The crystallization of recombinant nitrophorin 7 originating from the bloodsucking insect R. prolixus is described. The crystals diffracted to 1.8 Å resolution.
Keywords: nitrophorins, Rhodnius prolixus, β-barrel proteins, haem, membrane
Abstract
Nitrophorins (nitric oxide transport proteins) are haemproteins originating from the blood-feeding insect Rhodnius prolixus. They consist of an eight-stranded β-barrel, which classifies them into the lipocalin family. Nitrophorin 7 (NP7) and the E27V mutant protein NP7(E27V) were crystallized at 277 K using the vapour-diffusion method with PEG as the precipitating agent. Data sets for wild-type NP7 and NP7(E27V) were collected to 1.80 Å resolution from single crystals at 100 K using synchrotron radiation. The crystals belonged to space group P21, with unit-cell parameters a = 38, b = 67, c = 39 Å, β = 117°. The crystal contained one molecule per asymmetric unit, with a Matthews coefficient (V M) of 2.11 Å3 Da−1; the solvent content was estimated to be 41.8%.
1. Introduction
Nitrophorins (NPs) comprise a unique class of ferrihaem proteins originating from the blood-feeding insect Rhodnius prolixus (Lehane, 2005 ▶). Four nitrophorins, designated NP1–4, have been isolated from the insect saliva (Ribeiro et al., 1993 ▶; Andersen et al., 2005 ▶) and have subsequently been recombinantly expressed (Andersen et al., 1997 ▶; Andersen & Montfort, 2000 ▶). Another nitrophorin, NP7, has recently been established from a cDNA library and recombinantly expressed (Andersen et al., 2004 ▶; Knipp, Zhang et al., 2007 ▶). The major biological function of these proteins is the transport and delivery of NO from the insect saliva to the host tissue, where it acts as a vasodilator and a blood-coagulation inhibitor (Ribeiro & Francischetti, 2003 ▶; Knipp & He, 2011 ▶). NO transport is accomplished through the binding of NO to a haem iron and subsequent release inside the host tissue. The protein experiences a significant pH change from the acidic pH of the saliva (between 5 and 6; Soares et al., 2006 ▶) to that of the blood plasma (∼7.4), which decreases the affinity for NO, for example from >109 M −1 (pH 5.5) to 4 × 106 M −1 (pH 7.5) for NP7 (Knipp, Yang et al., 2007 ▶). Furthermore, it has recently been established that ferrihaem NPs, at least in vitro, are able to produce NO from NO2 −, which is a unique feature among ferrihaem proteins (He & Knipp, 2009 ▶; He et al., 2010 ▶).
In NPs the haem b cofactor is located inside an eight-stranded β-barrel, which is unusual for a haemprotein (Weichsel et al., 1998 ▶). The haem iron is coordinated by a His residue and the sixth coordination site is open for the binding of various ligands including the native ligands NO and histamine (Walker, 2005 ▶). The protein fold has been classified as a lipocalin-type fold (Weichsel et al., 1998 ▶), which is very common in the proteome and is typically found in proteins that bind lipophilic molecules (Flower et al., 2000 ▶). Another common characteristic of the lipocalins is the presence of disulfide bridges, of which the nitrophorins exhibit two (Knipp et al., 2011 ▶).
