection in vivo: a proof-of-concept study. Lancet. 2005;366(9485):549–555. doi: 10.1016/S0140-6736(05)67098-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg V. Manual on Uncertainties, Graphing, and the Vernier Caliper. 2010 http://www.rit.edu/cos/uphysics/uncertainties/Uncertaintiespart2.html.
- Liszewski MK, Graf EH, Mexas AM, Yu JJ, Migueles SA, O'Doherty U. Symposium on Antiviral Drug Resistance; Richmond, Virginia, USA. 2009. [Google Scholar]
- Liszewski MK, Yu JJ, O'Doherty U. Detecting HIV-1 integration by repetitive-sampling Alu-gag PCR. Methods. 2009;47(4):254–260. doi: 10.1016/j.ymeth.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Humeau L, Slepushkin V, Binder G, Yu Q, Slepushkina T, Chen Z, Merling R, Davis B, Chang YN, Dropulic B. Safe two-plasmid production for the first clinical lentivirus vector that achieves >99% transduction in primary cells using a one-step protocol. J Gene Med. 2004;6(9):963–973. doi: 10.1002/jgm.593. [DOI] [PubMed] [Google Scholar]
- Mighell AJ, Markham AF, Robinson PA. Alu sequences (Minireview) FEBS Letters. 1997;417:1–5. doi: 10.1016/s0014-5793(97)01259-3. [DOI] [PubMed] [Google Scholar]
- Nara PL, Fischinger PJ. Quantitative infectivity assay for HIV-1 and -2. Nature. 1988;332:469–470. doi: 10.1038/332469a0. [DOI] [PubMed] [Google Scholar]
- Nettles RE, Kieffer TL, Simmons RP, Cofrancesco J, Jr, Moore RD, Gallant JE, Persaud D, Siliciano RF. Genotypic resistance in HIV-1-infected patients with persistently detectable low-level viremia while receiving highly active antiretroviral therapy. Clin Infect Dis. 2004;39(7):1030–1037. doi: 10.1086/423388. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Cross GD, Callaway CS, McDougal JS. In vitro infection of human monocytes with human T lymphotropic virus type III/lymphadenopathy-associated virus (HTLV-III/LAV) J Immunol. 1986;137(1):323–329. [PubMed] [Google Scholar]
- Nottet HS, van Dijk SJ, Fanoy EB, Goedegebuure IW, de Jong D, Vrisekoop N, van Baarle D, Boltz V, Palmer S, Borleffs JC, Boucher CA. HIV-1 can persist in aged memory CD4+ T lymphocytes with minimal signs of evolution after 8.3 years of effective highly active antiretroviral therapy. J Acquir Immune Defic Syndr. 2009;50(4):345–353. doi: 10.1097/QAI.0b013e318197eb04. [DOI] [PubMed] [Google Scholar]
- O'Doherty U, Swiggard WJ, Jeyakumar D, McGain D, Malim MH. A sensitive, quantitative, assay for Human Immunodeficiency Virus type 1 integration. J Virol. 2002;76(21):10942–10950. doi: 10.1128/JVI.76.21.10942-10950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Doherty U, Swiggard WJ, Malim MH. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J Virol. 2000;74(21):10074–10080. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski MA, Chun T-W, Justement SJ, Motola I, Spinelli MA, Adelsberger J, Ehler LA, Mizell SB, Hallahan CW, Fauci AS. Both memory and CD45RA(+)/CD62L(+) naive CD4(+) T cells are infected in human immunodeficiency type 1-infected individuals. Journal of Virology. 1999;73:6430–6435. doi: 10.1128/jvi.73.8.6430-6435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persaud D, Siberry GK, Ahonkhai A, Kajdas J, Monie D, Hutton N, Watson DC, Quinn TC, Ray SC, Siliciano RF. Continued production of drug-sensitive human immunodeficiency virus type 1 in children on combination antiretroviral therapy who have undetectable viral loads. J Virol. 2004;78(2):968–979. doi: 10.1128/JVI.78.2.968-979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson TC, Kieffer TL, Ruff CT, Buck C, Gange SJ, Siliciano RF. Intrinsic stability of episomal circles formed during Human Immunodeficiency Virus Type-1 replication. Journal of Virology. 2002;76(8):4138–4144. doi: 10.1128/JVI.76.8.4138-4144.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramratnam B, Mittler JE, Zhang L, Boden D, Hurley A, Fang F, Macken CA, Perelson AS, Markowitz M, Ho DD. The decay of the latent reservoir of replication-competent HIV-1 is inversely correlated with the extent of residual viral replication during prolonged anti-retroviral therapy. Nature Medicine. 2000;6:82–85. doi: 10.1038/71577. [DOI] [PubMed] [Google Scholar]
