NahF is a salicylaldehyde dehydrogenase that is involved in the naphthalene-degradation pathway, converting salicylaldehyde into salicylate. The subcloning, expression, purification and preliminary X-ray diffraction studies at 2.42 Å resolution of P. putida G7 NahF are reported.
Keywords: Pseudomonas putida G7, naphthalene degradation, salicylaldehyde dehydrogenase, NahF
Abstract
Pseudomonas putida G7 is one of the most studied naphthalene-degrading species. The nah operon in P. putida, which is present on the 83 kb metabolic plasmid NAH7, codes for enzymes involved in the conversion of naphthalene to salicylate. The enzyme NahF (salicylaldehyde dehydrogenase) catalyzes the last reaction in this pathway. The nahF gene was subcloned into the pET28a(TEV) vector and the recombinant protein was overexpressed in Escherichia coli Arctic Express at 285 K. The soluble protein was purified by affinity chromatography followed by gel filtration. Crystals of recombinant NahF (6×His-NahF) were obtained at 291 K and diffracted to 2.42 Å resolution. They belonged to the hexagonal space group P6422, with unit-cell parameters a = b = 169.47, c = 157.94 Å. The asymmetric unit contained a monomer and a crystallographic twofold axis generated the dimeric biological unit.
1. Introduction
Polycyclic aromatic hydrocarbons (PAHs) are a group of aromatic hydrocarbons composed of two or more fused rings and include naphthalene, anthracene, phenanthrene, pyrene and benzo[a]pyrene amongst others (Haritash & Kaushik, 2009 ▶). PAHs from natural and anthropogenic sources are widely distributed in the environment (Kanaly & Harayama, 2000 ▶). These compounds have a high stability and a low solubility in water, which contribute to their persistence in the environment (Samanta et al., 2002 ▶; Haritash & Kaushik, 2009 ▶).
Exposure to naphthalene has been associated with several toxic manifestations in humans and laboratory animals, with the lens of the eye and the lungs being most sensitive (Stohs et al., 2002 ▶). Additionally, this compound has been reclassified as a possible human carcinogen by the US Environmental Protection Agency (EPA, 2004 ▶) and the International Agency for Research on Cancer (IARC, 2002 ▶). It has recently been suggested that naphthalene undergoes metabolic activation to 1,2-naphthoquinone, which reacts with DNA, leading to the formation of apurinic sites on DNA and depurinating DNA adducts (Saeed et al., 2009 ▶).
Moreover, a number of studies have attested to the carcinogenic and mutagenic properties of more complex PAHs (Park et al., 2006 ▶; Spink et al., 2008 ▶; Topinka et al., 2008 ▶), suggesting the need for further work on the elimination of these compounds from the environment. Bioremediation might be a potential strategy to achieve this purpose (Kanaly & Harayama, 2000 ▶). Briefly, this strategy involves the use of microorganisms or enzymes capable of metabolizing toxic compounds, such as PAHs, into substances that have little or no toxicity (Holliger et al., 1997 ▶).
Bacterial strains such as those from the genera Mycobacterium, Corynebacterium, Flavobacterium, Rhodococcus, Bacillus and particularly Pseudomonas that are able to degrade aromatic hydrocarbons have been widely described (Cerniglia, 1984 ▶; Annweiler et al., 2000 ▶; Kanaly & Harayama, 2000 ▶; Arun et al., 2008 ▶). Species of the genus Pseudomonas are metabolically versatile and use a large number of organic compounds as carbon sources, including aromatic hydrocarbons such as naphthalene (Rosselló-Mora et al., 1994 ▶).
There are several reports in the literature of complete nucleotide sequences coding for enzymes involved in naphthalene-degradation pathways in bacteria. The chromosome of P. stutzeri AN10 and plasmids from P. putida ND6, P. putida NCIB 9816-4 and P. putida G7 contain the genes of this pathway (Bosch et al., 1999 ▶; Dennis & Zylstra, 2004 ▶; Li et al., 2004 ▶).
