Abstract
Methylglyoxal (MG) and related alpha-oxoaldehydes react with proteins, lipids and DNA to give rise to covalent adducts known as advanced glycation end-products (AGEs). Elevated levels of AGEs have been implicated in the pathological complications of diabetes, uremia, Alzheimers disease, and possibly cancer. There is therefore widespread interest in developing sensitive methods for the in vivo measurement of AGEs as prognostic biomarkers and for treatment monitoring. The two diastereomeric MG-DNA adducts of N2-(1-carboxyethyl)-2'-deoxyguanosine (CEdG) are the primary glycation products formed in DNA; however, accurate assessment of their distribution in vivo has not been possible since there is no readily available quantitative method for CEdG determination in biological samples. In order to address these issues we have developed a sensitive and quantitative LC-ESI-MS/MS assay using the stable isotope dilution method with an 15N5-CEdG standard. Methods for CEdG determination in urine or tissue extracted DNA are described. Changes in urinary CEdG in diabetic rats in response to oral administration of the AGE inhibitor LR-90 are used to demonstrate the potential utility of the method for treatment monitoring. Both stereoisomeric CEdG adducts were detected in a human breast tumor and normal adjacent tissue at levels of 3–12 adducts/107 dG, suggesting that this lesion may be widely distributed in vivo. Strategies for dealing with artifactual adduct formation due to oxoaldehyde generation during DNA isolation and enzymatic workup procedures are described.
Introduction
The highly reactive electrophile methylglyoxal (MG)1 is a major environmental breakdown product of carbohydrates (1). It is also generated biochemically during glycolysis via elimination of phosphate from the common enediol intermediate resulting from deprotonation of dihydroxyacetone phosphate and glyceraldehyde 3-phosphate (2). Additional endogenous sources include the catabolism of threonine and the P450 mediated oxidation of ketone bodies (3). It is also formed during the oxidative breakdown of DNA and RNA under acidic conditions (4). Methylglyoxal is present at micromolar levels in many foods and most living organisms, and is a probable mutagen in vivo (5). It reacts readily with nucleophilic moieties on proteins, lipids and DNA to produce covalent adducts known as advanced glycation end-products (AGEs) (6, 7). Protein AGEs have been well characterized, and these highly modified proteins have been proposed to play a role in the various pathologies associated with diabetes, cancer, aging, and Alzheimers disease (7–9). The first clear correlation between abnormal levels of a protein-AGE and a human disease (diabetes) was described nearly 40 years ago for the hemoglobin HbA1c adduct (10). Since then, hemoglobin HbA1c has become a commonly used biomarker for the diagnosis and treatment monitoring of diabetes (11–13). Accordingly, there is continued interest in the development of novel, more sensitive assays for the quantitative measurement of biomolecule-derived AGEs to complement and extend the clinical biomarker repertoire, as well as to assist in elucidating their role in pathology.
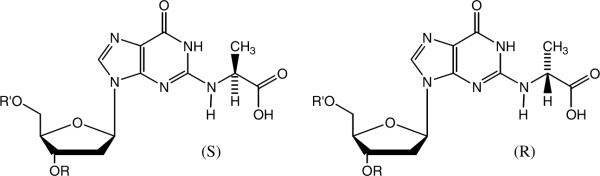
Approximately a dozen protein-AGEs have been characterized and LC-MS/MS methods have been described for their quantitative measurement. Choosing an appropriate protein-AGE biomarker for evaluating the glycation status of a particular target tissue or organ is complicated by unequal protein-AGE distributions across different tissues, varying adduct stabilities, and the limited availability of stable isotope standards for quantification (7, 14). Glycation adducts of DNA may have potential as biomarkers since all nucleated cells contain the same DNA content and should reflect the relative level of MG in the target tissue. Reaction of double stranded DNA with MG or glucose in vitro produces primarily N2-carboxyethyl-2'-deoxyguanosine (CEdG) as a diastereomeric mixture (Figure 1), suggesting that it is the likely major adduct formed in vivo (15, 16). This implies that CEdG might be a useful biomarker for monitoring oxoaldehyde-induced stress in response to enhanced glycolytic flux or environmental exposure to MG.
Figure 1.
The two CEdG diastereomers formed from reaction of MG with dG.
However, correlative studies of CEdG have been hindered by a lack of quantitative analytical methods. We have developed a sensitive LC-ESI-MS/MS method for the measurement of CEdG in urine or double-stranded DNA. Quantification is achieved by the stable isotope dilution method using synthetic 15N5-CEdG as an internal standard. We have measured urinary CEdG in normal and streptozoticin-induced diabetic rats, and have shown that adduct levels are significantly increased following the onset of hyperglycemia. LC-ESI-MS/MS was used to demonstrate a dose-dependent reduction in CEdG in response to administration of LR-90, an inhibitor of AGE formation. Measurement of CEdG from hydrolyzed and dephosphorylated double-stranded DNA was complicated by the fact that MG was present during the enzymatic workup. This was found to react with DNA during sample workup leading to artifactual overestimation of CEdG levels. In order to circumvent this problem, adventitious MG was sequestered by the addition of carbonyl scavengers such as aminoguanidine (AG) and D-penicillamine (D-P) prior to workup, resulting in stable and reproducible determinations.
Experimental Procedures
Materials and instrumentation
15N5-2'-deoxyguanosine was purchased from Silantes (Munich, Germany, lot # dG-N-0507-1/2); DL-glyceraldehyde (95%) and calf thymus DNA were from Sigma (St. Louis, MO), and ammonium acetate (1M, pH 7 solution) from Fluka (Buchs, Switzerland). Phosphate salts were A.C.S. reagent grade and purchased from J.T. Baker (Phillipsburg, NJ). HPLC grade CH3CN was purchased from Fisher Scientific (Fair Lawn, NJ). All water was purified to a resistivity of 18.2 MΩ using a Nanopure Diamond system by Barnstead International (Dubuque, IA). Solid phase extractions were performed using 1 ml strata-X-C cation mixed mode cartridges (Phenomenex, Torrance CA). Nuclease P1 was purchased from US Biologicals (Swampscott, MA). Phosphodiesterase II from bovine spleen and alkaline phosphatase from bovine intestinal mucosa was purchased from Sigma-Aldrich. HPLC separations were performed using a Hewlett-Packard Series 1100 Liquid Chromatography system equipped with a diode-array detector. Ultraviolet spectra were collected on an Ultrospec 3000 pro (Amersham Biosciences, Piscataway, NJ). Mass analysis of synthetic 15N5-CEdG was performed using a Thermo Finnigan LTQ-FT linear ion trap mass spectrometer (San Jose, CA) in the Mass Spectrometry-Proteomics Core Facility of the City of Hope.
LC-MS/MS analyses of CEdG in biological samples were carried out using a Micromass Quattro Ultima Triple Quadrupole Mass Spectrometer (Beverly, MA) interfaced to an Agilent 1100 Capillary HPLC system (Palo Alto, CA) equipped with a Synergi C18 analytical column (4μ, 150 × 2.0 mm; Phenomenex, Torrance, CA). 1H NMR spectra were recorded at 400 MHz on a VNMRS spectrometer (Varian, Inc., Palo Alto, CA) in the Synthesis and Biopolymer Core Facility of the City of Hope. 1D and 2D NMR data was processed using the Spinworks shareware program (version 2.5.5), copyright 1999–2006 by Kirk Marat and availible from the University of Manitoba website at http://www.umanitoba.ca/chemistry/nmr/spinworks/.
