Abstract
Glycine N-methyltransferase (GNMT) is a key regulatory enzyme in methyl group metabolism. In mammalian liver it reduces S-adenosylmethionine levels by using it to methylate glycine, producing N-methylglycine (sarcosine) and S-adenosylhomocysteine. GNMT is inhibited by binding two molecules of 5-methyltetrahydrofolate (mono- or polyglutamate forms) per tetramer of the active enzyme. Inhibition is sensitive to the status of the N-terminal valine of GNMT and to polyglutamation of the folate inhibitor. It is inhibited by pentaglutamate form more efficiently compared to monoglutamate form. The native rat liver GNMT contains an acetylated N-terminal valine and is inhibited much more efficiently compared to the recombinant protein expressed in E. coli where the N-terminus is not acetylated. In this work we used a protein crystallography approach to evaluate the structural basis for these differences. We show that in the folate-GNMT complexes with the native enzyme, two folate molecules establish three and four hydrogen bonds with the protein. In the folate-recombinant GNMT complex only one hydrogen bond is established. This difference results in more effective inhibition by folate of the native liver GNMT activity compared to the recombinant enzyme.
Keywords: Glycine N-methyltransferase, folate, crystal structure, inhibition
1. Introduction
The enzyme glycine N-methyltransferase (GNMT) is a key element in the regulation of the concentration of S-adenosylmethionine (AdoMet) in mammalian tissues (for review see [1]). GNMT transfers a methyl group from AdoMet to glycine producing S-adenosylhomocysteine (AdoHcy) and sarcosine. GNMT is a tetramer of identical 32 kDa subunits. Studies of humans with missense mutations and GNMT knockout mice (obtained by gene targeting) showed that a decrease or complete absence of GNMT activity leads to highly elevated concentrations of the critically important metabolites methionine and AdoMet in plasma and liver. The high level of AdoMet leads to damage of the liver and liver diseases including hepatocellular carcinoma in GNMT knockout mice. Thus, it is critical to regulate activity of GNMT in response to the changing metabolic state. The activity of GNMT is regulated by binding of specific inhibitor, 5-methyltetrahydrofolate. A regulatory scheme in which GNMT acts with 5-methyltetrahydrofolate to modulate high levels of AdoMet was discussed in detail earlier [1].
To obtain insight into the mechanism of regulation of GNMT activity by folate we performed a crystallographic study of the GNMT-folate complex with rat recombinant GNMT expressed in E. coli and with 5-CH3-H4PteGlu1 (monoglutamate) [2]. The essential elements of the GNMT-folate interactions were established in that study, however some important points remain to be addressed. First, GNMT prepared from rat liver differs from the recombinant protein expressed in E. coli by the status of the N-terminal valine. In rat liver protein the N-terminal valine is acetylated but in the recombinant protein it is not [3–4]. No other differences in post-translational modification of recombinant and rat liver proteins were found [4]. Secondly, all active folate cofactors in living organisms are polyglutamated with different number of glutamyl residues attached [5]. In the rat liver the most abundant form of folates is the pentaglutamate (5-CH3-H4PteGlu5).
Both native liver and recombinant GNMTs are inhibited by 5-CH3-H4PteGlu1 and 5-CH3-H4PteGlu5 and inhibition is much more effective by the pentaglutamate form than by the monoglutamate form. Moreover, inhibition of the native enzyme is much more efficient compared to recombinant enzyme by both forms of folate [6].
To investigate the structural basis for the difference in sensitivity to inhibition by the native and recombinant enzymes we solved the crystal structures of the native enzyme complexed with both mono- and pentaglutamate forms of 5-methyltetrahydrofolates.
2. Materials and methods
2.1 Chemicals
All chemicals for buffers and microbiological media, S-adenosylmethionine, S-adenosylhomocysteine and Sephacryl S-200 resin were from Sigma. Ceramic hydroxylapatite and Bio-Gels were from Bio-Rad. [3H-CH3]-S-Adenosylmethionine was from NEN Radiochemicals. (6S)-5-CH3-H4PteGlu1 was a gift from EPROVA (Switzerland). To prepare natural (6S)-5-CH3-H4PteGlu5 the mixture of (6R/6S)-5-CH3-H4PteGlu5 purchased from Schircks Laboratories (Switzerland) was separated by binding to GNMT, removal of non-bound folate (presumably the 6R-form) by using Spin-Columns and release of free folate by heating the GNMT-folate complex at +80 °C in presence of 60 mM β-mercaptoethanol as was done in [6].
