Apolipoprotein A-IV crystals consisted of a long unit-cell edge (540 Å) with a high mosaic spread, making them intractable for X-ray diffraction analysis. Extreme dehydration in 60% PEG 3350 was utilized as a post-crystallization treatment as well a screening method to significantly sharpen the mosaic spread and increase the overall resolution of diffraction.
Keywords: apolipoproteins, dehydration, long unit cell, reflection elongation
Abstract
Apolipoproteins are the protein component of high-density lipoproteins (HDL), which are necessary for mobilizing lipid-like molecules throughout the body. Apolipoproteins undergo self-association, especially at higher concentrations, making them difficult to crystallize. Here, the crystallization and diffraction of the core fragment of apolipoprotein A-IV (apoA-IV), consisting of residues 64–335, is presented. ApoA-IV64–335 crystallized readily in a variety of hexagonal (P6) morphologies with similar unit-cell parameters, all containing a long axis of nearly 550 Å in length. Preliminary diffraction experiments with the different crystal morphologies all resulted in limited streaky diffraction to 3.5 Å resolution. Crystal dehydration was applied to the different morphologies with variable success and was also used as a quality indicator of crystal-growth conditions. The results show that the morphologies that withstood the most extreme dehydration conditions showed the greatest improvement in diffraction. One morphology in particular was able to withstand dehydration in 60% PEG 3350 for over 12 h, which resulted in well defined intensities to 2.7 Å resolution. These results suggest that the approach of integrating dehydration with variation in crystal-growth conditions might be a general technique to optimize diffraction.
1. Introduction
Human apolipoprotein A-IV (apoA-IV) is the largest of the exchangeable apolipoproteins. Primarily made in the human gut, the most commonly appreciated role of apoA-IV is as a component of lipid transport and chylomicron assembly. As a prominent component of high-density lipoproteins (HDL), apoA-IV has been proposed to participate in the removal of excess cholesterol from peripheral cells, such as those in the coronary artery wall, and to return it to the liver for catabolism. ApoA-IV has also been proposed to function as an antioxidant (Qin et al., 1998 ▶; Ostos et al., 2001 ▶), a satiety factor (Tso & Xu, 2003 ▶) and a mediator of gut inflammation (Vowinkel et al., 2004 ▶), and has recently been linked to β-amyloid clearance in the brain (Cui et al., 2011 ▶).
One of the fundamental questions in lipoprotein structural biology relates to how the exchangeable apolipoproteins transition from a lipid-free state to a lipid-bound state. Apolipoproteins are highly helical and are thought to adopt a helical bundle in the free state and to undergo self-association into higher order oligomers (Weinberg & Spector, 1985 ▶; Garai & Frieden, 2010 ▶; Vitello & Scanu, 1976 ▶). This feature has hindered high-resolution structural studies and few structures of apolipoproteins are available (Borhani et al., 1997 ▶; Hatters et al., 2005 ▶; Mei & Atkinson, 2011 ▶). In the lipid-bound state, the evidence supports a general model in which two molecules wrap around a discoidal lipid bilayer (Segrest et al., 1999 ▶; Wu et al., 2007 ▶; Bhat et al., 2005 ▶). Exactly how the lipid-free protein transitions into a discoidal particle is also unknown. This transition mechanism is pivotal to understanding reverse cholesterol transport, in which in vivo apolipoproteins interact with ATP-binding cassette transporter ABCA1 and become lipidated, forming a nascent HDL. Nascent HDL proceeds through the bloodstream and absorbs excess lipids from peripheral cells for removal. To gain insight into this important mechanism, we pursued the crystal structure of a proteolytically stable fragment of human apoA-IV (residues 64–335) in its dimer configuration (Tubb et al., 2008 ▶). In our attempt to determine the structure of apoA-IV64–335, dehydrating the crystals with extreme polyethylene glycol (PEG) concentrations was critical to obtaining well defined diffraction images.
2. Methods
2.1. Purification of apoA-IV dimer
ApoA-IV64–335 was identified as a core domain through limited proteolysis (Tubb et al., 2008 ▶) and selected for crystallization. ApoA-IV64–335 was expressed in Escherichia coli BL21 (DE3) and purified as described previously (Tubb et al., 2009 ▶) with the addition of size-exclusion chromatography to isolate the dimer species. Size exclusion was carried out with a HiLoad S200 16/60 column using a buffer consisting of 25 mM Tris pH 8.5, 150 mM NaCl. Fractions that correlated with the dimer molecular weight were pooled and concentrated. The protein concentration was determined using the Bradford assay. SeMet-derivative ApoA-IV64–335 was expressed following Studier’s protocol (Studier, 2005 ▶) and purified as previously stated.