Synthesis and characterization of 15N5-CEdG
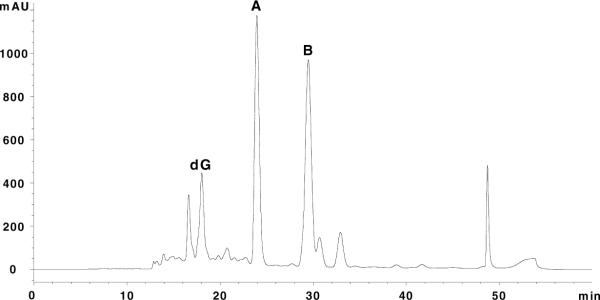
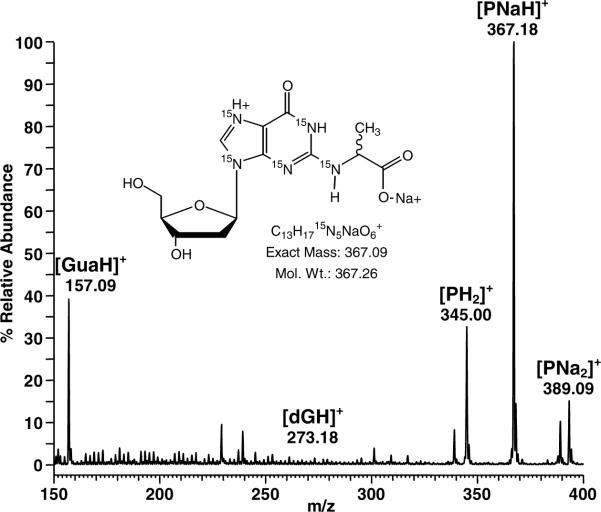
DL-Glyceraldehyde was used to generate MG in situ via guanine catalyzed dehydration (17). DL-Glyceraldehyde (9.5 mg) was added to 10 mg of 15N5-labeled dG, 12.3 mg potassium dihydrogen phosphate, and 24.0 mg disodium hydrogen phosphate in 87.7 μL H2O. The heterogeneous reaction mixture was vortexed and placed in a heat block at 40°C. Reactions were worked up following complete dissolution of solids (~14–17 days) yielding a yellow-red viscous solution. Products were purified by HPLC in 10–15 μL aliquots on a 10 × 50mm Waters XTerra MS C18 2.5μ column using a (Et)3NH4OAc (50 mM, pH 7)/CH3CN gradient. The CH3CN concentration was raised from 0 to 4.0% in the first 5 minutes, from 4.0 to 6.5% over 30 minutes; held at 6.5% for 5 minutes, then raised to 90% to wash residual material off the column. Diastereomers CEdG-A and B eluted at 24 and 29 minutes, respectively (Figure 2). Fractions were lyophilized to dryness prior to resuspension in 18.2 MΩ H2O. Concentrations of stock solutions were calculated by UV using a molar extinction coefficient of 12,300 @ 255 nm. Full scan UV spectra are provided in the Supporting Information. Mass analyses of 15N5-CEdG diastereomers were conducted using a Thermo-Finnigan LTQ FT ion trap mass spectrometer in the positive ion mode. A full scan MS for CEdG-A is shown in Figure 3. The most intense ion was observed for the sodiated peak, C13H17 15N5NaO6+: m/z 367.18 (obs), m/z 367.09 (calc). 1H NMR assignments for CEdG-A: 1H NMR (400 MHz, d6-DMSO, 18°C) δ 10.60 (s, 1H, N1-H), δ 7.93 (s, 1H, C8-H), δ 6.76 (d, 1H, C2-NH), δ 6.12 (dd, 1H, C1'-H), δ 5.30 (d, 1H, C3'-OH), δ 4.89 (vbr, 1H, C5'-OH), δ 4.36 (m, 1H, C2-NH-CH), δ 4.32 (m, 1H, C4'-H), δ 3.81 (m, 1H, C3'-H), δ 3.50 (ddd, 2H, C5'-H2), δ 2.64 (ddd, 1H, C2'-H), δ 2.18 (ddd, 1H, C2'-H), δ 1.39 (d, 3H, C2-NH-CH-CH3). 13C NMR assignments for CEdG-A: (100.5 MHz, d6-DMSO, 18°C) δ 174.1(C2-NH-CH-COOH), δ 156.3 (C6), δ 151.5 (C2), δ 149.9 (C4), δ 136.1 (C8), δ 117.1 (C5), δ 87.6 (C3'), δ 82.9 (C1'), δ 70.8 (C4'), δ 61.7 (C5'), δ 49.0 (C2-NH-CH), Δ ~39.5 (C2'), δ 17.7 (C2-NH-CH-CH3). 1H1H and 13C NMR assignments for CEdG-B are nearly identical to the A isomer and are provided in the Supporting Information.
Figure 2.
A representative HPLC chromatogram of the reaction of 15N5-dG with dl-glyceraldehyde. Peaks A and B correspond to the two diastereomers of 15N5-CEdG.
Figure 3.
Full scan positive ion ESI-MS spectrum for 15N5-CEdG diastereomer peak A.
Stability studies of CEdG in acidic solution
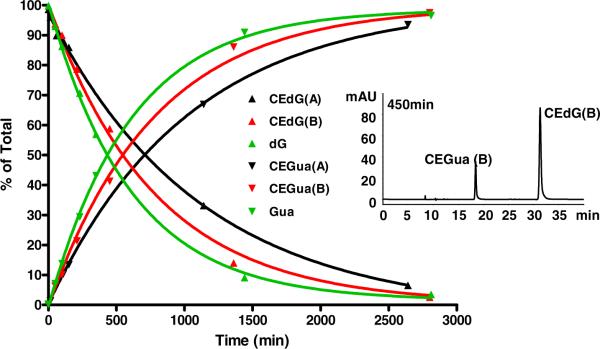
A 1.25 mM solution of CEdG-A, B or dG in 100 μL of 1M AcOH (pH 2.4) was stirred at 37°C. 10 μl aliquots were removed periodically and added to 40 μL of 2M TEAA (pH 7.0). HPLC product analyses were performed using an Alltech HS HyperPrep 100 BDS C18 8μ column. A gradient of 0 to 4% CH3CN over 5 min was followed by 6.5% CH3CN over 30 min. TEAA (pH 7) was kept constant at 50 mM. The ratio of free base (CEG or G) to intact nucleoside (CEdG or dG) was calculated by integration of the corresponding HPLC peaks (see inset in Figure 4). The CEG free base was identified as Peak A by ESI-MS in the negative ion mode. C8H8O3N5, observed : m/z 222.064; calculated : m/z 222.063.
Figure 4.
