Abstract
Photoactivation of rhodopsin in lipid bilayers results within milliseconds in a metarhodopsin I (MI) – metarhodopsin II (MII) equilibrium that is very sensitive to the lipid composition. It has been well established that lipid bilayers that are under negative curvature elastic stress from incorporation of lipids like phosphatidylethanolamines (PE) favor formation of MII, the rhodopsin photointermediate that is capable of activating G protein. Furthermore, formation of the MII state is favored by negatively charged lipids like phosphatidylserine and by lipids with longer hydrocarbon chains that yield bilayers with larger membrane hydrophobic thickness. Cholesterol and rhodopsin-rhodopsin interactions from crowding of rhodopsin molecules in lipid bilayers shift the MI – MII equilibrium towards MI. A variety of mechanisms seems to be responsible for the large, lipid-induced shifts between MI and MII: adjustment of the thickness of lipid bilayers to rhodopsin and adjustment of rhodopsin helicity to the thickness of bilayers, curvature elastic deformations in the lipid matrix surrounding the protein, direct interactions of PE headgroups and polyunsaturated hydrocarbon chains with rhodopsin, and direct or lipid-mediated interactions between rhodopsin molecules.
Keywords: rhodopsin, metarhodopsin I, metarhodopsin II, lipid-protein interaction, membranes, elastic energy, hydrophobic mismatch, docosahexaenoic acid, NMR, protein oligomerization
1. Introduction
Despite recent progress with expression, purification, crystallization and structural studies of several G protein-coupled membrane receptors (GPCR) of class A, rhodopsin, the mammalian dim-light receptor, remains the best-studied GPCR. The initial events of scotopic visual transduction take place in the disk membranes located in the rod outer segment (ROS) of rod cells. Each mammalian ROS consists of a stack of 1,000–2,000 distinct disks enclosed by the plasma membrane. The disks are formed from evaginations of the plasma membrane and move up the length of the rod cell as the disks age [1]. Rhodopsin is the major protein component (>90%) of a given disk and occupies approximately on third of its area [2]. The average disk membrane phospholipid composition consists of approximately 44% phosphatidylcholine (PC), 41% phosphatidylethanolamine (PE), 13% phosphatidylserine (PS), and 2% phosphatidylinositol (PI) [3]. The disk membrane contains a high proportion of the polyunsaturated fatty acid DHA in PS, PE, and PC [4 ]. Cholesterol concentration decreases from 30 mol% of all phospholipids to about 5 mol% as the disks age [5, 6]. Lipid distribution is asymmetric with a preferential location of PE and PS in the outer leaflet [7].
Rhodopsin has seven transmembrane helices (TM) that are connected by cytoplasmic and extracellular loops as well as an eights amphipathic cytoplasmic helix that follows TM7 and runs parallel to the membrane surface. The integral membrane receptor consists of the opsin apoprotein and the chromophore 11-cis retinal which is covalently bound by a protonated Schiff base to Lys 296 in TM7 and acts in the dark as a strong inverse agonist to constrain rhodopsin in the inactive conformation with a visible light adsorption maximum at λmax=500 nm [8, 9].
Absorption of a photon generates the moderately strong agonist, all-trans retinal, in situ and leads to the formation of a spectrally distinguishable equilibrium between a heterogeneous population of G protein-binding competent metarhodopsin II (MII, λmax=380 nm) and an inactive metarhodopsin I (MI, λmax=478 nm) [10]. MI formation occurs within a few microseconds and involves a series of fast transformations mainly occurring near the retinal binding pocket so that its overall conformation is very similar to dark-adapted rhodopsin [11]. In contrast, MII formation takes place on the timescale of milliseconds and is characterized by larger conformational changes taking place outside the protein photochemical core that are the consequence of three sequential events (i) the deprotonation of the retinal Schiff base and protonation of its complex counterion [12, 13], (ii) an outward tilt of TM6 [14, 15], and (iii) the proton uptake by a glutamic acid residue in the microdomain that forms the ionic lock between TM3 and TM6 [13]. Those structural changes alter the cytoplasmic receptor surface to allow G protein binding. MII activates multiple copies of the G protein transducin (Gt) in succession [16], setting off a biochemical amplification cascade that ultimately results in vivo in the signaling of second order neurons and the visual signal. In rod outer segments, the reaction ends with rhodopsin phosphorylation by rhodopsin kinases that allow arrestin binding and prevent further activation of Gt [17]. Eventually, the photolyzed chromophore is released and opsin is formed. In a physiological setting, 11-cis-retinal is metabolically supplied during the visual cycle and the 11-cis-retinal-bound dark adapted state is regenerated [18].
The influence of lipid bilayer properties on rhodopsin photoactivation has been studied for the past 25 years by the laboratories of Litman and Mitchell [19, 20], Brown [21, 22], and more recently by us. The aim of this review is to summarize those data and to provide a mechanistic interpretation of results.
2. Experimental observations
The MI-MII equilibrium is sensitive to:
Bilayer curvature elastic stress. Addition of PE to the lipid matrix generates negative curvature elastic stress in lipid monolayers and shifts the MI-MII equilibrium towards MII with increasing PE concentrations [21, 23–25].
Structure and dynamics of annular lipids. Lipids with a positively charged amino group like PE or methylated PE, or lipids with polyunsaturated acyl chains like docosahexaenoic acid (DHA) favor formation of MII by additional mechanisms unrelated to curvature elastic stress. Rhodopsin function is influenced by direct interactions of PE headgroups and DHA chains with rhodopsin [24].
Membrane thickness. With increasing thickness of bilayers composed of di-monounsaturated PCs, the MI-MII equilibrium is shifted toward MII for lipids with 14–18 carbon atoms per chain. The equilibrium shifts back toward MI for lipids with 20–24 carbon atoms per chain [26]. For a series of PCs with a perdeuterated, saturated sn-1 hydrocarbon chain and a monounsaturated sn-2 chain, both with 14–20 carbons per chain, the MI-MII equilibrium shifts steadily toward MII with increasing acyl chain length [27]. The lipids adjust to the length mismatch to rhodopsin by stretching or compressing hydrocarbons chains as detected by 2H NMR order parameter measurements. The crossover from stretching to compression occurs at a bilayer hydrophobic thickness of 27 Å [28]. Furthermore, an increase of the helicity of rhodopsin with increasing hydrocarbon chain length was detected by circular dichroism (CD) suggesting that the length of transmembrane helices adjusts to bilayer thickness [27].
