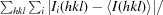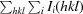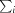The crystal structure at 1.0 Å resolution of the cellulose-binding CBM3b from the major scaffoldin subunit ScaA of A. cellulolyticus is described.
Keywords: CBM3b, cellulose-binding module, major scaffoldin subunit, ScaA, cellulosome, Acetivibrio cellulolyticus
Abstract
The carbohydrate-binding module (CBM) of the major scaffoldin subunit ScaA of the cellulosome of Acetivibrio cellulolyticus is classified as a family 3b CBM and binds strongly to cellulose. The CBM3b was overexpressed, purified and crystallized, and its three-dimensional structure was determined. The structure contained a nickel-binding site located at the N-terminal region in addition to a ‘classical’ CBM3b calcium-binding site. The structure was also determined independently by the SAD method using data collected at the Ni-absorption wavelength of 1.48395 Å and even at a wavelength of 0.97625 Å in a favourable case. The new scaffoldin-borne CBM3 structure reported here provides clear evidence for the proposition that a family 3b CBM may be accommodated in scaffoldin subunits and functions as the major substrate-binding entity of the cellulosome assembly.
1. Introduction
The cellulosome is a multi-protein complex consisting of cellulolytic and hemi-cellulolytic enzymes that has mainly been described in anaerobic bacteria (Lamed et al., 1983 ▶; Bayer, Shimon et al., 1998 ▶; Béguin & Lemaire, 1996 ▶; Felix & Ljungdahl, 1993 ▶; Karita et al., 1997 ▶; Ding et al., 1999 ▶). The cellulosomal enzymes are attached to a large multi-modular noncatalytic subunit called scaffoldin (Gerngross et al., 1993 ▶). The scaffoldin contains a single cellulose-specific carbohydrate-binding module (CBM) that is responsible for targeting the multi-enzyme complex to the crystalline cellulose substrate (Bayer, Shimon et al., 1998 ▶).
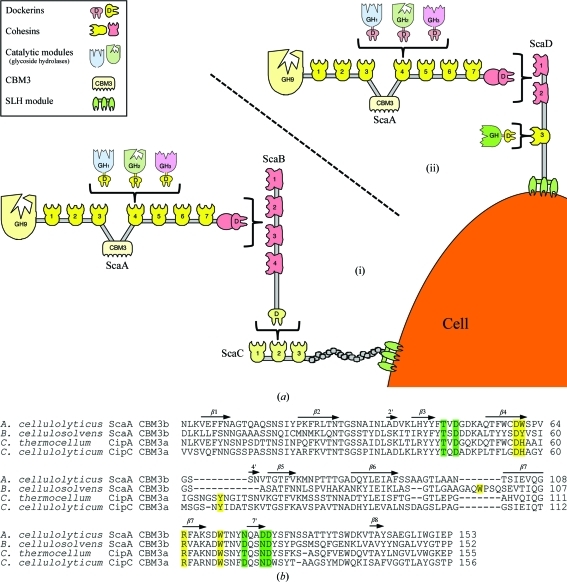
Acetivibrio cellulolyticus is a cellulosome-producing anaerobic mesophilic bacterium that is known for its efficient degradation of crystalline cellulose (Khan, 1980 ▶; Saddler et al., 1980 ▶; Saddler & Khan, 1981 ▶). The complicated cellulosome system of this bacterium (Fig. 1 ▶) is capable of housing up to 96 cellulose-degrading dockerin-containing enzymes (Xu et al., 2004 ▶), which all rely upon the substrate-binding abilities of the ScaA CBM3b.
Figure 1.
The scaffoldin-borne family 3b CBM of the A. cellulolyticus cellulosome. (a) Schematic representation of the proposed cell-surface disposition of the cellulosomal components of A. cellulolyticus. The intermolecular cellulosome interactions in this bacterium can be one of two types. (i) Enzyme-borne type I dockerins recognize the type I cohesins of ScaA, the ScaA type II dockerin binds to the type II cohesins of ScaB, the ScaB dockerin binds to the cohesins of ScaC and an SLH module on ScaC serves to attach the entire assembly to the cell surface. (ii) Enzyme-borne type I dockerins recognize the type I cohesins of ScaA and the ScaA type II dockerin binds to one of the two type II cohesins of ScaD. Additionally, a single type I dockerin-containing enzyme binds to the single type I cohesin of ScaD. (b) Sequence alignment of A. cellulolyticus ScaA CBM3b with sequences of other scaffoldin-borne family 3 CBMs: B. cellulosolvens ScaA CBM3b (PDB entry 2xbt; Yaniv et al., 2011 ▶), C. thermocellum CipA CBM3a (PDB entry 1nbc; Tormo et al., 1996 ▶) and C. cellulolyticum CipC CBM3a (PDB entry 1g43; Shimon et al., 2000 ▶). The alignment was performed using ClustalW v.2.0.12 (Larkin et al., 2007 ▶). Secondary-structural elements (β-strands) corresponding to the A. cellulolyticus ScaA CBM3b structure are indicated as arrows and numbered. Proposed cellulose-binding amino-acid residues are shown in yellow. Putative calcium-binding residues are shown in green. The numbering of the A. cellulolyticus ScaA CBM3b sequence is according to the observed electron density.
