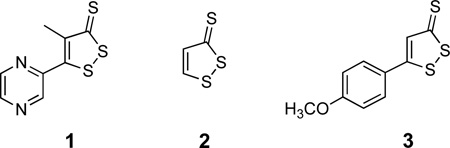
Abstract
Dithiolethiones upregulate the expression of cancer-preventive proteins via modification of thiol residues in the Keap1-Nrf2 transcription factor complex. In addition to Keap1-Nrf2, dithiolethiones have the potential to modify a variety of cysteine-containing proteins in the cell. Such “off target” reactions could contribute to either side effects or cancer-preventive efficacy. Evidence is presented here that cancer chemopreventive dithiolethiones inactivate protein tyrosine phosphatases via covalent, but thiol-labile, modification of active site residues. This observation may explain a number of previously reported biological cellular responses to dithiolethiones.
1. Introduction
Dithiolethiones are potent cancer-preventive agents.1,2 These compounds trigger upregulation in the expression of genes that encode a host of cancer-preventive phase II metabolic enzymes.2,3 This is driven primarily by activation of the transcription factor Nrf2 (nuclear factor-E2-related factor 2).3,4 Nrf2 exists in the cytosol complexed with a repressor protein known as Keap1 (Kelch-like ECH-associated protein 1). Keap1 acts as a sensor protein that responds to a variety of stresses. Modification of key thiol residues on Keap1 by either alkylating agents or reactive oxygen species (ROS) leads to increased levels of Nrf2 in the nucleus.3,4 The exact mechanism by which dithiolethiones activate Keap1-Nrf2 remains under investigation, but at least two possibilities have been considered. Dithiolethiones are thiol-reactive compounds that may directly modify cysteine thiol residues on Keap1.5 Alternatively, the reaction of thiols with dithiolethiones leads to the generation of ROS,6 which have the potential to modify cysteine residues on Keap1.
It is clear that the Keap1-Nrf2 complex is a central cellular target in the cancer-preventive activity of dithiolethiones. Nonetheless, dithiolethiones are thiol-reactive compounds that have the potential to modify a variety of cysteine-containing proteins in the cell. Such “off target” reactions could contribute to either side effects or cancer-preventive efficacy. The modification of protein thiol residues by dithiolethiones may be especially important when it occurs in the context of enzymes that contain critical catalytic cysteine residues. Along these lines, it may be important to consider the reactions of dithiolethiones with protein tyrosine phosphatases.
Protein tyrosine phosphatases (PTPs) operate alongside protein tyrosine kinases (PTKs) to regulate a variety of critical mammalian signal transduction pathways.7 PTKs catalyze the addition of phosphoryl groups to tyrosine residues on target proteins while PTPs catalyze their hydrolytic removal.8 PTPs contain a crucial active site cysteine residue with an abnormally low pKa value of about 5.7 (versus about 8.5 for a typical cysteine thiol).9 Thus, the catalytic cysteine residue (Cys215 in the numbering of the archetypal enzyme PTP1B) exists predominantly in the nucleophilic thiolate form (RS−) at physiological pH.9–13 Thiol-reactive chemicals can inactivate PTPs through selective modification of the exceptionally reactive active site cysteine.10,14–16 In addition, reactive oxygen species such as superoxide radical (O2•−) and hydrogen peroxide (H2O2) can inactivate PTPs via oxidation of the catalytic cysteine residue.17–21 Inactivation of PTPs can yield profound biological effects by modulating the activity of various signal transduction pathways.7
Interestingly, treatment of cells with various dithiolethiones alters the activity of a number of signal transduction pathways that are regulated by tyrosine phosphorylation, including the ERK1/2 and NF-κB pathways.22,23 We reasoned that inactivation of PTPs could contribute to modulation of these important signal transduction pathways by dithiolethiones. Indeed, here we report that oltipraz and other 1,2-dithiole-3-thiones inactivate protein tyrosine phosphatases under physiologically-relevant conditions.