2.2. Crystallization
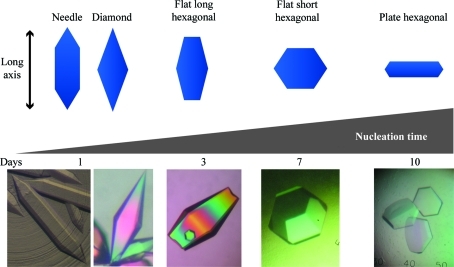
Initial crystal screening was performed with the commercial screens Index (Hampton Research), JCSG+ Suite (Qiagen) and Protein Complex Suite (Qiagen 130715) using a Crystal Phoenix Liquid Handling System (Art Robbins Instruments) to set up 96-well sitting-drop vapor-diffusion experiments. Multiple plates with various protein concentrations ranging from 0.7 to 45 mg ml−1 were analyzed for crystallization. A number of conditions produced crystals with different hexagonal morphologies. Each condition was optimized by hanging-drop vapor diffusion in a 24-well Linbro plate and further refined in sitting drops in nine-well batch plates (Rayment, 2002 ▶). In general, apoA-IV64–335 crystallized in a batch experiment by mixing protein with a 1:1 solution of 22–28% PEG 3350, 0.2–0.4 M ammonium sulfate pH 8.5. Minor alterations in the final crystallization mixture, including the protein concentration, resulted in different nucleation times and numerous crystal morphologies (Fig. 1 ▶).
Figure 1.
Different hexagonal crystal morphologies of apoA-IV64–335. Morphologies are arranged from left to right based on increasing nucleation time and are a product of slight variations in the crystallization solution. The shortest nucleation time creates a long crystal, termed a needle. As the nucleation time increases from 1 to 10 d, other hexagonal morphologies arise. The vertical direction of the scheme indicates the position of the long axis (c axis) relative to the crystal morphology.
3. Results and discussion
3.1. Crystal diffraction and dehydration
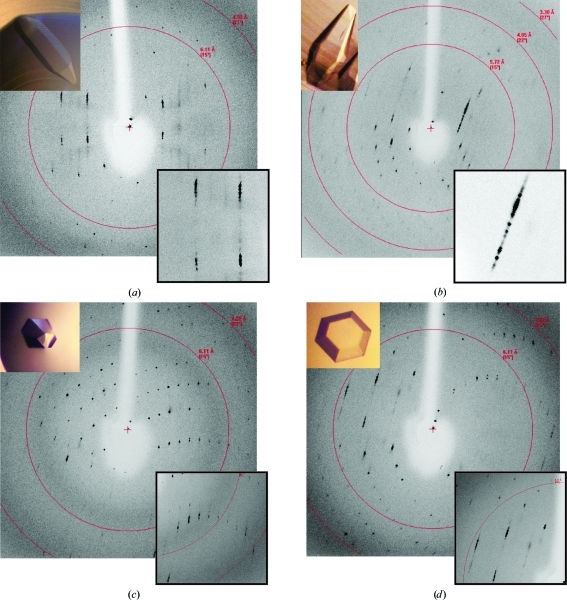
Crystals of varying morphology were mounted in a quartz capillary and tested for diffraction quality at room temperature. This revealed that although the crystals differed in morphology, they appeared to have similar lattices. Based on initial indexing, all morphologies belonged to space group P6, with approximately similar unit-cell parameters of a = b = 73.9, c = 544.7 Å, α = β = 90, γ = 120° (Fig. 2 ▶). Unfortunately, the array of hexagonal crystals only diffracted to 3.5 Å resolution and all had elongated streaky diffraction maxima. Since the unit cell is particularly long, high mosaicity resulted in significant overlap of intensities, which prohibited integration. Owing to the long axis, the hexagonal face had to be aligned perpendicular to the beam (Fig. 1 ▶). Because of this restriction, the hexagonal plate crystals were not pursued since they lacked a side dimension that could be properly oriented. Despite this, mounting crystals with the long axis as close as possible to the rotation axis still resulted in significant overlap of diffraction intensities, even with an oscillation angle of 0.15° or less. Overall, these characteristics rendered the apoA-IV64–335 crystals incompatible with high-resolution structure determination.
