Abstract
Mutations within the human TREX1 3' exonuclease are associated with Aicardi-Goutières Syndrome (AGS) and familial chilblain lupus (FCL). Both AGS and FCL are autoimmune diseases that result in increased levels of interferon alpha and circulating antibodies to DNA. TREX1 is a member of the endoplasmic reticulum (ER)-associated SET complex and participates in granzyme A-mediated cell death to degrade nicked genomic DNA. The loss of TREX1 activity may result in the accumulation of double-stranded DNA (dsDNA) degradation intermediates that trigger autoimmune activation. The X-ray crystal structures of the TREX1 wt apoprotein, the dominant D200H, D200N and D18N homodimer mutants derived from AGS and FCL patients, as well as the recessive V201D homodimer mutant have been determined. The structures of the D200H and D200N mutant proteins reveal the enzyme has lost coordination of one of the active site metals, and the catalytic histidine (H195) is trapped in a conformation pointing away from the active site. The TREX1 D18N and V201D mutants are able to bind both metals in the active site, but with inter-metal distances that are larger than optimal for catalysis. Additionally, all of the mutant structures reveal a reduced mobility in the catalytic histidine, providing further explanation for the loss of catalytic activity. The structures of the mutant TREX1 proteins provide insight into the dysfunction relating to human disease. Additionally, the TREX1 apoprotein structure together with the previously determined wild type substrate and product structures allow us to propose a distinct mechanism for the TREX1 exonuclease.
Keywords: TREX1, autoimmune disease, protein structure, enzyme mechanism, lupus, exonuclease
1. Introduction
The TREX1 3’ exonuclease is a member of the DnaQ family of enzymes, and catalyzes the major 3’ exonuclease activity in mammalian cells [1–2]. The exact cellular role of TREX1 remains unclear, but several potential functions have been identified. These include degradation of nicked genomic DNA during Granzyme A cell death [3], elimination of single-stranded DNA (ssDNA) derived from endogenous retroelements as a part of the interferon-stimulatory DNA response [4], and the disposal of aberrant ssDNA replication intermediates [5–6]. TREX1 interacts with the SET protein as a member of the endoplasmic reticulum (ER)-associated SET complex [3]. The 440 kDa SET complex contains proteins such as the endonuclease NM23H1, the apurinic endonuclease APE1, and the DNA bending protein HMG-2. During granzyme A-mediated cell death, the serine protease granzyme A enters a target cell and proteolyses certain components of the complex, which results in the translocation of the SET complex into the nucleus. Subsequently, the NM23H1 nuclease generates single-stranded nicks in the genomic DNA. TREX1 binds the 3' termini generated at these nicks and fully degrades the DNA, ensuring cell death [3]. It has been proposed that this TREX1 function is important to avoid the accumulation of host DNA after cell death that could trigger an autoimmune response. Alternatively, there is evidence that TREX1 acts as a negative regulator of the interferon stimulatory DNA response pathway by degrading single- and double-stranded DNA (ssDNA, dsDNA) resulting from replication of endogenous retroelements or DNA repair intermediates [4, 6]. Removal of this cytosolic DNA prevents a potent cell-intrinsic antiviral response activated through cytosolic DNA receptors.
Genetic studies have linked mutations in the TREX1 gene with a spectrum of autoimmune diseases including Aicardi-Goutières syndrome (AGS), familial chilblain lupus (FCL), systemic lupus erythematosus (SLE), and retinal vasculopathy and cerebral leukodystrophy (RVCL) [7–14]. Additionally, TREX1 deficient mice develop inflammatory myocarditis, strengthening the association with autoimmune disease [15]. The sites of mutations causing autoimmune diseases in humans are located throughout the TREX1 gene; affecting areas of the protein including the active site, the dimer interface, and the C-terminal domain. Mutations within the TREX1 active site have been linked to both AGS and FCL. AGS is a genetically determined encephalopathy exhibiting recessive inheritance, although isolated cases of dominant AGS have been identified. The symptoms of AGS mimic congenital viral infection without evidence of an infectious agent [16–18]. These patients exhibit a severe phenotype involving the calcification of basal ganglia and white matter in the brain that negatively impacts motor and social development [18–20]. FCL is a dominant form of lupus erythematosus [9, 11, 21], which shares significant phenotypic overlap with AGS including increased levels of interferon alpha and circulating antinuclear antibodies to dsDNA or ssDNA, suggesting a common pathogenesis.
The TREX1 protein is a stable homodimer in solution, and the crystal structure revealed that the active sites are positioned at opposite outer edges of the same face of the dimer [22–23]. The TREX1 protein contains four highly conserved acidic residues (DEDD) within the active site that are characteristic of the DnaQ family of exonucleases. These residues are required for coordinating two Mg2+ ions that are necessary for catalysis. The presence of a conserved histidine residue in the TREX1 active site places the protein within the DEDDh subgroup of the DnaQ family. The histidine is thought to contribute to deprotonation of a water molecule and to promoting nucleophilic attack on the scissile phosphate during catalysis [24–27]. The TREX1 protein has several additional distinct structural features that likely contribute to its cellular function. A polyproline type II (PPII) helix is positioned adjacent to the active site and dimer interface in each TREX1 protomer. The PPII helix has a putative role in mediating contacts with other proteins in the cell, and may be important for its interaction with the SET complex. The TREX1 protein also has an extended C-terminal region that is involved in the sub-cellular localization of TREX1 on the cytosolic side of the endoplasmic reticulum [3, 12]. Finally, a flexible loop that is proposed in DNA binding is located adjacent to each active site [26–27].
Multiple mechanisms of dysfunction underlie the observed clinical phenotypes in patients with TREX1 mutations, reflective of the fact that the mutations result in a broad range of catalytic activities, yet all result in similar human pathologies. Our previous biochemical data has begun to provide insight into the disease etiology for dominant TREX1-mediated FCL and AGS, but the mechanism of recessive AGS remains elusive. We have proposed a mechanism for biological dysfunction of dominant TREX1 mutants in which competitive binding of DNA nicks by the catalytically inactive dominant mutants precluding access by active TREX1 [28–29]. In order to better understand the effects of disease causing mutations on the structure and function of the TREX1 enzyme, we have determined the X-ray crystal structures of four mutant TREX1 proteins identified in patients with autoimmune disease, as well as the structure of the wild type TREX1 apoprotein. The D18N, D200N and D200H, TREX1 proteins are dominant mutants associated with FCL and AGS. The V201D mutant TREX1 is a recessive AGS mutation. All of these mutations are located in or directly adjacent to the TREX1 active site and display a spectrum of catalytic activities ranging from 25% of wild type activity to virtually inactive enzyme. The role of the D18 and D200 residues is to coordinate the two divalent metal ion cofactors necessary for catalysis. The structures of the mutant TREX1 proteins demonstrate a fundamental role for conformational movement of the catalytic histidine during nucleic acid hydrolysis. This reveals unique characteristics of two metal ion catalysis in the context of the DEDDh subgroup of the DnaQ family of exonucleases that rationalizes substrate trapping in FCL and dominant AGS. The structure of the TREX1 apoprotein, reveals conformational differences in the active site dependent upon the presence or absence of substrate. Additionally, in the structure of the D200H TREX1 mutant we observe an alternate conformation of the conserved catalytic residue, E20, which forms an interaction with an arginine from the opposing protomer. This data provides new insight into possible substrate release during the catalytic cycle, and for the first time provides a rationale for the unique dimeric architecture of the TREX1 protein.