2.2. GNMT preparation
Rat liver GNMT was prepared as reported earlier [6]. Briefly, rat liver (Pel-Freez Biologicals) was homogenized in homogenization buffer with concentration of the components as follows: 20 mM Tris-HCl pH 7.8, 1 mM EDTA, 2 mM PMSF, 14 mM β-mercaptoethanol and 500 μl of Protease Inhibitor Cocktail (Sigma)/100 ml. After centrifugation of homogenate GNMT was precipitated from the supernatant between 35–50% of ammonium sulfate saturation. The pelleted proteins were solubilized, desalted and loaded on an ion-exchange column (DE-52, Beckman) equilibrated with 20 mM Tris-HCl pH 7.8 containing 14 mM β-mercaptoethanol. GNMT was eluted using the same buffer containing 30 mM NaCl. After changing the buffer to 2 mM Na-phosphate, pH 7.5, the GNMT sample was applied to a ceramic hydroxylapatite Type I (Bio-Rad) column equilibrated with 2 mM Na-phosphate, pH 7.5 containing 14 mM β-mercaptoethanol. GNMT of about 95–97% purity was eluted from the column by 30 mM Na-phosphate, pH 8.0. The protein purity was further increased to at least 98% by size-exclusion chromatography on a Sephacryl S-200 column equilibrated with 50 mM Tris pH 7.5, 200 mM NaCl, 1 mM EDTA and 14 mM β-mercaptoethanol. All purification steps were performed at +3–4° C.
2.3. Crystallization
The GNMT-folate monoglutamate complex was crystallized by the sitting drop method at room temperature. The protein (4–6 mg/ml concentration in 20 mM Tris-HCl, pH 7.5, 25 mM NaCl, 5 mM Tris(2-carboxyethyl)phosphine (TCEP)) was incubated with 1–20 mM folate for 1 hour prior to the crystallization. The protein-folate complex was mixed with the reservoir solution (20% PEG 3350, 0.2 M Na-fluoride or Ca-acetate and 100 mM Tris-HCl, pH 7.5). The crystals of the GNMT-folate complexes appeared in a few hours and grew to the maximum dimensions in 4–6 days. The shape of the crystals for the complex was as shown in Fig. 1A.
Figure 1. Crystals of rat liver GNMT.
A: GNMT - 5-CH3-H4PteGlu complex; B: GNMT - 5-CH3-H4PteGlu5 complex; C: GNMT apoprotein.
A GNMT-folate pentaglutamate complex crystallized in the similar conditions as the GNMT-folate monoglutamate but the shape of the crystals was different (Fig. 1 B). Interestingly, crystals of GNMT-folate monoglutamate and GNMT-pentaglutamate belong to different space groups. The crystals of complex with monoglutamate belongs to the primitive monoclinic P21 space group with a = 59.19 Å, b = 83.79 Å, c = 134.44 Å and β = 91.32°. Crystals of the complex with pentaglutamate belong to the centered monoclinic C2 space group with a = 193.04 Å, b = 61.05 Å, c = 146.35 Å and β = 128.92°.
In contrast, crystals of the liver GNMT apoprotein grown from the above mentioned crystallization conditions appear only as tiny needles unsuitable for X-ray data collection (Fig. 1C) preventing us from comparison of the crystal structure of the apo form of the enzyme with structures of both folate bound complexes.
2.4. X-ray data collection
Diffraction data were collected at 100 K at SER-CAT beamline 22-BM, Advanced Photon Source, Argonne National Laboratory, Illinois using a MAR 225 CCD detector. Data were processed and scaled using HKL2000 package [7]. Data collection and data processing statistics are summarized in Table 1.
Table 1.
Data collection and refinement statistics
| GNMT-5-CH3-THF-Glu | GNMT-5CH3-THF-Glu5 | |
|---|---|---|
| Wavelength (Å) | 1.000 | 1.000 |
| Resolution (Å) | 2.00 | 2.50 |
| Space group | P21 | C2 |
| Cell dimensions | ||
| a (Å) | 59.19 | 193.04 |
| b (Å) | 83.79 | 61.05 |
| c (Å) | 134.44 | 146.35 |
| β (°) | 91.32 | 128.92 |
| Unique reflections | 83512 | 46441 |
| Rsym1,2 (%) | 8.4 (61.0) | 8.1 (56.5) |
| Completeness (%) | 92.9 (84.5) | 99.7 (98.5) |
| Redundancy | 6.2 (6.2) | 7.4 (7.0) |
| I/σ(I) | 22.2 (2.6) | 34.1 (3.0) |
| Refinement | ||
| Resolution range (Å) | 31.19 – 2.00 | 48.22 – 2.50 |
| Reflections used in refinement | 73600 | 43978 |
| σ cutoff used in refinement | none | none |
| R/R free3 (%) | 17.81/24.07 | 22.78/27.98 |
| No. of refined atoms | ||
| Protein | 8997 | 8926 |
| Waters | 725 | 88 |
| Folate | 132 | 66 |
| Other ligands | 33 | 11 |
| R.m.s. deviations | ||
| Bonds lengths (Å) | 0.023 | 0.016 |
| Bonds angles, ° | 2.017 | 1.551 |
| Average B factor (Å2) | ||
| Protein | 31.6 | 58.5 |
| Folate4 | 50.2 | 88.4 |
Values in parentheses are for the highest-resolution shell.