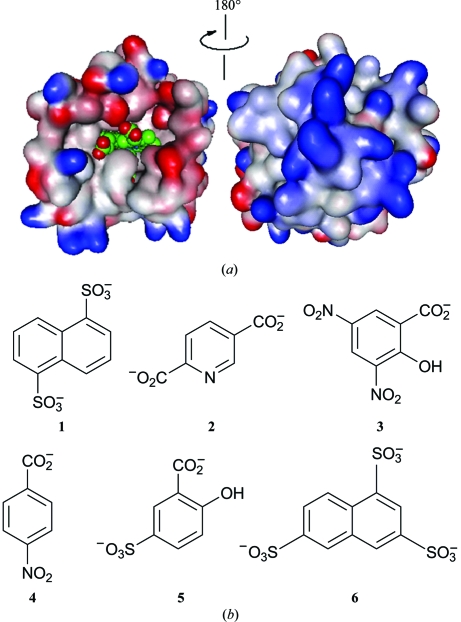
While in the cases of NP1, NP2 and NP4 crystal structures have been determined with numerous ligands bound to the iron [for example, NP2 (Andersen & Montfort, 2000 ▶), NP1, NP1(FeIII–CN−), NP1(FeIII–histamine) (Weichsel et al., 1998 ▶) and NP4(FeIII–NO2 −) (He et al., 2010 ▶)], attempts to crystallize NP7 have failed to date. However, NP7 is a particularly interesting case among the NPs because it is the only type that attaches to cell membranes (Andersen et al., 2004 ▶; Knipp, Zhang et al., 2007 ▶). This interaction is accomplished by a positively charged surface opposite the haem-binding cleft, which is evident from homology modelling (Knipp, Yang et al., 2007 ▶) in combination with surface plasmon resonance experiments (Andersen et al., 2004 ▶). The binding to negatively charged phospholipid surfaces is accomplished via this surface structure (see Fig. 1 ▶ a). The positively charged surface is established by a spatial clustering of Lys residues, which leads to a marked increase in the pI of NP7 (calculated as 8.9) compared with those of NP1–4 (calculated as between 6.1 and 6.3) (Knipp, Yang et al., 2007 ▶).
Figure 1.
(a) Representation of the electrostatic surface potential of the homology model of NP7 (Knipp, Yang et al., 2007 ▶). Blue indicates positive charge potential and red indicates negative charge potential. The haem is indicated in van der Waals representation (green). The electrostatic surface potential was calculated and the figure was prepared with Swiss-PdbViewer (Guex & Peitsch, 1997 ▶) and the figure was rendered with POV-Ray (http://www.povray.org/). (b) Reagents from the commercial Silver Bullets screen (Hampton Research) that successfully promoted crystallization of wt NP7 and NP7(E27V).
Structural information is key to detailed understanding of the NP7–membrane interaction. Therefore, on the basis of this information, a novel crystallization screen which takes the strong charge dipolar nature of NP7 into account was successfully applied. The screen was performed using wild-type (wt) NP7 and the partly characterized E27V mutant protein NP7(E27V) (Yang, Zhang et al., 2009 ▶). Glu27 is a unique residue in the NP7 haem pocket which is surprisingly directed towards the hydrophobic site of the haem cofactor. All other NPs instead have a Val residue at this position. It has previously been shown that mutation of Glu27 to Val has a great influence on the properties of the haem; for example, it determines the orientation of the haem in the pocket (Yang, Knipp et al., 2009 ▶; Yang, Zhang et al., 2009 ▶). In order to understand the role of Glu27 in NP7, crystallization of NP7(E27V) is required.
2. Materials and methods
2.1. Purification
wt NP7 and NP7(E27V) were expressed in Escherichia coli in inclusion bodies and the protein was reconstituted using previously reported methods (Knipp, Zhang et al., 2007 ▶; Yang, Zhang et al., 2009 ▶). All purification procedures were carried out at 277 K. In brief, upon insertion of the haem cofactor the protein was purified using a size-exclusion column (Sephacryl HR 100, 26 × 600 mm, GE Healthcare) with the running buffer 200 mM sodium acetate/acetic acid pH 5.5, 2% glycerol. A second chromatographic step using Ca2+-loaded Chelating Sepharose (GE Healthcare) with gradient elution (0–40 mM CaCl2) was then applied (Knipp, work to be published). The protein preparations were judged by SDS–PAGE to be >90% pure. Proteins were also subjected to MALDI–TOF MS to confirm the correct molecular masses (calculations consider the presence of two Cys–Cys disulfides): calculated for [wt NP7 + H]+ 20 969, observed 20 966 ± 20; calculated for [NP7(E27V) + H]+ 20 939, observed 20 939 ± 20. Proteins were kept at 253 K in 200 mM sodium acetate/acetic acid, 10%(v/v) glycerol pH 5.0 until use.