2. Materials and Methods
2.1 Protein Expression and Purification
The mouse TREX1 enzymes used for crystallization contain the catalytic core residues (1–242) of the protein. This C-terminal truncation was expressed as a fusion with an N-terminal polyhistidine sequence followed by maltose binding protein (MBP) as described previously [22]. Briefly, the expression vector containing the MBP-TREX1 fusion was transformed into BL21*(DE3) Rosetta II cells (Novagen) for overexpression. The cells were grown to an A600=0.5 and induced with 1 mM isopropyl-β-D-thiogalactopyranoside for 10 minutes at 37°C and quickly cooled on ice to 16 °C. Cells were allowed to grow for 18 hours at 16 °C. The MBP-TREX1 fusion was bound to amylose resin, washed thoroughly, and cleaved overnight by incubation with PreScission Protease (GE Life Sciences) at 4 °C to remove the polyhistidine tag and MBP protein. After cleavage, the mTREX1 protein was collected in the column flow-through and dialyzed against 50 mM Tris-HCl pH 7.5, 50 mM NaCl, 10% glycerol and 1 mM EDTA and purified to homogeneity using phosphocellulose chromatography.
2.2 Protein Crystallization and X-ray Data Collection
The TREX1 mutant proteins and the wild type TREX1 apoprotein were crystallized using the sitting drop vapor diffusion technique. All TREX1 proteins were dialyzed into 20 mM MES (pH 6.5), 50 mM NaCl. Substrate complex was formed by incubating the protein with a four-nucleotide ssDNA (5'-GACG, purchased from Operon) in a molar ratio of 1:2 and 5 mM calcium chloride (5 mM magnesium chloride in the case of the D200N mutant). TCEP-HCl pH 8.0 was added to a final concentration of 1 mM to each protein solution prior to placing in the crystallization tray. Two µl protein complex at 5 mg/ml TREX1 was mixed with an equal volume of reservoir solution and placed on a bridge above 500 µl of the reservoir solution. Optimized crystals of the wild type TREX1 apoprotein grew in 0.25 M tri-sodium citrate dihydrate and 20% PEG 3350. Optimized crystals of the D18N complex were obtained by microseeding into 0.15 M MES pH 6.5, 19% PEG 4000 and 10% ethylene glycol and grown at 15°C. Crystals of the D200N mutant complex were obtained by microseeding into 0.1 M MES pH 5.5, 0.075 M NaCl, 12% PEG 3350 and 5% 1,4-butanediol and grown at 15°C. The D200H complex was obtained by microseeding into 0.1 M MES pH 6.0, 16% PEG 4000, 2% 1,4-butanediol and grown at 30°C. Crystals of the V201D protein in complex with ssDNA were grown at 30°C in 16% PEG 3350, 0.1 M NaI and 5% 1,4-butanediol. All crystals grew within one week. Prior to data collection all crystals were dipped into reservoir solution containing 20% 1,4-butanediol or (for D18N) 20% glycerol in preparation for cryo-cooling. Crystals were mounted on a nylon loop and flash cooled to 100 K in a stream of liquid nitrogen.
2.3 Phasing and Refinement
The X-ray data were collected using CuKα radiation on a MicroMax 007 generator and a Saturn 92 CCD detector (Rigaku). Intensity data were processed using the programs d*TREK or HKL2000 [30–31]. The D18N, D200N, and D200H TREX1 mutants in complex with ssDNA belong to the P21 spacegroup. The TREX1 V201D- ssDNA complex and the wild type TREX1 apoprotein belong to the P212121 space-group (Table 1). Phases for the data were obtained by maximum likelihood molecular replacement using the program PHASER [32] and the TREX1 dimer (PDB ID: 2OA8), including the α7–α8 loop, as the search model (protein only). The TREX1 apoprotein and mutant models were built in the program COOT [33] following composite omit procedures and the structures refined using the programs CNS [34], Refmac5 [35], and Phenix.refine [36]. Translation/libration/screw (TLS) refinement was utilized to independently define subgroups within the apoprotein structure and to further refine their directions of movement as individual rigid bodies [37–38]. The inspection of clashes and stereochemical parameters was carried out using the program MolProbity [39]. All structure figures were generated in the program Pymol [40]. Coordinates have been deposited into the protein databank for the structures of TREX1 apoprotein, D200N, D200H, V201D and D18N TREX1 proteins.
Table 1.
Crystallographic data and refinement statistics
| TREX1 | Apoprotein | D18N | D200H | D200N | V201D |
|---|---|---|---|---|---|
| Space group | P212121 | P21 | P21 | P21 | P212121 |
| Unit cell, Å | a=64.2, | a=64.0, | a=65.2, | a=64.9, | a=70.5, |
| b=85.7, | b=58.8, | b=57.9, | b=56.6, | b=86, | |
| c=100.3 | c=66.5 | c=68.3 | c=68.1 | c=92.8 | |
| α,β,γ=90° | α,γ=90° | α,γ=90° | α,γ=90.0° | α,β,γ=90° | |
| β=110.3° | β=108° | β=108° | |||
| Molecules/asymmetric unit | 2 | 2 | 2 | 2 | 2 |
| Wavelength, Å | 1.54 | 1.54 | 1.54 | 1.54 | 1.54 |
| Resolution, Å | 29-1.95 | 43-2.55 | 23-2.3 | 33-2.3 | 47-1.75 |
| Completeness, % | 99.7 (97.1) | 96.6 (95.1) | 99.5 (99.5) | 99.7 (99.6) | 99 (91.2) |
| Rmerge, %a | 5.7 (27.6) | 10 (23) | 7.2 (29.3) | 9.0 (34.6) | 6.8 (24.0) |
| Mean I/σ | 16 (4.1) | 10.9 (4.5) | 8.8 (3.7) | 13.8 (4.0) | 11.9 (3.4) |
| Average redundancy | 6.7 | 6.9 | 3.5 | 7 | 5.7 |
| R-factor, %b | 20.4 | 20.9 | 21.5 | 21.9 | 17.2 |
| Rfree, %b | 23.7 | 26.2 | 27.5 | 28.7 | 20.3 |
| Average Wilson B-factor, Å2 | 24.7 | 26.9 | 33.8 | 36.5 | 19 |
| Root mean square deviation bond lengths, Å | 0.007 | 0.008 | 0.005 | 0.008 | 0.010 |
| Root mean square deviation bond angles, ° | 1.069 | 1.291 | 1.002 | 1.273 | 1.279 |
Statistics for outer resolution shell are given in parentheses.
Rmerge = Σ|I-〈I〉|/ΣI where I is the observed intensity and 〈I〉 is the average intensity.
R factor = Σ||Fo|-|Fc||/Σ|Fo|. Rfree is the same as R, but calculated with 5% of the reflections that were never used in crystallographic refinement.