Time course product profiles of the reaction of dG and the A and B stereoisomers of CEdG with 1 M AcOH at 37°C. The inset shows the HPLC chromatogram of the reaction of CEdG-B at 450 min.
Animal Studies
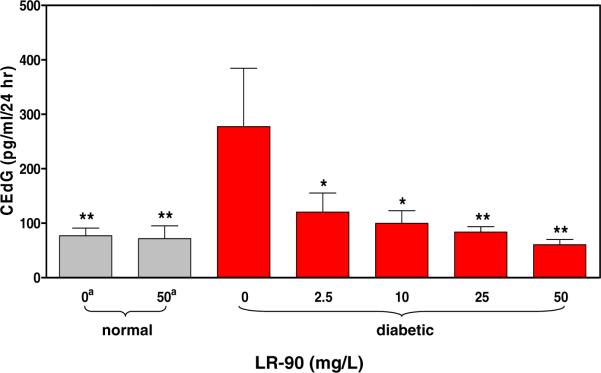
All animal studies were carried out in compliance with the policies outlined in NIH Publication No. 85-23 “Guide for the Care and Use of Laboratory Animals”. Male Sprague-Dawley rats were rendered diabetic by injection of streptozoticin and maintained as previously described (18). The AGE inhibitor LR-90 was administered ad libitum at concentrations ranging from 2.5–50 mg/L. Rats were housed in metabolic cages and urine was collected over a 24 h period with several drops of toluene to inhibit microbial growth. Urine samples were stored at −80°C prior to LC-MS/MS analysis for CEdG or 8-oxo-dG. The data in Figure 5 represent 3 replicates from n different animals: non-diabetic controls, n=6; non-diabetic treated with 50 mg/L LR-90, n=5; diabetic control, n=3. For diabetic rats treated with varying doses of LR-90: 2.5 mg/L, n=4; 10 mg/L, n=5; 25mg/L, n=6; 50 mg/L, n=8.
Figure 5.
Quantitation of CEdG in normal (grey) and diabetic (red) Sprague-Dawley rats. aOrdinate values represent ad libitum concentrations of the AGE inhibitor drug LR-90 (mg/L). *P <0.05 and **P <0.01 vs untreated diabetic animals (Bonferonni's test).
Urine sample preparation
CEdG was concentrated from urine by solid phase extraction. A 1 ml strata-X-C cartridge was pre-conditioned by the sequential addition of 1 ml MeOH/CH3CN (1:1) followed by 2 × 1 ml 2% H3PO4. Then 15 N5-CEdG was added as an internal standard (final concentration 5 μg/ml), the sample was acidified with 10 μl of 86% H3PO4, and finally 0.4 mL of urine was introduced via suction filtration. The cartridge was then washed with sequential additions of 1 ml 0.1% H3PO4 and 1 ml MeOH and then dried under vacuum for 1 minute. Finally, CEdG and 15N5-CEdG containing fractions were eluted from the cartridge with 1 mL 3% NH4OH in MeOH:CH3CN (2:8 v/v). The eluent was evaporated to dryness in a centrifugal concentrator and reconstituted with 200 μl 0.1% formic acid prior to LC-MS/MS injection.
Preparation of mononucleosides from DNA
Calf thymus or tissue-extracted DNA (100 μg) was dissolved in 80 μL of autoclaved 18.2 MΩ H2O containing 20 μL of sodium acetate (100 mM, pH 5.5), 20 μL of 1× TBE, 1.5 μL of 50 mM ZnCl2, and 2.37 μL of a 100 mM AG or D-P stock solution. DNA was denatured at 95°C for 5 min on a PCR heating block and then brought to 4°C for 5 min. After equilibration to 45°C, 1.5 μL of 10 U/μL nuclease P1 was added. Alkaline phosphatase (4 μL of 8 U/μL), 1 U of bovine phosphodiesterase, and 14μL of 100 mM CaCl2 were added after 1 h, and the hydrolysis/dephosphorylation was continued for another 7 hrs. DNA concentrations were determined by UV spectroscopy (1 OD260=50 μg/ml) and samples were stored at −80°C prior to MS analyses. A 5 μL aliquot of digest was diluted to 200 μL and used for quantitation of 2-deoxyguanosine by HPLC integration using a Beckman C-18 reverse phase (25 cm × 4.6 mm) column (Fullerton, CA). Separation was achieved isocratically using a mobile phase of 6% MeOH, 0.1% acetic acid in water.
DNA isolation from human tissue
Breast tumor and adjacent normal tissue was obtained from the frozen tumor bank of the City of Hope Pathology Core. A pea-sized section (~100 mg) of tissue was minced and suspended in 1.2 mL of digestion buffer (100 mM NaCl, 10 mM Tris HCl, pH 8, 25 mM EDTA, pH 8, 0.5% SDS, 0.2 mg/mL proteinase K, 10 mM D-penicillamine) and incubated at 50°C in a water bath for 12–18 h. DNA was then extracted using an equivalent volume of phenol/chloroform/isoamyl alcohol (25:24:1). The aqueous fraction was separated and 0.5 volumes of ammonium acetate and 2 volumes of 100% ethanol were added. The DNA was spooled, washed twice with 70% ethanol, pelleted, and resuspended in autoclaved 18.2 MΩ water. The enzymatic hydrolysis was carried out as described above.
LC-ESI-MS/MS
Quantification of CEdG was performed using a validated LCMS/MS method. Measurement of 8-oxo-dG was performed as previously described (19). CEdG and 15N5-CEdG (internal standard) were synthesized and purified as described above. Measurements were performed using an Agilent 1100 Capillary LC system (Agilent Technologies, Palo Alto, CA) in line with a Micromass Quattro Ultima Triple Quadrupole Mass Spectrometer (Micromass, Beverly, MA) operating in positiveion mode. The detector settings were as follows: capillary voltage, 3.5 kV; cone voltage, 18 V; collision cell voltage, 11 V; source temperature, 125°C; desolvation temperature, 350°C; cone gas flow, 150 liter/h; and desolvation gas flow, 620 liter/h. The mass transitions monitored for CEdG and 15N5-CEdG were 340.3→224.3 and 345.4→229.4 respectively (MS2 spectra for m/z 340 and 345 are provided in the Supporting Information). HPLC was accomplished using isocratic conditions with a mobile phase of 15% aqueous MeOH with 0.1% formic acid on a Prodigy ODS C-18 (25 cm × 2.0 mm × 5 micron) column (Phenomenex, Torrance, CA). The flow rate was 0.2 ml/min with a total run time of 30 min. The retention times for CEdG diastereomers A and B using these conditions were 9.3 and 16 min, respectively. The lower limit of quantitation for CEdG, defined as a peak height of ≥ 5× baseline noise, was 0.1 ng/ml in the starting solution or 0.2 pg on column. For urine analyses and calf thymus DNA digests, calibration curves were constructed using 0.75, 1.5, 3, 6, 12, 24, and 48 ng/mL of synthetic CEdG in urine or in blank nucleoside digestion buffer. For human breast tissues, CEdG concentrations used for calibration were 0.19, 0.38, 0.75, 1.5, 3, and 6 ng/mL. Linearity of the calibration curves were demonstrated with R-squared values of ≥ 0.996. Inter- and intra-day accuracy of the assay across the range of the standard curve was established to be 96% and 94% of target concentrations, respectively. The assay was also determined to be unbiased with both inter- and intra-day precision within ± 6%. Quantification of 2′-deoxyguanosine (dG) was performed by HPLC integration of DNA digests as described above and final values were expressed as CEdG/107dG.