Family 3 of the CBMs is a large family of carbohydrate-binding proteins that includes over 350 representatives from more than 60 different bacteria (data taken from the Carbohydrate Active Enzymes website; Cantarel et al., 2009 ▶). Historically, the division of the family into subgroups (a, b and c) was based on minor sequence differences combined with the fact that the known scaffoldin-borne CBM3s (subgroup a) from four different clostridia could be differentiated by the existence of a distinctive Trp-containing loop (Bayer, Morag et al., 1998 ▶). Subfamilies b and c lacked this loop and were further differentiated by the lack of the standard aromatic binding residues in subfamily c.
Here, we report the high-resolution X-ray structure of the scaffoldin-borne A. cellulolyticus ScaA CBM3b, which represents the second cellulosomal CBM3b structure to be published, following that of Bacteroides cellulosolvens (Yaniv et al., 2011 ▶).
2. Materials and methods
2.1. Cloning of the CBM3b from ScaA of A. cellulolyticus
A DNA fragment encoding CBM3b corresponding to residues 973–1121 of the full-length ScaA cellulosomal scaffoldin (GenBank accession No. AAF06064) was amplified by PCR from A. cellulolyticus ATCC 33288 genomic DNA, isolated as described by Murray & Thompson (1980 ▶), using the primers 5′-CCCATATGAACTTGAAAGTTGAGTTCTTCAATG-3′ and 5′-GAATTCTTAAGGTTCTATACCCCAGATCAAGCC-3′. The PCR product was inserted into pET-28a(+) (Novagen, Madison, Wisconsin, USA) via NdeI and EcoRI to generate the expression plasmid. The obtained construct contained an N-terminal hexahistidine (His) tag and thrombin cleavage site.
2.2. Protein expression and purification
Escherichia coli BL21 (DE3) harbouring the expression plasmid was grown under aerobic conditions at 310 K until the culture reached an A 600 of 0.6. The temperature was then decreased to 298 K for an additional 16 h. The recombinant ScaA CBM3b containing an N-terminal His tag was first isolated by metal-chelate affinity chromatography using Ni–IDA resin (Rimon Biotech, Israel) according to the manufacturer’s recommended protocol.
The His tag was cleaved off by thrombin protease (Novagen, EMD Chemicals Inc., San Diego, California, USA) following the manufacturer’s instructions.
After thrombin cleavage, the cleaved His tag was removed from the preparation by metal-chelate affinity chromatography. For this purpose, the solution was incubated with Ni–IDA resin and the suspension was decanted into a column. The flowthrough fluid (containing the cleaved protein and thrombin) was collected and finally purified by fast protein liquid chromatography (FPLC) using a Superdex 75 16/60 column (GE Healthcare) and an ÄKTAprime system (GE Healthcare). The protein was concentrated using Centriprep YM-3 centrifugal filter devices (Amicon Bioseparation, Millipore, Billerica, Massachusetts, USA) and the protein concentration was determined by measuring the UV absorbance at 280 nm. The purified protein solution consisted of 30 mg ml−1 protein, 150 mM Tris–HCl pH 7.5, 300 mM NaCl and 0.05% sodium azide.
2.3. Crystallization, data collection and processing
Crystals were grown at 293 K by the hanging-drop vapour-diffusion method (McPherson, 1982 ▶). The first crystals appeared in condition No. 39 of Crystal Screen from Hampton Research, consisting of 2.0 M ammonium sulfate, 0.1 M HEPES pH 7.5 and 2%(w/v) polyethylene glycol 400. The initial crystallization condition was optimized and the best crystals were obtained by setting up crystallization drops consisting of 1.9 M ammonium sulfate, 0.1 M HEPES pH 7.5 and 0.5%(w/v) polyethylene glycol 400. In addition, crystals were grown from these conditions supplemented with NiCl2 (0.2 M final concentration).