3. Results and Discussion
The TREX1 D18N, D200H, and V201D mutant proteins associated with disease (Figure 1) were co-crystallized with ssDNA and Ca2+, and the D200N mutant was co-crystallized with ssDNA and Mg2+. Co-crystallization of some of the TREX1 mutants with calcium was necessary to prevent degradation of substrate DNA. Replacement of the Mg2+ ion with other divalent metals is a strategy that has been successfully employed in the past to trap complexes of nucleases with DNA for structure determination [22, 41–47]. Data were collected to 1.95 Å, 2.55 Å, 2.35 Å, 2.3 Å, and 1.75 Å resolution for the wild type apoprotein, D18N, D200H, D200N, and V201D structures, respectively. All structures were determined by molecular replacement (MR), using the TREX1 dimer (PDB ID: 2OA8) protein only as the search model. After MR, good electron density was observed for most residues in the TREX1 dimer in each mutant structure and in the wild type apoprotein structure. In addition, electron density for the ssDNA was observed in the active site of all of the mutant structures indicating that the observed decrease in activity in these mutants is not due to an inability to bind substrate. Final R/Rfree at the conclusion of refinement for the X-ray crystallographic models of the dimeric TREX1 apoprotein and mutants are 20.4% / 23.7% for the apoprotein, 23.1% / 28.4% for D18N, 21.5% / 27.5% for D200H, 21.9% / 27.9%for D200N, and 19.5% / 22.6% for V201D. Further refinement statistics are listed in Table 1.
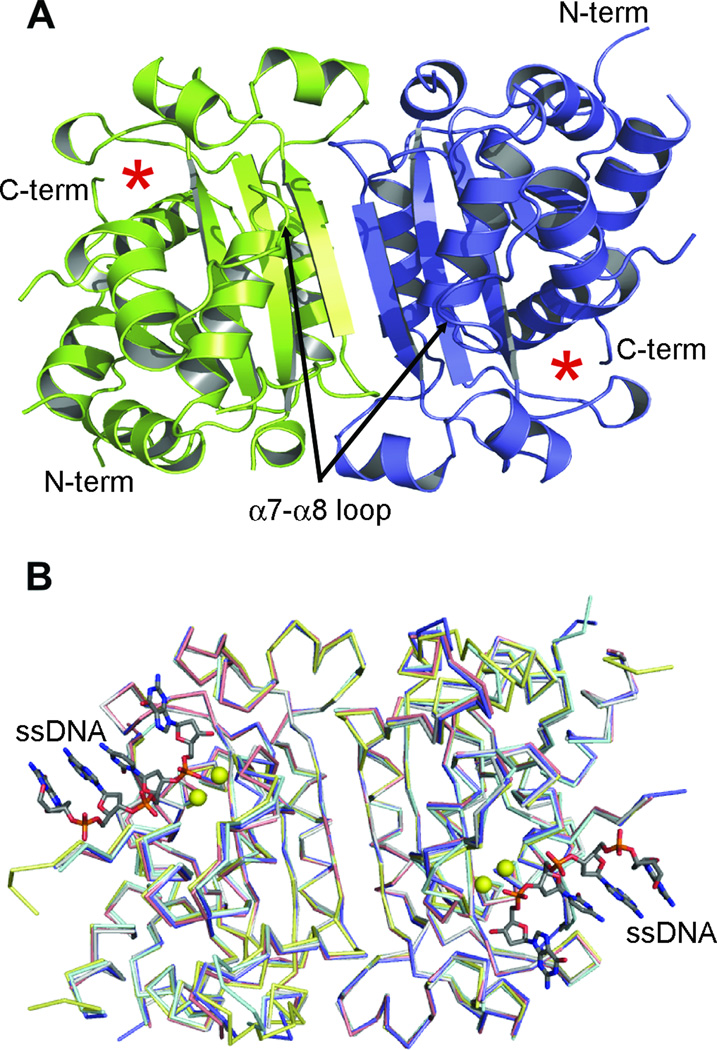
Figure 1. TREX1 structures.
(A) Ribbon diagram of the apo-TREX1 dimer, with one protomer in blue and the other in green. The active site of each monomer is indicated by asterisks. (B) Superposition of the TREX1-DNA complex (slate) with the D18N (cyan), D200N (gray), D200H (salmon) and V201D (yellow) mutant TREX1 structures shows the mutant proteins maintain a similar overall structure (overall RMSD 0.37–0.62). The major structural differences being in the coordination of the divalent metal ions and positioning of the catalytic residue, H195 (see Figures 2 & 3). TREX1 structures shown as Cα traces for clarity, divalent metals shown as yellow spheres and DNA as stick models.
Active site rearrangements in TREX1 apoprotein and disease mutant structures
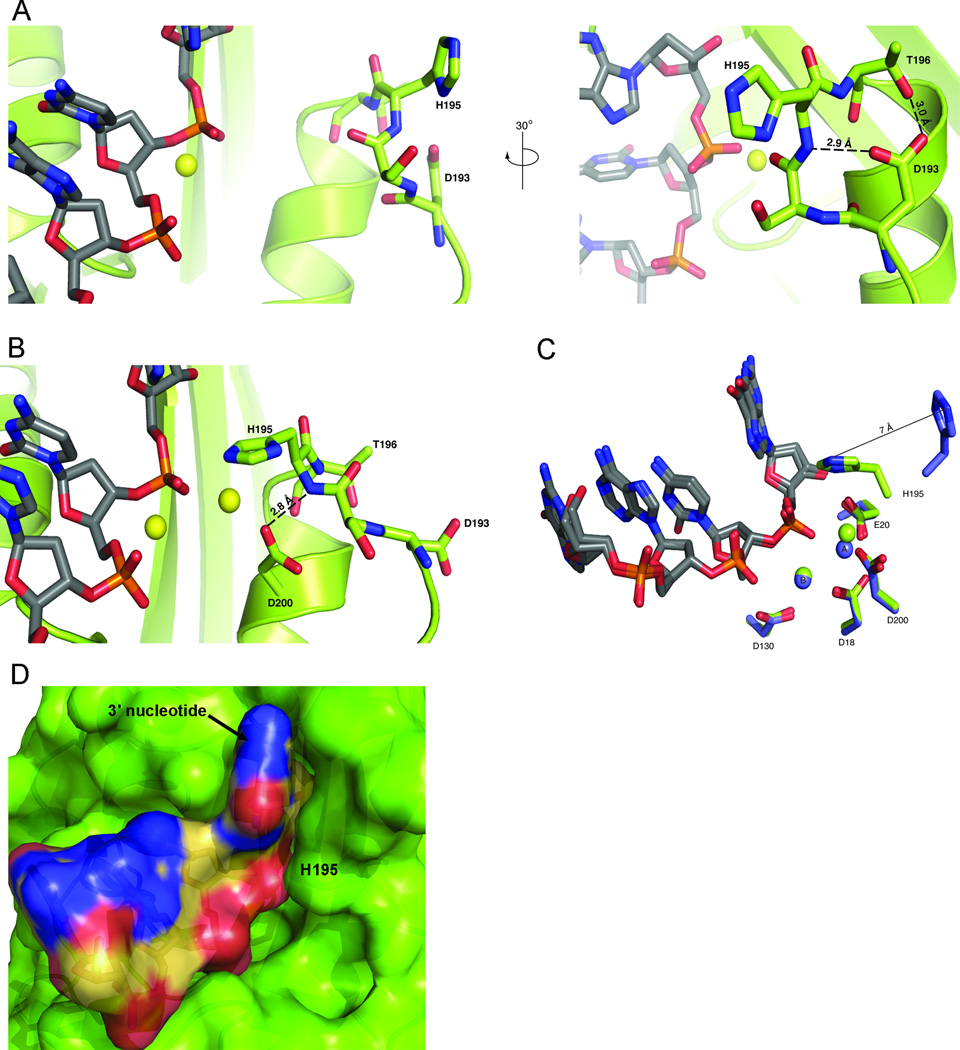
The structures of the mutant TREX1 proteins fit into two distinct groups based on the conformation of residue H195 and the number of metal ions coordinated in the active sites. Each of the TREX1 mutant proteins adopts the same overall α/β fold as the wild type TREX1 enzyme (RMSD 0.37–0.62 Å) (Figure 1). The positions of Mg2+ in the two-metal-ion mechanism for phosphoryl hydrolysis have the canonical names of a and b. Metal ion a is required for directing the inline attack of the scissile phosphate by an activated water molecule while metal ion b facilitates DNA binding and stabilization of the penta-covalent transition state [41, 48]. The structural roles for residues D18 and D200 are to coordinate divalent metal at position a, and residue D18 additionally coordinates metal at position b. In the structures of the D200H and D200N mutants associated with AGS, the conformation of the loop containing the catalytic H195 is oriented with the histidine pointing away from the active site in each monomer of the TREX1 dimer, and there is electron density only for a single metal ion in position b (Figure 2). This outward conformation of H195 is stabilized by hydrogen bonds from D193 to the backbone amide of H195 and the side chain hydroxyl of T196 (Figure 3). In contrast, the structures of the D18N and V201D TREX1 mutants reveal H195 is oriented towards the active site in both protomers of the dimer and electron density is present for both metal ions (Figure 2). Amino acid V201 is on the back side of the α-helix containing the adjacent metal-coordinating D200 residue. The inward conformation of the catalytic histidine is a movement of about 7 Å from the position of the outward conformation, and it is stabilized by hydrogen bonds from the amide nitrogen of H195 to a carboxylate oxygen of D200 and from the carbonyl oxygen of D193 to the side chain hydroxyl of T196 (Figure 3).