- Ramratnam B, Ribeiro R, He T, Chung C, Simon V, Vanderhoeven J, Hurley A, Zhang L, Perelson AS, Ho DD, Markowitz M. Intensification of antiretroviral therapy accelerates the decay of the HIV-1 latent reservoir and decreases, but does not eliminate, ongoing virus replication. J Acquir Immune Defic Syndr. 2004;35(1):33–37. doi: 10.1097/00126334-200401010-00004. [DOI] [PubMed] [Google Scholar]
- Sakai H, Kawamura M, Sakuragi J, Sakuragi S, Shibata R, Ishimoto A, Ono N, Ueda S, Adachi A. Integration is essential for efficient gene expression of human immunodeficiency virus type 1. Journal of Virology. 1993;67(3):1169–1174. doi: 10.1128/jvi.67.3.1169-1174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey M, Triques K, Kuritzkes DR, Stevenson M. In vivo evidence for instability of episomal human immunodeficiency virus type 1 cDNA. J Virol. 2005;79(8):5203–5210. doi: 10.1128/JVI.79.8.5203-5210.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Siliciano RF. Viral reservoirs, residual viremia, and the potential of highly active antiretroviral therapy to eradicate HIV infection. J Allergy Clin Immunol. 2008;122(1):22–28. doi: 10.1016/j.jaci.2008.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano JD, Siliciano RF. Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol Biol. 2005;304:3–15. doi: 10.1385/1-59259-907-9:003. [DOI] [PubMed] [Google Scholar]
- Stevenson M. HIV-1 pathogenesis. Nat Med. 2003;9(7):853–860. doi: 10.1038/nm0703-853. [DOI] [PubMed] [Google Scholar]
- Stevenson M, Haggerty S, Lamonica CA, Meier CM, Welch SK, Wasiak AJ. Integration is not necessary for expression of human immunodeficiency virus type 1 protein products. Journal of Virology. 1990a;64(5):2421–2425. doi: 10.1128/jvi.64.5.2421-2425.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson M, Stanwick TL, Dempsey MP, Lamonica CA. HIV-1 replication is controlled at the level of T cell activation and proviral integration. European Molecular Biology Organization Journal. 1990b;9:1551–1560. doi: 10.1002/j.1460-2075.1990.tb08274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiggard WJ, Baytop C, Yu JJ, Dai J, Li C, Schretzenmair R, Theodosopoulos T, O'Doherty U. Human immunodeficiency virus type 1 can establish latent infection in resting CD4+ T cells in the absence of activating stimuli. J Virol. 2005;79(22):14179–14188. doi: 10.1128/JVI.79.22.14179-14188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swingler S, Mann AM, Zhou J, Swingler C, Stevenson M. Apoptotic killing of HIV-1-infected macrophages is subverted by the viral envelope glycoprotein. PLoS Pathog. 2007;3(9):1281–1290. doi: 10.1371/journal.ppat.0030134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin NH, Learn GH, Holte SE, Wang Y, Melvin AJ, McKernan JL, Pawluk DM, Mohan KM, Lewis PF, Mullins JI, Frenkel LM. Evidence that low-level viremias during effective highly active antiretroviral therapy result from two processes: expression of archival virus and replication of virus. J Virol. 2005;79(15):9625–9634. doi: 10.1128/JVI.79.15.9625-9634.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiskerchen M, Meusing MA. Identification and characterization of a temperature-sensitive mutant of human immunodeficiency virus type 1 by alanine scanning mutagenesis of the integrase gene. Journal of Virology. 1995;69:597–601. doi: 10.1128/jvi.69.1.597-601.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JJ, Wu TL, Liszewski MK, Dai J, Swiggard WJ, Baytop C, Frank I, Levine BL, Yang W, Theodosopoulos T, O'Doherty U. A more precise HIV integration assay designed to detect small differences finds lower levels of integrated DNA in HAART treated patients. Virology. 2008;379(1):78–86. doi: 10.1016/j.virol.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.