Abstract
Dendritic cells (DC) direct the magnitude, polarity and effector function of the adaptive immune response. DC express toll-like receptors (TLR), antigen capturing and processing machinery, and costimulatory molecules, which facilitate innate sensing and T cell activation. Once activated, DC can efficiently migrate to lymphoid tissue and prime T cell responses. Therefore, DC play an integral role as mediators of the immune response to multiple pathogens. Elucidating the molecular mechanisms involved in DC activation is therefore central in gaining an understanding of host response to infection. Unfortunately, technical constraints have limited system-wide ‘omic’ analysis of human DC subsets collected ex vivo. Here we have applied novel proteomic approaches to human myeloid dendritic cells (mDCs) purified from 100 milliliters of peripheral blood to characterize specific molecular networks of cell activation at the individual patient level, and have successfully quantified over 700 proteins from individual samples containing as little as 200,000 mDCs. The proteomic and network readouts after ex vivo stimulation of mDCs with TLR3 agonists is measured and verified using flow cytometry.
Keywords: Myeloid dendritic cells, Proteomics, Polyinosinic:polycytidylic acid, Innate Immunity, Flow Cytometry
1. Introduction
Dendritic cells (DC) are essential antigen presenting cells (APC) that effect the innate and adaptive immune response (Banchereau and Steinman, 1998). DC derived from peripheral blood have been characterized by cell surface phenotype and divided into 2 main subsets: myeloid dendritic cells and plasmacytoid dendritic cells (mDC and pDC). They both possess toll-like receptors (TLR), antigen capturing and processing machinery, and costimulatory molecules, which allow them to act as professional APC (Banchereau et al., 2000; Jarrossay et al., 2001; Ito et al., 2002). mDCs express TLR1-6, 8, and 10 providing selectivity in response to specific pathogen associated molecular pattern signals (Liu, 2005). Therefore, TLR3 agonists such as polyinosinic:polycytidylic acid (polyI:C) can selectively activate mDC. Once activated, DC can migrate to lymphoid tissue and prime T cell responses (Banchereau et al., 2000). Elucidating the molecular mechanisms responsible for DC activation is essential towards developing a better understanding of their role and utility in combating disease (Ueno et al, 2010.).
In addition to overall mechanistic questions related to mDC function overall, important questions as to attenuations of function of mDCs against backgrounds of chronic infections and immune perturbations such as seen in HIV or HCV is very important for understanding disease progression and guiding clinical practice. A systems-wide proteomic comparison at the individual patient level to monitor alterations of DC function in disease and identify pathways that can be targeted for therapy would permit patient stratification far beyond current medical practice. Unfortunately, technical limitations have hindered omic-scale studies of DCs collected ex vivo due to insufficient sample yield from standard peripheral blood preparations. To overcome these limitations, most studies have utilized monocyte derived DC (Mo-DC) cultured in vitro (Wang et al., 2008; Buschow et al., 2010; Luber et al., 2010). While these studies have provided important insights with respect to the molecular mechanisms at play during DC activation, Mo-DC are thought to differ in important biological respects from mDC (Horlock et al., 2007). Recently, advanced quantitative proteomic techniques have been applied to relatively small amounts of mouse splenic DC subsets, resulting in over 5,000 proteins identified from several million cells (Luber et al., 2010). This study utilized a label free proteomic technique which qualifies a peptide sequence (and associates it with its parent protein) by tandem mass spectrometry (MS/MS) identification. Peptide species are quantified by ion intensity, while individual peptides are grouped across samples based on precise mass and retention time measurements (Chelius and Bondarenko, 2002; Wang et al., 2003; Bantscheff et al., 2007). The thousands of peptides thus grouped are analyzed by well-established techniques that have been developed for analyzing high-dimensional data and permitted the variations in protein expression for different DC subsets to be established for the first time.
We have capitalized on these techniques to develop a sensitive and reproducible ex vivo proteomic analysis of human mDCs freshly purified from peripheral blood of individual donors. In this analysis, we performed two independent studies of label free expression of mDC which were stimulated with the TLR3 ligand poly(I:C) to test reproducibility of proteomic analysis of stimulation and response in mDCs. Importantly, we have developed a reproducible method which can analyze the proteome of DC subsets collected ex vivo for as few as 200,000 DC cells and reliably detect proteomic changes due to immune activation and/or chronic infection.
2. Materials and Methods
2.1. Samples
Venous blood samples were collected from healthy control subjects (age 25–53 (median41), 57% female, no known chronic viral infection or other medical illness). The label free study contained n = 6 biological replicates which were used in a paired design that had two treatment groups. The flow cytometry validation analysis contained n=7 biological samples. All study subjects provided written informed consent for phlebotomy in accordance with protocols approved by the institutional review boards for human studies at University Hospitals of Cleveland and Cleveland VA Medical Center.