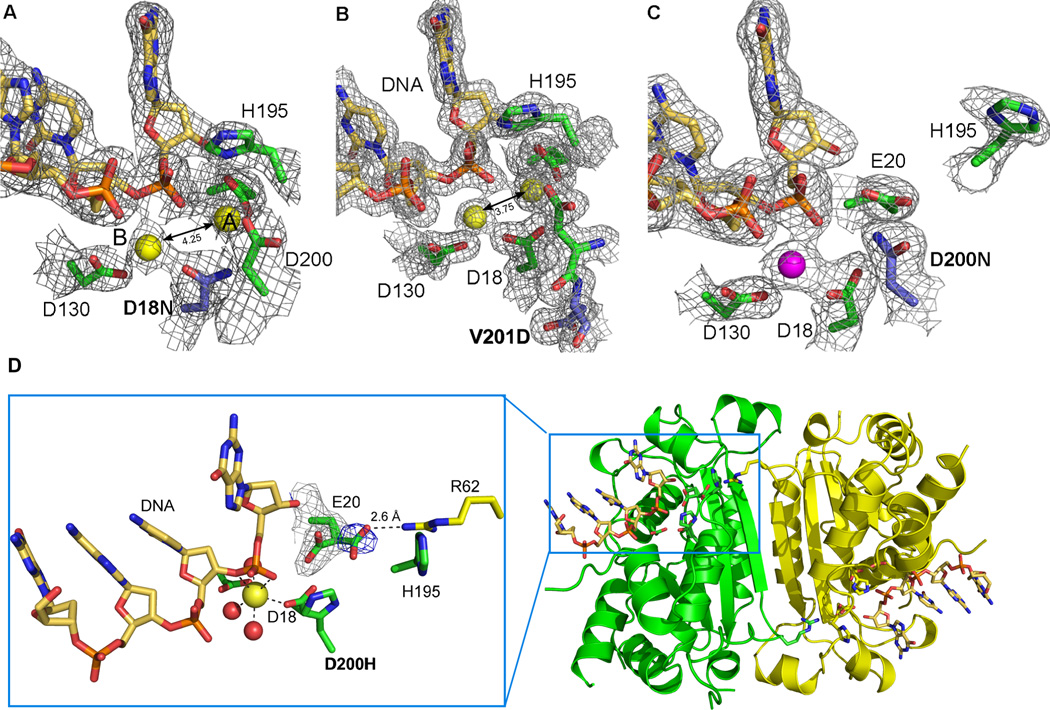
Figure 2. Active site morphology of TREX1 mutants.
The structures of the mutant TREX1 proteins fit into two distinct groups based on the number of metal ions coordinated and the position of residue H195 in the active sites. 2Fo-Fc electron density (1σ) for the D18N-DNA and V201D-DNA mutant TREX1 structures (A and B) reveal the catalytic H195 is pointing toward the active site and both divalent calcium ions (yellow spheres, canonical position A and B labeled) are present. However, the 4.25 Å and 3.75 Å interatomic distances for these metal ions is greater than the 3.5 Å distance needed for optimal catalytic activity. In contrast, the D200N-DNA and D200H-DNA structures (C and D) have the H195 oriented away from the active site and only a single divalent ion coordinated by the protein-DNA complex (Mg2+ shown as magenta sphere). The mutated residues in the AGS and FCL mutant active sites are highlighted in blue. (D) An alternate conformation of residue E20 is seen in the active site of the D200H mutant TREX1 structure (3.5 σ Fo-Fc density shown in blue) that allows an interaction with amino acid R62 from the opposing protomer in the dimer. This intersubunit interaction may play a role in substrate release and provides the first structural rationale for the dimeric nature of the TREX1 enzyme.
Figure 3. Stabilization of the inward and outward conformations of H195.
(A) When H195 adopts the outward conformation from the active site, the flexible loop containing the residue is stabilized by two hydrogen bonds from the carboxylate oxygens of residue D193 to the side chain oxygen of T196 and the backbone amide nitrogen of H195. Two orientations from the TREX1 D200H structure, rotated by 30° are shown. (B) The inward conformation of H195 into the active site is stabilized by hydrogen bonds from a carboxylate oxygen of D200 to the backbone amide of H195, as well as the hydroxyl oxygen of T196 to the backbone carbonyl oxygen of D193. The inward H195 conformation shown is from the α7-α8 loop of the TREX1 V201D mutant structure. Protein residues are shown in green, ssDNA as gray sticks, and divalent metal ions as yellow spheres. (C) Residue H195 adopts two distinct structural conformations that are displaced by 7 Å (PDB ID: 2OA8). (D) Surface representation of ssDNA (colored by atom) bound in V201D mutant TREX1 active site (green). The inward conformation of H195 (labeled) helps surround the 3' nucleotide of the substrate.
The dynamic conformations of H195 observed in the current TREX1 structures is likely important for catalysis. The orientation of the catalytic histidine appears to be influenced by the presence of metal ion at position a in the active site. In the structures of the D18N and V201D mutant TREX1 proteins, the exclusive orientation of H195 pointing into the active site suggests that the binding of metal ion a contributes to the inward orientation of H195 in the active site and that the loss of this metal in the D200N and D200H only supports the observed outward orientation of H195 (Figure 2). The effect of the metal must be indirect since there are no direct interactions between the metal and histidine. Interestingly in our previous structure of the wild type TREX1-ssDNA complex, H195 is observed in both the inward and outward conformations within the two protomers, implying that only one half of the dimer may be active at a time, and that both conformations are necessary in the catalytic cycle [22]. This is a distinction from the mutant structures that all have H195 locked into a single conformation and bolsters the idea that this residue must be mobile during catalysis. Furthermore, the alternating conformations between active and inactive states supports the possibility of communication across the dimer interface that may coordinate alternating catalytic activity between protomers of the TREX1 dimer. The idea of inter-protomer communication has other supporting evidence. Cooperativity within the homologous, dimeric TREX2 exonuclease has been demonstrated by kinetic analysis [26], suggesting an important role for communication across the dimer interface in the catalytic function of TREX exonucleases. Additionally, biochemical analysis of the disease causing R114H TREX1mutation reveals the R114 residue in one monomer contributes to catalysis of the opposing monomer [49]. The catalytic histidine (H195) has been previously proposed to play a role in the deprotonation of a water molecule coordinated by metal a to generate the nucleophile to carry out phosphodiester bond cleavage [44, 50]. Mutagenesis of the catalytic histidine within the DEDDh exonucleases TREX1, TREX2, and RNase T abolishes exonuclease activity [23, 26–27, 51]. Quantum mechanical/molecular mechanical studies of the catalytic mechanism of the structurally similar epsilon subunit of E. coli DNA polymerase III indicate that a proton is transferred from the nucleophilic water to the catalytic histidine, further supporting a role for the histidine in phosphoryl hydrolysis [25].