The Gram-negative bacterium P. putida G7 is among the best studied naphthalene-degrading species. The genes associated with naphthalene metabolism are localized on NAH7, an 83 kilobase plasmid (Yen & Gunsalus, 1982 ▶). Other species also have naphthalene catabolic plasmids that are quite similar to NAH7: pWW60 derivatives from P. putida strain NCIB 9816 (Yang et al., 1994 ▶), pDTG1 from P. putida strain NCIB 9816-4 (Dennis & Zylstra, 2004 ▶) and pKA1 from P. fluorescens strain 5R (Habe & Omori, 2003 ▶).
In P. putida G7 the naphthalene-oxidation genes are organized into two operons under salicylate control. The first operon, which is associated with the upper naphthalene-degradation pathway, includes the genes nahAaAbAcAdBCDEF, which code for the conversion of naphthalene to salicylate, while the second operon includes the genes nahGTHINLOMKJ coding for the lower pathway that includes the oxidation of salicylate via the catechol meta-cleavage pathway (Yen & Gunsalus, 1985 ▶).
The first step in naphthalene catabolism is the incorporation of molecular oxygen into the aromatic nucleus of naphthalene to produce cis-1,2-dihydroxy-1,2-dihydronaphthalene (Fig. 1 ▶). This reaction is catalyzed by the naphthalene dioxygenase (NDO) complex, which is composed of four proteins: ferredoxin reductase (NahAa), ferredoxin (NahAb) and two iron–sulfur proteins (NahAc and NahAd). The four subsequent reactions are catalyzed by cis-naphthalene dihydrodiol dehydrogenase (NahB), 1,2-dihydroxynaphthalene dioxygenase (NahC), 2-hydroxy-2H-chromene-2-carboxylate isomerase (NahD) and trans-o-hydroxybenzylidenepyruvic hydratase-aldolase (NahE), resulting in the formation of salicylaldehyde (Davies & Evans, 1964 ▶).
Figure 1.
Naphthalene-degradation upper pathway. The upper pathway is composed of NahAaAbAcAd (naphthalene dioxygenase complex; NDO), cis-naphthalene dihydrodiol dehydrogenase (NahB), 1,2-dihydroxynaphthalene dioxygenase (NahC), 2-hydroxy-2H-chromene-2-carboxylate isomerase (NahD), trans-o-hydroxybenzylidenepyruvic hydratase-aldolase (NahE) and salicylaldehyde dehydrogenase (NahF). The main substrates and products for these enzymes are also depicted. (1) Naphthalene, (2) cis-1,2-dihydroxy-1,2-dihydronaphthalene, (3) 1,2-dihydroxynaphthalene, (4) 2-hydroxy-2H-chromene-2-carboxylic acid, (5) trans-o-hydroxybenzylidenepyruvic acid, (6) salicylaldehyde, (7) pyruvate and (8) salicylic acid (adapted from Peng et al., 2008 ▶).
The enzyme NahF (salicylaldehyde dehydrogenase; EC 1.2.1.65; 483 amino acids) catalyzes the last reaction of the upper pathway converting salicylaldehyde to salicylate (Davies & Evans, 1964 ▶; Eaton & Chapman, 1992 ▶). This enzyme belongs to the aldehyde dehydrogenase (ALDH) superfamily, the members of which oxidize a wide variety of aliphatic and aromatic aldehydes using NAD+ or NADP+ as a cofactor.
Despite the fact that aldehyde dehydrogenases may share a similar scaffold and modes of oligomerization (dimers or tetramers; Perozich, Nicholas, Lindahl et al., 1999 ▶), substrate specificity dictates some of the differences between these enzymes. The salicylaldehyde dehydrogenases, for example, are classified as a family of ALDHs or aromatic ALDHs that oxidize relatively specific aromatic xenobiotics found in the environment (Perozich, Nicholas, Wang et al., 1999 ▶). Also belonging to this family are the benzaldehyde dehydrogenase from P. putida (Yeung et al., 2008 ▶) and vanillin dehydrogenase from Pseudomonas sp. strain HR199 (Priefert et al., 1997 ▶).