2. Results and Discussion
2.1. Inactivation of PTP1B by oltipraz
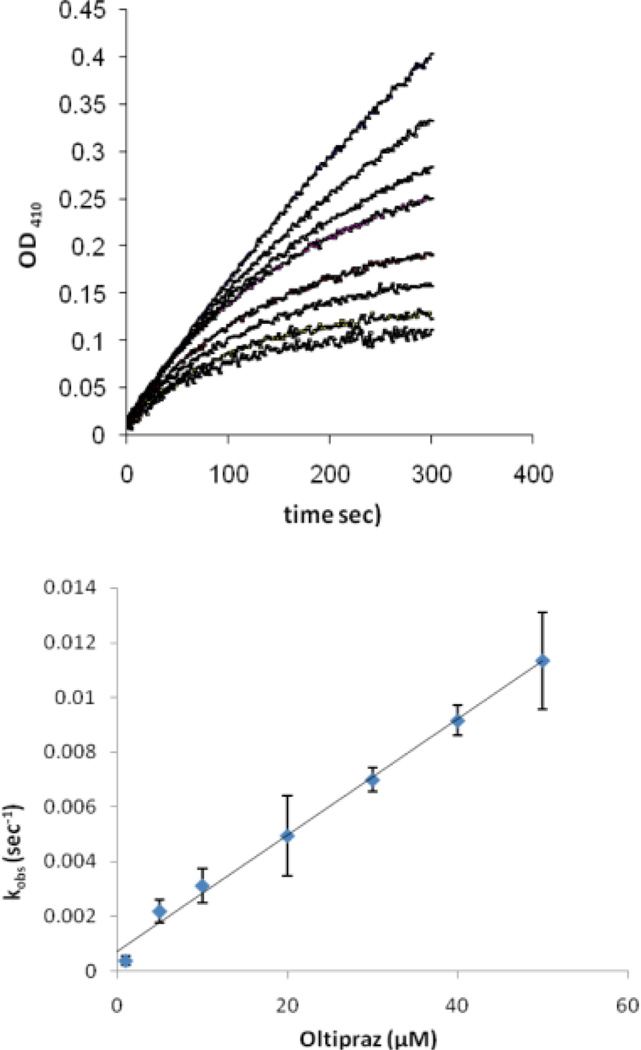
We utilized the catalytic subunit (amino acids 1–322) of recombinant human PTP1B to investigate whether oltipraz (1) can inactivate a protein tyrosine phosphatase. This enzyme is an archetypal member of the protein tyrosine phosphatase family.9 Various concentrations of oltipraz were incubated with the enzyme in the presence of the substrate p-nitrophenolphosphate (pNPP). The progress curves obtained in this manner clearly reveal time-dependent loss of enzyme activity (Figure 1). Each curve was fit to obtain the apparent rate of inactivation at each oltipraz concentration as described in the Experimental Section. A plot of these observed rate constants versus oltipraz concentration yields a line (Figure 1) indicative of a second-order reaction between oltipraz and the enzyme. The slope of the line yields a rate constant of 1970 ± 170 M−1 s−1 for this inactivation process. By way of comparison, the reaction 2-mercaptoethanol thiolate with oltipraz occurs with a rate constant of 2 M−1 s−1.24 It is possible that general acid catalysis by residues such as Asp181 or stabilization of a developing negative charge by residues such as Arg 211 in the phosphate binding pocket of the enzyme accelerate the reaction of the active site thiolate residue with oltipraz.
Figure 1.
Inactivation of PTP1B by oltipraz. Above: progress curves for the inactivation of PTP1B by oltipraz. Thiol-free PTP1B (12.5 nM final) was added to a solution of oltipraz (0, 5, 10, 20, 40, and 50 µM from top to bottom curves) in tris (50 mM), bis-tris (50 mM), sodium acetate (100 mM), DETPAC (10 mM), and 1% DMF, containing the substrate p-nitrophenyl phosphate (pNPP, 20 mM) at 23 °C. Below: A plot of observed rate of inactivation (kobs) versus oltipraz concentration. The apparent second order rate of enzyme inactivation was calculated as described in the Experimental Section.
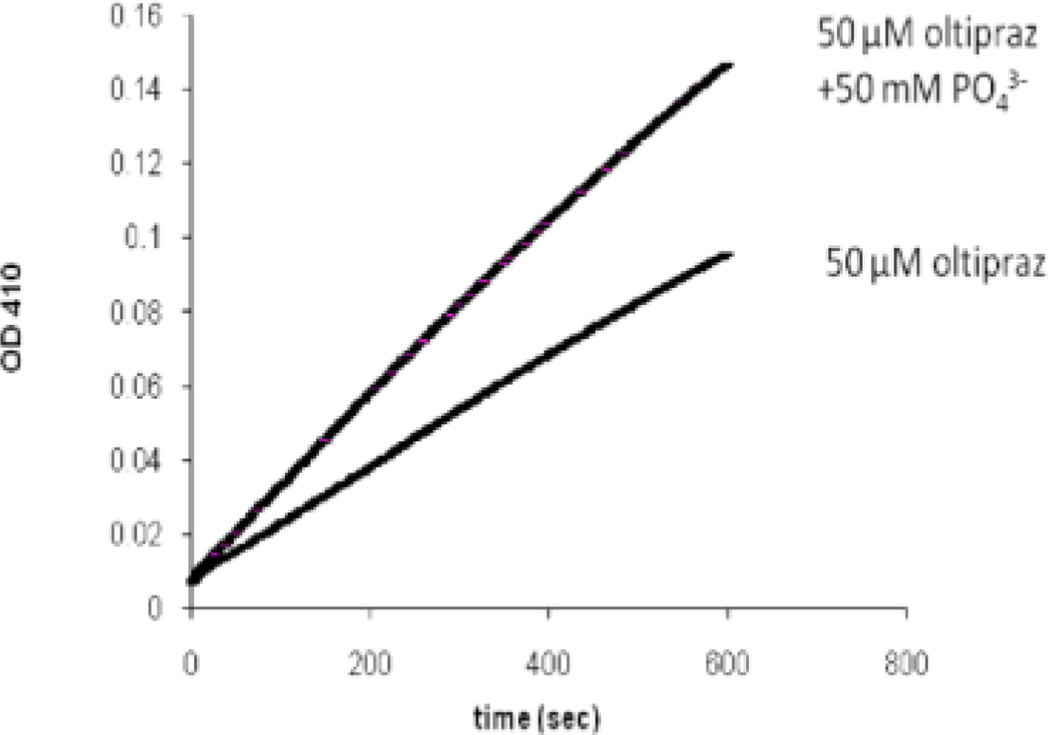
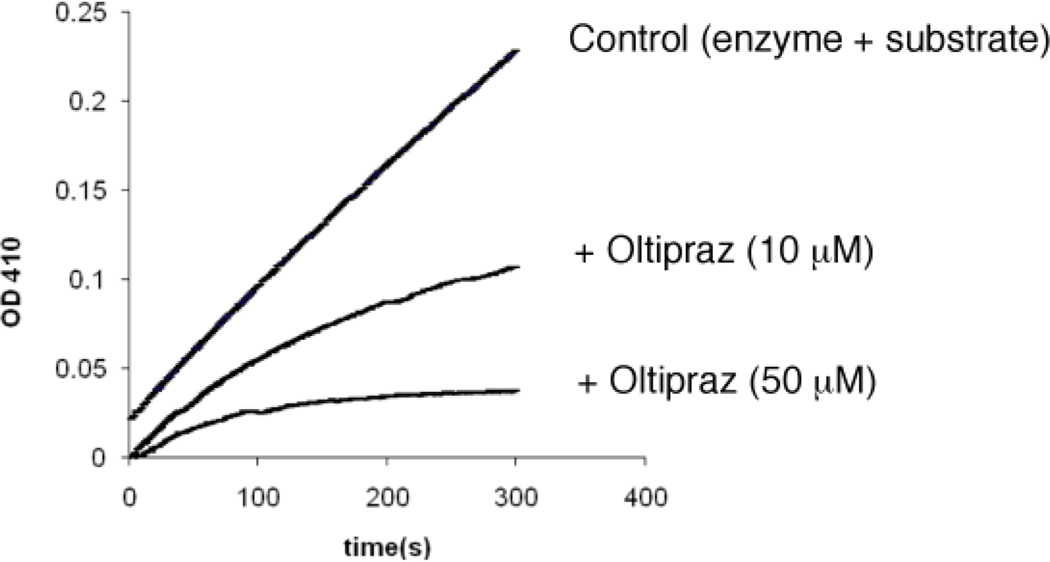