Figure 2.
Diffraction patterns of various crystal morphologies mounted in a quartz capillary: (a) needle, (b) diamond, (c) flat long hexagonal and (d) flat short hexagonal. In all cases a long unit cell is evident with streaky reflections. This results in significant overlap of the reflections and data that are unusable for high-resolution diffraction studies.
Since capillary mounting confirmed that the mosaic spread of the diffraction was a consequence of intrinsic disorder of the crystals, we tried to improve crystal packing by varying the crystallization conditions, including testing different additives and detergents. Despite these efforts, the quality of the diffraction did not improve. Similarly, annealing of cryogenically cooled crystals by blocking the cold stream for short periods did not improve the diffraction. We next turned to crystal dehydration, which has been shown to improve the quality of diffraction in a number of cases (Heras & Martin, 2005 ▶). A number of procedures for dehydrating crystals have been described that range from simply exposing the crystal-containing drop to air to sequential transfer of a cover slip to higher precipitant concentrations. The most suitable method for the dehydration of apoA-IV64–335 was to harvest crystals into a nine-well depression plate containing ∼50 µl mother liquor followed by incremental increases in the concentration of PEG 3350. This was accomplished by removing 25% of the mother liquor and replacing it with the dehydration solution [mother liquor + 30–60%(w/v) PEG 3350]. Crystals were allowed to equilibrate for 15–60 min before repeating. These incremental steps were repeated until 90–95% of the well was composed of the dehydration solution, with a final step in which the crystal was transferred with a nylon loop into 100% dehydration solution. At each step, the crystals were evaluated visually for stress fractures and maintainance of the ability to polarize light. It was apparent that crystals greater than 200 µm in the longest dimension suffered more during the dehydration procedure. Small crystals behaved well but had a significantly lower maximum resolution in diffraction. Crystals between 100 and 150 µm in the longest dimension provided an optimal balance between size and stress from dehydration as determined visually.
Initially, common cryoprotectants, various salts and different molecular-weight PEGs were tested as possible dehydration solutions, but ultimately the most successful conditions were similar to the mother liquor. These included PEG 3350 concentrations of 30–60%, with a lower range of ammonium sulfate concentrations (0.05–0.1 M) and a decrease in pH to around 6.5–7.0. Since different morphologies withstood dehydration to different extents, the overall best solution for each form was a variation of this general solution (Table 1 ▶). The needle-shaped crystals were the most sensitive to dehydration and could only withstand 5% incremental increases in PEG. Flat short and flat long hexagonal crystals also required 5% incremental increases in PEG, but were slightly better at enduring higher concentrations of PEG. Diamond-shaped crystals withstood the harshest dehydration conditions and could be directly transferred from the mother liquor to a solution of 70% PEG 3350 without visible defects in the crystal.
Table 1. Maximum dehydration solution for the various crystal forms and the transfer method needed to reach the level of dehydration.
| Dehydration solution | Transfer method | |
|---|---|---|
| Needle | 30% PEG 3350, 0.2 M ammonium sulfate, 0.1 M Tris pH 8.5 | Incremental steps |
| Flat long | 40% PEG 3350, 0.1 M Tris pH 8.5 | Incremental steps |
| Flat short | 35% PEG 3350, 0.1 M Tris pH 8.5 | Incremental steps |
| Diamond | 60% PEG 3350 | Direct |
Once we had determined the maximum level of PEG that each crystal morphology was able to withstand (as determined from the visual appearance of the crystals), the impact of dehydration on the diffraction quality was analyzed. Furthermore, we also varied the incubation time that the crystal was exposed to the final dehydrating solution from 15 min to overnight. In all cases, the high content of PEG 3350 served as the cryoprotectant and diffraction intensities were recorded at 93 K. Improved diffraction and significant changes in unit-cell parameters occurred after 2 h in the final dehydration solution, but incubation over 12 h produced a very reproducible improvement. Crystals of diamond morphology that were subjected to 12 h of dehydration remained free of crystal defects and consistently produced better diffraction data than the other forms.