2.2 mDC Isolation and poly(I:C) Stimulation
Peripheral blood mononuclear cells (PBMC) were prepared from 100 milliliters of fresh peripheral blood using Ficoll (Fisher Scientific, Hudson, NH). mDC were purified from PBMCs using CD1c (BDCA-1)+ dendritic cell isolation kit for human cells (Miltenyi Biotech, Auburn, CA), where CD19+ B cells are removed by negative selection, then mDCs are isolated by CD1c (BDCA-1) positive bead selection. Purified mDCs from 6 patients were split in two and incubated separately overnight in 96 well round bottom plate containing 5% human AB serum (Gemini Bio-Products, Woodland, CA). Samples were incubated in the presence or absence of 50 μg/mL poly(I:C) (Amersham Biosciences, Piscataway, NJ). mDC were removed from 96 well plates and were washed twice in sterile PBS before snap freezing cells for proteomic analysis.
2.3. mDC purity and activation by flow cytometric analysis
mDC were stained with anti-CCR7 PE-Cy7, anti-CD11c APC, (BD Biosciences, San Jose, CA). Flow cytometric analysis was performed on an LSRII flow cytometer (BD Biosciences) with FACSDiva Software (BD Biosciences) to establish baseline purity and analysis of CCR7 upregulation in response to poly(I:C) stimulation.
2.4. Label Free Protein Expression Studies
As outlined above, the six patient samples were split into two batches and incubated either with media or Poly(I:C) for 12 samples total. Each sample was lysed with a buffer of 1% Triton, 150mM NaCl, 20mM TRIS pH 8.0 and 0.1% protease inhibitor cocktail (Sigma-Aldrich, P2714, St.Louis, MO). The sample was incubated on ice for 1.5 hours and pulse-sonicated with a probe sonicator every 30 minutes. Following lysis, the samples were removed of detergent using the 2-D Clean-Up kit per manufacturer’s instructions (GE Healthcare Bio-Sciences, Piscataway, NJ) with the exception of the final solubilization step which was performed in 10 μL 8M urea. The samples were bath-sonicated on ice for 1 hour to resolubilize the protein pellet and 10 μL of Tris pH 8.0 was added to yield a final concentration of 4M urea. Subsequent to solubilization, total protein quantification of each sample was performed using BioRad protein assay per manufacturer’s instructions (BioRad Laboraories, Herucles, CA). Each sample was adjusted to 880 nanograms in 13 microliters with 50mM Tris pH 8.0. Dithiotheritol was added to a final concentration of 5mM and the samples were reduced at 37°C for 30 minutes and cooled to room temperature prior to alkylation with iodacetamide at a final concentration of 10mM for 30 minutes. A dual proteolytic digestion was performed with endopeptidase Lys C (Wako Chemicals, Richmond, VA) and trypsin with a final enzyme to protein ratio of 1:1 (w/w) for each protein. First, Lys C was added and incubated for 2 hours at 37°C and then subsequently adjusted to 2M urea with 50mM Tris pH 8.0 to accommodate the trypsin digestion which incubated overnight at 37°C.
One hundred nanograms of each sample were analyzed by LC/MS/MS. The samples were run in two separate batches with the order of sample injections randomized for the 6 samples in a batch. Separation of peptides via capillary liquid chromatography was performed using a Dionex Ultimate 3000 capillary LC system (Dionex Sunnyvale, CA). Mobile phase A (aqueous) contains 0.1% formic acid in 5% acetonitrile and mobile phase B (organic) contained 0.1% formic acid in 85% acetonitrile. Samples were trapped and desalted on-line in mobile phase A at 10 μL/min for 10 minutes using a Dionex PepMap 100, (300 μm × 5 mm). The sample was subsequently loaded onto a Dionex C18 PepMap (75 μm × 15 cm) reversed phase column with 5% mobile phase B. Separation was obtained by employing a gradient of 6% to 28% mobile B at 0.300 μL/min over 120 minutes. The column was washed at 99% mobile phase B for 10 minutes, followed by a re-equilibration at 100% A for 15 minutes. Mass spectrometry analyses of samples were performed using a LTQ-Orbitrap XL (Thermo, Waltham, MA). Positive mode electrospray was conducted using a nanospray source and the mass spectrometer (s) was operated at a resolution of 60,000. Quantitative and qualitative data were acquired using alternating full MS scan and MS/MS scans in normal mode. Survey data were acquired from m/z of 400 to 