Presence of the active site-directed competitive PTP1B inhibitor phosphate slows inactivation of the enzyme by oltipraz (Figure 2). For example, treatment of PTP1B with oltipraz (50 µM) for 15 min yields 70% inactivation while, in the presence of 50 mM sodium phosphate, only 50% inactivation is observed (the Ki for phosphate is 17 mM).25 Furthermore, the inactivation of PTP1B by oltipraz is not reversed upon gel filtration of the inactivated enzyme through G25 Sephadex to remove excess small molecules from the mixture. Taken together, the data suggests that oltipraz inactivates PTP1B via covalent modification of the active site.
Figure 2.
The reversible PTP inhibitor sodium phosphate protects PTP1B against inactivation by oltipraz. Thiol free PTP1B (0.5 µm) was incubated with oltipraz (50 µM) in a buffer composed of Tris (50 mM), bis-Tris (50 mM), sodium acetate (100 mM) and DETAPAC (10 mM) at pH 7.2) for 15 min at 23 °C (100 µL final volume). An aliquot (15 µL) was taken from the reaction mixture and assayed to measure the remaining enzyme activity using the substrate p-nitrophenol phosphate by monitoring the increase in absorbance at 410 nm due to the enzymatic release of p-nitrophenol.
2.2. Inactivation of PTP1B by oltipraz does not involve reactive oxygen species
Two mechanisms can be envisioned for the covalent modification of PTP1B by oltipraz. First, oltipraz may directly modify thiol residues in the protein. Alternatively, literature precedents indicate that reaction of thiols with oltipraz can lead to the production of reactive oxygen species (ROS) via the (unbalanced) reactions shown in Eqn 1.6,26 ROS have the potential to inactivate PTPs via oxidation of the active site thiol residue.17,18 Thus, we set out to examine whether reactions of oltipraz with solvent-exposed thiol residues on PTP1B leads to the generation of ROS that inactivate the enzyme. We found that the rate at which oltipraz inactivated PTP1B was not significantly altered by the presence of the HO• scavenger mannitol (1 mM). Similarly, the presence of the metal chelator diethylenetriaminepentaacetic acid (DETAPAC, 10 mM) had no significant effect on the inactivation process. DETAPAC sequesters traces of adventitious transition metal ions that otherwise have the potential to catalyze the conversion of hydrogen peroxide to hydroxyl radicals. Finally, addition of the peroxide-destroying enzyme catalase had no effect on the inactivation of PTP1B by oltipraz. Overall, these results suggest that the inactivation of PTP1B by oltipraz does not involve the generation of diffusible ROS.