Urine extracts or mononucleoside digests were spiked with 20 μL of 100 ng/mL 15N5-labeled CEdG and 10 μL of 86% phosphoric acid. Samples were then loaded onto strata-X-C cation mixed mode columns that had been pre-conditioned with MeOH/CH3CN (1:4) followed by 2% phosphoric acid. After sample loading, columns were washed with 0.1% phosphoric acid, followed by MeOH. Nucleosides were eluted with 3% ammonium hydroxide in MeOH/CH3CN (1:4) and evaporated to dryness in a centrifugal concentrator. Samples were reconstituted with 100 μL of 0.1% formic acid and analyzed directly by LC-MS/MS. Recovery of CEdG diastereomers and 15N5-CEdG from urine and mononucleoside digests was determined to be 85 +/- 0.9%.
Results
Synthesis and characterization of CEdG isotopomers
Isotopomers of CEdG were prepared by a modification of the method of Ochs and Severin (17). Reaction of 15N5-dG with (dl)-glyceraldehyde in phosphate buffer afforded the desired products as a ~1:1 mixture of diastereomers in ~ 60% yield. Unenriched CEdG diasteromers were prepared in an analogous manner. It has been proposed that the N2 amino group of dG catalyzes the dehydration of glyceraldehyde to yield the hemiacetal of MG in situ, which then reacts to provide CEdG either directly by condensation at N2 or alternatively via the rearrangement of an intermediate N1, N2 cyclic diol (15, 17). The two diastereomers of CEdG were readily resolved by HPLC and eluted at 24 and 29 minutes (Figure 2) on a C18 reverse phase column. In spite of significant differences in chromatographic retention times, both the proton and carbon NMR spectra for CEdG-A and B were essentially superimposable, with the chemical shift differential on the order of < 0.1 ppm for proton and < 1.0 ppm for carbon (see Supporting Information).
Mass analyses of the CEdG isotopomers were performed using a Thermo Finnigan LTQ ion trap mass spectrometer in the positive ion mode. The most intense signal in the parent ion spectrum of the isotopically enriched standard corresponded to the sodium salt of 15N5-CEdG at m/z 367 [PNaH]+ (Figure 3). The disodium salt [PNa2]+ and the dihydro adduct [PH2]+ were also observed at m/z 389 and 345, respectively. Collision induced dissociation of the m/z 367 parent ion gave rise primarily to the sodiated base ion [BNaH]+ at m/z 251 (see Supporting Information). The observed isotopic distribution for C13H1715N5NaO6 was found to be in good agreement with the calculated values (Supporting Information).
Stability of CEdG to acid-catalyzed depurination and sidechain isomerization
The chemical stability of CEdG was examined as an important criterion for evaluating its suitability as a quantitative biomarker. Purified stereoisomers of synthetic CEdG were subjected to acidic conditions (1 M AcOH at 37°C) and the extent of released free base and diastereomer interconversion was monitored by HPLC as a function of time. Analogous experiments were performed for dG and the results are presented in Figure 4. The approximate half-lives for depurination were 750 and 500 min for the A and B isomers respectively, whereas dG was observed to be less stable, with a half-life of 440 min under these conditions. No racemization of the sidechain stereocenter was detected during acidic hydrolysis, i.e., no interconversion of CEdG isomers A and B was observed.
Urinary CEdG measurement in diabetic rats
A diabetic animal model was used to examine the relationship between glycemic status and CEdG levels. Rats rendered diabetic by streptozoticin (STZ) treatment have been previously shown to possess elevated MG relative to normal controls and thus appeared likely to exhibit an increased burden of CEdG adducts (20). The effect of a novel AGE inhibitor, LR-90, was also examined (21, 22). The results of these experiments are shown in Figure 5. Analyses of urine from non-diabetic control animals collected over a 24 hr period revealed mean CEdG levels of 77 pg/ml (Figure 5). The induction of diabetes increased the level of excreted CEdG by ~ 4 fold. Administration of LR-90 to diabetic rats ad libitum at a dose corresponding to 2.5 mg/L resulted in a 2.3 fold decrease in CEdG titer. Increasing concentrations of LR-90 led to a dose dependent reduction in CEdG, and at 25 mg/L the adduct level in urine was comparable to that of non-diabetic animals. In contrast, administration of LR-90 at doses up to 50 mg/L in normal controls had no significant effect on CEdG levels. We also measured 8-oxo-dG as an indicator of oxidative stress in normal and diabetic rats; however, excreted 8-oxo-dG in diabetic animals was not statistically different (P>0.05) from controls (data not shown).
CEdG in calf thymus DNA
Commercial grade calf thymus DNA was used as a model substrate for developing a protocol for CEdG quantitation in double-stranded DNA. DNA was hydrolyzed and dephosphorylated by sequential addition of nuclease P1, alkaline phosphatase and phosphodiesterase; then mononucleosides were concentrated by solid phase extraction prior to LC-MS/MS analyses. The results of these experiments are shown in Figure 6. Initial determinations yielded values of CEdG in the range of 60–66 CEdG/106 dG. These surprisingly high levels suggested that some CEdG may have been formed artifactually during the hydrolysis and dephosphorylation. We hypothesized that additional CEdG may have been formed due to the release of MG from the protein reagents used in the workup during prolonged incubation. Proteins can bind MG reversibly, and it has been suggested that up to 90% of cellular MG may be sequestered in this manner (23). In order to prevent additional reactions of adventitiously generated MG with DNA, carbonyl scavenging agents AG or D-P were added prior to DNA digestion and dephosphorylation (24). These reagents sequester MG and other alpha-oxoaldehydes by forming stable cyclic aminotriazine and thiazolidine derivatives respectively (25). Concentrations of AG from 0.5 to 50 mM were added to prior to workup, and CEdG levels were measured in order to determine the optimal concentration required to achieve stable, reproducible levels (data not shown). The addition of 10 mM AG prior to sample processing resulted in a modest but significant drop in adduct levels (45–50 CEdG/106 guanines) in calf thymus DNA, suggesting that ~ 15 CEdG/106 guanines were formed as a direct result of the hydrolysis and dephosphorylation protocol.
Figure 6.
LC-ESI-MS/MS measurements of CEdG diastereomers in calf thymus DNA subjected to various workup procedures. Hydrolyzed samples correspond to DNA treated with nuclease P1/alkaline phosphatase/phosphodiesterase as described in Materials and Methods. Calf thymus DNA samples were also reacted with proteinase K (Extracted) prior to hydrolysis. Levels of CEdG were measured in the presence or absence of carbonyl scavenger AG.