Rsym = Σ|Ii − <Ii> |/ΣIi, where Ii is the intensity of the ith observation and <Ii> is the mean intensity of the reflection.
R = Σ||Fo| − |Fc||/Σ|Fo|, where Fo and Fc are the observed and calculated structure factors amplitudes. Rfree is calculated using 4.8 and 4.9 % of reflections omitted from the refinement for the GNMT-monoglutamate and GNMT-pentaglutamate complex, respectively.
Specifically bound folate molecules.
2.5. Structure solution and refinement
The crystal structures of both GNMT-folate complexes were solved by the molecular replacement procedure. A tetramer of the recombinant GNMT (PDB ID: 2IDK) was used as the search model in both cases. The positioned models of GNMT-folate complexes were then refined using the maximum likelihood refinement in REFMAC [8] with the TLS parameters generated by the TLSMD server [9]. TLS tensors were analyzed and anisotropic B-factors were derived with TLSANL program [10]. Two-fold non-crystallographic symmetry restraints and bulk solvent corrections were applied during the refinement. The program Coot [11] was used to build the models throughout the refinement. Refinement statistics for both complexes are listed in Table 1. Details of refinement of each of the structures follow.
GNMT-folate monoglutamate complex
The difference Fourier revealed the presence of two folate monoglutamate molecules in the intersubunits area in the tetramer of GNMT. Additional two nonspecifically bound folate monoglutamate molecules and three Tris molecules were located in the difference map and modeled according to the electron density shape. The final model consists of protein residues A1–A225, A232–A292, B1–B226, B233–B292, C1–C224, C233–C292, D1–D225 and D233–D292. Alternate conformations have been built for residues Ser182 (monomer A), His58 (monomer C) and Ser80 (monomer C). A total of 725 water molecules have been added into the final model.
GNMT-folate pentaglutamate complex
Two molecules of folate pentaglutamate are located in the intersubunits area as in the case of the GNMT-folate monoglutamate complex. However, no electron density for the pentaglutamate tail of the molecule was observed indicating that this part of the ligand molecule is very flexible. Consequently, the polyglutamate tail has not been modeled. The final model consists of protein residues A1–A224, A234–A292, B1–B225, B233–B292, C1–C224, C234–C292, D1–D223, D234–D292, two folates pentaglutamate molecules, one Tris molecule, one covalently bound molecule of β-mercaptoethanol in two alternate conformations and 88 water molecules.
2.6. AdoMet binding
Binding of AdoMet by both preparations of GNMT was determined based on protein fluorescence quenching upon AdoMet binding as done previously [12]. In these experiments the fluorescence emission spectra of GNMT in the presence of different amount of AdoMet excited by 285 nm light were registered on a Varian Eclipse fluorescence spectrophotometer. The concentration of AdoMet at which the maximal quenching was observed was considered to be the saturation concentration. Plotting of the loss of intensity of fluorescence at 343 nm (spectrum maximum) versus AdoMet concentration permitted calculation of the value of S0.5 which was used as a measure of AdoMet binding. All experiments were done in 0.1 M Tris-HCl, pH 7.5 in presence of 5 mM β-mercaptoethanol with GNMT concentration of 0.095 mg/ml at ambient temperature. The experimental data were fitted using the Hill equation with normalized quenching of protein fluorescence.
2.7. Other methods
Protein concentration was routinely determined by the BCA method (BCA Protein Assay Kit, Pierce) according to the manufacturer’s protocol with bovine serum albumin as a standard. Protein purity was determined by standard SDS electrophoresis with scanning of the Coomassie stained gel on Bio-Rad Gel/Chemi Doc system and Quantity One software. Absorbance spectra were recorded on a double beam Shimadzu UV-2401 Spectrophotometer.