| (Eqn 1) |
2.3. Inactivation of PTP1B by oltipraz is reversed by treatment with dithiothreitol
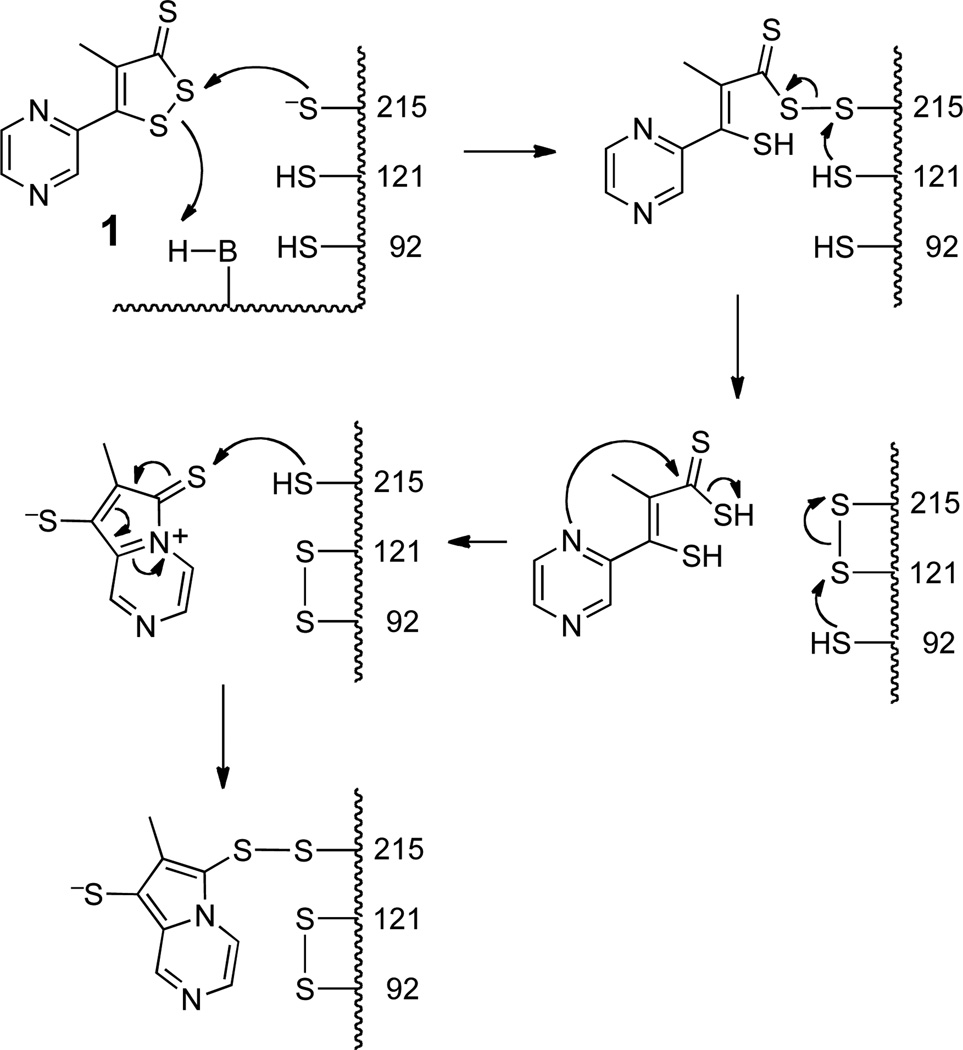
The inactivation of PTP1B by oltipraz (50 µM oltipraz, 1 hr, > 97% enzyme inactivation) can be reversed (75% recovery of activity) by treatment with dithiothreitol (100 mM, 1 h, 24 °C). It is noteworthy that activity does not return upon incubation with biologically-relevant concentrations of glutathione (1 mM, 1 h, 24 °C). This result suggests that inactivation of PTP1B by oltipraz involves covalent attachment of the compound to an active site residue via a thiol-labile chemical bond such as a disulfide. This suggests that the point of attachment might be the catalytic cysteine residue at position 215 in the enzyme.
2.4 Mass spectrometric analysis of oltipraz-inactivated PTP1B: evidence supporting covalent modification of the active site cysteine residue 215
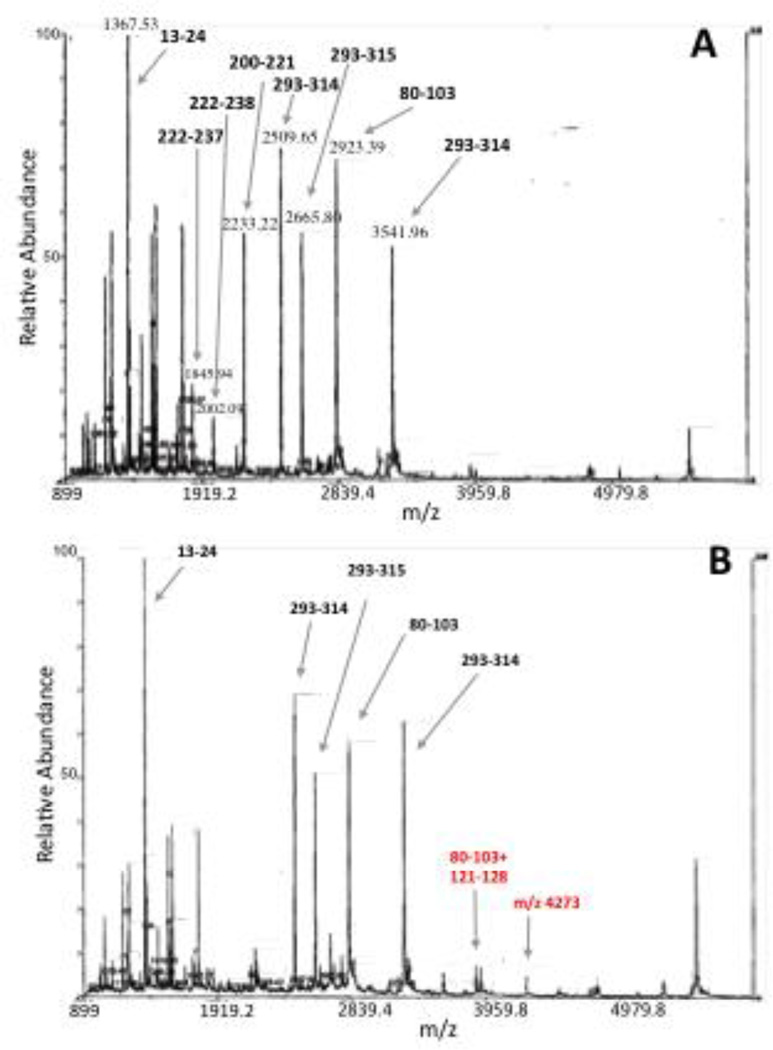
Mass spectrometric analysis was used to characterize the site(s) at which oltipraz modified PTP1B during the inactivation process. In these experiments, PTP1B was inactivated by oltipraz under conditions that leaves less than 10% enzyme activity remaining. Recognizing that dithiolethione adducts may be prone to reactions with thiols (as discussed above), we treated the inactivated enzyme with iodoacetamide to cap unmodified thiol residues in the protein (in its native form, the catalytic domain of PTP1B used in these studies contains six free cysteine residues and no disulfides). Samples of the proteins were then deposited on a polished stainless steel target with α-cyano-4-hydroxycinnamic acid (CHCA) and analyzed by MALDI-TOF-MS. Analysis of the control enzyme treated in this manner reveals reasonable coverage of the expected tryptic fragments (Figure 3A). A number of differences were observed in the mass spectrum of the oltipraz-inactivated enzyme. Perhaps most striking, the active site peptide consisting of amino acids 200–221 is completely missing from the spectrum of the inactivated enzyme. This strongly suggests that the active peptide has been modified by treatment of the enzyme with oltipraz. At the same time, significant new signals were observed in the mass spectrum of the oltipraz-inactivated protein at m/z 3887 and 4273. The species at m/z 4273 is in the general range expected for an oltipraz-modified active site peptide fragment composed of amino acids 200–237; however, even after repeated analyses we have been unable to assign a discrete structure to this signal.