Here, we report the subcloning and expression of recombinant NahF (6×His-NahF) from P. putida G7. To elucidate the structural motifs that may distinguish this enzyme from other dehydrogenases, we purified the recombinant protein and aimed to crystallize and collect preliminary diffraction data from 6×His-NahF crystals. As far as we are aware, this is the first crystallization report of an aromatic ALDH.
2. Materials and methods
2.1. Subcloning, protein expression and purification
The NAH7 plasmid from P. putida G7 (GenBank nucleotide entry NC_007926.1) was used as the polymerase chain reaction (PCR) template. The primers used to amplify the salicylaldehyde dehydrogenase gene from the NAH7 plasmid were nahF-F, 5′-CAT ATG AAG ACA AAA CTG TTT ATC AAC AAC-3′, and nahF-R, 5′-CTC GAG TTC AGA AGG GAT ATT GCT GCT-3′. The sites for restriction enzymes (NdeI and XhoI, respectively) are shown in bold.
The DNA fragments obtained were excised from the gel, purified and ligated into pGEM vector (Promega Corporation). Recombinant plasmid pGEM-nahF was used to transform Escherichia coli DH5a competent cells. Positive transformants were tested by restriction analysis with NdeI and XhoI and those presenting the nahF gene were propagated and used for construction of the expression vector. DNA fragments obtained from digestion of pGEM-nahF with NdeI and XhoI were ligated into pET28a(TEV) vector (Carneiro et al., 2006 ▶) previously digested with the same enzymes. pET28a(TEV) is a modified pET28a vector (Novagen) that adds a 6×His tag followed by a tobacco etch virus (TEV) protease-cleavage site at the N-terminus of the target protein (MGHHHHHHENLYFQGH–target protein). Electrocompetent E. coli BL21 Arctic Express (DE3) cells (Agilent Technologies) were transformed by electroporation using a MicroPulser Electroporation Apparatus (Bio-Rad Laboratories) with the recombinant plasmid pET28a(TEV)-nahF. Gene insertion was confirmed by colony PCR and sequencing using T7 primers in a MegaBACE instrument (GE Healthcare Life Sciences).
Transformed E. coli BL21 Arctic Express (DE3) cells were inoculated in 10 ml LB broth supplemented with 40 µg ml−1 gentamicin and 50 µg ml−1 kanamycin. Overnight cultures were transferred to 500 ml LB broth without antibiotics and cultivated at 303 K for 3 h. Expression was then induced by addition of IPTG to a final concentration of 0.5 mM and the culture was incubated at 285 K for 24 h at 200 rev min−1.
The bacterial cells were harvested by centrifugation and were resuspended in 20 ml lysis buffer [50 mM Tris–HCl pH 7.4, 1%(w/v) sucrose, 1%(v/v) Tween 20, 1%(v/v) glycerol, 0.25%(w/v) lysozyme]. Cells were ruptured by three cycles of freezing and thawing followed by three cycles of 30 s on/off sonication in an ice–water bath at 30% amplitude. Subsequently, debris was removed by centrifugation at 277 K for 30 min at 10 000g.
Imidazole was added to the clear supernatant to a final concentration of 30 mM and the solution was immediately loaded onto a 5 ml HisTrap HP affinity column (GE Healthcare Biosciences) pre-equilibrated with five column volumes of equilibration buffer (50 mM sodium phosphate buffer pH 7.4, 500 mM NaCl, 30 mM imidazole) and connected to an ÄKTAprime chromatography system (GE Healthcare Life Sciences). The column was washed at a flow rate of 1.0 ml min−1 with five column volumes of equilibration buffer. Subsequently, gradient elution with elution buffer (50 mM sodium phosphate buffer pH 7.4, 500 mM NaCl, 500 mM imidazole) at a flow rate of 5.0 ml min−1 was used to elute the retained proteins.