Since the extraction of DNA from biological samples requires extended reaction with proteinase K (up to 24 h), we investigated whether this treatment could also contribute to artifactual CEdG formation. Accordingly, calf thymus DNA was subjected to mock proteolysis using the procedure described in Materials and Methods prior to hydrolysis and workup in the absence of carbonyl scavenger. The data in Figure 6 revealed an increase in adduct levels significantly higher than those observed following hydrolysis alone, with values ranging from 80–100 CEdG/106 guanines. The addition of 10 mM AG in two aliquots prior to the mock lysis treatment and hydrolysis/dephosphorylation steps resulted in a drop in measured CEdG levels comparable to that observed previously for calf thymus DNA subjected only to the hydrolysis/dephosphorylation in the presence of AG. No apparent stereoisomer bias was detected in any of these samples, i. e., the ratio of CEdGA: CEdGB was not significantly different from 1:1.
Quantitation of CEdG in a human breast tumor and adjacent normal tissue
Many cancer cells in the hyopoxic tumor microenvironment primarily utilize glycolysis to meet their energetic demands. This glycolytic phenotype (Warburg effect) is characterized by constitutive cell surface expression of glucose transporter proteins such as GLUT-1, and forms the basis for the diagnostic use of 18FDG-PET in the imaging of breast and other cancers (26, 27). Enhanced glyocolytic flux suggests that breast tumors might exhibit abnormal levels of AGEs including CEdG. Accordingly we measured the levels of CEdG diastereomers in DNA extracted from a clinical breast tumor specimen as well as adjacent normal tissue. The data in Table 1 reveal some significant (P<0.05) differences in the levels of CEdG between tumor and normal tissue. Both stereoisomers were observed at ~ 3-fold higher levels in normal relative to tumor tissue (CEdG-A, P=0.02; CEdG-B, P=0.003). Within normal tissue, the levels of CEdGA and B were not significantly different (P=0.08), while in tumor there was a small bias favoring CEdG-A (P=0.03). Levels of CEdG in DNA extracted from either breast tumor or adjacent tissue in the absence of carbonyl scavenger were ~ 1.5–2.0× higher (data not shown); however, artifactual formation was inhibited by the addition of 10 mM D-penicillamine in two aliquots during both the cell lysis/DNA isolation and hydrolysis/dephosphorylation steps.
Table 1.
CEdG isomers from a human breast tumor and adjacent normal tissue.
| CEdG (fmol) | dG (fmol) | CEdG/107 dG | ||
|---|---|---|---|---|
| CEdG-A | Normal | 234 ± 24.9 | 1.91 × 108 | 12.3a ± 1.3 |
| Tumor | 247 ± 11.6 | 6.48 × 108 | 3.9b ± 0.2 | |
| CEdG-B | Normal | 151 ± 4.98 | 1.91 × 108 | 7.9c ± 0.3 |
| Tumor | 173 ± 6.64 | 6.48 × 108 | 2.7d ± 0.1 |
P=0.08 versus CEdG-B in normal tissue.
P=0.02 versus CEdG-A in adjacent normal tissue.
P=0.003 versus CEdQ-B in adjacent tumor tissue.
P=0.03 versus CEdG-A in tumor tissue.
Discussion
In spite of longstanding interest in the role of biopolymer glycation in human disease, no generally applicable method for the quantitative determination of CEdG has been described. A 32P post-labeling assay has been used to estimate endogenous levels of CEdG in human buccal epithelial cells of 2–3/107 nucleotides (28). However, although the post-labeling method offers potentially great advantages in sensitivity, a major drawback is that direct analyte verification is usually not possible. Moreover, post-labeling is prone to artifacts and false positives, and may lead to inaccurate estimation of adduct levels due to several factors including RNA contamination (29). An immunoaffinity-based method for the detection of CEdG using a polyclonal antibody coupled to a diode array HPLC platform has more recently been described by the Pischetsreider group (30). This approach was used to provide the first demonstration of CEdG in human urine and cultured smooth muscle cells. In some cases, peak identity was confirmed by LC-MS/MS, but quantitation was not practical owing to the imprecise nature of immunoaffinity chromatography. A monoclonal-based immunohistochemical detection method has also been reported and was used to demonstrate elevated levels of CEdG in aorta and kidney of diabetic patients relative to normal controls (31). However, antibody-based assays are primarily of value in qualitative and comparative determinations of adduct abundance. For quantitative determinations LC-ESI-MS/MS in conjunction with the isotope dilution method is rapidly becoming the standard approach for the quantitative analysis of DNA adducts (32, 33). Although structural analyses of DNA-AGEs by LC-ESI-MS/MS have been previously described and recently reviewed (34), to the best of our knowledge no reliable quantitative methods for CEdG measurement have appeared, presumably due to a lack of suitable isotopically enriched standards. To address this issue we synthesized 15N-enriched isotopomers of CEdG differing from the unlabelled adducts by 5 amu, which provided sufficient mass resolution for accurate and reproducible quantitation using the stable isotope dilution method.
The ability to simultaneously resolve and quantitate both diastereomers of CEdG provided two independent parameters for assessing DNA glycation levels within a single sample. The biological significance of the CEdG diastereomer ratio in vivo is presently unclear, but could reflect stereochemical biases in adduct repair or polymerase bypass. Examination of the CEdG stereoisomer distribution in vivo by LC-ESI-MS/MS would only be meaningful if the rate of stereochemical interconversion was negligible. Superseding the issue of stereochemical integrity is that of overall adduct stability. Loss of the CEG base from either stereoisomer during workup would result in the generation of abasic sites leading to an underestimation of true nucleoside adduct levels. This was of particular concern since CEdG has been purported to undergo depurination more readily than dG at elevated temperatures (35). We decided to quantify the extent of depurination and racemization by monitoring free base formation and isomer interconversion under acidic conditions at 37°C rather than at non-physiological temperatures. The data in Figure 4 suggest that the CEdG diastereomers possess similar stability, and are slightly more resistant to depurination under acidic conditions than dG. This fact, together with the prohibitive barrier to stereochemical interconversion, indicates that determination of CEdG diastereomer ratios may be plausibly used in quantitative biomarker studies.
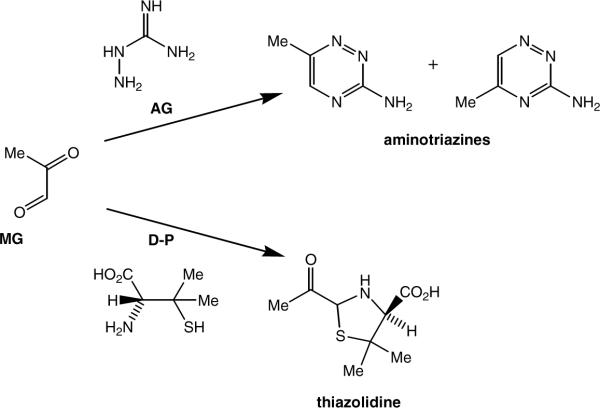
One important confounding factor in the quantitation of adducts resulting from oxidative or oxoaldehyde DNA modification is artifactual product formation during sample isolation and workup. For example, the problems surrounding the measurement of 8-oxo-dG using GC-MS and/or mildly oxidizing workup conditions have been detailed previously (36–38). In the case of CEdG adducts, the presence of MG during the workup could complicate the accurate determination of endogenous levels. The high background levels of CEdG detected in reagent grade calf thymus DNA prompted us to examine the effects of carbonyl scavenger addition prior to the enzymatic digestions. Scavengers such as AG and D-P react rapidly with MG and other oxoaldehydes to yield aminotriazines and thiazolidines respectively (Figure 7) which are relatively unreactive electrophiles (24, 25). D-penicillamine has been reported to react with MG 60χ faster than AG (24), and thus may be more advantageous for CEdG determinations requiring DNA isolation from complex tissue matrices.