Figure 3.
MADLI-MS of native (panel A) and oltipraz-inactivated PTP1B (panel B).
A peptide fragment in the range m/z 4000 could arise if modification of the active site cysteine residue 215 inhibits the usual tryptic cleavage at residue 221. The new peak observed at m/z 3887 in the oltipraz-treated sample corresponds to the [M+H]+ ion expected for the disulfide-crosslinked tryptic fragments 80–103 and 121–128. While not seen previously in the context of PTP1B, a disulfide “relay” leading to analogous disulfide-linked tryptic fragments has been observed following modification of the active site cysteine residue in the protein tyrosine phosphatase family members, SHP1 and SHP2.27 Scheme 1 shows a speculative mechanism by which reaction of PTP1B with oltipraz could lead to both modification of the active site cysteine and also disulfide formation between tryptic fragments 80–103 and 121–128. The proposed mechanism closely follows precedents for the reaction of oltipraz with low molecular weight thiols.5 Clearly, generation of the disulfide crosslink between tryptic fragments 80–103 and 121–128 is not quantitative because a strong signal for the 80–103 tryptic fragment remains in the spectrum. While the nature of the oltipraz-PTP adduct remains uncertain, the mass spectrometric data nonetheless is consistent with an inactivation mechanism involving modification of the active site cysteine residue in PTP1B by oltipraz.
Scheme 1.
2.5. Various dithiolethiones inactivate PTP1B
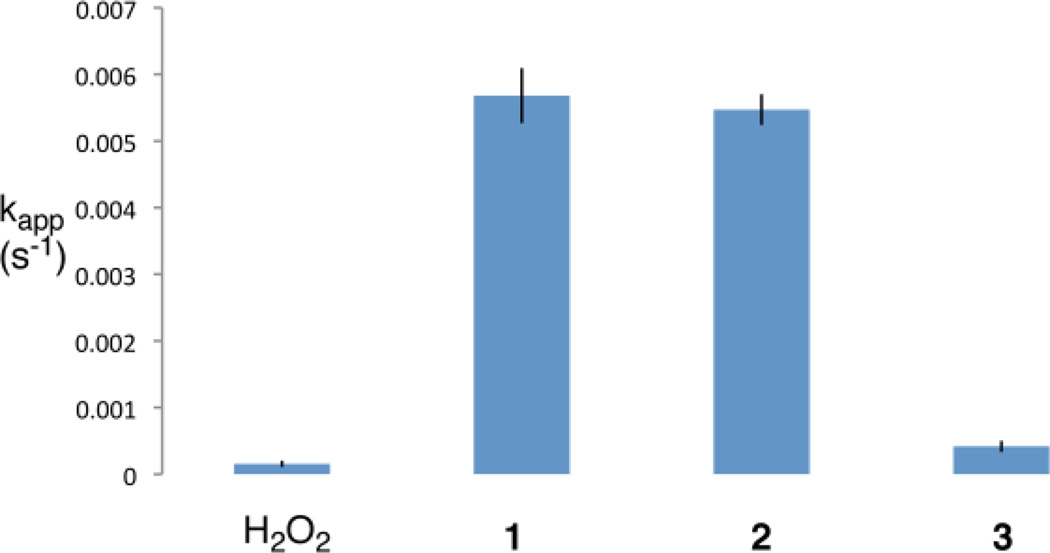
A wide variety of dithiolethiones possess chemopreventive activity.1 Thus, we felt it would be interesting to examine the ability of two additional, structurally varied dithiolethiones to inactivate PTP1B. We compared the ability of equal concentrations of oltipraz (1), dithiolethione (2), and anethole dithione (3) to inactivate the enzyme. As shown in Figure 4, we find that dithiolethione (2) is comparable to oltipraz in its ability to inactivate PTP1B. Anethole dithione is less potent than oltipraz, but is nonetheless comparable to the endogenous regulator of PTP activity, hydrogen peroxide,18 in its potency as an inactivator of PTP1B.
Figure 4.