Crystals were harvested from the crystallization drop using a MiTeGen Micromount and transferred for a short time into a cryo-stabilization solution mimicking the mother liquor supplemented with 18%(w/v) sucrose, 16%(w/v) glycerol, 16%(w/v) ethylene glycol and 4%(w/v) glucose. For data collection, crystals were mounted on MiTeGen MicroMounts, plunged into liquid nitrogen and transported to the synchrotron for data collection in a nitrogen stream at a temperature of 100 K produced by an Oxford Cryostream low-temperature generator (Cosier & Glazer, 1986 ▶).
Owing to the unexpected presence of the Ni atom in the structure, a total of three data sets were measured for this system on the ID29 and BM14 beamlines at the European Synchrotron Radiation Facility (ESRF), Grenoble, France. A Pilatus 6M detector and X-ray radiation of 0.97622 Å wavelength were used on the ID29 beamline for crystals 1 and 3 (Table 1 ▶). A MAR 325 CCD detector and X-ray radiation of 1.48395 Å wavelength (Ni absorption edge) were used on the BM14 beamline for crystal 2 (Table 1 ▶). Diffraction data sets, each consisting of 200 images with 1° oscillation per frame, were collected for crystals 1, 2 and 3. The data were indexed and integrated with DENZO and scaled with SCALEPACK as implemented in HKL-2000 (Otwinowski & Minor, 1997 ▶). During the scaling procedure in SCALEPACK, Friedel pairs of reflections were kept unmerged in order to preserve the anomalous diffraction signal for further use. Crystals 1 and 3 diffracted to 1.08 and 1.0 Å resolution, respectively, on beamline ID29 and belonged to the hexagonal space group P6122 or P6522. Data for crystal 2 were collected on the tunable beamline BM14 to a moderate resolution of 1.80 Å only, owing to the detector size and wavelength limitations, and were used for independent determination of the structure using single-wavelength anomalous dispersion (SAD) and as an opportunity to explore the phasing power of Ni anomalous scattering. The calculated Matthews coefficient (Matthews, 1968 ▶) was 2.24 Å3 Da−1 for all crystals and gave a solvent content of 45.2%, corresponding to the presence of one monomer in the asymmetric unit. Data-collection statistics are given in Table 1 ▶.
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Data collection | Crystal 1 | Crystal 2, Ni absorption edge | Crystal 3, crystallized with Ni |
|---|---|---|---|
| X-ray source | ESRF ID29 | ESRF BM14 | ESRF ID29 |
| Date of measurement | 23 February 2011 | 13 April 2011 | 25 June 2011 |
| Wavelength | 0.97622 | 1.48395 | 0.97622 |
| Temperature (K) | 100 | 100 | 100 |
| Resolution (Å) | 20.0–1.07 (1.09–1.07) | 50.0–1.80 (1.83–1.80) | 50.0–1.00 (1.02–1.00) |
| Space group | P6122 | P6122 | P6122 |
| Unit-cell parameters (Å) | a = b = 52.51, c = 193.98 | a = b = 52.26, c = 192.92 | a = b = 52.70, c = 194.16 |
| Total No. of reflections | 1291610 | 168927 | 679030 |
| Unique reflections | 71090 | 15364 | 86080 |
| Completeness (%) | 98.8 (95.9) | 99.3 (97.1) | 98.4 (82.20) |
| Mosaicity (°) | 0.10–0.13 | 0.33–0.44 | 0.11–0.13 |
| Wilson B factor (Å2) | 7.59 | 20.3 | 8.24 |
| Rmerge† | 0.086 (0.624) | 0.058 (0.446) | 0.056 (0.57) |
| 〈I/σ(I)〉 | 30.9 (2.2) | 43.6 (5.0) | 33.17 (1.61) |
| Refinement data | |||
| Protein residues | 153 | 153 | 153 |
| Water molecules | 294 | 173 | 311 |
| Rcryst (%) | 10.3 | 13.7 | 12.49 |
| Rfree (%) | 14.5 | 17.3 | 13.60 |
| R.m.s.d., bonds (Å) | 0.014 | 0.014 | 0.012 |
| R.m.s.d., angles (°) | 1.57 | 1.70 | 1.52 |
| Ramachandran favoured‡ (%) | 99.0 | 98.0 | 98.0 |
| Ramachandran outliers‡ (%) | 0.62 | 0.0 | 0.62 |
| Clashscore‡ | 7.31 | 8.92 | 7.14 |
| Average B factor (Å2) | 11.80 | 23.0 | 12.80 |
2.4. Structure determination and refinement
The structure of the A. cellulolyticus CBM3b crystal was determined by molecular replacement using the data from crystal 1 employing MOLREP (Vagin & Teplyakov, 2010 ▶) as implemented in CCP4 (Winn et al., 2011 ▶), using the atomic coordinates of Clostridium thermocellum CBM3a (PDB entry 1nbc; Tormo et al., 1996 ▶), which exhibits 53% sequence identity, as a search model. Molecular-replacement attempts in the two alternative enantiomer space groups (P6122 and P6522) clearly supported P6122 as the correct space group with CC = 0.415 and RF = 0.539 (CC = 0.259 and RF = 0.608 for space group P6522).