Rhodopsin-rhodopsin interactions. For rhodopsin in POPC or SDPC bilayers, the MI-MII equilibrium is shifted towards MI with increasing rhodopsin concentration [26, 28, 29]. This shift is correlated with a reduced fraction of perturbed lipids per rhodopsin detected by 2H NMR [28] as well as an increase of Förster resonance energy transfer (FRET) efficiency between labeled rhodopsin molecules suggesting that the shift toward MI is related to rhodopsin oligomerization [26]. The onset of oligomerization depends on the degree of mismatch of hydrophobic thickness between the lipid matrix and rhodopsin [28, 30–32] .
Electric membrane surface potential. Increasing concentrations of negatively charged phosphatidylserine (PS) at neutral pH or the lowering of pH shift the MI-MII equilibrium toward MII [33].
Cholesterol concentration. Addition of cholesterol to PC membranes shifts the MI-MII equilibrium towards MI [34–36].
3. Rhodopsin-lipid interaction
In this section we attempt interpretation of the experimental results presented above. How are the properties of membranes linked to the MI-MII equilibrium? The free energy of MI or MII in a lipid matrix is the sum of the protein’s intrinsic free energy plus the energy of the surrounding lipid domain perturbed by lipid-protein interactions. The position of the MI-MII equilibrium depends on the free energy difference between MI and MII. Rhodopsin resides with higher probability in the state that has lower free energy. Readers that want to learn more about the thermodynamics of the MI-MII equilibrium in a lipid matrix are referred to publications by the Brown laboratory [23, 25].
For all integral membrane proteins, transmembrane helices tend to be formed by mostly hydrophobic amino acids. The matching of the hydrophobic length of transmembrane helices to the thickness of the hydrophobic core of the bilayer is a general principle of membrane organization that must be met [37–40]. Exposure of hydrophobic sections of lipids or protein to water is energetically highly unfavorable and, therefore, prevented by structural adjustments. They include (i) stretching, compressing, or tilting of the fluid hydrocarbon chains of lipids in the lipid matrix surrounding the protein, (ii) extension or contraction of transmembrane helices, (iii) tilting of transmembrane helices either over their entire length, or tilting of helix segments that are separated by a kink, and (iv) a reduction of the lipid-exposed protein surface by protein oligomerization. The adjustments in the lipid matrix to the protein may also generate monolayer curvature [23], e.g. as a result of a tilt of transmembrane helices. Hydrophobic coupling between lipids and proteins and the resulting bilayer elastic deformations are interpreted by continuum elastic theory of lipid bilayers [40–42].
However, the influence of the lipid matrix on protein function cannot be entirely reduced to membrane elastic deformations [43]. Intuition, experimental results [44, 45], as well as molecular simulations [46, 47] strongly suggest that the lipids nearest to the protein participate in pointed interactions with protein segments via (i) hydrogen bonds between lipid headgroups and polar residues on the protein [24], (ii) electrostatic interactions between lipid charges and dipoles in the lipid-water interface and charged amino acids [48] , (iii) π- π interactions between double bonds in lipid hydrocarbon chains and aromatic sidechains on the protein [49], and (iv) cation-π-interactions between positively charged headgroups and aromatic sidechains [50]. Those interactions are unique for the first layer of lipids surrounding the protein, and they can not be treated by the formalism of elastic bilayer deformations that is based on interactions of lipids with each other and with water. In combination with structural differences between MI and MII that expose different regions of the protein to the lipid matrix, they may shift the MI-MII equilibrium as well.
3.1. Lipid-rhodopsin hydrophobic mismatch
3.1.1. Adjustment of bilayers to rhodopsin
The experiments on membranes with variable thickness revealed that rhodopsin prefers residing in bilayers with a hydrophobic thickness of 27 Å [28]. If bilayers are thinner (negative hydrophobic mismatch), the protein raises order parameters of lipids, suggesting that lipids near the protein stretch their hydrocarbon chains to match the hydrophobic length of the protein. If membranes are thicker (positive hydrophobic mismatch), the lipids near the protein have hydrocarbon chains with lower order, suggesting that bilayer thickness near the protein is reduced.
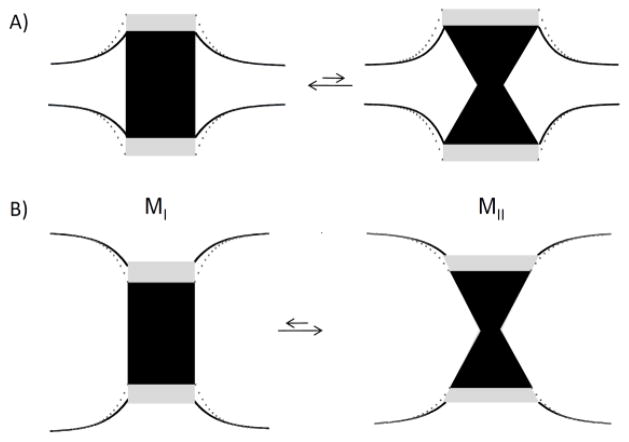
Why does increasing bilayer hydrophobic thickness favor MII? Bilayers with lower hydrophobic thickness than rhodopsin have a negative curvature in their lipid monolayers near the protein, while bilayers that are thicker have positive monolayer curvature (see Fig. 1). As it was discussed, the transition from MI to MII involves an outward movement of TM6 [14] and the elongation of TM5 [15], which generates negative curvature in monolayers near the protein. Thinner bilayers like MMPC (14:0–14:1n5-PC) experience a further increase in curvature stress from negative curvature while thicker bilayers like EEPC (20:0–20:1n9-PC) experience a reduction of stress from positive curvature (see Fig. 1). The reduction of curvature stress with increasing thickness of PC bilayers upon MII formation shifts the MI-MII equilibrium towards MII as it is observed experimentally.
Fig. 1.
Schematic presentation of the rhodopsin MI-MII equilibrium in a bilayer with (A) small hydrophobic thickness as in MMPC (14:0-14:1n-5-PC) and (B) large hydrophobic thickness as in EEPC (20:0-20:1n-9-PC). The structural transition from MI to MII is shown as change of shape from cylindrical (MI) to conical (MII). (A) Rhodopsin in thin bilayers has reduced helicity, probably reflecting a reduction in length of transmembrane helices (transition from grey to black). The bilayer near rhodopsin adjusts to the length of hydrophobic transmembrane helices by stretching and tilting of lipid hydrocarbon chains. Lipid monolayers near the protein have negative radii of curvature. (B) Rhodopsin in thick bilayers adjusts to bilayer hydrophobic thickness with an increase in helicity (transition from black to grey). Furthermore, the bilayer adjusts to rhodopsin with a reduction of thickness. Lipid monolayers near rhodopsin have positive radii of curvature.