1600 and up to 8 precursors based on intensity, were interrogated by MS/MS per switch. Two micro scans were acquired for every precursor interrogated and MS/MS was acquired as centroid data. The instrument was mass calibrated immediately before the analysis using the instrument protocol. The raw data for each run were extracted via Rosetta Elucidator to provide MS/MS peak lists for identification and intensity based profile peak lists for quantification. The MS/MS peak lists were subsequently searched by Mascot version 2.2.0 (Matrix Science London, UK). The database used was the human International Protein Index (IPI) (68020 sequences). Search settings were as follows: trypsin enzyme specificity, mass accuracy window for precursor ion, 10 ppm; mass accuracy window for fragment ions, 0.8 Daltons; variable modification, including only carbamidomethylation of cysteines and oxidation of methionine. Peptide and protein assignments were made using peptide and protein teller algorithms (Keller et al., 2002; Nesvizhskii et al., 2003) in Rosetta Elucidator with a false discovery rate of 1% (Rosetta Biosoftware, Seattle, WA). In addition, to provide additional confidence in the assignments we considered proteins that had > 2 peptides matching the above criteria to be confirmed assignments while proteins identified with one peptide which met the above criteria were highlighted as tentative assignments. Automated differential quantification of peptides in a set of samples was accomplished with Rosetta Elucidator as previously described (Neubert et al., 2008; Chan et al., 2009). Estimated protein intensities for each sample were determined by summing the peptide intensities for individual proteins. In addition, a Student’s T-test was performed comparing protein intensities across media and poly(I:C) stimulated groups and p values were reported for these comparisons. Overall protein fold changes were determined by dividing the median protein intensity for each peptide in each treatment group.
2.5. Data Pre-Processing
ANOVA models require at least the following two basic assumptions: the normality distribution of variable measurements (peptide expressions) and the homoscedasticity between sample groups (LCMS runs/observations) in multi-group designs. Since expression levels from high-throughput data do not generally follow these assumptions, a transformation is often required. We applied here a log-transformation, followed by a joint adaptive mean-variance regularization procedure (Dazard and Rao, 2010; Dazard and Rao, 2011), now available as a R package called “MVR”. This procedure simultaneously overcomes the issues of lack of degrees of freedom and variance-mean dependency in this type of data where the number of variables is ≫ greater than the number of samples.
2.6. Power Analysis
We performed power analysis to determine a sample size per group that would assure a certain power for variables with a fold change of 2.5 between stimulated and non-stimulated groups. After variance stabilization and normalization of the data (see sub-section 2.5 above), the usual distributional assumptions of test statistics are satisfied. Power was calculated for a regular two-sided paired t-test statistic, for a range of estimated standard deviations in common to all peptides σ̂0.50–0.65, and for a range of values of the estimated proportion of non-differentially expressed peptides π̂0 ≈ 0.85–0.95, defined and estimated in (Storey et al., 2004), while controlling the 0 Positive-False Discovery Rate at or below 5% pFDR ≤ α = 0.05. Under the above assumptions and estimations, results show that the experimental design can achieve up to ~ 65 – 93% power with a sample size of n = 6 independent biological replicates per experimental group (Supplemental Figure 1).
2.7. Ingenuity Pathway Analysis
Following statistical analysis, significant proteins from both studies (P value < = 0.05 and a fold change > 2.5) and their corresponding estimated fold changes were imported into Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA, USA). Once imported, the software utilized biomedical literature and existing protein interaction databases to annotate cellular locations and elucidate biological networks within the uploaded protein lists. (www.ingenuity.com).