Inactivation of PTP1B by H2O2 and various dithiolethiones. Compounds (25 µM) were incubated with thiol-free PTP1B (12.5 nM final concentration) in a quartz cuvette containing the chromogenic substrate pNPP (20 mM) in Tris (50 mM), bis-Tris (50 mM), sodium acetate (100 mM), DETAPAC (10 mM), and 1% dimethylformamide at pH 7.2 and 23 °C. The apparent rate of inactivation was obtained by non-linear fitting of the inactivation progress curves to a first-order reaction as described in the Experimental Section. The results shown are the averages of three experiments.
2.6 Inactivation of SHP2 by oltipraz
The human genome encodes for approximately 80 catalytically active PTPs.7 These enzymes possess structurally homologous active sites that contain a nucleophilic cysteine residue. With this in mind, we felt it would be interesting to examine whether oltipraz inactivates a PTP enzyme other than PTP1B. Accordingly, we examined the phosphatase SHP2 which is involved in mammalian T-cell antigen and growth factor signaling.28 We find that the inactivation of SHP2 by oltipraz occurs about two-fold faster than the analogous inactivation of PTP1B (Figure 5). The inactivation of SHP2 by oltipraz is inhibited in the presence competitive inhibitors such as vanadate and arsenate, suggesting that the inactivation involves reaction of the chemical at the active site of the enzyme. Treatment of the inactivated enzyme with dithiothreitol leads to recovery of enzyme activity similar to the results described above for PTP1B. The results indicate that the dithiolethione oltipraz is a general inactivator of PTP enzymes.
Figure 5.
Progress curves for the inactivation of SHP2 by oltipraz. Thiol-free SHP2 was added to a solution of oltipraz (10 or 50 µM) in tris (50 mM), bis-tris (50 mM), sodium acetate (100 mM), DETAPAC (10 mM), and 1% dimethylformamide, containing the substrate (p-NPP, 20 mM) at 23 °C. The remaining enzyme activity was determined as a function of time by monitoring the release of p-nitrophenol at 410 nm. Fitting of the data to the equation for a first order reaction reveals an observed rate of inactivation of the enzyme by 10 µM oltipraz to be 5.2 × 10−3 s−1 while 50 µM yields an observed inactivation rate constant of 2.1 × 10−2 s−1. For comparison, the kobs values for the inactivation of PTP1B by oltipraz at 10 µM and 50 µM in the presence of p-NPP (20 mM) are 3.1 × 10−3 s−1 and 1.1 × 10−2 s−1, respectively.
3. Conclusion
Our results clearly show that the chemopreventive agent oltipraz inactivates protein tyrosine phosphatases. The inactivation of PTP1B occurs with a rate constant of 1970 M−1 s−1. This rate constant suggests that clinically-relevant concentrations of oltipraz (1–20 µM)2 could cause efficient inactivation of PTP enzymes in vivo. We find that other cancer-preventive dithiolethiones also inactivate PTP1B. Mechanistic studies in the context of oltipraz suggest that the inactivation reaction involves modification of the active site cysteine residue via a thiol-labile linkage such as a disulfide. Such a mechanism is consistent with chemical model reactions involving the reaction of small-molecule thiols with oltipraz.5
The inactivation of PTPs by dithiolethiones reported here may be relevant to a number of the cellular responses observed for this class of chemopreventive agents. First, recent studies have shown that “PTP1B deficiency or inhibition delays ErbB2-induced mammary tumorigenesis”.29 Our findings offer the possibility that inactivation of cellular PTP1B could contribute to the cancer-preventive properties of oltipraz and other dithiolethiones. Second, dithiolethiones result in activation of Nrf2 via upregulation of the ERK1/2 signaling pathways.23 In separate studies, it has been shown that inactivation of PTPs by vanadate or hydrogen peroxide leads to activation of ERK1/2 pathways.30 In light of these literature precedents, the results presented here suggest that inactivation of PTPs could contribute to the ability of dithiolethiones to activate Nrf2 via the ERK1/2 signaling pathway. At the same time, is important to note that regulation of the Nrf2/Keap1 system is complex and that phosphorylation of some tyrosine residues in these proteins inhibits activation of the Nrf2 transcription factor.31 Third, oltipraz displays antiangiogenic properties.32 Other studies have shown that inhibition of the enzyme SHP2 blocks angiogenesis in the same HUVEC cell line used in the earlier study.33 These literature reports, taken together with our findings, suggest the possibility that the antiangiogenic properties of oltipraz could stem from its ability to inactivate SHP2. Finally, oltipraz activates the transcription factor NF-κB.22 Phosphorylation of the inhibitor protein IκB-α at tyrosine 42 leads to activation of NF-κB.34 Significantly, inhibition of cellular PTPs was shown to elevate tyrosine phosphorylation levels on IκB-α and activate NF-κB via this mechanism.34 Our results offer the possibility that inhibition of PTPs contributes to the activation of NF-κB by oltipraz.