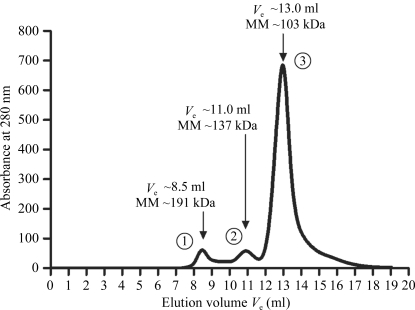
The eluted fractions containing the recombinant NahF (6×His-NahF, 499 amino acids, 54 kDa) from the affinity chromatography were extensively dialyzed against 20 mM sodium phosphate buffer pH 7.4 containing 20 mM NaCl. Recombinant NahF was further purified on a Superdex 200 gel-filtration column (GE Healthcare) previously equilibrated with 20 mM sodium phosphate buffer pH 7.4 with 20 mM NaCl and eluted with the same solution (Fig. 2 ▶). The peak fractions containing purified 6×His-NahF were analyzed by a dynamic light-scattering (DLS) experiment and the main peak fractions (peak 3, Fig. 2 ▶) were pooled and concentrated using a dialysis membrane covered with PEG 8000 powder until a final protein concentration of 15 mg ml−1 was achieved. This final 6×His-NahF protein solution was used in crystallization trials.
Figure 2.
Typical size-exclusion chromatography chromatogram obtained with recombinant NahF. The elution of recombinant NahF in peak 3 (the main peak) corresponds to the profile of a 103 kDa protein and is in good agreement with a dimeric form of 6×His-NahF (108 kDa). The elution volume (V e) and molecular mass (MM) associated with each peak (1, 2 and 3) are shown.
2.2. Dynamic light-scattering and crystallization assays
The oligomerization state of 6×His-NahF at 2.5 mg ml−1 was assessed by a DLS experiment using a DynaPro instrument (Wyatt Technology). The sample tested consisted of elution fractions corresponding to peak 3 from the size-exclusion chromatography (Fig. 2 ▶).
Initial crystallization attempts were carried out at 291 K using 24-well plates and the hanging-drop vapour-diffusion method. Drops consisted of 1 µl final 6×His-NahF protein solution and 1 µl of the screening solutions from Crystal Screen and Crystal Screen 2 (Hampton Research) and were equilibrated against 1 ml reservoir solution (screening solution). Initial crystals were only observed in condition No. 5 from Crystal Screen 2, consisting of 2.0 M ammonium sulfate and 5%(v/v) 2-propanol. The condition was optimized by grid screening of the ammonium sulfate concentration and the addition of sodium acetate–acetic acid buffer and β-nicotinamide adenine dinucleotide hydrate (NAD+; Sigma–Aldrich) to the crystallization condition. The flower-shaped crystals reached a maximum dimension of 500 µm after 2 d in 2 µl drops equilibrated against 1 ml reservoir solution (Fig. 3 ▶).
Figure 3.
Flower-shaped crystals of recombinant NahF. Crystals were obtained using 1.5 M ammonium sulfate, 5%(v/v) 2-propanol, 5 mM NAD+ in 100 mM sodium acetate–acetic acid buffer pH 5.0.
2.3. Data collection, processing and phase determination
Prior to data acquisition, a flower-shaped 6×His-NahF crystal was carefully broken and a smaller piece (a single ‘petal’) was harvested using a nylon loop (Hampton Research). The ‘petal’ was then transferred for a few seconds into a 10 µl drop of a cryogenic solution formed by the addition of 1 µl ethylene glycol to 9 µl reservoir solution. The crystal was then flash-cooled to 100 K in a cold nitrogen stream and used for data acquisition. X-ray diffraction images were recorded on a MAR CCD 165 detector using the D03B-MX1 beamline (Polikarpov et al., 1998 ▶) at the Laboratório Nacional de Luz Síncrotron (LNLS), Brazil. Each image was exposed for 60 s with a rotation range of 0.5°.