The structure of the TREX1 apoprotein provides further evidence for the conformational malleability of H195. The TREX1 apoprotein (Figure 1) maintains the same overall fold as the TREX1 protein in complex with DNA [22] (RMSD 0.66 Å), however, a significant difference between the two structures exists in the active site of the enzyme where the α7–α8 loop that contains the catalytic His195 is disordered in the absence of DNA binding. There is weak electron density for the backbone of this loop in the TREX1 apoprotein structure, but no density for the side chains of residues S194 and H195; indicative of a dynamic movement of the H195 loop that becomes ordered during DNA binding. The transition of residue H195 from a disordered to ordered state upon DNA binding combined with the observation that it has two distinct structural conformations supports the idea that there is deliberate and directed movement of H195 that is required during the catalytic cycle.
While the conformation of residue H195 is significant for catalysis, the positions of the divalent metal ions are also essential. In the structures of the D18N and V201D TREX1 mutant proteins, the H195 residue is oriented inward towards the active site to assist catalysis, yet these proteins display only 1/ 160000 and ¼ relative catalytic activity, respectively, to wild type TREX1 [11, 22]. Within both of these structures electron density is present for metal ions in both positions a and b (Figure 2). However, the metal ions in the two protomers of the D18N TREX1 dimer are separated by 4.18 and 4.45 Å. This distance is consistent with the 4.5 Å inter-magnesium distance that Nowotny et al. [45] observed in the structure of the D132N-Mg2+ RNase H protein (D132 is a bridging aspartate in RNase H), which is a catalytically inactive enzyme, and analogous to the TREX1 D18N mutation. Metal ion position is an important factor in two metal ion catalysis, and it has been proposed that metal a moves toward metal ion b during catalysis to bring the nucleophile close enough to the scissile phosphate to promote catalysis [52–53]. In both the substrate and product complex structures of the TREX1 protein [22] electron density for two metal ions is present in the enzyme active site and the metal ion spacing is similar to the inter-metal distance of 3.5 Å that has been proposed as the metal spacing for an active nuclease [45, 54–55]. In the TREX1 substrate complex, the distance between metal ions is an average 3.4 Å [22]. When the metal ions are separated by 4 Å or more, as observed in the TREX1 D18N structure, metal a is likely unable to help the nucleophile bridge the distance to the phosphorous. The metal ion spacing is between 3.7 and 3.8 Å in both protomers of the V201D dimer (Figure 2). This is closer to ideal but probably still suboptimal and likely explains the higher level of nuclease activity in the V201D versus the D18N mutant [22]. The caveat to this interpretation is that the V201D and D18N structures contain calcium as the divalent ion rather than magnesium. Using calcium was necessary to prevent catalysis and trap the complex for crystallization. There are two points, however, that provide confidence that the observed positions of the divalent metals in the structures of the mutant proteins are relevant. First, our previous structures of the wild type TREX1 protein in complex with nucleic acid using calcium or manganese show these metals are properly positioned and spaced for catalysis (Supplemental Figure 1). Secondly, the degree of misalignment of the metal in each of the current mutant structures correlates with the loss of catalytic activity for these mutants in our biochemical experiments [22].
Another very interesting phenomenon that we observe with the D200H mutant structure is an alternate conformation of the amino acid E20 in the active site (Figure 2). Residue E20 is one of the conserved acidic residues critical for coordination of divalent metal in position a. The active site of the D200H TREX1 has a metal ion only at position b. After the first round of crystallographic refinement of this structure, a region of positive Fo-Fc electron density (3.5 σ) appeared adjacent to the carbon-γ of E20. We modeled an alternate conformation of the amino acid into the density, which refined to an occupancy of about 30%. The carboxylate group in the alternate conformation of E20 is rotated away from the active site towards the dimer interface and places it 2.6 Å from residue R62 of the opposing protomer in the dimer. The conformational change of this amino acid side chain appears to be linked to the loss of metal ion at position A, and likely has significant consequences for catalysis. Loss of at least one of the coordinating metals ions in the active site is thought to be necessary for product release after catalysis [45, 47, 52, 56]. This process might be facilitated by the reorientation of the E20 side chain through an interaction with R62, which would reduce the coordination of metal ion a and effectively release it. Amino acid R62 of each protomer stretches across the dimer interface, positioning the guanidinium group adjacent to the active site of the opposing protomer. It was previously noted that residue R62 is highly conserved in all species of TREX enzymes [27] and its position is nearly identical in all of the TREX1 structures including the D200H mutant. The structural conservation of this residue coupled with the observation that it interacts with the active site of the opposing protomer provides a possible biological rationale for the dimeric structure of the TREX1 exonuclease.
Consequences of TREX1 mutations for disease
The D18N, D200N and D200H mutations in TREX1 cause dominant disease in humans, meaning patients with a single mutated allele display the disease phenotype. In vitro assays confirm the homodimer mutant enzymes are essentially catalytically inactive on either ssDNA or dsDNA [11, 22, 28]. However, heterozygous individuals have the formal possibility of expressing both wild type and mutant TREX1 enzyme and the potential to create wild type-mutant heterodimers in cells. The recombinantly expressed D18 and D200 heterodimers, containing one wild type and one mutant protomer in the dimer, are about 50% active on ssDNA [28]. Most notably, they display no activity on a nicked dsDNA plasmid substrate, and both the homo- and heterodimer D18 and D200 mutants inhibit the activity of wild type TREX1 on a dsDNA substrate. This ability of either the TREX1 homodimer or heterodimer mutant proteins to inhibit the wild type enzyme at sub-stoichiometric concentrations is likely connected to the dominant genetics of the autoimmune dysfunction associated with these TREX1 mutations. One explanation for the inhibition of wild type TREX1 activity by the dominant mutant proteins is that they bind the substrate and do not readily release it; therefore, blocking access to the DNA 3' termini by wild type enzyme. Residues D18 and D200 participate in coordination of metal a, which in turn is necessary for phosphoryl hydrolysis. Metal a is absent from the D200N/H mutant structures and is coordinated with a significantly larger inter-metal ion distance in the D18 mutant structure than is observed in the TREX1 substrate complex. The structures of the dominant D18N, D200H, and D200N TREX1 proteins reveal DNA substrate is bound by the mutants, but is not able to undergo hydrolysis, therefore trapping them in a non-productive complex. In contrast, the structure of the recessive V201D mutant TREX1 protein shows the active site morphology is the most similar to wild type of any of the mutant proteins we analyzed. It has only minor distortions in metal ion spacing that are consistent with its relatively higher in vitro catalytic activity, and strengthens the notion that the V201D mutation results in a different mechanism of disease than the dominant mutants.