2.8. Structural data
Atomic coordinates and structure factors have been deposited to the Protein Data Bank with the accession codes: 3THR and 3THS.
3. Results
3.1. Overall structures
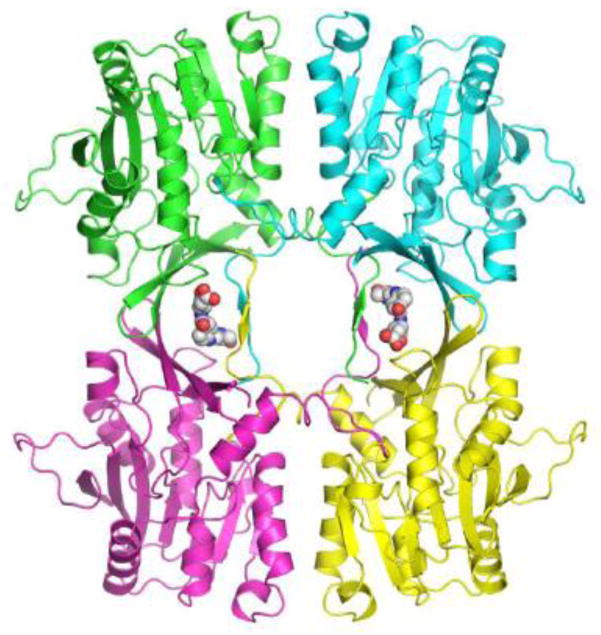
The asymmetric unit in both native liver GNMT-folate complexes is represented by a tetramer in the so-called closed conformation (Fig. 2) as was observed in the GNMT-folate complex formed by the recombinant protein and 5-CH3-H4PteGlu1 [2]. The comparison of liver GNMT-folate complexes with that of recombinant GNMT showed that the protein conformation in all three complexes is essentially the same with the root mean square deviations for the equivalent CA atoms less than 1 Å. The crystal structure of the recombinant GNMT-folate monoglutamate complex was solved at the 2.55 Å resolution and folate molecules were quite disordered in the structure with high average B-factors of 93.2 Å2. Such high B-factors could be explained by the fact that no strong H-bonds between the folate and protein molecules have been observed. Folate molecules are stabilized inside the tetramer by mainly hydrophobic interactions. Crystals of the rat liver GNMT-folate monoglutamate complex reported here diffract to significantly better resolution (2.0 Å) and folate monoglutamate molecules have B-factors of 50.2 Å2 which are much closer to the protein B-factors. In addition to two folate monoglutamate molecules located in the intersubunit areas, two non-specifically bound folate monoglutamate molecules were found in electron density maps on the surface of the tetramer stabilizing crystal packing. No non-specifically bound folate molecules were found in case of the GNMT-pentaglutamate complex.
Figure 2. Cartoon representation of the tetramer of GNMT in the native rat liver GNMT-folate monoglutamate complex.
Protein subunits are shown in different colors. Bound folate molecules are shown as space filled models.
3.2. Folate-protein interactions and folate conformations
Elucidation of interaction of the natural folate form (pentaglutamate) with native rat liver GNMT and the possible difference in interaction of folate with native and recombinant GNMT was a principal goal of this project.
The folates in various folate binding proteins have been found in different conformations, which depend on the specific interactions with protein molecules [13–16]. The interaction of 5-CH3-H4PteGlu in its mono- and pentaglutamate forms with GNMT protein molecules follows the general rule for folate-protein interactions: the pterin and p-aminobenzoyl rings are buried into the more hydrophobic interior of the protein with the negatively charged glutamate tail exposed to the surface of the protein globule. The best assignment of the folate in the electron density in GNMT- folate complexes in this work was achieved when the folate was built in an extended conformation in both the monoglutamate and pentaglutamate forms. Interestingly, the two molecules of the non-specifically trapped 5-CH3-H4PteGlu1 are in the bent conformation.