In addition, combination of the high-resolution data from crystal 1 with the data collected at the wavelength corresponding to the peak of Ni anomalous dispersion from crystal 2 permitted crystal structure solution by SAD as implemented in the SHELXC/D/E set of programs (Sheldrick, 2010 ▶). The excellent quality of the diffraction data from crystal 3 (grown in the presence of Ni) permitted determination of the structure using SAD. This could be accomplished without experimental optimization of the anomalous signals of Ni2+ and Ca2+ ions (i.e. collection of data at the Ni or Ca absorption edge).
Model refinement was performed using REFMAC5 (Murshudov et al., 1997 ▶) as implemented in CCP4 (Winn et al., 2011 ▶) and PHENIX (Adams et al., 2010 ▶) and was assessed using Coot (Emsley et al., 2010 ▶). Water molecules were added by PHENIX, utilizing the strength of the peaks in a difference electron-density map and hydrogen-bond patterns, checked by Coot and modified manually if necessary. H atoms were refined in the riding mode for crystals 1 and 3.
The structures were validated using the MolProbity suite (Chen et al., 2010 ▶) as implemented in PHENIX. Data-collection and refinement statistics are summarized in Table 1 ▶. The structures have been deposited in the Protein Data Bank with codes 3zqw, 3zuc and 3zu8.
2.5. X-ray fluorescence measurements
X-ray fluorescence (XRF) spectra (Leonard et al., 2009 ▶) of the crystals were measured on the ID29 beamline at ESRF in order to analyze the metal content of the crystals (Garcia et al., 2006 ▶). Crystalline samples were mounted on MiTeGen MicroMounts as described for X-ray diffraction analysis. A silicon drift diode detector coupled with X-flash MMAX signal-processing units (Xflash 5010, Bruker AXS Microanalysis, Germany) was used. Incident beam energies were 12.7 keV. The collected spectra were analyzed using the PyMCA program (Sole et al., 2007 ▶).
3. Results
3.1. Overview
We determined the X-ray structure of the CBM3b module from the cellulosomal scaffoldin ScaA of A. cellulolyticus to 1.07 Å resolution from crystal 1 (Table 1 ▶) by molecular replacement. Independently, the structure was determined twice by the SAD method: once using data from crystals 1 and 2 and again using data from crystal 3. The 153 residues of the CBM3b form an antiparallel β-sandwich fold similar to that of other family 3 CBMs (Fig. 2 ▶). The ‘bottom’ β-sheet of the β-sandwich is formed by antiparallel strands 1, 2, 7 and 4, while the ‘top’ β-sheet is formed by antiparallel strands 5, 6, 3 and 8. Both the N- and the C-termini are located in the ‘top’ region (Fig. 2 ▶). The first three residues Gly-Ser-His are remnants of the thrombin cleavage site from the His tag.
Figure 2.
Structural features of A. cellulolyticus ScaA CBM3b. (a) Overall structure. Cartoon representation of the major secondary-structural elements (numbered according to sequence). The N- and C-termini are indicated. Calcium and nickel ions are shown as spheres (blue and orange, respectively). The solvent-accessible surface calculated using PyMOL is shown. (b) Space-filling representation of the proposed cellulose-binding region. Upper view; residues forming the cellulose-binding strip are coloured by element (carbon, yellow; oxygen, red; nitrogen, blue) and numbered. (c) Lateral view presenting the planar face that is free from topographic obstacles that could interfere in the formation of interactions with the glucose rings of the cellulose.