3.1.2. Adjustment of rhodopsin helicity to bilayer thickness
The adjustment of bilayer thickness to the protein is only part of the story. The increase in rhodopsin helicity with increasing bilayer thickness indicates that the protein adjusts to the bilayer as well [27]. We suspect that the number of turns per transmembrane helix increases with increasing bilayer thickness. This is not surprising since helices tend to form or unwind from the ends where helical structures are less stable because they are not as well supported by intrahelical hydrogen bonds as in the center of a helix. Therefore, with increasing bilayer thickness, and under conditions where oligomerization can be ruled out (see below), two events are taking place: (i) the lipid matrix tends to adjust its thickness to the protein and (ii) the protein adjusts its structure to the thickness of the lipid matrix. Structural plasticity of rhodopsin smoothes out interactions between the protein and the lipid matrix. Since it was reported that MII formation results in an elongation of TM5 [15], we suspect that the increase of helicity of rhodopsin with increasing bilayer thickness itself shifts the MI-MII equilibrium toward MII.
3.2. Membrane curvature elastic deformation vs. direct lipid-rhodopsin interactions
3.2.1. Membrane curvature elasticity
The critical outward movement of TM6 and the subsequent elongation of TM5 upon MII formation seem to be also responsible for the strong shift of the MI-MII equilibrium towards MII by lipids that generate negative curvature elastic stress in bilayers. The shape change upon MII formation relieves a fraction of the stress stored in the membrane which in turn lowers the free energy of MII. Our recent data show a nice quantitative agreement between measured coefficients of curvature elasticity for the lipid series DOPE-Me2, DOPE-Me1, DOPE and shifts in the MI-MII equilibrium [24] as predicted by the Flexible Surface Model (FSM) proposed by Brown and coworkers [23, 25].
However, the discrepancy between theoretical predictions for shifts in the MI-MII equilibrium and experiments for several other lipids indicates that the change of free energy due to a release of membrane curvature stress is not the sole determinant of the conformational energetics of rhodopsin photointermediates. There are additional energetic contributions that depend on the presence of lipids with certain properties in the lipid matrix, e.g. lipids with positively charged amino groups as well as polyunsaturated hydrocarbon chains. We assigned them to direct interactions between lipids and the protein, i.e. interactions that take place only between annular lipids and rhodopsin [24]. The interactions appear to be sufficiently weak and transient such that effects on the MI-MII equilibrium scale with the mole fraction of lipids in the membrane. What could be the nature of those interactions that contribute to shifts in the MI-MII equilibrium?
3.2.2. Hydrogen Bonding
One of those additional interactions seems to stem from the ability of lipids to form a hydrogen bond with rhodopsin. That was established by comparing MI-MII equilibria in POPC, DOPC, DOPE-Me2, DOPE-Me1, and DOPE bilayers. The lipids DOPE-Me2, DOPE-Me1, and DOPE have in common that their amino group may form a hydrogen bond with the protein, while POPC and DOPC may not. If the ability of lipids to form a hydrogen bond with rhodopsin is a critical energetic contribution to the MI-MII equilibrium, then the shifts in the equilibrium between DOPE-Me2, DOPE-Me1 and DOPE bilayers should strictly follow predictions from membrane elastic theory and a jump should occur between DOPC and DOPE-Me2 bilayers. This was confirmed experimentally [24], suggesting that the propensity of lipid amino groups to establish a hydrogen bond with rhodopsin shifts the MI-MII equilibrium toward MII.
3.2.3. Polyunsaturated Hydrocarbon Chains
The influence of polyunsaturated hydrocarbon chains on the MI-MII equilibrium was quantified by replacing the di-monounsaturated DOPC and DOPE with the mixed-chain saturated/polyunsaturated SDPC and SDPE [24]. How do membranes rich in polyunsaturated DHA differ from membranes with less unsaturated hydrocarbon chains? Curvature elasticity and the spontaneous radius of curvature of SDPE monolayers were measured at our lab and determined to be close to values of DOPE [51] which eliminates membrane curvature elastic stress as cause for the shift toward MII. NMR studies as well as quantum chemical calculations and molecular dynamic simulations revealed a high level of conformational flexibility of DHA chains [52, 53]. By analyzing 13C relaxation data, we showed that even the DHA chains near rhodopsin isomerize on a timescale of 1–100 ps, and that those chains explore their entire conformational space within 10 ns [54]. The quantum chemical and molecular mechanical calculations demonstrated that this flexibility is caused by extremely low potential barriers in vinyl bonds for changes of dihedral bond angles [53, 55]. We speculate that the low potential barriers permit the polyunsaturated chains to better adjust to the structure of the MII photointermediate, shifting the MI-MII equilibrium toward MII.
In crystallographic studies on occasion well structured segments of lipid molecules near rhodopsin are detected [56]. However, in our NMR studies conducted on fluid bilayers, all annular lipid molecules remain highly mobile and are in rapid exchange with lipids further away from the protein [54]. Because of limits in sensitivity, we may not exclude existence of a small number of immobilized lipids per rhodopsin molecule that could remain undetected. However, functional experiments on rhodopsin reconstituted into binary lipid mixtures of variable composition suggest as well that rapid exchange with lipids in the bulk of the matrix occurs [24].
3.4. Membrane electrical surface potential
Formation of an ionic lock between TM3 and TM6 requires that Glu134 in the ERY motif of TM3 is deprotonated which is favored at pH of 7 or higher [13, 15, 25, 57]. A lower pH favors the protonated state of Glu134, equivalent to an open ionic lock as in MII. The connection between pH, the concentration of negatively charged lipids in bilayers and increased formation of MII was studied and quantitatively interpreted by the Brown laboratory [33]. Negatively charged lipids in the bilayer attract the positively charged hydronium ions from the cytoplasm which lowers the pH near the surface and favors the protonated state of Glu134 as in MII.