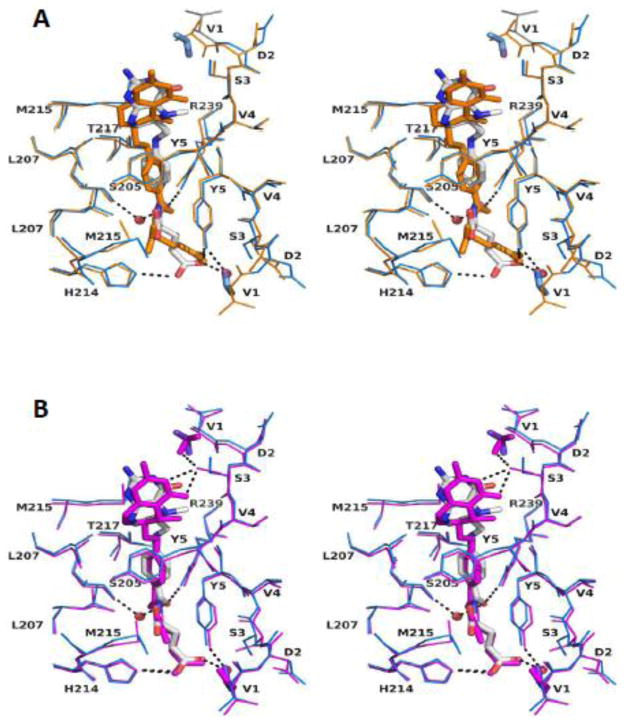
The specific interactions of folates with native liver and recombinant GNMTs are shown in Fig. 3A and B and listed in Table 2. For both complexes the folate binding sites are located in the same areas but the exact interactions are slightly different. In both complexes two molecules of 5-CH3-H4PteGlu monoglutamate interact with all four GNMT subunits in both binding sites through interactions with mostly, but not entirely, the same residues.
Figure 3. A stereo figure of superposition of the folate binding sites.
A: Rat liver GNMT-folate monoglutamate (blue) and recombinant GNMT-folate monoglutamate (orange) complexes.
B: Rat liver GNMT-folate monoglutamate (blue) and rat liver GNMT-folate pentaglutamate (magenta) complexes. N-terminal acetyl groups are shown in stick representation. Only one folate binding site (site A) is shown.
Table 2.
Amino acid residues in rat liver and recombinant GNMT interacting with folates.
| Protein and Folate | Folate molecule A | Folate molecule B |
|---|---|---|
| Recombinant with monoglutamate |
6 Hydrophobic contacts: Met215A, Leu207A, Ser3B, Tyr5B, Leu207C, Met215C |
8 Hydrophobic contacts: Thr7A, Met215A, Leu207A, Ser3B, Tyr5B, Leu207C, Met215C, His214D |
| One H-bond: PABA-Tyr5 | One H-Bond: PABA-Tyr5 | |
|
| ||
| Liver with monoglutamate |
8 Hydrophobic contacts: Thr7A, Leu207B, Met215B, Thr217B, Ser3C, Val4C, Tyr5C, Leu207D |
8 Hydrophobic contacts: Leu207A, Val4B, Tyr5B, Leu207C, Met215C, Thr217C, Tyr5D, Thr7D |
|
3 H-bonds: 1-Arg239B, 2-His214D, 3-H2O-Tyr5 and Ser205B |
3 H-bonds: 1-Arg239C, 2-Ser3B, 3-H2O-Arg145C and Ser205C |
|
|
| ||
| Liver with pentaglutamate |
7 Hydrophobic contacts: Tyr5A, Leu207B, Thr217B, Arg239B, Val4C, Tyr5C, Leu207D |
6 Hydrophobic contacts: Leu207A, Met215A, Val4B, Tyr5B, Leu207C, Tyr5D |
|
3 H-bonds: (1+1)- bonds-Ser3C, 3-His214D |
4 H-bonds: 1-Ser3B, 2-Ser146C, (1+1)-Arg239C |
|
In the recombinant protein folate molecules interact with protein molecule by mainly hydrophobic interactions. One molecule interacts with Ser3B, Tyr5B, Met215A, Leu207A, Leu207C and Met215C. The second folate molecule interacts through the same residues plus Thr7A and His214D. The total number of hydrophobic contacts for the two folates is 15. Both folate molecules interact with protein by one hydrogen bond of a glutamate residue of folate with the hydroxyl group of Tyr5 of GNMT.
Although the interactions of the folates with the native protein are very similar to those with the recombinant protein there are some differences. Both folate molecules interact with the native protein through almost the same number of hydrophobic contacts, eight of them for each folate molecules: folate A makes contacts with Thr7A, Leu207B, Met215B, Thr217B, Ser3C, Val4C, Tyr5C and Leu207D and folate B makes contacts with Leu207A, Val4B, Tyr5B, Leu207C, Met215C, Thr217C, Tyr5D and Thr7D. The most distinctive feature in the folate-native GNMT interaction is the number and position of hydrogen bonds. Folate molecule A makes H-bonds directly with the side chain NH of the guanidinium group of Arg239B and the NE2 of His214D and with the hydroxyl groups of Tyr5A and Ser205B via water molecules. The folate molecule B establishes H-bonds directly with the side chain of Ser3B, the side chain NH of Arg239C and with the carbonyl oxygen of Asp145C and hydroxyl group of Ser205C via water molecules.