Two-metal-ion mediated nucleic acid hydrolysis and the TREX1 mechanism
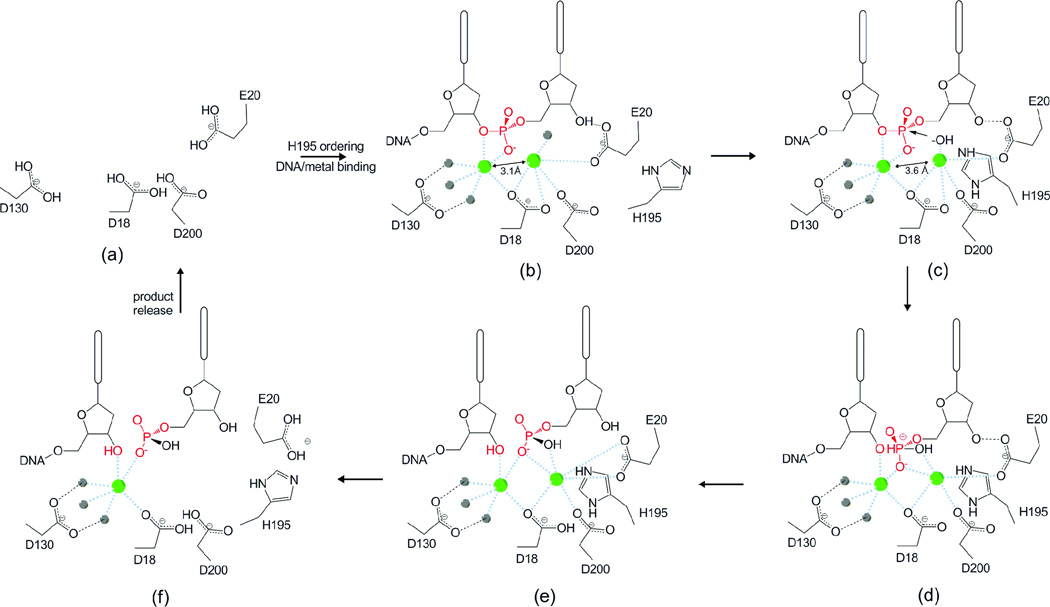
Together, our wild type and mutant structure data of the TREX1 proteins provide us with direct information for most steps in the catalytic degradation of single-stranded DNA (ssDNA). This enables us to propose a detailed catalytic mechanism for TREX1 (Figure 4). Based on our apo enzyme structure and previously determined wt TREX1 in complex with ssDNA, the TREX1 reaction begins with the binding of metal and ssDNA in the enzyme active site (Figure 4a–b), which results in the ordering of the H195 loop. In one protomer, H195 flips into the active site to abstract a proton from the lone water molecule coordinating metal a and displacing it towards the scissile phosphate (Figure 4c)[57]. The structures of the wtTREX1-ssDNA complex and the V201D mutant show the motion of the activated water molecule towards the substrate results in the shift in metal a diagonally away from metal b and towards the substrate. This positions metal a within 3.5 Å of the scissile phosphate as observed in the structure of the wt substrate complex. Nucleophilic attack on the scissile phosphate ensues due to the placement of the hydroxide anion within the ~2–2.5 Å of the scissile phosphate that is required for nucleophilic attack to occur [45, 51]. This motion of metal a is different from that proposed for metal a towards metal b to promote catalysis in RNase H, but may be explained by differences in active site architecture or the fact that metal a is penta-coordinated in TREX1 and hexa-coordinated in RNase H [45, 52]. Metal b participates in the stabilization of the penta-covalent transition state (Figure 4d). Once the scissile bond is broken, H195 flips back out of the active site, releasing the product and allowing the alignment of the new 3’-terminal nucleotide for excision. This movement of H195 also provides a physical opening of the active site relative to the closed conformation which securely surrounds the 3' nucleotide (Figure 3). Product release may be facilitated by the movement of residue E20 through an interaction with R62 of the opposing protomer and subsequent loss of divalent metal in the active site (Figure 4f). TREX1 is a non-processive enzyme; therefore, the substrate and/or metals will be lost from the active site at the end of the catalytic cycle. Rebinding of the DNA 3' end along with metal ions must occur for catalysis to continue. Other DEDDh subgroup exonucleases likely follow a similar mechanism involving an active and resting state that are defined by both the inter-metal distance and the position of the catalytic histidine. Because of the possibility of communication across the dimer interface, it is important to address the significance of dimerization on the catalytic mechanism of TREX1. We propose that the TREX1 3’ exonuclease and other similar homodimeric nucleases such as TREX2 catalyze the nucleotide excision reaction via a mechanism where the catalytic histidine (H195 in TREX1) acts as a switch between the active and resting states of the enzyme and that the protomers alternate between these two states as they degrade substrate. The cumulative structural data suggest a mechanism where the two protomers alternate back and forth between an active and resting state. This is corroborated by the dual conformation of H195 in the wild type-substrate complex (PDB ID: 2OA8) and the shift in the metal ion spacing from 3.1 Å when H195 is out of the active site (resting) to 3.6 Å when H195 moves into the active site (active) in this structure. It is also consistent with the active and resting states that were previously proposed for two metal ion catalysis based solely on the inter-metal ion distance [52].
Figure 4. Proposed mechanism of the TREX1 exonuclease.
(a) In the absence of DNA and metal binding H195 in the active site is disordered. (b) Binding of DNA and metal ions (green spheres) orders residue H195 along with (c) a shift of divalent metal ions to inter-atomic spacing of ~3.6 Å. H195 contributes to deprotonation of a water molecule (gray sphere) above divalent metal ion a. (d) The activated nucleophile attacks the scissile phosphate (highlighted in red). The transition state (e) is stabilized by the divalent ions. Dissociation of the cleaved nucleotide monophosphate product and DNA may be facilitated by rotation of amino acids E20 and H195 away from the active site (f) allowing for the release of divalent metal and product from the enzyme in a distributive fashion.
4. Conclusions
The structures of the TREX1 mutant proteins provide new information about the defects in DNA degradation that result in disease. We have observed mobility of the catalytic residue H195 in the active site which correlates with substrate and metal binding as well as catalytic activity. Proper positioning of the two divalent metal ions in the active site also plays an essential role in catalysis. Both of these elements are disrupted by the dominant D18N, D200N, and D200H TREX1 mutant proteins, which trap the TREX1-DNA in a catalytically inactive complex. These structures reveal a possible mechanism for disease with the dominant alleles. Additionally, we observed an interaction of residue R62 of one protomer with E20 in the active site of the other protomer that points to a possible mechanism for substrate release and provides a structural rationale for the dimeric nature of the TREX1 enzyme. Finally, the cumulative information we have gained from the structures of the TREX1 proteins in progressive states of catalysis has allowed us to propose a general mechanism for the TREX enzymes. Unraveling of the mechanistic properties of the TREX1 protein is essential in the continued pursuit of understanding the link between degradation of nucleic acids and chronic activation of the immune system.
Highlights.
-
>
In this study we determine the crystal structures of TREX1 mutants linked to autoimmune disease.
-
>
The structures reveal changes in the architecture of the active site that explain loss of catalytic activity.
-
>
The structures also reveal conformational movement that is required for catalytic activity.
-
>
Based on our cumulative work we propose a mechanism for catalysis for TREX1.
-
>
The structures provide insight into dysfunction relating to human disease.
Supplementary Material
Acknowledgements
We thank Min Ae Lee-Kirsch for sharing unpublished data on TREX1 AGS alleles.