3.5. Rhodopsin-rhodopsin interactions
Functional studies on model bilayers show beyond doubt that at the physiological ratio of 70 lipids per rhodopsin, the rhodopsin molecules interact with each other, either by formation of oligomers or via interactions mediated by superposition of lipid domains that surround each rhodopsin molecule (see Fig. 2). Those interactions shift the MI-MII equilibrium toward MI [26, 29]. The rhodopsin concentration of onset of interactions depends on membrane hydrophobic thickness. Any deviation of hydrophobic thickness from 27 Å shifts formation of rhodopsin-rhodopsin contacts toward lower rhodopsin concentrations [28]. The high sensitivity of rhodopsin interactions to rhodopsin concentration and to lipid properties suggests that affinities of rhodopsin molecules for each other are modest. It highlights the importance of lipid-protein interaction for rhodopsin-rhodopsin contacts.
Fig. 2.
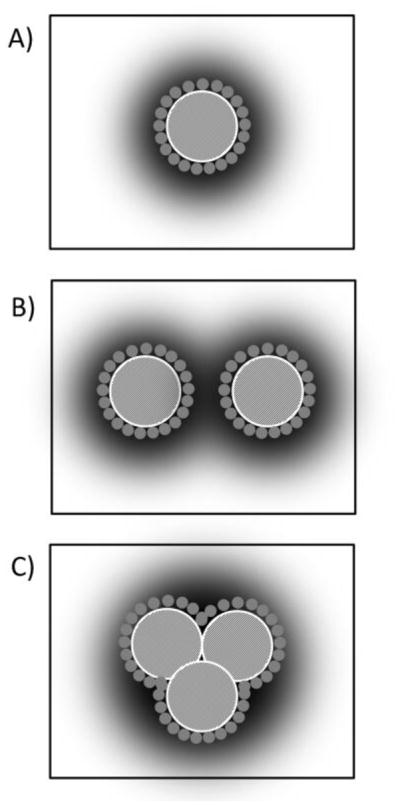
Schematic presentation of rhodopsin-rhodopsin interactions triggered by crowding of rhodopsin molecules in the lipid matrix. At low rhodopsin concentrations (A) rhodopsin (crosshatched circle) exists preferentially as monomers. The first layer of lipids near the protein (small grey circles) influences protein function via direct interactions with rhodopsin. Furthermore, rhodopsin is surrounded by a domain of lipid molecules (shaded area) that is deformed elastically to adjust the lipid matrix to the protein. Lipids in the first layer, in the lipid domain surrounding the protein, and in the bulk of the lipid matrix are in rapid exchange on the timescale of microseconds. At high rhodopsin concentrations, the rhodopsin molecules interact with each other via (B) a superposition of domains with elastically perturbed lipid layers or (C) via rhodopsin oligomerization. Oligomerization reduces the fraction of lipids per rhodopsin that is perturbed by lipid-protein interactions. The latter is detected by NMR experiments on samples with variable rhodopsin concentration.
Most likely, the shift in the MI-MII equilibrium from crowding of rhodopsin molecules in model membranes depends on both direct associations between rhodopsin molecules as in oligomers and lipid-mediated interactions. The oligomerization of GPCR and its functional implications has received a lot of attention lately. For rhodopsin, atomic force microscopy suggested existence of dimers and oligomers in isolated ROS disk membranes [58]. However, this has not stopped the debate on the existence of rhodopsin oligomers since microscopy results could have been influenced by sample preparation procedures [59]. A recent study reported that rhodopsin in ROS is loosely packed in the center of disks [2]. Furthermore, other studies have questioned the functional role of oligomerization by demonstrating that a rhodopsin monomer is a fully functional unit [60].
The following questions need to be address on oligomerization: what are the properties of rhodopsin sites where protein-protein interactions occur and how do they depend on lipid-protein interaction? The lipid-exposed surface of rhodopsin molecules appears to be heterogeneous about its circumference. The poly- and monounsaturated lipids in the lipid matrix interact preferentially with a limited number of sites on rhodopsin that are specific for mono- or polyunsaturated hydrocarbon chains [44, 45]. Coarse-grained molecular simulations (CG-MD) suggest a localized adaptation of the membrane bilayer to rhodopsin that is most pronounced near TM2, 4, and 7 [32]. This local membrane deformation appears to be a key factor defining the rate, extent, and orientational preference of rhodopsin-rhodopsin associations. The possibility that rhodopsin molecules may influence each other functionally without direct interactions needs to be better investigated as well [61]. Over which distances do rhodopsin-induced perturbations of the lipid matrix decay and how does this decay length depend on lipid composition?
Experiments on various bilayers conducted as a function of rhodopsin concentration strongly suggest that rhodopsin-rhodopsin interactions are of functional importance. Clearly, more experiments are needed to determine the underlying causes for the observed shifts in function.
3.6. Cholesterol
Addition of cholesterol to membranes shifts the MI-MII equilibrium towards MI [36, 62]. The Litman laboratory related the cholesterol effect to a tighter packing of lipid hydrocarbon chains which generates a less permissive environment for the formation of MII [62]. But other factors may contribute as well. Bilayer thickness increases with increasing cholesterol concentration (compare to sections 3.1 and 3.5) and the lateral area compressibility coefficient and elastic bending modulus increase altering the energy of membrane elastic deformations [63]. The cholesterol-induced increase of membrane stiffness increases the decay length of protein-induced perturbations in the lipid matrix [64], effectively creating a larger rhodopsin-lipid domain. The latter increases the range of lipid-mediated interactions between rhodopsin monomers (compare to section 3.5).
Furthermore, cholesterol-containing lipid monolayers have lowest energy at a negative radius of curvature, suggesting that cholesterol induces negative curvature elastic stress in lipid bilayers [65]. But membranes under negative curvature elastic stress shift the MI-MII equilibrium towards MII (see section 3.2.1) which is not generally the case for cholesterol.
Additionally, cholesterol may shift the MI-MII equilibrium through direct interactions with rhodopsin. Direct interactions between rhodopsin and cholesterol were detected by fluorescence energy transfer studies [66] and evidence for preferred sites on rhodopsin for interaction with cholesterol was found in molecular simulations [46]. Existence of a specific cholesterol binding site was also established for the human β2-adrenergic receptor [67]. Furthermore, it was reported that cholesterol thermally stabilizes rhodopsin [68]. However, the role of such direct interactions for rhodopsin function remains uncertain.