Two molecules of 5-CH3-H4PteGlu5 bind to native GNMT through essentially the same hydrophobic interactions as the monoglutamate form with occupation by the pterin-PABA parts of molecules of the same binding sites on the protein. There was no electron density in the folate-GNMT complex to assign to the pentaglutamate tails of both folate molecules, probably due to the highly flexible properties of such a structure. The essential interactions of one folate molecules (molecule A) are shown in the Fig. 3B. The folate molecule A makes hydrophobic contacts with seven residues: Tyr5A, Leu207B, Thr217B, Arg239B, Val4C, Tyr5C and Leu207D. This folate molecule is also bound through three hydrogen bonds, two of them between the pterine ring N3 atom and the side chain OH group of Ser3C and one bond between the glutamate part of folate and the NE2 of His214D. The folate molecule B establishes hydrophobic contacts with six protein residues: Leu207A, Met215A, Val4B, Tyr5B, Leu207C and Tyr5D. There are four H-bonds between the folate molecule and the side chain hydroxyl groups of Ser3B, Ser146C and the guanidinium group of Arg239C (two bonds).
4. Discussion
The data presented here show that native rat liver GNMT binds two molecules of 5-methyltetrahydrofolates, mono- and pentaglutamate at the same binding sites located in the intersubunits areas. The folate binding sites of GNMT are formed by substantial contributions by all four N-termini of all subunits. About of half of hydrophobic contacts and hydrogen bonds are established between folates and residues in the N-termini (Table 2). That means that any changes of amino acid residues in these parts of GNMT would lead to a change of interactions with folates either by loss of some contacts or by changing the microenvironment in the folate binding sites.
Despite an additional four glutamate residues in pentaglutamate form of the folate, it interacts with GNMT through essentially the same contacts as monoglutamate form, an indication that the most important interactions of folate with GNMT are achieved through interactions the monoglutamate part of folate. However, the much higher efficiency of inhibition of GNMT activity by pentaglutamates [6] suggests that additional interactions with the polyglutamyl part of folate are possible.
The N-terminal fragments of 10 residues of all subunits interact with each other not only in GNMT-folate complexes but also in the GNMT apoprotein tetramer where these fragments close access to active center for substrate, AdoMet and glycine [2, 17]. This implies that any changes in chemical structure of residues that increase N-termini interactions will result in increasing the energy required for access of substrates to the binding sites in active centers located deep inside the globular parts of each subunit. One such change is the transformation of the N-terminal valine from its neutral acetylated form in the native form to a positively charged state, in the recombinant protein. This change decreases the hydrophobicity of the amino terminus and therefore its interaction with globular parts of GNMT tetramer subunits.
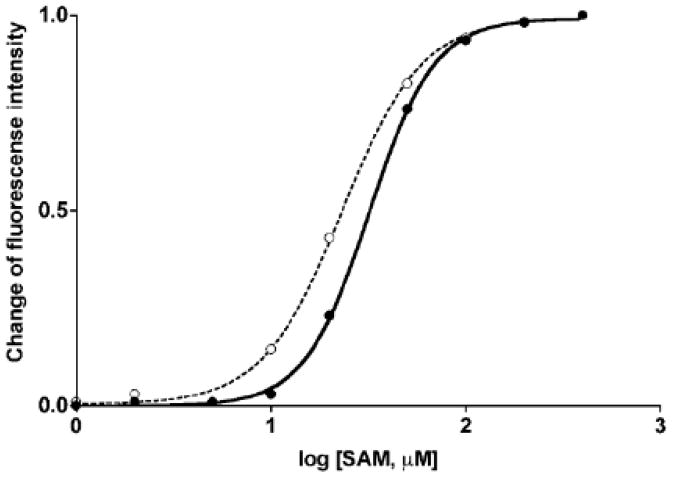
The importance of the N-terminal fragment of GNMT in determining its kinetic properties was established shortly after this enzyme was first purified from rat liver. It was found that removal of 7 N-terminal residues in rat liver GNMT resulted in the loss of cooperativity of AdoMet concentration on enzyme activity [12]. Later, it was shown that rat recombinant GNMT expressed in E. coli exhibited the same absence of cooperativity as N-terminal truncated enzyme [3]. Loss of cooperativity was attributed to the difference in the status of N-terminal valine which was acetylated in the native liver protein and non-acetylated in recombinant GNMT [3]. Moreover, binding of AdoMet to GNMT was consistent with that finding: the value of S0.5 for AdoMet for the native enzyme without the 7 N-terminal amino acid residues was found to be 13.9 μM by dialysis and 21.1 μM by fluorescence methods [12], which was lower compared to the native liver enzyme as of 42.7 μM by equilibrium dialysis and 50.1 μM by the fluorescence method [18]. Similarly, our data in this work showed that AdoMet is bound with significantly (P <0.001 as it calculated by GraphPad Prism) higher affinity to recombinant GNMT (S0.5 value of 23.1 μM) compared to liver enzyme (S0.5 value of 31.9 μM) with cooperativity coefficients of 2.1 and 2.6, respectively (Fig. 4). All these differences can be attributed to the status of the N-terminal valine as analysis of liver and recombinant GNMT by mass-spectrometry and proteomics tools showed that there was no other difference in post-translational modification of recombinant and rat liver protein [4].