3.2. Cellulose binding
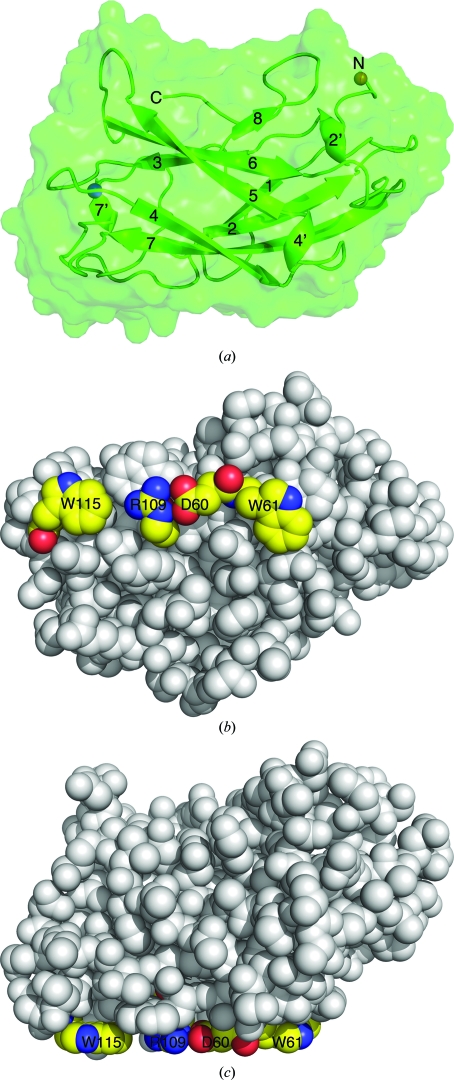
According to the accepted hypothesis (Tormo et al., 1996 ▶; Lehtiö et al., 2003 ▶; Ding et al., 2006 ▶), the cellulose-binding CBM3 module interacts with the glucose rings of cellulose via a conserved planar strip of aromatic residues located on the ‘bottom’ 1–2–7–4 sheet (Shimon et al., 2000 ▶).
In the A. cellulolyticus ScaA CBM3b, the aromatic residues forming the cellulose-binding array are Trp61 and Trp115. In addition, as seen in other scaffoldin-borne CBM3s from C. thermocellum, C. cellulolyticum and B. cellulosolvens, Asp60 and Arg109 of the A. cellulolyticus ScaA CBM3b form a salt bridge, creating a closed hydrogen-bonded ring capable of interacting with glucose rings that mimics an aromatic residue (Fig. 2 ▶).
Another requirement for cellulose binding is apparently that the cellulose-binding residues are on a planar face without topographic obstacles that enables proximity between the aromatic residues of the CBM3 and the glucose rings of the cellulose (Petkun et al., 2010 ▶). The ‘bottom’ 1–2–7–4 face of the A. cellulolyticus ScaA CBM3b (Fig. 2 ▶) is relatively flat and close proximity between the protein and its substrate can be achieved so that stacking interactions can be formed between the aromatic residue array and the glucose rings.
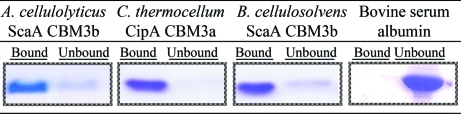
The cellulose-binding abilities of this CBM3 (Fig. 3 ▶) have been confirmed experimentally, demonstrating that even though there are only three cellulose-binding groups in the linear aromatic strip [compared with four groups in the scaffoldin-borne CBM3s of C. thermocellum (PDB entry 1nbc; Tormo et al., 1996 ▶), C. cellulolyticum (PDB entry 1g43; Shimon et al., 2000 ▶) and B. cellulosolvens (PDB entry 2xbt; Yaniv et al., 2011 ▶)] the resulting interaction with the glucose rings of the cellulose chains is stable enough for strong binding. It thus appears that three aromatic groups are sufficient to establish stable substrate–protein interactions.
Figure 3.
Cellulose-binding assay. Recombinant forms (all highly expressed in E. coli) of the designated CBM3s (20 µg) were mixed in predetermined amounts with 5 mg insoluble cellulose. The suspension was centrifuged and the pellet was washed (bound CBM3); the supernatant fluids (unbound CBM3) were collected, mixed with sample buffer and aliquots corresponding to 10% of the bound and unbound fractions were subjected to SDS–PAGE. Each experiment was repeated at least three times. The partition of the CBM3 bands between the bound and unbound states is shown. BSA was used as a negative control in all experiments.