Last but not least, cholesterol content of membranes is critical for the formation of lateral domains and clusters. The Litman laboratory has shown that rhodopsin has a preference for locating in the liquid disordered domains of a lipid matrix formed of saturated DPPC, polyunsaturated DDPC, and cholesterol [69]. On the other hand, clear evidence for existence of domains in the membranes of ROS disks has not been presented yet. The technique of detection of detergent resistant membranes (DRM) by solubilization with Triton X-100 at 4 oC was applied to ROS disks [70], but its value for the study of lateral organization in membranes is debated (see review by Albert and Boesze-Battaglia [71] and references therein).
Most likely, cholesterol shifts the MI-MII equilibrium via a combination of several of the mechanisms described above. More experimental data are required for a better understanding of the role of cholesterol in rhodopsin function.
4. Are those mechanisms transferable to other GPCR?
Recently, several crystal structures of rhodopsin-like GPCR (class A) were reported. Although the homology between their amino acid sequences is limited, the general structure of these GPCR and their mechanisms of activation are surprisingly similar. It appears that rhodopsin activation is representative for the entire family of class A GPCR. This raises the question if other GPCR will show a similar sensitivity to lipid composition in their function? Unfortunately, there are very few data for other GPCR available to make such a comparison meaningful.
Recently, we successfully reconstituted recombinant, peripheral cannabinoid receptor CB2 into a lipid matrix of controlled composition and investigated the dependence of G protein activation by the receptor on anionic lipid and cholesterol content of membranes [72]. While CB2 activity increased with PS concentration as found for rhodopsin, no changes in function with a raise of cholesterol content was observed. We speculate that the high sensitivity of rhodopsin to composition of the lipid matrix may be linked to the type of ligand that activates the GPCR. While 11-cis retinal is a strong inverse agonist that turns off all basal G protein activation by dark adapted rhodopsin, all-trans retinal seems to be a weak agonist that makes rhodopsin activation vulnerable to a variety of cofactors, the composition of the lipid matrix included. In difference to rhodopsin, other GPCR of class A are known to have a significant basal activity of G protein activation [73]. It is conceivable that basal activity of GPCR as well as the level of activation upon interaction with weakly binding ligands could be particularly sensitive to composition of the lipid matrix.
It must be pointed out that most experiments on reconstituted rhodopsin in model bilayers have limitations related to sample preparation. While rhodopsin molecules in ROS disks are oriented with the C-terminal end facing outside, reconstitution of rhodopsin by rapid dilution most likely produces proteoliposomes with rhodopsin of both orientations. Furthermore, while lipid composition of monolayers in ROS disk membranes is known to be asymmetric, the composition of monolayers in recombinant proteoliposomes is likely to be equal. Those differences in sidedness between ROS disk membranes and model systems may have consequences for lipid-lipid and lipid-protein interactions that need to be investigated in the future.
5. Conclusions
Functional studies on rhodopsin reconstituted into bilayers with well-characterized biophysical properties revealed that the equilibrium of rhodopsin MI-MII photointermediates responds to membrane hydrophobic thickness, curvature elastic stress, lipid order, negative electric surface potentials, direct interactions with rhodopsin via PE headgroups and polyunsaturated hydrocarbon chains, membrane elastic properties, and last but not least to direct or lipid-mediated interactions between rhodopsin molecules. A functional influence from rhodopsin-rhodopsin interactions was already observed at rhodopsin concentrations that are significantly lower than concentrations in rod outer segment disks. The latter is a reminder that concentration of integral proteins in the lipid matrix is typically very high. Every protein is surrounded by just a few layers of lipids that are somewhat perturbed by the presence of the protein. Even if protein molecules have low affinity for each other, because of their crowding in a lipid matrix an influence on protein function from oligomerization or lipid-mediated interactions seems almost unavoidable. This subject requires more attention. It may at least partially explain the frequently observed crosstalk between GPCR signaling pathways. Cholesterol is known to trigger formation of lipid domains and clusters. Therefore, cholesterol is likely to play a major role in regulation of lipid-mediated interactions between GPCR as well.
Highlights.
adjustment of the thickness of lipid bilayers to rhodopsin,
adjustment of rhodopsin helicity to the thickness of bilayers,
curvature elastic deformations in the lipid matrix surrounding the protein,
direct interactions of PE headgroups and polyunsaturated hydrocarbon chains with rhodopsin,
direct or lipid-mediated interactions between rhodopsin molecules.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute on Alcohol Abuse and Alcoholism, National Institutes of Health.
Abbreviations
- GPCR
G protein-coupled membrane receptors
- TM
transmembrane helix
- Gt
G protein transducin
- MI
metarhodopsin I
- MII
metarhodopsin II
- ROS
rod outer segments
- CD
circular dichroism,
- FRET
Förster resonance energy transfer
- EPR
electron paramagnetic resonance
- EM
electron microscopy
- CG-MD
coarse-grained molecular dynamic simulations
- FSM
Flexible Surface Model
- HII
inverse hexagonal phase
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PS
phosphatidylserine
- DHA
docosahexaenoic acid (22:6n-3)
- POPC
1-palmitoyl-2-oleolyl-sn-glycero-3-phosphocholine (16:0-18:1n-9-PC)
- DOPC
1,2-dioleoyl-sn-glycero-3-phosphocholine (18:1n-9-18:1n-9-PC)
- SDPC