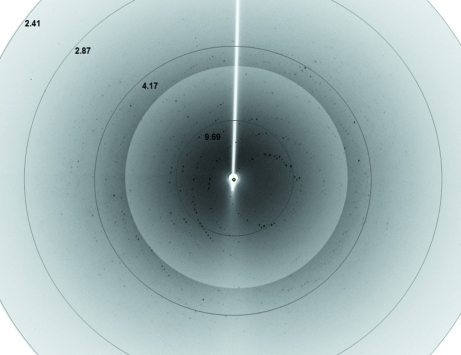
Diffraction images (Fig. 4 ▶) were processed using iMOSFLM (Battye et al., 2011 ▶) and SCALA (Evans, 2006 ▶) as distributed in the CCP4 package (Winn et al., 1994 ▶). Diffraction data statistics are shown in Table 1 ▶.
Figure 4.
A diffraction image from a broken piece of a flower-shaped 6×His-NahF crystal. Diffraction data were collected to 2.42 Å resolution. The diffraction pattern background around the centre of the image was altered to allow a better visualization of the reflections. Resolution shells are shown (in Å) close to their respective circles.
Table 1. Summary of diffraction data statistics for the 6×His-NahF crystal.
Values in parentheses are for the lowest/highest resolution shells.
| Wavelength (Å) | 1.608 |
| Temperature (K) | 100 |
| Crystal-to-detector distance (mm) | 100 |
| Rotation range per image (°) | 0.5 |
| Total rotation range (°) | 120 |
| Space group | P6422 |
| Unit-cell parameters (Å) | a = b = 169.47, c = 157.94 |
| Resolution range (Å) | 32.17–2.42 (32.17–7.64/2.55–2.42) |
| No. of observations | 710305 (23697/97367) |
| No. of unique reflections | 51558 (1811/7385) |
| Data completeness (%) | 99.9 (98.7/99.8) |
| 〈I/σ(I)〉 | 10.4 (34.8/4.0) |
| Multiplicity | 13.8 (13.1/13.2) |
| Rmerge† | 0.272 (0.062/0.651) |
| Average mosaicity (°) | 0.38 |
| Wilson B factor (Å2) | 18.8 |
| Matthews coefficient (Å3 Da−1) | 6.06 |
| Solvent content (%) | 79.7 |
R
merge = 
 , where Ii(hkl) is the ith intensity measurement of reflection hkl and 〈I(hkl)〉 is its average.
, where Ii(hkl) is the ith intensity measurement of reflection hkl and 〈I(hkl)〉 is its average.
The molecular-replacement (MR) method, as implemented in Phaser (McCoy et al., 2007 ▶), was used to solve the phase problem with a single monomer of the sheep liver cytosolic aldehyde dehydrogenase crystallographic structure (PDB entry 1bxs; Moore et al., 1998 ▶) as the search template. This protein template was chosen as its primary structure shows one of the highest identities (37%) to the NahF amino-acid sequence.
Preliminary 6×His-NahF model building was performed using the programs Buccaneer (Cowtan, 2006 ▶) and Coot (Emsley et al., 2010 ▶). Restrained refinement of the model was performed by the reciprocal-space refinement program REFMAC (Murshudov et al., 2011 ▶).
3. Results and discussion
The recombinant salicylaldehyde dehydrogenase from P. putida G7 was overexpressed in E. coli BL21 Arctic Express cells in a soluble form. Ni2+-affinity and size-exclusion chromatography (SEC) were used to obtain high-purity 6×His-NahF with a yield of approximately 40 mg purified protein per litre of culture. Several attempts to cleave the 6×His tag from the recombinant protein were unsuccessful and subsequent experiments were carried out with 6×His-NahF.
The biological unit of 6×His-NahF appears to be a dimer as suggested by the elution profile in the SEC chromatogram (Fig. 2 ▶). This finding is in good agreement with the dynamic light-scattering results, in which the monodisperse sample from SEC peak 3 (with a polydispersity of 14%) exhibits particles with a hydrodynamic radius of 4.7 nm (suggested protein molecular mass of 125 kDa).
Purified 6×His-NahF was concentrated to 15.0 mg ml−1 and flower-shaped crystals suitable for diffraction (maximum dimension of 500 µm; Fig. 3 ▶) were grown in 1.5 M ammonium sulfate, 5%(v/v) 2-propanol, 5 mM NAD+ in 100 mM sodium acetate–acetic acid buffer pH 5.0. Diffraction data collected to 2.42 Å resolution from a broken piece of a flower-shaped crystal indicated that 6×His-NahF crystallized in space group P6422, with unit-cell parameters a = b = 169.47, c = 157.94 Å (Table 1 ▶).