Figure 7.
Reactions of carbonyl scavengers AG and D-P with MG yield isomeric aminotriazines and 2-acylthiazolidine, respectively.
We suggest that MG bound reversibly to proteins (23) was predominantly responsible for the formation of DNA glycation artifacts observed during the isolation and workup of dsDNA. Extraction and workup procedures which expose DNA for extended periods to cell lysates and partially purified enzyme reagents increase the probability for the ex vivo formation of CEdG, necessitating the need for carbonyl scavengers. This hypothesis is consistent with a previously published report showing that MG-BSA conjugates prepared by incubating MG with BSA can be used as reagents to induce DNA damage in cultured mammalian cells (39).. The data in Figure 6 suggest that the addition of AG or D-P can largely eliminate artifactual CEdG formation. Minimizing exposure to proteins by shortening the enzymatic lysis and hydrolysis/dephosphorylation steps may also reduce the requirement for carbonyl scavengers.
We are currently examining a diverse array of tumor and corresponding control tissues in order to determine whether the trends noted in the breast cancer specimen are a general feature of tumors which display elevated levels of glycolysis. The finding of significantly lower CEdG in breast tumors relative to adjacent normal tissue can potentially be explained by the observation that glycolytic cancers possess lower levels of MG as a result of overexpression of the glyoxalase system. This highly evolutionarily conserved system consists of two non-homologous zinc metalloenzymes Glo1 and 2, which act sequentially to convert MG into lactate using reduced glutathione (GSH) as a catalytic cofactor (40, 41).
Glo1/2 are overexpressed ~ 3–5× in many breast cancers relative to normal mammary tissue, and enhanced expression of either one or both enzymes has also been observed in prostate, kidney, lung, colon, stomach, brain and ovarian cancers (42, 43). This is believed to be a metabolic adaptation to counter the pro-apoptotic effect of MG accumulation in glycolytic tumors. This phenomenon has formed the basis for the proposal to develop Glo1 inhibitors as cancer therapeutics (43, 44). One potential application of the quantitative LC-ESI-MS/MS method would be in monitoring the efficacy of glyoxalase inhibitors, which would be expected to induce a dose dependent increase in CEdG levels.
Methylglyoxal is a potent mutagen in mammalian cells, inducing G>T and G>C transversions, as well as a large number (50%) of multibase deletions (5). Since 89% of the base substitution mutations are observed at guanosine, and CEdG is the predominant adduct formed from reaction of MG with DNA, it appears likely that this pattern of transversions arises from CEdG. Evidence for this has recently been obtained via primer extension assays using oligonucleotide templates containing CEdG. (45). The presence of CEdG in DNA has also been shown to induce single-strand breaks, suggesting an alternative mechanism by which this adduct may contribute to genetic instability (46).
Embryonic hyperglycemia has been shown to result in a high frequency of congenital malformations leading to perinatal death; and transgenic (lacI) mouse models have implicated a prominent role for DNA damage and mutagenesis in glucose-associated embryopathy (47, 48). Conditions which result in the impairment of glucose regulation such as diabetes and metabolic syndrome have been shown to significantly increase the risk for cancers of the breast, liver, pancreas, colon, cervix and endometrium (49, 50). Although the mechanistic reasons for this are as yet unclear, it is conceivable that genetic instability induced by CEdG may be involved.
The availability of a quantitative LC-MS/MS method for the measurement of CEdG complements methods currently available for protein AGEs (7), and should allow for a more comprehensive evaluation of the role of nucleotide glycation in human metabolic disease.
Supplementary Material
Acknowledgement
Support from the City of Hope Comprehensive Cancer Center (P30 CA33572) and the California Breast Cancer Research Program for a pre-doctoral fellowship to D. Tamae (14GB-0162) are gratefully acknowledged.
Footnotes
Abbreviations: MG, methylglyoxal AGE, advanced glycation endproduct LC-ESI-MS/MS, liquid chromatography electrospray ionization tandem mass spectrometry CEdG, N2-carboxyethyl-2'-deoxyguanosine AG, aminoguanidine D-P, D-penicillamine.
Supporting Information Available: UV, 1H NMR and 13C NMR spectra for CEdG-A and B; MS2 and MS3 for 15N5-CEdG-A and B; and observed and calculated isotopic distributions for both diastereomers. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Nemet I, Varga-Defterdarovic L, Turk Z. Methylglyoxal in food and living organisms. Mol. Nutr. Food Res. 2006;50:1105–1117. doi: 10.1002/mnfr.200600065. [DOI] [PubMed] [Google Scholar]
- (2).Phillips SA, Thornalley PJ. The formation of methylglyoxal from triose phosphates. Investigation using a specific assay for methylglyoxal. Eur. J. Biochem. 1993;212:101–105. doi: 10.1111/j.1432-1033.1993.tb17638.x. [DOI] [PubMed] [Google Scholar]
- (3).Casazza JP, Felver ME, Veech RL. The metabolism of acetone in rat. J. Biol. Chem. 1984;259:231–236. [PubMed] [Google Scholar]
- (4).Chaplen FW, Fahl WE, Cameron DC. Detection of methylglyoxal as a degradation product of DNA and nucleic acid components treated with strong acid. Anal. Biochem. 1996;236:262–269. doi: 10.1006/abio.1996.0165. [DOI] [PubMed] [Google Scholar]
- (5).Murata-Kamiya N, Kamiya H, Kaji H, Kasai H. Methylglyoxal induces G:C to C:G and G:C to T:A transversions in the supF gene on a shuttle vector plasmid replicated in mammalian cells. Mutat. Res. 2000;468:173–182. doi: 10.1016/s1383-5718(00)00044-9. [DOI] [PubMed] [Google Scholar]