In conclusion, the data presented here suggests that the inactivation of protein tyrosine phosphatases may contribute to the biological activities of oltipraz and other cancer-preventive 1,2-dithiole-3-thiones. Oltipraz has been tested in humans and shows a reasonable safety profile.2 It is possible that appropriately substituted 1,2-dithiole-3-thiones could provide a platform for the development of nontoxic and therapeutically useful PTP inactivators. For example, the ability of oltipraz to inactivate PTP1B suggests that 1,2-dithiole-3-thiones might find utility in the treatment of type 2 diabetes.35
4. Experimental
4.1 General
All chemicals were of the highest grade available unless otherwise noted. Dithiothreitol (DTT), 2-mercaptoethanol (βME), sodium vanadate, mannitol, G-25 sephadex, dimethyl formamide (DMF), diethylenetriaminepentaacetic acid (DETAPAC), 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl) propane-1,3-diol (bis-tris), sodium acetate, tris(hydroxymethyl)aminomethane (tris), 4-nitrophenylphosphate (pNPP) and glutathione (GSH) were purchased from Sigma-Aldrich Company and oltipraz, 1,2-dithiol-3-thione, and anethole dithione were synthesized by Dr. Hong Zang as described previously.6
4.2. Inactivation of protein tyrosine phosphatases by oltipraz
PTP1B was isolated as described previously and thiol-free samples of the enzyme were prepared by gel filtration of the protein through G25 Sephadex immediately prior to use. Typical assays contained (in a final volume of 1.0 mL at 23 °C) tris (50 mM, pH 7.2), bis-tris (50 mM), sodium acetate (100 mM), DETAPAC (10 mM), DMF (1% v/v), 20 mM pNPP, dithiolethione (0–50 µM) and thiol-free20 enzyme (0.01–0.05 µM). The dithiolethiones were introduced to the assay mixture as stock solutions in DMF. Following addition of the enzyme, remaining catalytic activity was determined as a function of time by monitoring the release of p-nitrophenol at 410 nm. The apparent rates of inactivation at various concentrations of dithiolethione were determined by fitting the progress curves to the equation for a first order reaction. The apparent rates of inactivation were plotted versus inactivator concentration. The data afforded a good fit to a line and, according to the equation kobs= kinact[I]0/(1+[S]/Km) the slope of the line corresponds to kinact/(1+[S]/Km), where [I0] is the initial concentration of inactivator.35 The effects of added radical scavenging agents on the observed rate constants for inactivation were determined as described above and compared to control reactions containing no dithiolethione. In experiments that examined remaining enzyme activity following treatment with DTT or gel filtration, an aliqout of enzyme was added to a quartz cuvette containing p-NPP (20 mM). The remaining catalytic activity was determined by monitoring the rate release of p-nitrophenol at 410 nm.
4.3. Mass spectrometric analysis of inactivated PTP1B
After incubation, both solutions were passed through a Zeba mini centrifugal buffer exchange column. To the resulting mixtures, sequence grade modified trypsin (50 µL of a 1µg/50 µL solution in 50 mM ammonium bicarbonate) was added. The digestion was incubated at 37 °C for 18 h then quenched with TFA (5 µL of a 10% aqueous solution). An aliquot (20 µL) of each solution was transferred to a microcentrifuge tube, frozen in liquid nitrogen, and lyophilized to dryness. The residue was resuspended in water (10 µL) and again lyophilized. The final dried sample was reconstituted in acetonitrile/water/88% formic acid (5 µL of 700/290/10 v/v/v). For MALDI TOF/TOF MS analysis, a 0.6 µL portion of diluted sample was mixed with an equal volume of alpha-cyano-4-hydroxycinnamic acid (CHCA) matrix solution (5 mg CHCA/mL in 500/455/20/25 (v/v/v/v) acetronitrile/water/10% TFA/400 mM aqueous ammonium dihydrogen phosphate). Aliquots (0.4 µL) of the mixture were deposited on a polished stainless steel target. Crystallization of the mixture proceeded under ambient conditions. Mass spectra were acquired on an Applied Biosystems Inc. 4700 MALDI TOF/TOF mass spectrometer with a 355 nm Nd:YAG laser (200 Hz) in the positive ion delayed extraction reflector MS mode. The MS spectra (2000 laser shots summed/averaged) were acquired over the mass range 700–4000 Da. Each MS spectrum was re-calibrated internally using the masses (monoisotopic [M+H]+) for the PTP1B tryptic fragments observed at 1366.675 Da ([13–24]) and 2508.288 Da ([293–314]).
Acknowledgements
The authors are grateful to the National Institutes of Health (CA83925 and 119131 to KSG) for partial financial support of this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Maxiutenko YY, Libby AH, Joyner HH, Curphey TJ, MacMillan DL, Kensler TW, Roebuck BD. Carcinogenesis. 1998;19:1609–1615. doi: 10.1093/carcin/19.9.1609. [DOI] [PubMed] [Google Scholar]