Calculation of the Matthews coefficient suggests the presence of two protein molecules in the asymmetric unit. Surprisingly, all molecular-replacement attempts aiming to find a dimer in the asymmetric unit were unsuccessful. A good MR solution (translation-function Z score of 24.7) was only obtained when a monomer was expected in the asymmetric unit (V M = 6.06 Å3 Da−1; solvent content of almost 80%). This solution and the correct recombinant NahF amino-acid sequence were used as input files in the Buccaneer software (Cowtan, 2006 ▶) for automated model building. The resulting PDB file was manually adjusted in Coot (Emsley et al., 2010 ▶) and refined against the original data set in REFMAC (Murshudov et al., 1997 ▶), achieving R and R free values of 25.7% and 28.9%, respectively, prior to the modelling of water molecules.
Inspection of crystal packing shows that a solvent tunnel along the 64 screw axis of approximately 90 Å in diameter is responsible for the high solvent content of this crystal. Even though a monomer was observed in the asymmetric unit, the 6×His-NahF biological unit (a dimer) could be generated by a crystallographic twofold axis. The detailed structure of recombinant NahF will be published elsewhere.
Acknowledgments
We would like to thank Dr Masataka Tsuda and Dr Masahiro Sota for kindly providing us with the P. putida G7 strain. This work was supported by the Laboratório Nacional de Luz Síncrotron (LNLS; Brazilian Synchrotron Light Laboratory; Project 6970), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq; Project 482173/2010-6) and Fundação de Desenvolvimento à Pesquisa do Estado de Minas Gerais (FAPEMIG; Projects EDT-0185-0.00-07, Rede-170/08 and RDP-00174-10). JBC, DMAC and SLG are recipients of scholarships from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
References
- Annweiler, E., Richnow, H. H., Antranikian, G., Hebenbrock, S., Garms, C., Franke, S., Francke, W. & Michaelis, W. (2000). Appl. Environ. Microbiol. 66, 518–523. [DOI] [PMC free article] [PubMed]
- Arun, A., Raja, P. P., Arthi, R., Ananthi, M., Kumar, K. S. & Eyini, M. (2008). Appl. Biochem. Biotechnol. 151, 132–142. [DOI] [PubMed]
- Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. (2011). Acta Cryst. D67, 271–281. [DOI] [PMC free article] [PubMed]
- Bosch, R., García-Valdés, E. & Moore, E. R. (1999). Gene, 236, 149–157. [DOI] [PubMed]
- Carneiro, F. R., Silva, T. C., Alves, A. C., Haline-Vaz, T., Gozzo, F. C. & Zanchin, N. I. (2006). Biochem. Biophys. Res. Commun. 343, 260–268. [DOI] [PubMed]
- Cerniglia, C. E. (1984). Adv. Appl. Microbiol. 30, 31–71. [DOI] [PubMed]
- Cowtan, K. (2006). Acta Cryst. D62, 1002–1011. [DOI] [PubMed]
- Davies, J. I. & Evans, W. C. (1964). Biochem. J. 91, 251–261. [DOI] [PMC free article] [PubMed]
- Dennis, J. J. & Zylstra, G. J. (2004). J. Mol. Biol. 341, 753–768. [DOI] [PubMed]
- Eaton, R. W. & Chapman, P. J. (1992). J. Bacteriol. 174, 7542–7554. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- EPA (2004). Toxicological Review of Naphthalene http://oaspub.epa.gov/eims/eimscomm.getfile?p_download_id=501313.