Funding:
This work was supported by an American Heart Association predoctoral fellowship to SLB; American Heart Association Grant 10GRNT3650033 (TH); and National Institutes of Health Grant RO1 GM069962 (FWP). The funding sources had no role in the study design; in data collection; analysis and interpretation of the data; in writing the manuscript; or in the decision to submit for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement:
The authors declare they have no personal or financial relationships with other people or organization that could inappropriately influence this work.
References
- 1.Mazur DJ, Perrino FW. Identification and expression of the TREX1 and TREX2 cDNA sequences encoding mammalian 3'-->5' exonucleases. J Biol Chem. 1999;274:19655–19660. doi: 10.1074/jbc.274.28.19655. [DOI] [PubMed] [Google Scholar]
- 2.Hoss M, Robins P, Naven TJ, Pappin DJ, Sgouros J, Lindahl T. A human DNA editing enzyme homologous to the Escherichia coli DnaQ/MutD protein. Embo J. 1999;18:3868–3875. doi: 10.1093/emboj/18.13.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chowdhury D, Beresford PJ, Zhu P, Zhang D, Sung JS, Demple B, Perrino FW, Lieberman J. The Exonuclease TREX1 Is in the SET Complex and Acts in Concert with NM23-H1 to Degrade DNA during Granzyme A-Mediated Cell Death. Mol Cell. 2006;23:133–142. doi: 10.1016/j.molcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindahl T, Barnes DE, Yang YG, Robins P. Biochemical properties of mammalian TREX1 and its association with DNA replication and inherited inflammatory disease. Biochemical Society transactions. 2009;37:535–538. doi: 10.1042/BST0370535. [DOI] [PubMed] [Google Scholar]
- 6.Yang YG, Lindahl T, Barnes DE. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell. 2007;131:873–886. doi: 10.1016/j.cell.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 7.Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, van Bokhoven H, Brunner HG, Hamel BC, Corry PC, Cowan FM, Frints SG, Klepper J, Livingston JH, Lynch SA, Massey RF, Meritet JF, Michaud JL, Ponsot G, Voit T, Lebon P, Bonthron DT, Jackson AP, Barnes DE, Lindahl T. Mutations in the gene encoding the 3'–5' DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nature genetics. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- 8.de Vries B, Steup-Beekman G, Haan J, Bollen E, Luyendijk J, Frants R, Terwindt G, van Buchem M, Huizinga T, van den Maagdenberg A, Ferrari M. TREX1 Gene Variant in Neuropsychiatric Systemic Lupus Erythematosus. Annals of the rheumatic diseases. 2009 doi: 10.1136/ard.2009.114157. [DOI] [PubMed] [Google Scholar]
- 9.Gunther C, Meurer M, Stein A, Viehweg A, Lee-Kirsch MA. Familial Chilblain Lupus - A Monogenic Form of Cutaneous Lupus Erythematosus due to a Heterozygous Mutation in TREX1. Dermatology (Basel, Switzerland) 2009 doi: 10.1159/000222430. [DOI] [PubMed] [Google Scholar]
- 10.Hur JW, Sung YK, Shin HD, Park BL, Cheong HS, Bae SC. TREX1 polymorphisms associated with autoantibodies in patients with systemic lupus erythematosus. Rheumatology international. 2008;28:783–789. doi: 10.1007/s00296-007-0509-0. [DOI] [PubMed] [Google Scholar]
- 11.Lee-Kirsch MA, Chowdhury D, Harvey S, Gong M, Senenko L, Engel K, Pfeiffer C, Hollis T, Gahr M, Perrino FW, Lieberman J, Hubner N. A mutation in TREX1 that impairs susceptibility to granzyme A-mediated cell death underlies familial chilblain lupus. Journal of molecular medicine (Berlin, Germany) 2007;85:531–537. doi: 10.1007/s00109-007-0199-9. [DOI] [PubMed] [Google Scholar]
- 12.Lee-Kirsch MA, Gong M, Chowdhury D, Senenko L, Engel K, Lee YA, de Silva U, Bailey SL, Witte T, Vyse TJ, Kere J, Pfeiffer C, Harvey S, Wong A, Koskenmies S, Hummel O, Rohde K, Schmidt RE, Dominiczak AF, Gahr M, Hollis T, Perrino FW, Lieberman J, Hubner N. Mutations in the gene encoding the 3'–5' DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nature genetics. 2007;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- 13.Rice G, Newman WG, Dean J, Patrick T, Parmar R, Flintoff K, Robins P, Harvey S, Hollis T, O'Hara A, Herrick AL, Bowden AP, Perrino FW, Lindahl T, Barnes DE, Crow YJ. Heterozygous mutations in TREX1 cause familial chilblain lupus and dominant Aicardi-Goutieres syndrome. American journal of human genetics. 2007;80:811–815. doi: 10.1086/513443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richards A, van den Maagdenberg AM, Jen JC, Kavanagh D, Bertram P, Spitzer D, Liszewski MK, Barilla-Labarca ML, Terwindt GM, Kasai Y, McLellan M, Grand MG, Vanmolkot KR, de Vries B, Wan J, Kane MJ, Mamsa H, Schafer R, Stam AH, Haan J, de Jong PT, Storimans CW, van Schooneveld MJ, Oosterhuis JA, Gschwendter A, Dichgans M, Kotschet KE, Hodgkinson S, Hardy TA, Delatycki MB, Hajj-Ali RA, Kothari PH, Nelson SF, Frants RR, Baloh RW, Ferrari MD, Atkinson JP. C-terminal truncations in human 3'–5' DNA exonuclease TREX1 cause autosomal dominant retinal vasculopathy with cerebral leukodystrophy. Nature genetics. 2007;39:1068–1070. doi: 10.1038/ng2082. [DOI] [PubMed] [Google Scholar]
- 15.Morita M, Stamp G, Robins P, Dulic A, Rosewell I, Hrivnak G, Daly G, Lindahl T, Barnes DE. Gene-targeted mice lacking the Trex1 (DNase III) 3'-->5' DNA exonuclease develop inflammatory myocarditis. Molecular and cellular biology. 2004;24:6719–6727. doi: 10.1128/MCB.24.15.6719-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crow YJ, Livingston JH. Aicardi-Goutieres syndrome: an important Mendelian mimic of congenital infection. Developmental medicine and child neurology. 2008;50:410–416. doi: 10.1111/j.1469-8749.2008.02062.x. [DOI] [PubMed] [Google Scholar]
- 17.Jepps H, Seal S, Hattingh L, Crow YJ. The neonatal form of Aicardi-Goutieres syndrome masquerading as congenital infection. Early human development. 2008;84:783–785. doi: 10.1016/j.earlhumdev.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Stephenson JB. Aicardi-Goutieres syndrome (AGS) Eur J Paediatr Neurol. 2008;12:355–358. doi: 10.1016/j.ejpn.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 19.Crow YJ, Rehwinkel J. Aicardi-Goutieres syndrome and related phenotypes: linking nucleic acid metabolism with autoimmunity. Human molecular genetics. 