Since protein crystals typically grow within a range of precipitant and protein concentrations, minor adjustments in the growth parameters can alter the quality of crystal packing and diffraction. Differences in quality may not be apparent from visual examination of the crystal. Through our dehydration experiments, we observed trends in which crystals of the same morphology grown from slightly different conditions suffered different extents. This led us to utilize the dehydration process to evaluate and adjust crystal-growth parameters to select conditions that maximized the amount of dehydration crystals could withstand. For example, conditions that resulted in the formation of diamond-shaped crystals were optimized by adjusting the protein concentration and altering the ratio of protein to mother liquor mixed in batch experiments. In general, this technique might be widely applicable as a rapid way to assess different growth conditions, especially if the macromolecule crystallizes over a broad range of conditions.
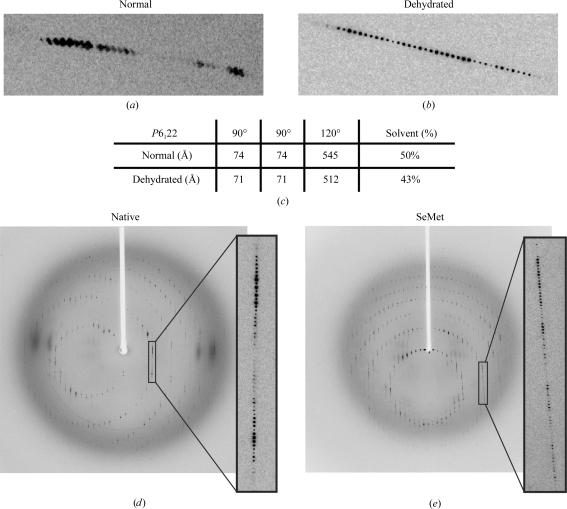
Optimized growth conditions produced diamond-shaped crystals that could withstand 70% PEG 3350, but over a period of hours in the nine-well plates this solution began to precipitate, making crystal harvesting impossible. Therefore, the diamond-shaped crystals were transferred directly into 60% PEG 3350, which produced strong diffraction to 2.7 Å resolution with a mosaic spread of 0.3° (down from ∼1.2°; Fig. 3 ▶ a). Data were collected from native and SeMet crystals dehydrated in 60% PEG 3350 for 12 h (Figs. 3 ▶ c and 3 ▶ d). During the dehydration process the unit-cell volume of the crystals decreased by 13.6%, which corresponded to a significant reduction of solvent content from 64.5 to 58.9% (Kantardjieff & Rupp, 2003 ▶; Fig. 3 ▶ c).
Figure 3.
Comparison of diffraction patterns before and after dehydration. (a) Diffraction pattern prior to dehydration and (b) that after dehydration. Dehydration results in reflections that are sharpened significantly with clear separation between intensities. (c) Unit-cell volume before and after dehydration. A significant reduction in unit-cell volume is observed on dehydration. This results in a 6.6% decrease in solvent content. (d, e) Full-frame diffraction images of native and SeMet diamond-shaped crystals after dehydration. Native data were collected at a wavelength of 1.03 Å with a crystal-to-detector distance of 320 mm, an oscillation range of 0.8° and a mosaicity of 0.26°. SeMet data were collected at three wavelengths corresponding to the low remote, inflection and the high remote with a crystal-to-detector distance of 380 mm, an oscillation range of 1° and a mosaicity ranging from 0.415° to 0.48°.
3.2. Structure determination and packing analysis
Native and SeMet data sets were indexed and integrated with XDS (Kabsch, 2010 ▶) and were determined to belong to space group P6122 , with unit-cell parameters a = b = 70.8, c = 512.4 Å, α = β = 90, γ = 120°. Scaling was performed with SCALA, which is part of the CCP4 suite (Winn et al., 2011 ▶). The highly automated program CRANK (Ness et al., 2004 ▶) was used to locate the Se atoms (AFRO/CRUNCH2; De Graaff et al., 2001 ▶), determine the initial phases (BP3; McCoy et al., 2005 ▶; Pannu & Read, 2004 ▶), perform density modification (SOLOMON; Abrahams & Leslie, 1996 ▶) and build an initial model (Buccaneer; Cowtan, 2006 ▶). The model was further refined using an iterative process with manual model building in Coot and refinement with autoBUSTER (Bricogne et al., 2011 ▶) using TLS parameters (Painter & Merritt, 2006a ▶,b ▶) and NCS (Smart et al., 2008 ▶). The structure of apoA-IV64–335 was determined to 2.7 Å resolution and will be published in a separate manuscript.