3.0 Results
3.1 Label Free Protein Expression
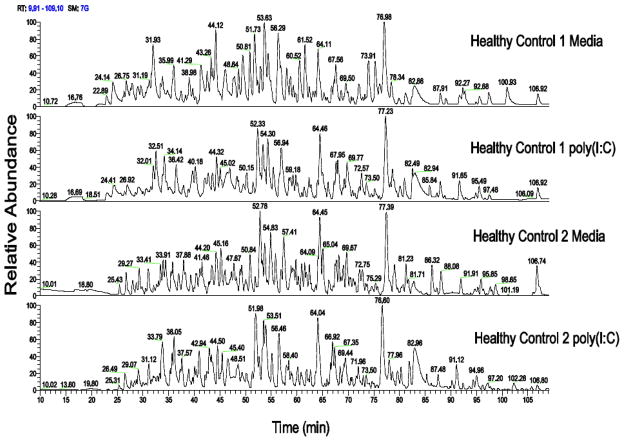
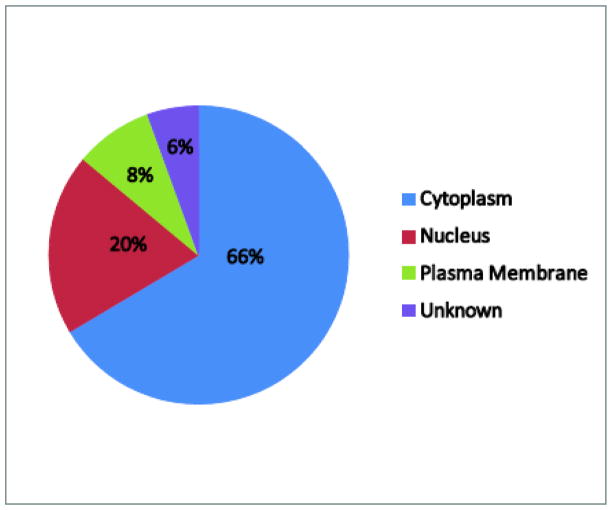
The sample preparation protocol we have developed provided sufficient protein recovery for proteomic analysis. Table 1 highlights the total protein concentration for all samples analyzed after cell lysis and detergent removal. The procedure consistently yielded greater than 1.5 microgram of total protein which is a sufficient amount for proteomic analysis via LC/MS/MS. For the two batch runs of 6 LC-MS samples, the samples were subsequently digested and analyzed by LC/MS/MS. Figure 1 highlights the chromatographic reproducibility in terms of intensity and retention time among two media and poly(I:C) samples. In addition, each label free study provided good proteomic coverage with over 2500 peptides detected and quantified which mapped to 679 and 574 proteins respectively for each of the two studies. Overall, a total of 725 non-redundant proteins were identified across the two studies. These identified proteins are shown in table S1. The label free method provided simultaneous measurements of both intra and extracellular proteins. The pie chart in Figure 2 displays the distribution of cellular location for proteins that were detected in both label free studies using annotations from IPA. As anticipated, many of the proteins we identified were cytoplasmic, with 66% of the proteins identified located in the cytoplasm. However, both nuclear and plasma membrane were also represented, comprising 20% and 8% of the total proteins identified, respectively.
Table 1.
Protein concentrations for each sample analyzed in both studies after sample preparation for label free analysis
| Sample | Label Free Study | Cell count | Protein Yield (total micorgrams) |
|---|---|---|---|
| 1 Media | 1 | 260,000 | 1.7 |
| 1 polyl:C | 1 | 260,000 | 2.4 |
| 55 Media | 1 | 375,000 | 2.4 |
| 55 polyl:C | 1 | 375,000 | 2.8 |
| 75 Media | 1 | 550,000 | 2.9 |
| 75 polyl:C | 1 | 550,000 | 2.1 |
| 42 Media | 2 | 475,000 | 1.8 |
| 42 polyl:C | 2 | 475,000 | 2.8 |
| 73 Media | 2 | 550,000 | 7.1 |
| 73 polyl:C | 2 | 550,000 | 2.9 |
| 103 Media | 2 | 950,000 | 2.5 |
| 103 polyl:C | 2 | 950,000 | 2.5 |
Figure 1.
mDC samples were freshly prepared from 2 separate healthy control subjects. Chromatograms are shown for these two individual mDC samples stimulated with poly(I:C) or media.
Figure 2.
Cellular location pie chart for non redundant proteins identified in both label free studies. Cellular locations were assigned for each protein using IPA annotations.
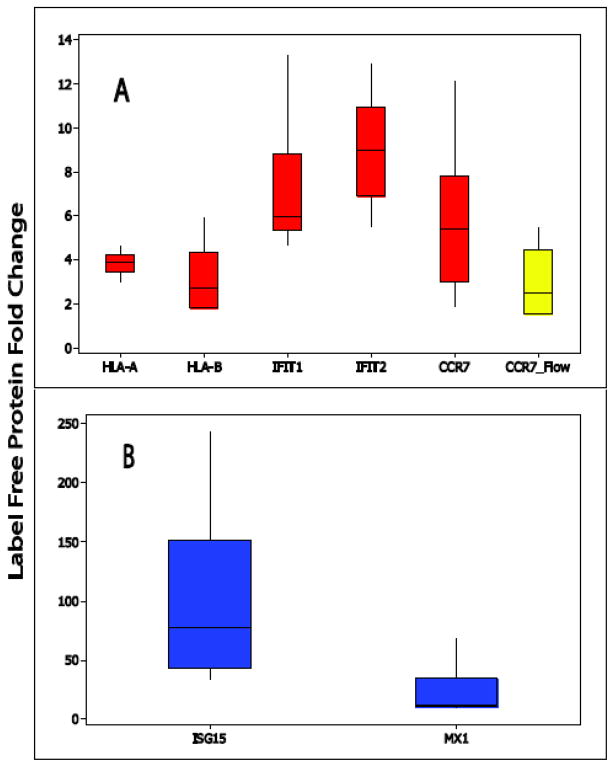
Importantly, reproducible fold changes were observed across immune-related proteins detected in both studies, suggesting the method measured reliable and consistent activation responses for mDC stimulation. This included C-C chemokine receptor 7 (CCR7) whose expression is up-regulated during DC maturation (Sallusto et al., 1999; Forster et al., 2008). CCR7 along with its ligands, CCL19 and CCL21 are important regulators in leuckocyte trafficking and cellular movement. (Forster et al., 2008). The label free studies detected up-regulation of greater than 5 fold in both CCR7 and CCL19 upon poly(I:C) stimulation. Figure 3 highlights the distribution of fold change for CCR7 as well as other selected immune-related proteins that were significantly changed due to DC activation and also highlights the distribution fold change for CCR7 analyzed via flow cytometry on seven additional (and independent) samples. We observed good agreement with regard to up-regulation across both platforms for CCR7 with a median fold change of 5.3 (error range 12.9 to 1.9) for label free proteomics and 2.3 (error range 5.5 to 1.5) for flow cytometry results.