- (6).Lo TW, Selwood T, Thornalley PJ. The reaction of methylglyoxal with aminoguanidine under physiological conditions and prevention of methylglyoxal binding to plasma proteins. Biochem. Pharmacol. 1994;48:1865–1870. doi: 10.1016/0006-2952(94)90584-3. [DOI] [PubMed] [Google Scholar]
- (7).Ahmed N, Thornalley PJ. Quantitative screening of protein biomarkers of early glycation, advanced glycation, oxidation and nitrosation in cellular and extracellular proteins by tandem mass spectrometry multiple reaction monitoring. Biochem. Soc. Trans. 2003;31:1417–1422. doi: 10.1042/bst0311417. [DOI] [PubMed] [Google Scholar]
- (8).Wautier JL, Schmidt AM. Protein glycation: a firm link to endothelial cell dysfunction. Circ. Res. 2004;95:233–238. doi: 10.1161/01.RES.0000137876.28454.64. [DOI] [PubMed] [Google Scholar]
- (9).Yao D, Taguchi T, Matsumura T, Pestell R, Edelstein D, Giardino I, Suske G, Ahmed N, Thornalley PJ, Sarthy VP, Hammes HP, Brownlee M. Methylglyoxal modification of mSin3A links glycolysis to angiopoietin-2 transcription. Cell. 2006;124:275–286. doi: 10.1016/j.cell.2005.11.024. [DOI] [PubMed] [Google Scholar]
- (10).Rahbar S, Blumenfeld O, Ranney HM. Studies of an unusual hemoglobin in patients with diabetes mellitus. Biochem. Biophys. Res. Commun. 1969;36:838–843. doi: 10.1016/0006-291x(69)90685-8. [DOI] [PubMed] [Google Scholar]
- (11).Norberg M, Eriksson JW, Lindahl B, Andersson C, Rolandsson O, Stenlund H, Weinehall L. A combination of HbA1c, fasting glucose and BMI is effective in screening for individuals at risk of future type 2 diabetes: OGTT is not needed. J. Intern. Med. 2006;260:263–271. doi: 10.1111/j.1365-2796.2006.01689.x. [DOI] [PubMed] [Google Scholar]
- (12).Edelman D, Olsen MK, Dudley TK, Harris AC, Oddone EZ. Utility of hemoglobin A1c in predicting diabetes risk. J. Gen. Intern. Med. 2004;19:1175–1180. doi: 10.1111/j.1525-1497.2004.40178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Rosenstock J, Sugimoto D, Strange P, Stewart JA, Soltes-Rak E, Dailey G. Triple therapy in type 2 diabetes: insulin glargine or rosiglitazone added to combination therapy of sulfonylurea plus metformin in insulin-naive patients. Diabetes Care. 2006;29:554–559. doi: 10.2337/diacare.29.03.06.dc05-0695. [DOI] [PubMed] [Google Scholar]
- (14).van Heijst JW, Niessen HW, Hoekman K, Schalkwijk CG. Advanced glycation end products in human cancer tissues: detection of Nepsilon-(carboxymethyl)lysine and argpyrimidine. Ann. N.Y. Acad. Sci. 2005;1043:725–733. doi: 10.1196/annals.1333.084. [DOI] [PubMed] [Google Scholar]
- (15).Frischmann M, Bidmon C, Angerer J, Pischetsrieder M. Identification of DNA adducts of methylglyoxal. Chem. Res. in Toxicol. 2005;18:1586–1592. doi: 10.1021/tx0501278. [DOI] [PubMed] [Google Scholar]
- (16).Papoulis A, al-Abed Y, Bucala R. Identification of N2-(1-carboxyethyl)guanine (CEG) as a guanine advanced glycosylation end product. Biochemistry. 1995;34:648–655. doi: 10.1021/bi00002a032. [DOI] [PubMed] [Google Scholar]
- (17).Ochs S, Severin T. Reaction of 2'-Deoxyguanosine with Glyceraldehyde. Liebigs Ann. Chem. 1994:851–853. [Google Scholar]
- (18).Rahbar S. Novel inhibitors of glycation and AGE formation. Cell biochemistry and biophysics. 2007;48:147–157. doi: 10.1007/s12013-007-0021-x. [DOI] [PubMed] [Google Scholar]
- (19).Besaratinia A, Bates SE, Synold TW, Pfeifer GP. Similar mutagenicity of photoactivated porphyrins and ultraviolet A radiation in mouse embryonic fibroblasts: involvement of oxidative DNA lesions in mutagenesis. Biochemistry. 2004;43:15557–15566. doi: 10.1021/bi048717c. [DOI] [PubMed] [Google Scholar]
- (20).Phillips SA, Mirrlees D, Thornalley PJ. Modification of the glyoxalase system in streptozotocin-induced diabetic rats. Effect of the aldose reductase inhibitor Statil. Biochem. Pharmacol. 1993;46:805–811. doi: 10.1016/0006-2952(93)90488-i. [DOI] [PubMed] [Google Scholar]
- (21).Figarola JL, Scott S, Loera S, Xi B, Synold T, Rahbar S. Renoprotective and lipid-lowering effects of LR compounds, novel advanced glycation end product inhibitors, in streptozotocin-induced diabetic rats. Ann. N. Y. Acad. Sci. 2005;1043:767–776. doi: 10.1196/annals.1333.089. [DOI] [PubMed] [Google Scholar]
- (22).Figarola JL, Scott S, Loera S, Tessler C, Chu P, Weiss L, Hardy J, Rahbar S. LR-90 a new advanced glycation endproduct inhibitor prevents progression of diabetic nephropathy in streptozotocin-diabetic rats. Diabetologia. 2003;46:1140–1152. doi: 10.1007/s00125-003-1162-0. [DOI] [PubMed] [Google Scholar]
- (23).Lo TW, Westwood ME, McLellan AC, Selwood T, Thornalley PJ. Binding and modification of proteins by methylglyoxal under physiological conditions. A kinetic and mechanistic study with N alpha-acetylarginine, N alpha-acetylcysteine, and N alpha-acetyllysine, and bovine serum albumin. J. Biol. Chem. 1994;269:32299–32305. [PubMed] [Google Scholar]
- (24).Wondrak GT, Cervantes-Laurean D, Roberts MJ, Qasem JG, Kim M, Jacobson EL, Jacobson MK. Identification of alpha-dicarbonyl scavengers for cellular protection against carbonyl stress. Biochem. Pharmacol. 2002;63:361–373. doi: 10.1016/s0006-2952(01)00915-7. [DOI] [PubMed] [Google Scholar]
- (25).Thornalley PJ, Yurek-George A, Argirov OK. Kinetics and mechanism of the reaction of aminoguanidine with the alpha-oxoaldehydes glyoxal, methylglyoxal, and 3-deoxyglucosone under physiological conditions. Biochem. Pharmacol. 2000;60:55–65. doi: 10.1016/s0006-2952(00)00287-2. [DOI] [PubMed] [Google Scholar]
- (26).Avril N, Menzel M, Dose J, Schelling M, Weber W, Janicke F, Nathrath W, Schwaiger M. Glucose metabolism of breast cancer assessed by 18F-FDG PET: histologic and immunohistochemical tissue analysis. J. Nucl. Med. 2001;42:9–16. [PubMed] [Google Scholar]
- (27).Bos R, van Der Hoeven JJ, van Der Wall E, van Der Groep P, van Diest PJ, Comans EF, Joshi U, Semenza GL, Hoekstra OS, Lammertsma AA, Molthoff CF. Biologic correlates of (18)fluorodeoxyglucose uptake in human breast cancer measured by positron emission tomography. J. Clin. Oncol. 2002;20:379–387. doi: 10.1200/JCO.2002.20.2.379. [DOI] [PubMed] [Google Scholar]