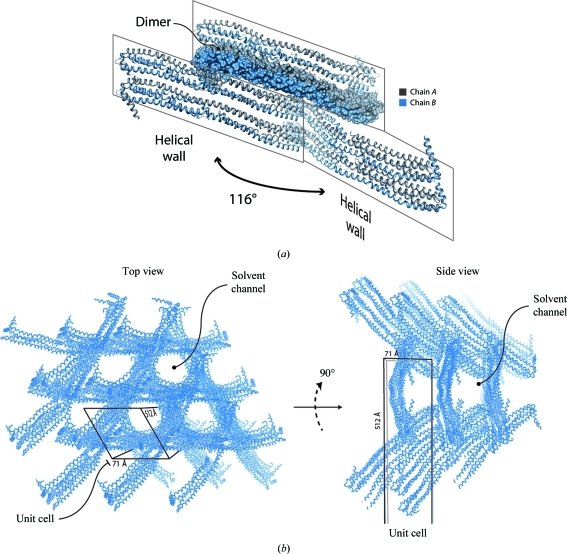
In the asymmetric unit there are two highly helical molecules of apoA-IV that undergo extensive domain swapping, forming an extensive dimer interface. This creates an elongated four-helix bundle that stretches over 170 Å in length. Two of these elongated helical bundles stack, forming the building blocks that pack vertically in a staggered fashion to generate a set of helical walls that intersect at an angle of 116° (Fig. 4 ▶ a). In between the intersecting helical walls are large solvent channels. This results in a honeycomb-like structure in directions parallel and perpendicular to the 512 Å unit-cell edge (Figs. 4 ▶ b and 4 ▶ b). It is possible that this honeycomb-like lattice, composed of highly helical intersecting walls, provides the strength to withstand the compression imposed during dehydration.
Figure 4.
Crystal-packing analysis of hexagonal apoA-IV64–335 crystals. (a) The asymmetric unit contains a dimer, chains A and B, that domain swaps through a long helix that extends the length of the molecule. Two dimer molecules stack vertically by a slight offset, forming a helical wall. This wall then intersects adjacent walls at 116°, creating a honeycomb-like lattice. (b) Top view of the honeycomb-like lattice looking down the long unit-cell dimension. Large solvent channels are the result of helices intersecting. (c) Side view of (b) looking perpendicular to the long axis.
It is interesting that the different morphologies responded differently to dehydration. One possibility is that the success of dehydration is correlated with different nucleation times or the rate at which the crystals grow (Fig. 1 ▶). Crystal morphologies that required longer nucleation and growth periods resulted in more limited dehydration success. Another possibility is that the success of dehydration could be dependent on the overall surface area to crystal volume ratio of the particular crystal morphology exposed to the dehydrating solution. Compared with the other crystal morphologies, the needles actually have the smallest ratio of surface area to crystal size (Fig. 1 ▶). Overall, we have shown that the technique of dehydration can vary significantly with different crystal morphologies. In addition, once the final crystallization conditions have been determined, dehydration can also be a useful technique to rapidly assess the various growth conditions prior to diffraction analysis.
References
- Abrahams, J. P. & Leslie, A. G. W. (1996). Acta Cryst. D52, 30–42. [DOI] [PubMed]
- Bhat, S., Sorci-Thomas, M. G., Alexander, E. T., Samuel, M. P. & Thomas, M. J. (2005). J. Biol. Chem. 280, 33015–33025. [DOI] [PubMed]
- Borhani, D. W., Rogers, D. P., Engler, J. A. & Brouillette, C. G. (1997). Proc. Natl Acad. Sci. USA, 94, 12291–12296. [DOI] [PMC free article] [PubMed]
- Bricogne, G., Blanc, E., Brandl, M., Flensburg, C., Keller, P., Paciorek, W., Roversi, P., Smart, O. S., Vonrhein, C. & Womack, T. O. (2011). BUSTER v.2.10.0 Cambridge: Global Phasing Ltd.