3.3. Metal ions
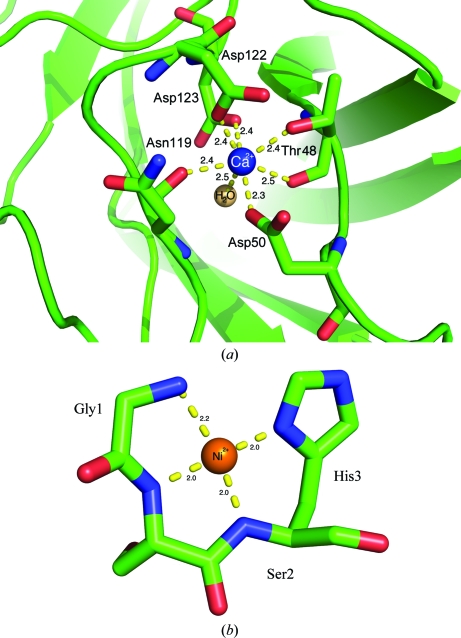
The X-ray structure revealed the presence of two heavy atoms in the ScaA CBM3b molecule manifested by two strong peaks in the anomalous difference Fourier density map (30.3σ and 14.57σ; the next peak is 6.5σ for the S atom of Cys59 as calculated by Coot). The second largest peak forms seven coordination bonds with bipyramidal geometry with O atoms in a calcium-binding site (Fig. 4 ▶ a), is conserved in CBMs with a β-sandwich fold (Hashimoto, 2006 ▶; Tormo et al., 1996 ▶) and can be assigned unambiguously as Ca2+. The square-planar coordination of the largest anomalous peak location and the presence of Ni ions in the purification procedure led to the presumption that this heavy atom could be Ni. The presence of an Ni atom in the crystals was verified by X-ray fluorescence (XRF) spectrum measurements (Thompson et al., 2001 ▶; Leonard et al., 2009 ▶) and was manifested by the presence of emission lines that are characteristic for nickel (Kα1 of 7.48 keV, Kβ1 of 8.26 keV). One possible source of this unexpected nickel may be Ni leakage during the Ni–IDA metal-chelate affinity chromatography procedure. Some of the nickel ions that form coordinative interactions within the IDA resin might have created alternative interactions with the protein residues.
Figure 4.
Metal-binding sites in A. cellulolyticus CBM3b. (a) Cartoon and stick representation of the Ca2+-binding site. Ca2+-coordinating residues are shown as sticks and numbered. Bond distances (Å) are shown. (b) Cartoon and stick representation of the Ni2+ pocket. Ni2+-coordinating residues are shown as sticks and numbered. Bond distances (Å) are shown
The nickel ion is located in a small pocket that is formed at the N-terminus of the protein by the extra residues Gly1-Ser2-His3 that are left after thrombin cleavage of the His tag. It displays square-planar geometry and forms four coordinative bonds to the main-chain N atoms and His3 Nδ (Fig. 4 ▶ b). Refinement of the structure with the first and the second data sets (see Table 1 ▶) revealed that the first three residues of the N-terminal part of the molecule are present in two states. One state was ordered and formed a complex with Ni. The other state was disordered and was not modelled.
A site-occupancy factor for the Ni2+ and the ordered state of the N-terminus was refined and converged to 0.53, 0.43 and 0.39 for crystal 1, crystal 2 and crystal 3, respectively (Table 1 ▶). Crystals 1 and 2 were from the same crystallization batch but were mounted and measured with a time difference of about six weeks. Crystal 3 was grown with supplementary Ni2+ in the crystallization conditions, yet the Ni occupancy in its structure was lower than that in crystals 1 and 2. This inability to increase the partial occupancy of the Ni during crystallization may imply that the mechanism of incorporation of Ni into the binding site of the IDA resin could not be reproduced in bulk solution.
In conclusion, we have shown that the structure of ScaA CBM3b from A. cellulolyticus can be determined using the anomalous signal of an Ni2+ ion with partial occupancy supplemented by the anomalous signal from the Ca2+ ion. As the Ni-binding residues remained after thrombin cleavage of the His tag and are therefore independent of the native protein sequence, this Ni phasing can be developed into a general approach for the determination of protein structures.
4. Discussion
The family 3 CBMs mediate one of two important biological functions in cellulase and cellulosome systems: (i) they confer the cellulose-binding property on the parent protein, whether scaffoldin or enzyme, as exemplified in subfamilies a and b, or (ii) they can serve to modify the character of the catalytic module of a parent enzyme, as exemplified in subfamily c. Members of subfamilies 3a and 3b of CBMs bind strongly to crystalline cellulose, whereas those of family 3c essentially fail to bind to cellulosic substrates. In some cases, members of subfamilies b and c show anomalies in their sequences and their precise contribution to the parent protein is currently obscure; these are referred to as CBM3b′ and CBM3c′ (Jindou et al., 2006 ▶).