1-stearoyl-2-docosahexaenoyl- sn-glycero-3-phosphocholine (18:0-22:6n-3-PC)
- DOPE-Me1
1,2-dioleoyl-sn-glycero-3-monomethyl-phosphoethanolamine (18:1n-9-18:1n-9-PE-Me1)
- DOPE-Me2
1,2-dioleoyl-sn-glycero-3-dimethyl-phosphoethanolamine (18:1n-9-18:1n-9-PE-Me2)
- DOPE
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (18:1n-9-18:1n-9-PE)
- SDPE
1-stearoyl-2-docosahexaenoyl-sn-glycero-3-phosphoethanolamine (18:0-22:6n-3-PE)
- MMPC
1-myristoyl-2-myristoleoyl-sn-glycero-3-phosphocholine (14:0-14:1n-5-PC)
- EEPC
1-eicosanoyl-2-eicosenoyl-sn-glycero-3-phosphocholine (20:0-20:1n-9-PC)
- DPPC
1,2-dipalmitoyl-sn-glycero-3-phosphocholine (16:0-16:0-PC)
- DDPC
1,2-didocosahexaenoyl-sn-glycero-3-phosphocholine (22:6n-3-22:6n-3-PC)
Glossary
- Curvature elastic stress
Lipid monolayers may bend to accommodate lipids with, e.g. smaller headgroups and wider hydrocarbon chains such as DOPE. In a bent conformation, those monolayers have lowest energy. When lipid monolayers form a bilayer, their ability to bend is limited by the apposing lipid monolayer. For simplicity, let’s assume that a bilayer is a flat sheet. Monolayers that have lowest energy when curved are now elastically deformed to be flat - they are under curvature elastic stress. The stress in such bilayers can be raised or reduced by integral membrane proteins, depending on their shape. A protein shaped like an hourglass is reducing stresses in bilayers composed of lipids like DOPE
- Hydrophobic mismatch
Lipid membranes have a hydrophobic core of hydrocarbon chains, and integral membrane proteins have sections with transmembrane orientation formed by amino acids with hydrophobic sidechains. A difference between the thickness of the bilayer hydrophobic core and the length of hydrophobic protein sections is called hydrophobic mismatch. Since exposure of hydrophobic residues to water is energetically costly, such mismatch results in either an elastic deformation of the membrane to match the length of hydrophobic protein sections and/or a change of protein conformation to match the thickness of the hydrophobic layer of membranes
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Young RW. In: The organization of vertebrate photoreceptor cells. Straatsma BR, Allen RA, Crescittelli F, editors. The Retina, University of California Press; Los Angeles: 1969. pp. 177–200. [Google Scholar]
- 2.Buzhynskyy N, Salesse C, Scheuring S. Rhodopsin is spatially heterogeneously distributed in rod outer segment disk membranes. J Mol Recognition. 2011;24:483–489. doi: 10.1002/jmr.1086. [DOI] [PubMed] [Google Scholar]
- 3.Boesze-Battaglia K, Albert AD. Phospholipid distribution among bovine rod outer segment plasma membrane and disk membranes. Exp Eye Res. 1992;54:821–823. doi: 10.1016/0014-4835(92)90040-y. [DOI] [PubMed] [Google Scholar]
- 4.Albert AD, Young JE, Paw Z. Phospholipid fatty acyl spatial distribution in bovine rod outer segment disk membranes. Biochim Biophys Acta. 1998;1368:52–60. doi: 10.1016/s0005-2736(97)00200-9. [DOI] [PubMed] [Google Scholar]
- 5.Boesze-Battaglia K, Hennessey T, Albert AD. Cholesterol heterogeneity in bovine rod outer segment disk membranes. J Biol Chem. 1989;264:8151–8155. [PMC free article] [PubMed] [Google Scholar]
- 6.Boesze-Battaglia K, Albert AD. Cholesterol modulation of photoreceptor function in bovine retinal rod outer segments. J Biol Chem. 1990;265:20727–20730. [PubMed] [Google Scholar]
- 7.Miljanich GP, Nemes PP, White DL, Dratz EA. The asymmetric transmembrane distribution of phosphatidylethanolamine, phosphatidylserine, and fatty acids of the bovine retinal rod outer segment disk membrane. J Membr Biol. 1981;60:249–255. doi: 10.1007/BF01992562. [DOI] [PubMed] [Google Scholar]
- 8.Hargrave PA, Hamm HE, Hofmann KP. Interaction of rhodopsin with the G-protein, transducin. Bioessays. 1993;15:43–50. doi: 10.1002/bies.950150107. [DOI] [PubMed] [Google Scholar]
- 9.Palczewski K. G protein-coupled receptor rhodopsin. Ann Rev Biochemistry. 2006;75:743–767. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wald G. Molecular basis of visual excitation. Nature. 1968;219:800–807. doi: 10.1038/219800a0. [DOI] [PubMed] [Google Scholar]
- 11.Schertler GFX. Structure of rhodopsin and the metarhodopsin I photointermediate. Current Opinion Struct Biol. 2005;15:408–415. doi: 10.1016/j.sbi.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 12.Longstaff C, Calhoon RD, Rando RR. Deprotonation of the Schiff-base of rhodopsin is obligate in the activation of the G-protein. Proc Natl Acad Sci USA. 1986;83:4209–4213. doi: 10.1073/pnas.83.12.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahalingam M, Martinez-Mayorga K, Brown MF, Vogel R. Two protonation switches control rhodopsin activation in membranes. Proceed Nat Acad Sci USA. 2008;105:17795–17800. doi: 10.1073/pnas.0804541105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Procees Nat Acad Sci USA. 2008;105:7439–7444. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choe HW, Kim YJ, Park JH, Morizumi T, Pai EF, Krauss N, Hofmann KP, Scheerer P, Ernst OP. Crystal structure of metarhodopsin II. Nature. 2011;471:651–U137. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 16.Kuhn H, Bennett N, Michelvillaz M, Chabre M. Interactions between photo-excited rhodopsin and GTP-binding protein - kinetic and stoichiometric analyses from light-scattering changes. Proceed Natl Acad Sci USA. 1981;78:6873–6877. doi: 10.1073/pnas.78.11.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilden U, Hall SW, Kuhn H. Phosphodiesterase activation by photoexcited rhodopsin is quenched when rhodopsin is phosphorylated and binds the intrinsic 48-kDa protein of rod outer segments. Proceed Natl Acad Sci USA. 1986;83:1174–1178. doi: 10.1073/pnas.83.5.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kefalov VJ, Cornwall MC, Crouch RK. Occupancy of the chromophore binding site of opsin activates visual transduction in rod photoreceptors. J Gen Physiol. 1999;113:491–503. doi: 10.1085/jgp.113.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Litman BJ, Mitchell DC. Rhodopsin structure and function. In: Lee AG, editor. Rhodopsin and G-protein linked receptors, part A. Vol. 2. JAI Press; Greenwich, CT: 1996. pp. 1–32. [Google Scholar]