Figure 3.
Box-Whiskers plots of fold change for selected immune related proteins across all samples analyzed in both studies. Panel A highlights proteins with an average fold change of <10 and panel B highlights protein with >15 fold change (stimulated versus non-stimulated). The yellow box-whisker plot highlights flow cytometry fold change (stimulated versus non-stimulated) for a selected protein, CCR-7.
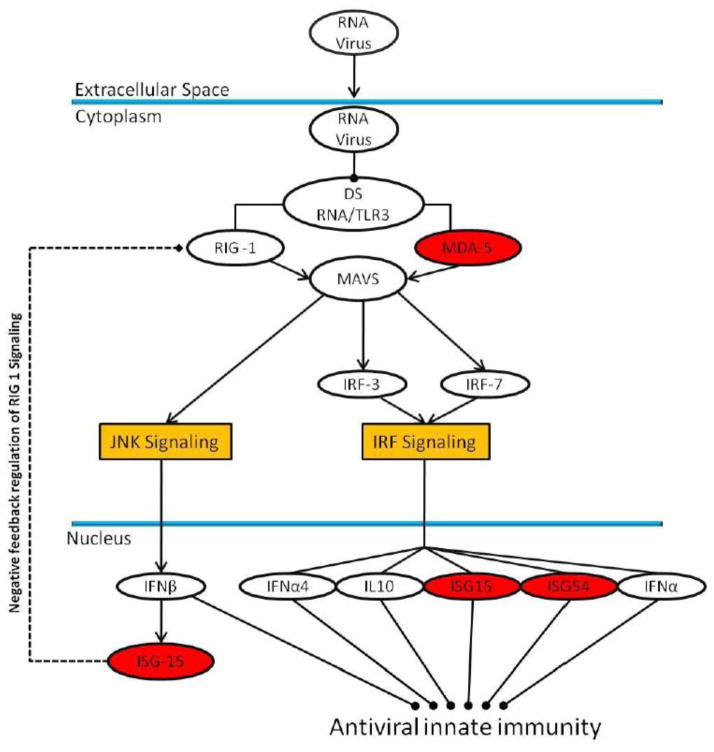
Using the statistical cutoffs described in the methods section, a total of 60 proteins were identified as having a significant fold change and were subsequently imported into IPA for pathway analysis. Table 2 highlights the fold change for selected immune activated proteins identified in this analysis. Overall, 25 proteins were mapped to 9 relevant immunologic pathways. These pathways included: dendritic cell maturation, interferon signaling, antigen presentation pathway, calveolar-mediated endocytosis, activation of interferon regulatory factor (IRF) signaling by pattern recognition receptors, cross-talk between dendritic and natural killer cells and communication between innate and adaptive immune cells. Figure 4 highlights one example pathway, activation of IRF signaling by pattern recognition receptors and the proteins identified as changing in our analysis.
Table 2.
Selected proteins that change in abundance due to poly(I:C) stimulation. FC S/M column represents fold change values from stimulated (poly(I:C)) versus non stimulated (media) comparison.