- Cowtan, K. (2006). Acta Cryst. D62, 1002–1011. [DOI] [PubMed]
- Cui, Y., Huang, M., He, Y., Zhang, S. & Luo, Y. (2011). Am. J. Pathol. 178, 1298–1308. [DOI] [PMC free article] [PubMed]
- Garai, K. & Frieden, C. (2010). Biochemistry, 49, 9533–9541. [DOI] [PMC free article] [PubMed]
- Graaff, R. A. G. de, Hilge, M., van der Plas, J. L. & Abrahams, J. P. (2001). Acta Cryst. D57, 1857–1862. [DOI] [PubMed]
- Hatters, D. M., Peters-Libeu, C. A. & Weisgraber, K. H. (2005). J. Biol. Chem. 280, 26477–26482. [DOI] [PubMed]
- Heras, B. & Martin, J. L. (2005). Acta Cryst. D61, 1173–1180. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kantardjieff, K. A. & Rupp, B. (2003). Protein Sci. 12, 1865–1871. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. (2005). Acta Cryst. D61, 458–464. [DOI] [PubMed]
- Mei, X. & Atkinson, D. (2011). J. Biol. Chem. 286, 38570–38582. [DOI] [PMC free article] [PubMed]
- Ness, S. R., de Graaff, R. A. G., Abrahams, J. P. & Pannu, N. S. (2004). Structure, 12, 1753–1761. [DOI] [PubMed]
- Ostos, M. A., Conconi, M., Vergnes, L., Baroukh, N., Ribalta, J., Girona, J., Caillaud, J. M., Ochoa, A. & Zakin, M. M. (2001). Arterioscler. Thromb. Vasc. Biol. 21, 1023–1028. [DOI] [PubMed]
- Painter, J. & Merritt, E. A. (2006a). Acta Cryst. D62, 439–450. [DOI] [PubMed]
- Painter, J. & Merritt, E. A. (2006b). J. Appl. Cryst. 39, 109–111.
- Pannu, N. S. & Read, R. J. (2004). Acta Cryst. D60, 22–27. [DOI] [PubMed]
- Qin, X., Swertfeger, D. K., Zheng, S., Hui, D. Y. & Tso, P. (1998). Am. J. Physiol. 274, H1836–H1840. [DOI] [PubMed]
- Rayment, I. (2002). Structure, 10, 147–151. [DOI] [PubMed]
- Segrest, J. P., Jones, M. K., Klon, A. E., Sheldahl, C. J., Hellinger, M., De Loof, H. & Harvey, S. C. (1999). J. Biol. Chem. 274, 31755–31758. [DOI] [PubMed]
- Smart, O. S., Brandl, M., Flensburg, C., Keller, P. A., Paciorek, W., Vonrhein, C., Womack, T. O. & Bricogne, G. (2008). Abstr. Annu. Meet. Am. Crystallogr. Assoc., Abstract TP139, p. 117.
- Studier, F. W. (2005). Protein Expr. Purif. 41, 207–234. [DOI] [PubMed]
- Tso, P. & Xu, G. (2003). Wei Sheng Yan Jiu, 32, 67. [PubMed]
- Tubb, M. R., Silva, R. A., Fang, J., Tso, P. & Davidson, W. S. (2008). J. Biol. Chem. 283, 17314–17323. [DOI] [PMC free article] [PubMed]
- Tubb, M. R., Smith, L. E. & Davidson, W. S. (2009). J. Lipid Res. 50, 1497–1504. [DOI] [PMC free article] [PubMed]
- Vitello, L. B. & Scanu, A. M. (1976). J. Biol. Chem. 251, 1131–1136. [PubMed]
- Vowinkel, T., Mori, M., Krieglstein, C. F., Russell, J., Saijo, F., Bharwani, S., Turnage, R. H., Davidson, W. S., Tso, P., Granger, D. N. & Kalogeris, T. J. (2004). J. Clin. Invest. 114, 260–269. [DOI] [PMC free article] [PubMed]
- Weinberg, R. B. & Spector, M. S. (1985). J. Biol. Chem. 260, 14279–14286. [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Wu, Z., Wagner, M. A., Zheng, L., Parks, J. S., Shy, J. M., Smith, J. D., Gogonea, V. & Hazen, S. L. (2007). Nature Struct. Mol. Biol. 14, 861–868. [DOI] [PubMed]