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Habe, H. & Omori, T. (2003). Biosci. Biotechnol. Biochem. 67, 225–243. [DOI] [PubMed]
- Haritash, A. K. & Kaushik, C. P. (2009). J. Hazard. Mater. 169, 1–15. [DOI] [PubMed]
- Holliger, C., Gaspard, S., Glod, G., Heijman, C., Schumacher, W., Schwarzenbach, R. P. & Vazquez, F. (1997). FEMS Microbiol. Rev. 20, 517–523. [DOI] [PubMed]
- IARC (2002). IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, Vol. 82. Lyon: IARC.
- Kanaly, R. A. & Harayama, S. (2000). J. Bacteriol. 182, 2059–2067. [DOI] [PMC free article] [PubMed]
- Li, W., Shi, J., Wang, X., Han, Y., Tong, W., Ma, L., Liu, B. & Cai, B. (2004). Gene, 336, 231–240. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Moore, S. A., Baker, H. M., Blythe, T. J., Kitson, K. E., Kitson, T. M. & Baker, E. N. (1998). Structure, 6, 1541–1551. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Park, S.-Y., Lee, S.-M., Ye, S.-K., Yoon, S.-H., Chung, M.-H. & Choi, J. (2006). Toxicol. Lett. 167, 27–33. [DOI] [PubMed]
- Peng, R.-H., Xiong, A.-S., Xue, Y., Fu, X.-Y., Gao, F., Zhao, W., Tian, Y.-S. & Yao, Q.-H. (2008). FEMS Microbiol. Rev. 32, 927–955. [DOI] [PubMed]
- Perozich, J., Nicholas, H., Lindahl, R. & Hempel, J. (1999). Adv. Exp. Med. Biol. 463, 1–7. [DOI] [PubMed]
- Perozich, J., Nicholas, H., Wang, B.-C., Lindahl, R. & Hempel, J. (1999). Protein Sci. 8, 137–146. [DOI] [PMC free article] [PubMed]
- Polikarpov, I., Perles, L. A., de Oliveira, R. T., Oliva, G., Castellano, E. E., Garratt, R. C. & Craievich, A. (1998). J. Synchrotron Rad. 5, 72–76. [DOI] [PubMed]
- Priefert, H., Rabenhorst, J. & Steinbüchel, A. (1997). J. Bacteriol. 179, 2595–2607. [DOI] [PMC free article] [PubMed]
- Rosselló-Mora, R. A., Lalucat, J. & García-Valdés, E. (1994). Appl. Environ. Microbiol. 60, 966–972. [DOI] [PMC free article] [PubMed]
- Saeed, M., Higginbotham, S., Gaikwad, N., Chakravarti, D., Rogan, E. & Cavalieri, E. (2009). Free Radic. Biol. Med. 47, 1075–1081. [DOI] [PMC free article] [PubMed]
- Samanta, S. K., Singh, O. V. & Jain, R. K. (2002). Trends Biotechnol. 20, 243–248. [DOI] [PubMed]
- Spink, D. C., Wu, S. J., Spink, B. C., Hussain, M. M., Vakharia, D. D., Pentecost, B. T. & Kaminsky, L. S. (2008). Toxicol. Appl. Pharmacol. 226, 213–224. [DOI] [PMC free article] [PubMed]
- Stohs, S. J., Ohia, S. & Bagchi, D. (2002). Toxicology, 180, 97–105. [DOI] [PubMed]
- Topinka, J., Marvanová, S., Vondrácek, J., Sevastyanova, O., Nováková, Z., Krcmár, P., Pencíková, K. & Machala, M. (2008). Mutat. Res. 638, 122–132. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yang, Y., Chen, R. F. & Shiaris, M. P. (1994). J. Bacteriol. 176, 2158–2164. [DOI] [PMC free article] [PubMed]
- Yen, K.-M. & Gunsalus, I. C. (1982). Proc. Natl Acad. Sci. USA, 79, 874–878. [DOI] [PMC free article] [PubMed]
- Yen, K.-M. & Gunsalus, I. C. (1985). J. Bacteriol. 162, 1008–1013. [DOI] [PMC free article] [PubMed]
- Yeung, C. K., Yep, A., Kenyon, G. L. & McLeish, M. J. (2008). Biochim. Biophys. Acta, 1784, 1248–1255. [DOI] [PubMed]