| FC S/M | Primary Protein Name | Protein Description |
|---|---|---|
| 5.7 | IPI :IPI00011549.1 | TRAF1 TNF receptor-associated factor 1 |
| 5.8 | IPI:IPI00295400.1 | WARS Tryptophanyl-tRNA synthetase, cytoplasmic |
| 5.8 | IPI:IPI00027687.1 | CCR7 C-C chemokine receptor type 7 |
| 6.9 | IPI:IPI00018300.2 | IFIT1 Interferon-induced protein with tetratricopeptide |
| 7.2 | IPI:IPI00005577.5 | IFIH1 Isoform 2 of Interferon-induced helicase C domain (MDA-5) |
| 8.2 | IPI:IPI00303726.3 | IFITM2 Interferon-induced transmembrane protein 2 |
| 8.3 | IPI:IPI00157365.2 | NUB1 Isoform 1 of NEDD8 ultimate buster 1 |
| 8.9 | IPI:IPI00847322.1 | SOD2 manganese superoxide dismutase isoform A precursor |
| 8.9 | IPI:IPI00028564.1 | GBP1 Interferon-induced guanylate-binding protein 1 |
| 8.9 | IPI:IPI00018298.3 | IFIT2 Interferon-induced protein with tetratricopeptide |
| 9.0 | IPI:IPI00291463.4 | RSAD2 Radical S-adenosyl methionine domain-containing p |
| 9.7 | IPI:IPI00217049.4 | OAS2 Isoform p69 of 2′-5′-oligoadenylate synthase 2 |
| 9.8 | IPI:IPI00028096.3 | IDO1 Indoleamine 2,3-dioxygenase |
| 15.0 | IPI:IPI00303726.3 | IFITM3 Interferon-induced transmembrane protein 3 |
| 16.1 | IPI:IPI00167949.6 | MX1 Interferon-induced GTP-binding protein Mx1 |
| 17.9 | IPI:IPI00647246.1 | ISG20 Isoform 1 of Interferon-stimulated gene 20 kDa pr |
| 27.3 | IPI:IPI00024254.3 | IFIT3 Interferon-induced protein with tetratricopeptide |
| 69.8 | IPI:IPI00375631.6 | ISG15 Interferon-induced 17 kDa protein |
| 2398.5 | IPI:IPI00479775.1 | CCL19 Chemokine (C-C motif) ligand 19 |
Figure 4.
IRF signaling pathway that was generated from pathway analysis of proteins with significant fold changes across both studies. The proteins highlighted in red were detected in our analysis and had a significant increase in abundance upon stimulation.
4.0 Discussion
As mentioned above, in vivo proteomic studies of DC have been hindered due to insufficient sample and purity. This was primarily due to their scarcity in peripheral blood and the lack of unique surface markers. Recent discoveries of new surface markers coupled with advances in commercially available immunomagnetic selection have enabled improved isolation and purification of DC subsets including mDC. However, DCs are very rare in perhipheral blood, comprising only 1% of circulating peripheral blood mononuclear cells (PBMCs), and even with improved isolations techniques average mDC yield from a standard blood draw (100 milliters or less) range from approximately 200,000–800,000 cells. In addition, quantitative proteomics requires cell lysis and detergent removal subsequent to mass spectrometry analysis with most procedures experiencing some degree of protein loss during this step. Therefore, good sample preparation and ultra-sensitive quantitative techniques are essential to analyzing human peripheral blood DCs. The methods described above have successfully addressed these challenges and have provided a robust, reproducible platform from which to probe DC subsets collected from 100 mL of peripheral blood. This proteomic technique can be applied to smaller blood collections of less than 100 milliliters however its success will be dependent upon the number of DCs collected from the sample rather than the volume of the draw itself.
In addition, the method quantified extracellular and intracellular proteins simultaneously without the need for antibodies or additional treatment of the cells. Moreover, the ability to measure over 700 proteins from DC subsets enables the application of pathway analysis tools such as IPA which can identify molecular networks and monitor relevant immuno-biological pathways across states of immune activation. One such example that was described above was interferon signaling. Poly(I:C) is a synthetic double stranded RNA that is a potent inducer of TLR3 signaling and subsequent interferon (IFN) production (Field et al., 1967). Upon poly(I:C) recognition, TLR3 elicits an activation cascade via the adaptor molecule Toll-IL-1 receptor domain containing adaptor molecule 1 (TICAM-1) which results in activation of transcription factors including interferon regulatory factor 3 (IRF-3) and NF-κB. This leads to production of type 1 interferons and subsequent interferon inducible proteins. In addition, other cytosolic dsRNA receptors such retinoic acid-inducible gene-I (RIG-I) and melanoma differentiation associated antigen 5 (MDA-5) recognize poly(I:C) and induce IRF signaling in mice (Gitlin et al., 2006; Kato et al., 2006). Moreover, recent work investigating the role of TLR3 and MDA5 have suggested that both are essential to produce interferon in response to poly(I:C) in human mDCs (Perrot et al., 2010). Interestingly, we have identified a significant increase of MDA5 in poly(I:C) stimulated DCs as well as in some of the corresponding interferon-inducible proteins, suggesting engagement of MDA5 in response to poly(I:C). Figure 3 highlights the magnitude of change for inducible proteins including interferon-induced tetratricopeptide protein 1 and 2 (IFIT1 and IFIT2), interferon-induced GTP-binding protein (MX1) and interferon-induced 17kDa protein (ISG15). The magnitude of change was over 5 fold for these proteins with a robust increase of approximately 70 fold for ISG15 upon stimulation. ISG15 is an ubiquitin-like protein which is up-regulated in cells upon IFN treatment and conjugates to proteins, a process commonly referred to as ISGylation (Skaug and Chen, 2010). Studies in mouse models of viral infection have suggested that ISG15 plays an important role in mammalian antiviral immunity (Lenschow et al., 2007). In this work, ISG−/− mice experienced increase susceptibility to influenza, herpes virus type 1 and Sindbis virus infection. Mechanistic studies have suggested ISG15 targets viral proteins and ISGylation of these proteins may impair viral replication and impairment of viral protein function (Skaug and Chen, 2010). Interestingly, ISG15 mRNA has been shown to have a strong up-regulation upon poly(I:C) stimulation in mDC, resulting in increased abundance of free ISG15 protein and subsequent high molecular weight ISG15 conjugates 12 hours post-stimulation (Ebstein et al., 2009). It is suspected these modifications may be important in proteasomal degradation and antigen presentation in mDC.