Figure 4. Binding of AdoMet by liver and recombinant GNMT.
Binding of AdoMet registered by the quenching of fluorescence emission upon AdoMet binding. Closed circles- native liver GNMT, open circles- recombinant GNMT.
Crystallographic studies showed that movement of the N-termini of four subunits in GNMT tetramer is an essential part of the mechanism of glycine methylation [17]. AdoMet is bound to the active sites deep within each subunit of the tetramer. When AdoMet is bound to all subunits, the N-terminal domains of all subunits which had blocked entrance to the active sites in the apoenzyme, have moved to allow entrance and exit from the active site. The N-terminal fragments are then exposed to solvent. Other crystallographic studies showed also that in the absence of AdoMet two molecules of 5-CH3-H4PteGlu1 are bound to intersubunit areas of tetrameric GNMT and interact with all four N-termini of GNMT [2].
In this case of binding of folate all four N-termini of all subunits “clamp” around the folate molecules and this results not only in cooperativity in AdoMet binding with liver GNMT apoprotein but in significant inhibition of GNMT activity. This inhibition is obviously dependent on the strength of interaction. The latter is much higher in the case of the native enzyme compared to recombinant GNMT due to more hydrogen bonds between folate molecules and protein (Table 2). The folate pentaglutamate most likely interacts with the apoprotein through the polyglutamate (not modeled in presented structure) which additionally increases affinity and therefore inhibition efficiency.
The location of acetyl groups of acetylated Val1 in relation to the position of bound folates is quite interesting. Although no hydrogen bonds between the acetyl groups and folates are observed in the crystal structures, acetyl groups from two different GNMT monomers are located close (4 – 6.5 A) to both the pterin and the glutamate tail parts of the folate molecules, making “clamps” around the folate molecule (Fig. 2). Such structural feature may restrict access to the folate binding site. There are also hydrogen bonds between the acetyl group and OH group of Ser3 that could help to position Ser3 for hydrogen bonded interactions with pterin rings of folate molecules.
The study of importance of N-terminal valine modification on interaction of folate with GNMT apoprotein and inhibition of enzyme activity presented in this paper is a part of elucidation of function of this enzyme in vivo. It is known [4] that several serine residues in rat GNMT are phosphorylated. One of those residues is Ser9. The position of this residue might influence the interaction of N-terminal fragments of GNMT with folate binding. This means that phosphorylation of Ser9 could be another way for modulation of GNMT activity.
This modification could be used differently in different tissues. GNMT is expressed in significant amounts only in the liver, pancreas, kidney and prostate. Although post-translational modification of GNMT in kidney, pancreas and prostate it is not known it may be possible that that phosphorylation of N-terminal residues in different tissues vary and could be used in regulation of activity of GNMT in tissue-specific way.
The results presented here, although specific to GNMT, add further arguments for the importance of differences between proteins from natural sources and recombinant proteins for the studies of their function.
Highlights.
We crystallized rat liver glycine N-methyltransferase with folate inhibitors.
We compared structures of these complexes with that formed by recombinant protein.
Folates establish more hydrogen bonds with native liver protein.
This explains more efficient inhibition of liver glycine N-methyltransferase.
Acknowledgments
This work has been supported by grants DK15289 and DK080010 of the U.S. Public Health Service to C. W. and by Louisiana Governor’s Biotechnology Initiative to M.E.
The authors thank Vanderbilt University Center for Structural Biology for access to Biomolecular Crystallography Facilities.
The authors thank EPROVA (Switzerland) for free samples of folate.
Data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-BM beamline at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38.