2.2. Crystallization
The purified proteins were concentrated to 5 mg ml−1 and dialyzed against Milli Q water. The crystallization of wt NP7 and NP7(E27V) from R. prolixus was carried out using the sitting-drop vapour-diffusion method at 277 K. Initial crystallization screening was performed using a Crystal Phoenix (Art Robbins Instruments, California, USA) liquid-handling system. The screening kits used were Silver Bullets HT, PEG/Ion and PEG/Ion 2 (Hampton Research, California, USA). The protein solution and the buffer were dispensed with 0.2 µl droplets each. For the NP7(E27V) sample, Silver Bullets HT pH 7.8 was used. After 4–6 d, red-coloured crystals were obtained. Crystals suitable for diffraction experiments were obtained under the following conditions: 25%(w/v) PEG 3350, 0.1 M bis-tris propane pH 7.8, 0.02 M HEPES pH 6.8, 0.03%(w/v) 1,5-naphthalenedisulfonic acid disodium salt (1); 0.33%(w/v) 2,5-pyridinedicarboxylic acid (2); 0.33%(w/v) 3,5-dinitrosalicylic acid (3). For the crystal of NP7(E27V) the conditions were 25%(w/v) PEG 3350, 0.1 M bis-tris propane pH 7.8, 0.02 M HEPES pH 6.8, 0.25%(w/v) 2,6-naphthalenedisulfonic acid disodium salt (1); 0.25%(w/v) 4-aminobenzoic acid (4); 0.25%(w/v) 5-sulfosalicylic acid dihydrate (5); 0.25%(w/v) naphthalene-1,3,6-trisulfonic acid trisodium salt hydrate (6). The structures of the numbered compounds are shown in Fig. 1 ▶(b). The dimensions of the wt NP7 and NP7(E27V) crystals were typically 0.1 × 0.02 × 0.01 and 0.3 × 0.01 × 0.01 mm, respectively (Fig. 2 ▶).
Figure 2.
Crystals of NP7 from R. prolixus: (a) wt NP7 crystals, (b) NP7(E27V) crystals.
2.3. Data collection and analysis
The crystals used for data collection were directly taken from the initial screen batches described above. In order to collect data at cryogenic temperature, crystals that had been soaked in cryoprotectant (15% glycerol in the crystallization buffer) were cooled in liquid nitrogen and mounted on the goniostat under a nitrogen-gas stream at 100 K. A data set was collected on beamline BL14.2 at BESSY II (Berlin, Germany). The detector was a Rayonix MX-225 (Rayonix, Illinois, USA). Complete data sets for both wt and E27V mutant NP7 were collected to 1.80 Å resolution at an X-ray wavelength of 0.91841 Å. For each data set, 180 frames of 3.2 s exposure time and 1.0° oscillation were collected. The flux of the X-ray beam was approximately 3 × 1011 photons s−1. The distance between the crystal and the detector was maintained at 180 mm for wt NP7 and at 170 mm for NP7(E27V). Diffraction images were indexed and integrated using the program XDS (Kabsch, 2010 ▶). Scaling was carried out using XSCALE (Kabsch, 2010 ▶). Molecular replacement and initial refinement were carried out using the programs MOLREP (Vagin & Teplyakov, 2010 ▶) and REFMAC5 (Murshudov et al., 2011 ▶), respectively, from the CCP4 program package (Winn et al., 2011 ▶). The data-collection conditions and the results obtained are summarized in Table 1 ▶.
Table 1. X-ray data-collection statistics for wt NP7 and NP7(E27V).
Values in parentheses are for the highest resolution shell.
| wt NP7 | NP7(E27V) | |
|---|---|---|
| Wavelength (Å) | 0.91841 | 0.91841 |
| Space group | P21 | P21 |
| Unit-cell parameters (Å, °) | a = 38.13, b = 66.70, c = 38.93, α = 90.0, β = 116.7, γ = 90.0 | a = 38.23, b = 66.88, c = 38.71, α = 90.0, β = 116.6, γ = 90.0 |
| Resolution (Å) | 32.79–1.80 (1.85–1.80) | 34.63–1.80 (1.85–1.80) |
| No. of observed reflections | 60743 (4503) | 60943 (4524) |
| No. of unique reflections | 16059 (1185) | 15776 (1166) |
| Rmrgd-F† | 0.062 (0.310) | 0.083 (0.433) |
| Completeness (%) | 98.7 (97.9) | 96.8 (96.0) |
| 〈I/σ(I)〉 | 25.2 (6.6) | 20.4 (4.4) |
R mrgd-F is the quality of the amplitudes (F) in the scaled data set (see Diederichs & Karplus, 1997 ▶).