- 20.Mitchell DC, Litman BJ. Modulation of receptor signaling by phospholipid acyl chain unsaturation. In: Mostofsky DI, Yehuda S, Salem N Jr, editors. Fatty acids: from neuronal membranes to physiological and behavioral function. Humana Press; Totowa N.J: 2001. pp. 41–62. [Google Scholar]
- 21.Brown MF. Modulation of rhodopsin function by properties of the membrane bilayer. Chem Phys Lipids. 1994;73:159–180. doi: 10.1016/0009-3084(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 22.Brown MF. Influence of nonlamellar-forming lipids on rhodopsin. In: Epand RM, editor. Curr Topics Membranes. Vol. 44. Academic Press; San Diego: 1997. pp. 285–356. [Google Scholar]
- 23.Botelho AV, Gibson NJ, Thurmond RL, Wang Y, Brown MF. Conformational energetics of rhodopsin modulated by nonlamellar-forming lipids. Biochemistry. 2002;41:6354–6368. doi: 10.1021/bi011995g. [DOI] [PubMed] [Google Scholar]
- 24.Soubias O, Teague WE, Hines KG, Mitchell DC, Gawrisch K. Contribution of Membrane Elastic Energy to Rhodopsin Function. Biophys J. 2010;99:817–824. doi: 10.1016/j.bpj.2010.04.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibson NJ, Brown MF. Lipid headgroup and acyl chain composition modulate the MI-MII equilibrium of rhodopsin in recombinant membranes. Biochemistry. 1993;32:2438–2454. doi: 10.1021/bi00060a040. [DOI] [PubMed] [Google Scholar]
- 26.Botelho AV, Huber T, Sakmar TP, Brown MF. Curvature and hydrophobic forces drive oligomerization and modulate activity of rhodopsin in membranes. Biophys J. 2006;91:4464–4477. doi: 10.1529/biophysj.106.082776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soubias O, Niu SL, Mitchell DC, Gawrisch K. Lipid-rhodopsin hydrophobic mismatch alters rhodopsin helical content. J Am Chem Soc. 2008;130:12465–12471. doi: 10.1021/ja803599x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soubias O, Teague WE, Hines KG, Gawrisch K. Rhodopsin-rhodopsin oligomerization in model lipid bilayers - functional implications. Biophys J. 2011;100:525a–526a. [Google Scholar]
- 29.Niu SL, Mitchell DC. Effect of packing density on rhodopsin stability and function in polyunsaturated membranes. Biophys J. 2005;89:1833–1840. doi: 10.1529/biophysj.105.061812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryba NJ, Marsh D. Protein rotational diffusion and lipid/protein interactions in recombinants of bovine rhodopsin with saturated diacylphosphatidylcholines of different chain lengths studied by conventional and saturation-transfer electron spin resonance. Biochemistry. 1992;31:7511–7518. doi: 10.1021/bi00148a011. [DOI] [PubMed] [Google Scholar]
- 31.Pearson LT, Chan SI, Lewis BA, Engelman DM. Pair distribution functions of bacteriorhodopsin and rhodopsin in model bilayers. Biophys J. 1983;43:167–174. doi: 10.1016/S0006-3495(83)84337-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Periole X, Huber T, Marrink SJ, Sakmar TP. G protein-coupled receptors self-assemble in dynamics simulations of model bilayers. J Am Chem Soc. 2007;129:10126–10132. doi: 10.1021/ja0706246. [DOI] [PubMed] [Google Scholar]
- 33.Gibson NJ, Brown MF. Role of phosphatidylserine in the MI-MII equilibrium of rhodopsin. Biochem Biophys Res Commun. 1991;176:915–921. doi: 10.1016/s0006-291x(05)80273-6. [DOI] [PubMed] [Google Scholar]
- 34.Straume M, Litman BJ. Influence of cholesterol on equilibrium and dynamic bilayer structure of unsaturated acyl chain phosphatidylcholine vesicles as determined from higher order analysis of fluorescence anisotropy decay. Biochemistry. 1987;26:5121–5126. doi: 10.1021/bi00390a034. [DOI] [PubMed] [Google Scholar]
- 35.Straume M, Litman BJ. Equilibrium and dynamic bilayer structural properties of unsaturated acyl chain phosphatidylcholine-cholesterol-rhodopsin recombinant vesicles and rod outer segment disk membranes as determined from higher order analysis of fluorescence anisotropy decay. Biochemistry. 1988;27:7723–7733. doi: 10.1021/bi00420a022. [DOI] [PubMed] [Google Scholar]
- 36.Mitchell DC, Straume M, Miller JL, Litman BJ. Modulation of metarhodopsin formation by cholesterol-induced ordering of bilayer lipids. Biochemistry. 1990;29:9143–9149. doi: 10.1021/bi00491a007. [DOI] [PubMed] [Google Scholar]
- 37.Mall S, Sharma RP, East JM, Lee AG. Lipid-protein interactions in the membrane: Studies with model peptides. Faraday Discussions. 1998:127–136. doi: 10.1039/a809299k. [DOI] [PubMed] [Google Scholar]
- 38.Killian JA. Hydrophobic mismatch between proteins and lipids in membranes. Biochim Biophys Acta. 1998;1376:401–416. doi: 10.1016/s0304-4157(98)00017-3. [DOI] [PubMed] [Google Scholar]
- 39.Lee AG. How lipids affect the activities of integral membrane proteins. Biochim Biophys Acta. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 40.Andersen OS, Koeppe RE. Bilayer thickness and membrane protein function: An energetic perspective. Ann Rev Biophys Biomol Struct. 2007;36:107–130. doi: 10.1146/annurev.biophys.36.040306.132643. [DOI] [PubMed] [Google Scholar]
- 41.Helfrich W. Elastic properties of lipid bilayers: Theory and possible experiments. Z Naturforsch. 1973;28c:693–703. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- 42.Hamm M, Kozlov MM. Tilt model of inverted amphiphilic mesophases. Europ Phys J B. 1998;6:519–528. [Google Scholar]
- 43.Gawrisch K, Soubias O, Mihailescu M. Insights from biophysical studies on the role of polyunsaturated fatty acids for function of G-protein coupled membrane receptors. Prostaglandins Leukotrienes Essent Fatty Acids. 2008;79:131–134. doi: 10.1016/j.plefa.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soubias O, Teague WT, Gawrisch K. Evidence for specificity in lipid-rhodopsin interactions. J Biol Chem. 2006;281:33233–33241. doi: 10.1074/jbc.M603059200. [DOI] [PubMed] [Google Scholar]
- 45.Soubias O, Gawrisch K. Probing specific lipid-protein interaction by saturation transfer difference NMR spectroscopy. J Am Chem Soc. 2005;127:13110–13111. doi: 10.1021/ja0538942. [DOI] [PubMed] [Google Scholar]