Abbreviations
- GNMT
glycine N-methyltransferase
- AdoMet
S-adenosylmethionine
- AdoHcy
S-adenosylhomocysteine
- 5-CH3-H4PteGlu1
5-methyltetrahydropteroyl monoglutamate
- 5-CH3-H4Pte-Glu5
5-methyltetrahydropteroyl pentaglutamate.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Luka Z, Mudd SH, Wagner C. Glycine N-methyltransferase and regulation of S-adenosylmethionine levels. J Biol Chem. 2010;284:22507–22511. doi: 10.1074/jbc.R109.019273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luka Z, Pakhomova S, Loukachevitch LV, Egli M, Newcomer ME, Wagner C. 5-methyltetrahydrofolate is bound in intersubunit areas of rat liver folate-binding protein glycine N-methyltransferase. J Biol Chem. 2006;282:4069–4075. doi: 10.1074/jbc.M610384200. [DOI] [PubMed] [Google Scholar]
- 3.Ogawa H, Gomi T, Takata Y, Date T, Fujioka M. Recombinant expression of rat glycine N-methyltransferase and evidence for contribution of N-terminal acetylation to co-operative binding of S-adenosylmethionine. Biochem J. 1997;327:407–412. doi: 10.1042/bj3270407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luka Z, Ham AJ, Norris JL, Yeo EJ, Yermalitsky V, Glenn B, Caprioli RM, Liebler DC, Wagner C. Identification of phosphorylation sites in glycine N-methyltransferase from rat liver. Protein Sci. 2006;15:785–794. doi: 10.1110/ps.051906706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blakley RL. The Biochemistry of Folic Acid and Related Pterines. Wiley; New York, NY: 1969. [Google Scholar]
- 6.Luka Z, Loukachevitch LV, Wagner C. Acetylation of N-terminal valine of glycine N-methyltransferase affects enzyme inhibition by folate. Biochim Biophys Acta. 2008;1784:1342–1346. doi: 10.1016/j.bbapap.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation Mode. Meth Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 8.Bailey S. The ccp4 Suite – Programs for Protein Crystallography. Acta Crystallogr D-Biological Crystallography. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 9.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. Journal of Applied Crystallography. 2006;39:109–111. [Google Scholar]
- 10.Howlin B, Butler AA, Moss DS, Harris GW, Driessen HPC. Tslanl – Tls Parameter Analysis Program for Segmented Anisotropic Refinement of Macromolecular Structures. Journal of Applied Crystallography. 1993;26:622–624. [Google Scholar]
- 11.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 12.Konishi K, Fujioka M. Rat liver glycine methyltransferase. Cooperative binding of S-adenosylmethionine and loss of cooperativity by removal of a short NH2-terminal Segment. J Biol Chem. 1988;263:13381–13385. [PubMed] [Google Scholar]
- 13.Matthews DA, Alden RA, Bolin JT, Freer ST, Hamlin R, Xuong N, Kraut J, Poe M, Williams M, Hoogsteen K. Dihydrofolate reductase: x-ray structure of the binary complex with methotrexate. Science. 1977;197:452–455. doi: 10.1126/science.17920. [DOI] [PubMed] [Google Scholar]
- 14.Hyatt DC, Maley F, Montfort WR. Use of strain in a stereospecific catalytic mechanism: crystal structures of Escherichia coli thymidylate synthase bound to FdUMP and methylenetetrahydrofolate. Biochemistry. 1997;36:4585–4594. doi: 10.1021/bi962936j. [DOI] [PubMed] [Google Scholar]
- 15.Pejchal R, Sargeant R, Ludwig ML. Structures of NADH and CH3-H4folate complexes of Escherichia coli methylenetetrahydrofolate reductase reveal a spartan strategy for a ping-pong reaction. Biochemistry. 2005;44:11447–11457. doi: 10.1021/bi050533q. [DOI] [PubMed] [Google Scholar]
- 16.Evans JC, Huddler DP, Hilgers MT, Romanchuk G, Matthews RG, Ludwig ML. Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase. Proc Natl Acad Sci USA. 2004;01:3729–3736. doi: 10.1073/pnas.0308082100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takata Y, Huang Y, Komoto J, Yamada T, Konishi K, Ogawa H, Gomi T, Fujioka M, Takusagawa F. Catalytic mechanism of glycine N-methyltransferase. Biochemistry. 2003;42:8394–8402. doi: 10.1021/bi034245a. [DOI] [PubMed] [Google Scholar]
- 18.Konishi K, Fujioka M. Chemical modification of a functional arginine residue of rat liver glycine methyltransferase. Biochemistry. 1987;26:8496–502. doi: 10.1021/bi00399a069. [DOI] [PubMed] [Google Scholar]