Early studies (Tormo et al., 1996 ▶) indicated that the short 4′ β-strand was distinctive of the CBM3a′s, which suggested that the additional aromatic residue provides a stronger attachment to the crystalline cellulose substrate than those of enzyme-borne CBM3b′s that lack this insert. However, subsequent sequencing studies revealed the existence of CBM3b′s in the cellulosomal scaffoldins of A. cellulolyticus and B. cellulosolvens (Ding et al., 1999 ▶, 2000 ▶), thus implying that this simplistic view is perhaps more complicated. Intriguingly, the recently reported crystal structure of the CBM3b from B. cellulosolvens (Yaniv et al., 2011 ▶) revealed an alternative insert which occurs between β-strands 6 and 7 and provides an alternative compensatory aromatic residue (Trp) to the planar aromatic strip. The unresolved question therefore was whether a crystal structure of the simple scaffoldin-borne CBM3b of A. cellulolyticus, which lacks such an insert, would reveal a compensatory aromatic residue.
The structure of CBM3b from ScaA of A. cellulolyticus provides evidence that the relationship between the family 3a and 3b CBMs is not necessarily connected to a precise function of its parent protein. The appearance of a CBM in a given scaffoldin is thus not restricted to the CBM3a group and a CBM3b may also be located in a scaffoldin subunit and function as the major substrate-binding entity of the cellulosome.
We now have two confirmed cases of scaffoldin-borne CBM3s that belong to subgroup b rather than subgroup a. Moreover, various deviant forms of subgroup b CBMs (Petkun et al., 2010 ▶; Jindou et al., 2006 ▶) have been shown to lack important cellulose-binding residues in their sequences and also to lack carbohydrate-binding ability. In view of the increased sequencing of bacterial genomes and the hundreds of CBM3 sequences now appearing in the databases, these findings raise the question as to whether the historical classification of subgroups a, b and c is still valid for the family 3 CBMs or whether a new and more accurate method of classification (based on sequence homology, functional studies and three-dimensional structure) should be adopted for clearer differentiation between the subgroups.
Supplementary Material
PDB reference: CBM3b of ScaA, 3zqw
PDB reference: 3zuc
PDB reference: 3zu8
Acknowledgments
We thank the ESRF, Grenoble, France for use of the macromolecular crystallographic data-collection facilities and the ID29 and BM14 staff for their assistance. This research was supported by the Israel Science Foundation (ISF; Grant No. 293/08). EAB holds The Maynard I. and Elaine Wishner Chair of Bio-Organic Chemistry at the Weizmann Institute of Science.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Bayer, E. A., Morag, E., Lamed, R., Yaron, S. & Shoham, Y. (1998). Carbohydrases from Trichoderma reesei and Other Microorganisms, edited by M. Claeyssens, W. Nerinckx & K. Piens, pp. 39–65. London: The Royal Society of Chemistry.
- Bayer, E. A., Shimon, L. J., Shoham, Y. & Lamed, R. (1998). J. Struct. Biol. 124, 221–234. [DOI] [PubMed]
- Béguin, P. & Lemaire, M. (1996). Crit. Rev. Biochem. Mol. Biol. 31, 201–236. [DOI] [PubMed]
- Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V. & Henrissat, B. (2009). Nucleic Acids Res. 37, D233–D238. [DOI] [PMC free article] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Cosier, J. & Glazer, A. M. (1986). J. Appl. Cryst. 19, 105–107.