- 46.Grossfield A, Feller SE, Pitman MC. A role for direct interactions in the modulation of rhodopsin by omega-3 polyunsaturated lipids. Proceed Natl Acad Sci USA. 2006;103:4888–4893. doi: 10.1073/pnas.0508352103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feller SE, Gawrisch K, Woolf TB. Rhodopsin exhibits a preference for solvation by polyunsaturated docosohexaenoic acid. J Am Chem Soc. 2003;125:4434–4435. doi: 10.1021/ja0345874. [DOI] [PubMed] [Google Scholar]
- 48.Tsui FC, Sundberg SA, Hubbell WL. Distribution of charge on photoreceptor disc membranes and implications for charged lipid asymmetry. Biophys J. 1990;57:85–97. doi: 10.1016/S0006-3495(90)82509-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yau WM, Wimley WC, Gawrisch K, White SH. The preference of tryptophan for membrane interfaces. Biochemistry. 1998;37:14713–14718. doi: 10.1021/bi980809c. [DOI] [PubMed] [Google Scholar]
- 50.Gaede HC, Yau WM, Gawrisch K. Electrostatic contributions to indole-lipid interactions. J Phys Chem B. 2005;109:13014–13023. doi: 10.1021/jp0511000. [DOI] [PubMed] [Google Scholar]
- 51.Teague WE, Fuller NL, Rand RP, Gawrisch K. Polyunsaturated lipids in membrane fusion events. Cell Mol Biol Lett. 2002;7:262–264. [PubMed] [Google Scholar]
- 52.Eldho NV, Feller SE, Tristram-Nagle S, Polozov IV, Gawrisch K. Polyunsaturated docosahexaenoic vs docosapentaenoic acid - Differences in lipid matrix properties from the loss of one double bond. J Am Chem Soc. 2003;125:6409–6421. doi: 10.1021/ja029029o. [DOI] [PubMed] [Google Scholar]
- 53.Feller SE, Gawrisch K, MacKerell AD. Polyunsaturated fatty acids in lipid bilayers: Intrinsic and environmental contributions to their unique physical properties. J Am Chem Soc. 2002;124:318–326. doi: 10.1021/ja0118340. [DOI] [PubMed] [Google Scholar]
- 54.Soubias O, Gawrisch K. Docosahexaenoyl chains isomerize on the subnanosecond time scale. J Am Chem Soc. 2007;129:6678–6679. doi: 10.1021/ja068856c. [DOI] [PubMed] [Google Scholar]
- 55.Rabinovich AL, Ripatti PO. On the conformational, physical properties and functions of polyunsaturated acyl chains. Biochim Biophys Acta. 1991;1085:53–62. doi: 10.1016/0005-2760(91)90231-6. [DOI] [PubMed] [Google Scholar]
- 56.Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 57.Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, Hofmann KP, Ernst OP. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–U430. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 58.Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. Atomic-force microscopy: Rhodopsin dimers in native disc membranes. Nature. 2003;421:127–128. doi: 10.1038/421127a. [DOI] [PubMed] [Google Scholar]
- 59.Chabre M, Cone R, Saibil H. Biophysics - Is rhodopsin dimeric in native rods? Nature. 2003;426:30–31. doi: 10.1038/426030b. [DOI] [PubMed] [Google Scholar]
- 60.Ernst OP, Gramse V, Kolbe M, Hofmann KP, Heck M. Monomeric G protein-coupled receptor rhodopsin in solution activates its G protein transducin at the diffusion limit. Proceed Natl Acad Sci USA. 2007;104:10859–10864. doi: 10.1073/pnas.0701967104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Phillips R, Ursell T, Wiggins P, Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459:379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Niu SL, Mitchell DC, Litman BJ. Manipulation of cholesterol levels in rod disk membranes by methyl-beta-cyclodextrin: effects on receptor activation. J Biol Chem. 2002;277:20139–20145. doi: 10.1074/jbc.M200594200. [DOI] [PubMed] [Google Scholar]
- 63.Needham D, McIntosh TJ, Evans E. Thermomechanical and transition properties of dimyristoylphosphatidylcholine/cholesterol bilayers. Biochemistry. 1988;27:4668–4673. doi: 10.1021/bi00413a013. [DOI] [PubMed] [Google Scholar]
- 64.Nielsen C, Andersen OS. Inclusion-induced bilayer deformations: Effects of monolayer equilibrium curvature. Biophys J. 2000;79:2583–2604. doi: 10.1016/S0006-3495(00)76498-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen Z, Rand RP. The influence of cholesterol on phospholipid membrane curvature and bending elasticity. Biophys J. 1997;73:267–276. doi: 10.1016/S0006-3495(97)78067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Albert AD, Young JE, Yeagle PL. Rhodopsin-cholesterol interactions in bovine rod outer segment disk membranes. Biochim Biophys Acta. 1996;1285:47–55. doi: 10.1016/s0005-2736(96)00145-9. [DOI] [PubMed] [Google Scholar]
- 67.Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien EYT, Velasquez J, Kuhn P, Stevens RC. A specific cholesterol binding site is established by the 2.8 angstrom structure of the human beta(2)-adrenergic receptor. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Albert AD, Boeszebattaglia K, Paw Z, Watts A, Epand RM. Effect of cholesterol on rhodopsin stability in disk membranes. Biochim Biophys Acta. 1996;1297:77–82. doi: 10.1016/0167-4838(96)00102-1. [DOI] [PubMed] [Google Scholar]
- 69.Polozova A, Litman BJ. Cholesterol dependent recruitment of di22:6-PC by a G protein-coupled receptor into lateral domains. Biophys J. 2000;79:2632–2643. doi: 10.1016/S0006-3495(00)76502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seno K, Kishimoto M, Abe M, Higuchi Y, Mieda M, Owada Y, Yoshiyama W, Liu H, Hayashi F. Light- and guanosine 5 '-3-O-(thio)triphosphate-sensitive localization of a G protein and its effector on detergent-resistant membrane rafts in rod photoreceptor outer segments. J Biol Chem. 2001;276:20813–20816. doi: 10.1074/jbc.C100032200. [DOI] [PubMed] [Google Scholar]
- 71.Albert AD, Boesze-Battaglia K. The role of cholesterol in rod outer segment membranes. Prog Lipid Res. 2005;44:99–124. doi: 10.1016/j.plipres.2005.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kimura T, Yeliseev AA, Vukoti K, Gawrisch K. Influence of membrane composition on function of human peripheral cannabinoid receptor CB2. Biophys J. 2010;98:290a. [Google Scholar]
- 73.Kobilka BK, Deupi X. Conformational complexity of G-protein-coupled receptors. Trends Pharmacol Sci. 2007;28:397–406. doi: 10.1016/j.tips.2007.06.003. [DOI] [PubMed] [Google Scholar]