3. Results and discussion
Both wt NP7 and NP7(E27V) were successfully overexpressed in E. coli and purified using two chromatographic steps. The second chromatographic purification step is mandatory to obtain the degree of purity required for crystallization. Crystals of NP7 were obtained at 277 K using the sitting-drop vapour-diffusion method with PEG as the precipitating agent. In the early stages of the crystallization attempts we found no crystals using the PEG/Ion screening kits. However, crystals were obtained in the presence of the Silver Bullets HT compounds shown in Fig. 1(b) ▶. All of the compounds are monoanions, dianions or trianions and thus can compensate for the high positive surface-charge density that is responsible for the interaction between the negatively charged phospholipid membrane and NP7.
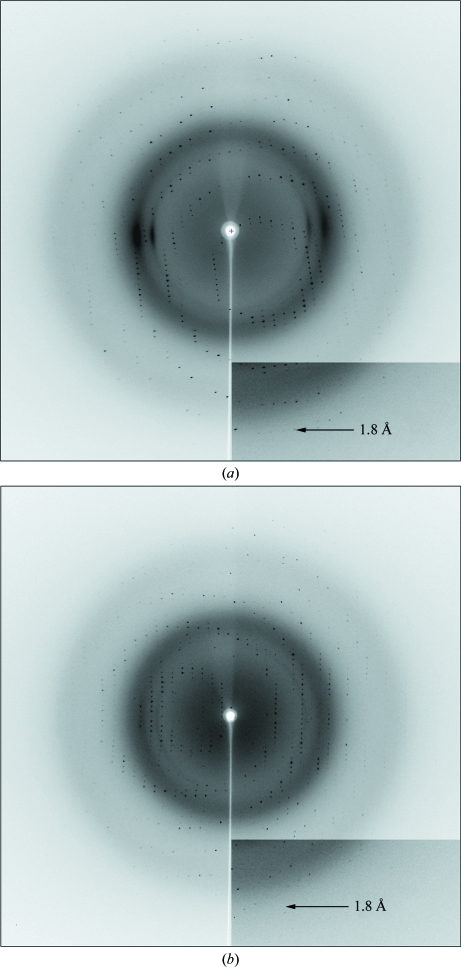
The crystals of both wt NP7 and NP7(E27V) diffracted to 1.80 Å resolution (Fig. 3 ▶) and, like those of other nitrophorins (Andersen & Montfort, 2000 ▶; Weichsel et al., 1998 ▶; He et al., 2010 ▶), belonged to space group P21. The unit-cell parameters of wt NP7 were a = 38.13, b = 66.70, c = 38.93 Å, β = 116.7°. The calculated Matthews coefficient (V M) of 2.11 Å3 Da−1 with a solvent content of 41.8% indicates the presence of one molecule in the asymmetric unit. The NP7(E27V) crystals had similar unit-cell parameters as those of wt NP7 (see Table 1 ▶).
Figure 3.
Diffraction patterns: (a) wt NP7, (b) NP7(E27V).
The molecular-replacement method was applied using the program MOLREP. The coordinates of NP2 from R. prolixus (PDB entry 2eu7; A. Weichsel, R. E. Berry, F. A. Walker & W. R. Montfort, unpublished work) were used as a search model. After calculation of the electron-density map using the molecular-replacement solution, the presence of one monomer in the asymmetric unit was confirmed. In both wt NP7 and NP7(E27V) the N-terminal amino acids (Pro2 and Gly3), which do not exist in NP1–4 (Knipp, Yang et al., 2007 ▶), were observed in the electron-density map. However, the initial residues Met0 and Leu1 could not be observed in the electron-density map. In contrast, the C-terminal region (Lys185) was confirmed. A total of 184 amino-acid residues were successfully confirmed. Model building and refinement are now in progress.