5.0 Conclusion
We have developed a method for robust and reproducible proteomic analysis of human dendritic cells. The method is amendable to ex vivo collections of mDC with as little as 200,000 DC enabling -omic scale analysis in individual samples collected from peripheral blood of individual human subjects. In addition, the method simultaneously measures hundreds of proteins across different cellular compartments without additional treatments to enable quantification. Our data suggest there are reliable and consistent protein profile changes upon DC activation, many of which are associated with immune activation. Finally, we have observed consistent up-regulation in the measurement of CCR7 using flow cytometry and label free protein expression providing cross-validation for both methods. The methods developed here provide a tool to evaluate the immunological response of DC in the context infection and disease.
Supplementary Material
Highlights.
Ultra sensitive quantitative method for proteomic analysis of myleiod dendritic cells purified from 100mL human peripherial blood
Molecular network analysis of PolyI:C stimulated myleiod dendritic cells
Validation of proteomic measures via flow cytometry
Acknowledgments
The authors would like to thank John C. Tilton for discussion and comments regarding this manuscript. In addition, we thank Sara Tomechko for mass spectrometry support and analysis. This work was funded by National Institute of Health grant P20-DA-026133, NIH R01 DK068361 and NIH R01 DK068361, the CWRU Center for AIDS Research Core facilities (AI 36219).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu YJ, Pulendran B, Palucka K. Immunobiology of dendritic cells. Annu Rev Immunol. 2000;18:767–811. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–52. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem. 2007;389:1017–31. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- Buschow SI, Lasonder E, van Deutekom HW, Oud MM, Beltrame L, Huynen MA, de Vries IJ, Figdor CG, Cavalieri D. Dominant processes during human dendritic cell maturation revealed by integration of proteome and transcriptome at the pathway level. J Proteome Res. 2010;9:1727–37. doi: 10.1021/pr9008546. [DOI] [PubMed] [Google Scholar]
- Chan EY, Sutton JN, Jacobs JM, Bondarenko A, Smith RD, Katze MG. Dynamic host energetics and cytoskeletal proteomes in human immunodeficiency virus type 1-infected human primary CD4 cells: analysis by multiplexed label-free mass spectrometry. J Virol. 2009;83:9283–95. doi: 10.1128/JVI.00814-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelius D, Bondarenko PV. Quantitative profiling of proteins in complex mixtures using liquid chromatography and mass spectrometry. J Proteome Res. 2002;1:317–23. doi: 10.1021/pr025517j. [DOI] [PubMed] [Google Scholar]
- Dazard J-E, Rao JS. JSM Proceedings. High-Dimensional Data Analysis and Variable Selection Section., Vol. IMS – JSM. American Statistical Association; Vancouver, BC. Canada: 2010. Regularized Variance Estimation and Variance Stabilization of High-Dimensional Data; pp. 5295–5309. [PMC free article] [PubMed] [Google Scholar]
- Dazard J-E, Rao JS. Joint Adaptive Mean-Variance Regularization and Variance Stabilization of High Dimensional Data. Comput Statist Data Anal. 2011 doi: 10.1016/j.csda.2012.01.012. (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebstein F, Lange N, Urban S, Seifert U, Kruger E, Kloetzel PM. Maturation of human dendritic cells is accompanied by functional remodelling of the ubiquitin-proteasome system. Int J Biochem Cell Biol. 2009;41:1205–15. doi: 10.1016/j.biocel.2008.10.023. [DOI] [PubMed] [Google Scholar]
- Field AK, Tytell AA, Lampson GP, Hilleman MR. Inducers of interferon and host resistance. II. Multistranded synthetic polynucleotide complexes. Proc Natl Acad Sci U S A. 1967;58:1004–10. doi: 10.1073/pnas.58.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster R, Davalos-Misslitz AC, Rot A. CCR7 and its ligands: balancing immunity and tolerance. Nat Rev Immunol. 2008;8:362–71. doi: 10.1038/nri2297. [DOI] [PubMed] [Google Scholar]
- Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci U S A. 2006;103:8459–64. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlock C, Shakib F, Mahdavi J, Jones NS, Sewell HF, Ghaemmaghami AM. Analysis of proteomic profiles and functional properties of human peripheral blood myeloid dendritic cells, monocyte-derived dendritic cells and the dendritic cell-like KG-1 cells reveals distinct characteristics. Genome Biol. 2007;8:R30. doi: 10.1186/gb-2007-8-3-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Amakawa R, Kaisho T, Hemmi H, Tajima K, Uehira K, Ozaki Y, Tomizawa H, Akira S, Fukuhara S. Interferon-alpha and interleukin-12 are induced differentially by Toll-like receptor 7 ligands in human blood dendritic cell subsets. J Exp Med. 2002;195:1507–12. doi: 10.1084/jem.20020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrossay D, Napolitani G, Colonna M, Sallusto F, Lanzavecchia A. Specialization and complementarity in microbial molecule recognition by human myeloid and plasmacytoid dendritic cells. Eur J Immunol. 2001;31:3388–93. doi: 10.1002/1521-4141(200111)31:11<3388::aid-immu3388>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–5. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–92. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Lenschow DJ, Lai C, Frias-Staheli N, Giannakopoulos NV, Lutz A, Wolff T, Osiak A, Levine B, Schmidt RE, Garcia-Sastre A, Leib DA, Pekosz A, Knobeloch KP, Horak I, Virgin HWt. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc Natl Acad Sci U S A. 2007;104:1371–6. doi: 10.1073/pnas.0607038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu Rev Immunol. 2005;23:275–306. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- Luber CA, Cox J, Lauterbach H, Fancke B, Selbach M, Tschopp J, Akira S, Wiegand M, Hochrein H, O’Keeffe M, Mann M. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity. 2010;32:279–89. doi: 10.1016/j.immuni.2010.01.013. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Neubert H, Bonnert TP, Rumpel K, Hunt BT, Henle ES, James IT. Label-free detection of differential protein expression by LC/MALDI mass spectrometry. J Proteome Res. 2008;7:2270–9. doi: 10.1021/pr700705u. [DOI] [PubMed] [Google Scholar]
- Perrot I, Deauvieau F, Massacrier C, Hughes N, Garrone P, Durand I, Demaria O, Viaud N, Gauthier L, Blery M, Bonnefoy-Berard N, Morel Y, Tschopp J, Alexopoulou L, Trinchieri G, Paturel C, Caux C. TLR3 and Rig-like receptor on myeloid dendritic cells and Rig-like receptor on human NK cells are both mandatory for production of IFN-gamma in response to double-stranded RNA. J Immunol. 2010;185:2080–8. doi: 10.4049/jimmunol.1000532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F, Palermo B, Lenig D, Miettinen M, Matikainen S, Julkunen I, Forster R, Burgstahler R, Lipp M, Lanzavecchia A. Distinct patterns and kinetics of chemokine production regulate dendritic cell function. Eur J Immunol. 1999;29:1617–25. doi: 10.1002/(SICI)1521-4141(199905)29:05<1617::AID-IMMU1617>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Skaug B, Chen ZJ. Emerging role of ISG15 in antiviral immunity. Cell. 2010;143:187–90. doi: 10.1016/j.cell.2010.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, ETJ, DS Strong control, conservative point estimation, and simultaneous conservative consistency of false discovery rates: A unified approach. J R Statist Soc. 2004;66:187–205. [Google Scholar]
- Ueno H, Schmitt N, Klechevsky E, Pedroza-Gonzalez A, Matsui T, Zurawski G, Oh S, Fay J, Pascual V, Banchereau J, Palucka K. Harnessing human dendritic cell subsets for medicine. Immunol Rev. 2010;234:199–212. doi: 10.1111/j.0105-2896.2009.00884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, Staniforth V, Chiao MT, Hou CC, Wu HM, Yeh KC, Chen CH, Hwang PI, Wen TN, Shyur LF, Yang NS. Genomics and proteomics of immune modulatory effects of a butanol fraction of echinacea purpurea in human dendritic cells. BMC Genomics. 2008;9:479. doi: 10.1186/1471-2164-9-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zhou H, Lin H, Roy S, Shaler TA, Hill LR, Norton S, Kumar P, Anderle M, Becker CH. Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Anal Chem. 2003;75:4818–26. doi: 10.1021/ac026468x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.