- Ding, S.-Y., Bayer, E. A., Steiner, D., Shoham, Y. & Lamed, R. (1999). J. Bacteriol. 181, 6720–6729. [DOI] [PMC free article] [PubMed]
- Ding, S.-Y., Bayer, E. A., Steiner, D., Shoham, Y. & Lamed, R. (2000). J. Bacteriol. 182, 4915–4925. [DOI] [PMC free article] [PubMed]
- Ding, S.-Y., Xu, Q., Mursheda, K. A., Baker, J. O., Bayer, E. A., Barak, Y., Lamed, R., Sugiyama, J., Rumbles, G. & Himmel, M. E. (2006). Biotechniques, 41, 435–440. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Felix, C. R. & Ljungdahl, L. G. (1993). Annu. Rev. Microbiol. 47, 791–819. [DOI] [PubMed]
- Garcia, J. S., Magalhães, C. S. & Arruda, M. A. (2006). Talanta, 69, 1–15. [DOI] [PubMed]
- Gerngross, U. T., Romaniec, M. P., Kobayashi, T., Huskisson, N. S. & Demain, A. L. (1993). Mol. Microbiol. 8, 325–334. [DOI] [PubMed]
- Hashimoto, H. (2006). Cell. Mol. Life Sci. 63, 2954–2967. [DOI] [PMC free article] [PubMed]
- Jindou, S., Xu, Q., Kenig, R., Shulman, M., Shoham, Y., Bayer, E. A. & Lamed, R. (2006). FEMS Microbiol. Lett. 254, 308–316. [DOI] [PubMed]
- Karita, S., Sakka, K. & Ohmiya, K. (1997). Rumen Microbes and Digestive Physiology in Ruminants, edited by R. Onodera, K. Itabashi, H. Ushida, H. Yano & Y. Sasaki, pp. 47–57. Tokyo: Japan Scientific Society.
- Khan, A. W. (1980). J. Gen. Microbiol. 121, 499–502.
- Lamed, R., Setter, E. & Bayer, E. A. (1983). J. Bacteriol. 156, 828–836. [DOI] [PMC free article] [PubMed]
- Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J. & Higgins, D. G. (2007). Bioinformatics, 23, 2947–2948. [DOI] [PubMed]
- Lehtiö, J., Sugiyama, J., Gustavsson, M., Fransson, L., Linder, M. & Teeri, T. T. (2003). Proc. Natl Acad. Sci. USA, 100, 484–489. [DOI] [PMC free article] [PubMed]
- Leonard, G., Solé, V. A., Beteva, A., Gabadinho, J., Guijarro, M., McCarthy, J., Marrocchelli, D., Nurizzo, D., McSweeney, S. & Mueller-Dieckmann, C. (2009). J. Appl. Cryst. 42, 333–335.
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McPherson, A. (1982). Preparation and Analysis of Protein Crystals, 1st ed. New York: Wiley.
- Murray, M. G. & Thompson, W. F. (1980). Nucleic Acids Res. 8, 4321–4325. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Petkun, S., Jindou, S., Shimon, L. J. W., Rosenheck, S., Bayer, E. A., Lamed, R. & Frolow, F. (2010). Acta Cryst. D66, 33–43. [DOI] [PubMed]
- Saddler, J. N. & Khan, A. W. (1981). Can. J. Microbiol. 27, 288–294. [DOI] [PubMed]
- Saddler, J. N., Khan, A. W. & Martin, S. M. (1980). Microbios, 28, 97–106. [PubMed]
- Sheldrick, G. M. (2010). Acta Cryst. D66, 479–485. [DOI] [PMC free article] [PubMed]
- Shimon, L. J. W., Pagès, S., Belaich, A., Belaich, J.-P., Bayer, E. A., Lamed, R., Shoham, Y. & Frolow, F. (2000). Acta Cryst. D56, 1560–1568. [DOI] [PubMed]
- Sole, V. A., Papillon, E., Cotte, M., Walter, P. & Susini, J. (2007). Spectrochim. Acta B, 62, 63–68.
- Stout, G. H. & Jensen, L. H. (1968). X-ray Structure Determination. A Practical Guide. London: Macmillan.
- Thompson, A. C., Attwood, D. T., Gullikson, E. M., Howells, M. R., Kim, K.-J., Kirz, J., Kortright, J. B., Lindau, I., Pianetta, P., Robinson, A. L., Scofield, J. H., Underwood, J. H., Williams, G. P. & Winik, H. (2001). X-ray Data Booklet, edited by A. C. Thompson & D. Vaughan. Berkeley: Lawrence Berkeley National Laboratory.
- Tormo, J., Lamed, R., Chirino, A. J., Morag, E., Bayer, E. A., Shoham, Y. & Steitz, T. A. (1996). EMBO J. 15, 5739–5751. [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Xu, Q., Barak, Y., Kenig, R., Shoham, Y., Bayer, E. A. & Lamed, R. (2004). J. Bacteriol. 186, 5782–5789. [DOI] [PMC free article] [PubMed]
- Yaniv, O., Shimon, L. J. W., Bayer, E. A., Lamed, R. & Frolow, F. (2011). Acta Cryst. D67, 506–515. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: CBM3b of ScaA, 3zqw
PDB reference: 3zuc
PDB reference: 3zu8