Acknowledgments
We are grateful to Johanna J. Taing and Yvonne Brandenburger (MPI for Bioinorganic Chemistry, Mülheim, Germany) for sample preparation. We thank Dr Koji Nishikawa (MPI for Bioinorganic Chemistry, Mülheim, Germany) for his help during data collection. We thank the staff of beamline BL14.2 at BESSY II (Helmholtz-Zentrum Berlin, Berlin, Germany) for their assistance during data collection. This work was financially supported by the Max Planck Society.
References
- Andersen, J. F., Champagne, D. E., Weichsel, A., Ribeiro, J. M., Balfour, C. A., Dress, V. & Montfort, W. R. (1997). Biochemistry, 36, 4423–4428. [DOI] [PubMed]
- Andersen, J. F., Gudderra, N. P., Francischetti, I. M. & Ribeiro, J. M. (2005). Arch. Insect Biochem. Physiol. 58, 97–105. [DOI] [PMC free article] [PubMed]
- Andersen, J. F., Gudderra, N. P., Francischetti, I. M., Valenzuela, J. G. & Ribeiro, J. M. (2004). Biochemistry, 43, 6987–6994. [DOI] [PMC free article] [PubMed]
- Andersen, J. F. & Montfort, W. R. (2000). J. Biol. Chem. 275, 30496–30503. [DOI] [PubMed]
- Diederichs, K. & Karplus, P. A. (1997). Nature Struct. Biol. 4, 269–275. [DOI] [PubMed]
- Flower, D. R., North, A. C. & Sansom, C. E. (2000). Biochim. Biophys. Acta, 1482, 9–24. [DOI] [PubMed]
- Guex, N. & Peitsch, M. C. (1997). Electrophoresis, 18, 2714–2723. [DOI] [PubMed]
- He, C. & Knipp, M. (2009). J. Am. Chem. Soc. 131, 12042–12043. [DOI] [PubMed]
- He, C., Ogata, H. & Knipp, M. (2010). Biochemistry, 49, 5841–5851. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Knipp, M. & He, C. (2011). IUBMB Life, 63, 304–312. [DOI] [PubMed]
- Knipp, M., Taing, J. J. & He, C. (2011). J. Inorg. Biochem. 105, 1405–1412. [DOI] [PubMed]
- Knipp, M., Yang, F., Berry, R. E., Zhang, H., Shokhirev, M. N. & Walker, F. A. (2007). Biochemistry, 46, 13254–13268. [DOI] [PMC free article] [PubMed]
- Knipp, M., Zhang, H., Berry, R. E. & Walker, F. A. (2007). Protein Expr. Purif. 54, 183–191. [DOI] [PMC free article] [PubMed]
- Lehane, M. J. (2005). The Biology of Blood-Sucking in Insects, 2nd ed. Cambridge University Press.
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Ribeiro, J. M. & Francischetti, I. M. (2003). Annu. Rev. Entomol. 48, 73–88. [DOI] [PubMed]
- Ribeiro, J. M., Hazzard, J. M., Nussenzveig, R. H., Champagne, D. E. & Walker, F. A. (1993). Science, 260, 539–541. [DOI] [PubMed]
- Soares, A. C., Carvalho-Tavares, J., Gontijo, N. de F., dos Santos, V. C., Teixeira, M. M. & Pereira, M. H. (2006). J. Insect Physiol. 52, 468–472. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Walker, F. A. (2005). J. Inorg. Biochem. 99, 216–236. [DOI] [PubMed]
- Weichsel, A., Andersen, J. F., Champagne, D. E., Walker, F. A. & Montfort, W. R. (1998). Nature Struct. Biol. 5, 304–309. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yang, F., Knipp, M., Berry, B., Shokhireva, K., Zhang, H. & Walker, F. A. (2009). J. Biol. Inorg. Chem. 14, 1077–1095. [DOI] [PMC free article] [PubMed]
- Yang, F., Zhang, H. & Knipp, M. (2009). Biochemistry, 48, 235–241. [DOI] [PubMed]