- 2.Kensler TW, Groopman JD, Sutter TR, Curphey TJ, Roebuck BD. Chem. Res. Toxicol. 1999;12:113–126. doi: 10.1021/tx980185b. [DOI] [PubMed] [Google Scholar]
- 3.Dinkova-Kostova AT, Holtzclaw WD, Kensler TW. Chem. Res. Toxicol. 2005;18:1779–1791. doi: 10.1021/tx050217c. [DOI] [PubMed] [Google Scholar]
- 4.Kern JT, Hannink M, Haas JF. Curr. Topics Med. Chem. 2007;7:972–978. doi: 10.2174/156802607780906825. [DOI] [PubMed] [Google Scholar]
- 5.Fluery MB, Largenon M, Barreau M, Vuilhorgne M. Tetrahedron. 1985;41:3705–3715. [Google Scholar]
- 6.Kim W, Gates KS. Chem. Res. Toxicol. 1997;10:296–301. doi: 10.1021/tx9601667. [DOI] [PubMed] [Google Scholar]
- 7.Tonks NK. Nature Rev. Cell. Biol. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 8.Hunter T. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Z-Y. Acc. Chem. Res. 2003;36:385–392. doi: 10.1021/ar020122r. [DOI] [PubMed] [Google Scholar]
- 10.Lohse DL, Denu JM, Santoro N, Dixon JE. Biochemistry. 1997;36:4568–4575. doi: 10.1021/bi963094r. [DOI] [PubMed] [Google Scholar]
- 11.Jackson MD, Denu JM. Chem. Rev. 2001;101:2313–2340. doi: 10.1021/cr000247e. [DOI] [PubMed] [Google Scholar]
- 12.Barford D, Flint AJ, Tonks K. Science. 1994;263:1397–1404. [PubMed] [Google Scholar]
- 13.Pedersen AK, Peters GH, Moller KB, Iverson LF, Kastrup JS. Acta Cryst. D. 2004;D60:1527–1534. doi: 10.1107/S0907444904015094. [DOI] [PubMed] [Google Scholar]
- 14.Seiner DR, Gates KS. Chem. Res. Toxicol. 2007;20:1315–1320. doi: 10.1021/tx700213s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu S, Yang H, He Y, Jiang Z-H, Kumar S, Wu L, Zhang Z-Y. J. Am. Chem. Soc. 2008;130:8251–8260. doi: 10.1021/ja711125p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arabaci G, Guo X-C, Beebe KD, Coggeshall KM, Pei D. J. Am. Chem. Soc. 1999;121:5085–5086. [Google Scholar]
- 17.Barrett WC, DeGnore JP, Keng Y-F, Zhang Z-Y, Yim MB, Chock PB. J. Biol. Chem. 1999;274:34543–34546. doi: 10.1074/jbc.274.49.34543. [DOI] [PubMed] [Google Scholar]
- 18.Denu JM, Tanner KG. Biochemistry. 1998;37:5633–5642. doi: 10.1021/bi973035t. [DOI] [PubMed] [Google Scholar]
- 19.Bhattacharya S, LaButti JN, Seiner DR, Gates KS. Bioorganic Med. Chem. Lett. 2008;18:5856–5859. doi: 10.1016/j.bmcl.2008.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LaButti JN, Chowdhury G, Reilly TJ, Gates KS. J. Am. Chem. Soc. 2007;129:5320–5321. doi: 10.1021/ja070194j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LaButti JN, Gates KS. Bioorg. Med. Chem. Lett. 2009;19:218–221. doi: 10.1016/j.bmcl.2008.10.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nho CW, O'Dwyer PJ. J. Biol. Chem. 2004;279:26019–26027. doi: 10.1074/jbc.M309022200. [DOI] [PubMed] [Google Scholar]
- 23.Manandar S, Cho J-M, Kim J-A, Kensler TW, Kwak M-K. Eur. J. Pharmacol. 2007;577:17–27. doi: 10.1016/j.ejphar.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 24.Carey KA, Kensler TW, Fishbein JC. Chem. Res. Toxicol. 2001;14:939–945. doi: 10.1021/tx0100340. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y-Z, Zhang Z-Y. Anal. Biochem. 1998;261:139–148. doi: 10.1006/abio.1998.2738. [DOI] [PubMed] [Google Scholar]
- 26.Velanyutham M, Villamena FA, Fishbein JC, Zweier JL. Arch. Biochem. Biophys. 2005;435:83–88. doi: 10.1016/j.abb.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 27.Chen C-Y, Willard D, Rudolph J. Biochemistry. 2009;48:1399–1409. doi: 10.1021/bi801973z. [DOI] [PubMed] [Google Scholar]
- 28.Neel BG, Gu H, Pao L. Trends Biochem. Sci. 2003;28:284–293. doi: 10.1016/S0968-0004(03)00091-4. [DOI] [PubMed] [Google Scholar]
- 29.Julien SG, Dubé N, Read M, Penney J, Paquet M, Han Y, Kennedy BP, Muller WJ, Tremblay ML. Nat. Genetics. 2007;39:338–346. doi: 10.1038/ng1963. [DOI] [PubMed] [Google Scholar]
- 30.Torres M, Forman HJ. Ann. NY Acad. Sci. 2002;973:345–348. doi: 10.1111/j.1749-6632.2002.tb04663.x. [DOI] [PubMed] [Google Scholar]
- 31.Jain AK, Mahajan S, Jaiswal AK. J. Biol. Chem. 2008;283:17712–17720. doi: 10.1074/jbc.M709854200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Ruggeri BA, Robinson C, Angeles T, Wilkinson J, IV, Clapper ML. Clin. Cancer Res. 2002;8:267–274. [PubMed] [Google Scholar]
- 33.Mannell H, Hellwig N, Gloe T, Plank C, Sohn H-Y, Groesser L, Walzog B, Pohl U, Krötz F. Vascular Res. 2007;45:153–163. doi: 10.1159/000110081. [DOI] [PubMed] [Google Scholar]
- 34.Imbert V, Rupec RA, Livolsi A, Pahl HL, Traenckner EB-M, Mueller-Dieckmann C, Farahifar D, Rossi B, Auberger P, Baeuerle PA, Peyron J-F. Cell. 1996;86:787–798. doi: 10.1016/s0092-8674(00)80153-1. [DOI] [PubMed] [Google Scholar]
- 35.Johnson TO, Ermolieff J, Jirousek MR. Nat. Rev. Drug Disc. 2002;1:696–709. doi: 10.1038/nrd895. [DOI] [PubMed] [Google Scholar]
- 36.Duranton J, Adam C, Bieth JG. Biochemistry. 1998;37:11239–11245. doi: 10.1021/bi980223q. [DOI] [PubMed] [Google Scholar]