- (28).Vaca CE, Nilsson JA, Fang JL, Grafstrom RC. Formation of DNA adducts in human buccal epithelial cells exposed to acetaldehyde and methylglyoxal in vitro. Chemico-Biological Interactions. 1998;108:197–208. doi: 10.1016/s0009-2797(97)00107-5. [DOI] [PubMed] [Google Scholar]
- (29).Godschalk RW, Maas LM, Kleinjans JC, Van Schooten FJ. Influences of DNA isolation and RNA contamination on carcinogen-DNA adduct analysis by 32P-postlabeling. Environ. Mol. Mutagen. 1998;32:344–350. [PubMed] [Google Scholar]
- (30).Schneider M, Georgescu A, Bidmon C, Tutsch M, Fleischmann EH, Popov D, Pischetsrieder M. Detection of DNA-bound advanced glycation end-products by immunoaffinity chromatography coupled to HPLC-diode array detection. Mol. Nutr. Food Res. 2006;50:424–429. doi: 10.1002/mnfr.200500217. [DOI] [PubMed] [Google Scholar]
- (31).Li H, Nakamura S, Miyazaki S, Morita T, Suzuki M, Pischetsrieder M, Niwa T. N2-carboxyethyl-2'-deoxyguanosine, a DNA glycation marker, in kidneys and aortas of diabetic and uremic patients. Kidney Int. 2006;69:388–392. doi: 10.1038/sj.ki.5000064. [DOI] [PubMed] [Google Scholar]
- (32).Singh R, Farmer PB. Liquid chromatography-electrospray ionization-mass spectrometry: the future of DNA adduct detection. Carcinogenesis. 2006;27:178–196. doi: 10.1093/carcin/bgi260. [DOI] [PubMed] [Google Scholar]
- (33).Koc H, Swenberg JA. Applications of mass spectrometry for quantitation of DNA adducts. J. Chromatogr. 2002;778:323–343. doi: 10.1016/s1570-0232(02)00135-6. [DOI] [PubMed] [Google Scholar]
- (34).Bidmon C, Frischmann M, Pischetsrieder M. Analysis of DNA-bound advanced glycation end-products by LC and mass spectrometry. J.Chromatogr. 2007;855:51–58. doi: 10.1016/j.jchromb.2006.11.033. [DOI] [PubMed] [Google Scholar]
- (35).Seidel W, Pischetsrieder M. DNA-glycation leads to depurination by the loss of N2-carboxyethylguanine in vitro. Cell. Mol. Biol. (Noisy-le-Grand, France) 1998;44:1165–1170. [PubMed] [Google Scholar]
- (36).Cadet J, D'Ham C, Douki T, Pouget JP, Ravanat JL, Sauvaigo S. Facts and artifacts in the measurement of oxidative base damage to DNA. Free Radical Res. 1998;29:541–550. doi: 10.1080/10715769800300581. [DOI] [PubMed] [Google Scholar]
- (37).Rodriguez H, Jurado J, Laval J, Dizdaroglu M. Comparison of the levels of 8-hydroxyguanine in DNA as measured by gas chromatography mass spectrometry following hydrolysis of DNA by Escherichia coli Fpg protein or formic acid. Nucleic Acids Res. 2000;28:E75. doi: 10.1093/nar/28.15.e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).ESCODD Comparison of different methods of measuring 8-oxoguanine as a marker of oxidative DNA damage. ESCODD (European Standards Committee on Oxidative DNA Damage) Free Radic Res. 2000;32:333–341. doi: 10.1080/10715760000300331. [DOI] [PubMed] [Google Scholar]
- (39).Schupp N, Schinzel R, Heidland A, Stopper H. Genotoxicity of advanced glycation end products: involvement of oxidative stress and of angiotensin II type 1 receptors. Ann. N. Y. Acad. Sci. 2005;1043:685–695. doi: 10.1196/annals.1333.079. [DOI] [PubMed] [Google Scholar]
- (40).Creighton DJ, Hamilton DS. Brief history of glyoxalase I and what we have learned about metal ion-dependent, enzyme-catalyzed isomerizations. Arch. Biochem. Biophys. 2001;387:1–10. doi: 10.1006/abbi.2000.2253. [DOI] [PubMed] [Google Scholar]
- (41).Thornalley PJ. Glutathione-dependent detoxification of alpha-oxoaldehydes by the glyoxalase system: involvement in disease mechanisms and antiproliferative activity of glyoxalase I inhibitors. Chemico-Biological Interactions. 1998;111–112:137–151. doi: 10.1016/s0009-2797(97)00157-9. [DOI] [PubMed] [Google Scholar]
- (42).Rulli A, Carli L, Romani R, Baroni T, Giovannini E, Rosi G, Talesa V. Expression of glyoxalase I and II in normal and breast cancer tissues. Breast Cancer Res. Treat. 2001;66:67–72. doi: 10.1023/a:1010632919129. [DOI] [PubMed] [Google Scholar]
- (43).Sakamoto H, Mashima T, Sato S, Hashimoto Y, Yamori T, Tsuruo T. Selective activation of apoptosis program by S-p-bromobenzylglutathione cyclopentyl diester in glyoxalase I-overexpressing human lung cancer cells. Clin. Cancer Res. 2001;7:2513–2518. [PubMed] [Google Scholar]
- (44).Kavarana MJ, Kovaleva EG, Creighton DJ, Wollman MB, Eiseman JL. Mechanism-based competitive inhibitors of glyoxalase I: intracellular delivery, in vitro antitumor activities, and stabilities in human serum and mouse serum. J. Med. Chem. 1999;42:221–228. doi: 10.1021/jm9708036. [DOI] [PubMed] [Google Scholar]
- (45).Cao H, Jiang Y, Wang Y. Stereospecific synthesis and characterization of oligodeoxyribonucleotides containing an N2-(1-carboxyethyl)-2'-deoxyguanosine. JACS. 2007;129:12123–12130. doi: 10.1021/ja072130e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Pischetsrieder M, Seidel W, Munch G, Schinzel R. N(2)-(1-Carboxyethyl)deoxyguanosine, a nonenzymatic glycation adduct of DNA, induces single-strand breaks and increases mutation frequencies. Biochem. Biophys. Res. Commun. 1999;264:544–549. doi: 10.1006/bbrc.1999.1528. [DOI] [PubMed] [Google Scholar]
- (47).Lee AT, Plump A, DeSimone C, Cerami A, Bucala R. A role for DNA mutations in diabetes-associated teratogenesis in transgenic embryos. Diabetes. 1995;44:20–24. doi: 10.2337/diab.44.1.20. [DOI] [PubMed] [Google Scholar]
- (48).Eriksson UJ, Wentzel P, Minhas HS, Thornalley PJ. Teratogenicity of 3-deoxyglucosone and diabetic embryopathy. Diabetes. 1998;47:1960–1966. doi: 10.2337/diabetes.47.12.1960. [DOI] [PubMed] [Google Scholar]
- (49).La Vecchia C, Negri E, Franceschi S, D'Avanzo B, Boyle P. A case-control study of diabetes mellitus and cancer risk. Br. J. Cancer. 1994;70:950–953. doi: 10.1038/bjc.1994.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Cowey S, Hardy RW. The metabolic syndrome: A high-risk state for cancer? Am. J. Pathol. 2006;169:1505–1522. doi: 10.2353/ajpath.2006.051090. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.