2009;18:R130–R136. doi: 10.1093/hmg/ddp293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rice G, Patrick T, Parmar R, Taylor CF, Aeby A, Aicardi J, Artuch R, Montalto SA, Bacino CA, Barroso B, Benko WS, Bergmann C, Bertini E, Biancheri R, Blair EM, Blau N, Bonthron DT, Briggs T, Brueton LA, Brunner HG, Burke CJ, Carr IM, Carvalho DR, Chandler KE, Christen HJ, Corry PC, Cowan FM, Cox H, D'Arrigo S, Dean J, De Laet C, De Praeter C, Dery C, Ferrie CD, Flintoff K, Frints SG, Garcia-Cazorla A, Gener B, Goizet C, Goutieres F, Green AJ, Guet A, Hamel BC, Hayward BE, Heiberg A, Hennekam RC, Husson M, Jackson AP, Jayatunga R, Jiang YH, Kant SG, Kao A, King MD, Kingston HM, Klepper J, van der Knaap MS, Kornberg AJ, Kotzot D, Kratzer W, Lacombe D, Lagae L, Landrieu PG, Lanzi G, Leitch A, Lim MJ, Livingston JH, Lourenco CM, Lyall EG, Lynch SA, Lyons MJ, Marom D, McClure JP, McWilliam R, Melancon SB, Mewasingh LD, Moutard ML, Nischal KK, Ostergaard JR, Prendiville J, Rasmussen M, Rogers RC, Roland D, Rosser EM, Rostasy K, Roubertie A, Sanchis A, Schiffmann R, Scholl-Burgi S, Seal S, Shalev SA, Corcoles CS, Sinha GP, Soler D, Spiegel R, Stephenson JB, Tacke U, Tan TY, Till M, Tolmie JL, Tomlin P, Vagnarelli F, Valente EM, Van Coster RN, Van der Aa N, Vanderver A, Vles JS, Voit T, Wassmer E, Weschke B, Whiteford ML, Willemsen MA, Zankl A, Zuberi SM, Orcesi S, Fazzi E, Lebon P, Crow YJ. Clinical and molecular phenotype of Aicardi-Goutieres syndrome. American journal of human genetics. 2007;81:713–725. doi: 10.1086/521373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hedrich CM, Fiebig B, Hauck FH, Sallmann S, Hahn G, Pfeiffer C, Heubner G, Lee-Kirsch MA, Gahr M. Chilblain lupus erythematosus-a review of literature. Clinical rheumatology. 2008;27:1341. doi: 10.1007/s10067-008-0975-0. [DOI] [PubMed] [Google Scholar]
- 22.de Silva U, Choudhury S, Bailey SL, Harvey S, Perrino FW, Hollis T. The crystal structure of TREX1 explains the 3' nucleotide specificity and reveals a polyproline II helix for protein partnering. J Biol Chem. 2007;282:10537–10543. doi: 10.1074/jbc.M700039200. [DOI] [PubMed] [Google Scholar]
- 23.Brucet M, Querol-Audi J, Serra M, Ramirez-Espain X, Bertlik K, Ruiz L, Lloberas J, Macias MJ, Fita I, Celada A. Structure of the dimeric exonuclease TREX1 in complex with DNA displays a proline-rich binding site for WW Domains. J Biol Chem. 2007;282:14547–14557. doi: 10.1074/jbc.M700236200. [DOI] [PubMed] [Google Scholar]
- 24.Kirby TW, Harvey S, DeRose EF, Chalov S, Chikova AK, Perrino FW, Schaaper RM, London RE, Pedersen LC. Structure of the Escherichia coli DNA polymerase III epsilon-HOT proofreading complex. J Biol Chem. 2006;281:38466–38471. doi: 10.1074/jbc.M606917200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cisneros GA, Perera L, Schaaper RM, Pedersen LC, London RE, Pedersen LG, Darden TA. Reaction mechanism of the epsilon subunit of E. coli DNA polymerase III: insights into active site metal coordination and catalytically significant residues. Journal of the American Chemical Society. 2009;131:1550–1556. doi: 10.1021/ja8082818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perrino FW, de Silva U, Harvey S, Pryor EE, Jr, Cole DW, Hollis T. Cooperative DNA binding and communication across the dimer interface in the TREX2 3'–5' exonuclease. J Biol Chem. 2008 doi: 10.1074/jbc.M803629200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perrino FW, Harvey S, McMillin S, Hollis T. The human TREX2 3' -> 5'-exonuclease structure suggests a mechanism for efficient nonprocessive DNA catalysis. J Biol Chem. 2005;280:15212–15218. doi: 10.1074/jbc.M500108200. [DOI] [PubMed] [Google Scholar]
- 28.Lehtinen DA, Harvey S, Mulcahy MJ, Hollis T, Perrino FW. The TREX1 double-stranded DNA degradation activity is defective in dominant mutations associated with autoimmune disease. J Biol Chem. 2008;283:31649–31656. doi: 10.1074/jbc.M806155200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fye JM, Orebaugh CD, Coffin SR, Hollis T, Perrino FW. TREX1 dominant mutations in lupus and Aicardi-Goutieres syndrome. J Biol Chem. 2011 doi: 10.1074/jbc.M111.276287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otwinowski W, Minor Z. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr D Biol Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 32.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of applied crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 35.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 36.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 37.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 38.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. Journal of Applied Crystallography. 2006;39:109–111. [Google Scholar]
- 39.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeLano WL. Delano Scientific. Palo Alto, California: 2002. The PyMol Molecular Graphics System. [Google Scholar]
- 41.Beese LS, Steitz TA. Structural basis for the 3'–5' exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. Embo J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brautigam CA, Aschheim K, Steitz TA. Structural elucidation of the binding and inhibitory properties of lanthanide (III) ions at the 3'–5' exonucleolytic active site of the Klenow fragment. Chem Biol. 1999;6:901–908. doi: 10.1016/s1074-5521(00)80009-5. [DOI] [PubMed] [Google Scholar]
- 43.Derbyshire V, Freemont PS, Sanderson MR, Beese L, Friedman JM, Joyce CM, Steitz TA. Genetic and crystallographic studies of the 3',5'-exonucleolytic site of DNA polymerase I. Science. 1988;240:199–201. doi: 10.1126/science.2832946. [DOI] [PubMed] [Google Scholar]
- 44.Hamdan S, Carr PD, Brown SE, Ollis DL, Dixon NE. Structural basis for proofreading during replication of the Escherichia coli chromosome. Structure (Camb) 2002;10:535–546. doi: 10.1016/s0969-2126(02)00738-4. [DOI] [PubMed] [Google Scholar]
- 45.Nowotny M, Yang W. Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. Embo J. 2006;25:1924–1933. doi: 10.1038/sj.emboj.7601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected] Nature. 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 47.Viadiu H, Aggarwal AK. The role of metals in catalysis by the restriction endonuclease BamHI. Nature structural biology. 1998;5:910–916. doi: 10.1038/2352. [DOI] [PubMed] [Google Scholar]
- 48.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci U S A. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Orebaugh CD, Fye JM, Harvey S, Hollis T, Perrino FW. The TREX1 Exonuclease R114H Mutation in Aicardi-Goutieres Syndrome and Lupus Reveals Dimeric Structure Requirements for DNA Degradation Activity. J Biol Chem. 2011 doi: 10.1074/jbc.M111.297903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Breyer WA, Matthews BW. Structure of Escherichia coli exonuclease I suggests how processivity is achieved. Nature structural biology. 2000;7:1125–1128. doi: 10.1038/81978. [DOI] [PubMed] [Google Scholar]
- 51.Zuo Y, Deutscher MP. Mechanism of action of RNase T. II. A structural and functional model of the enzyme. J Biol Chem. 2002;277:50160–50164. doi: 10.1074/jbc.M207707200. [DOI] [PubMed] [Google Scholar]
- 52.Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 53.De Vivo M, Dal Peraro M, Klein ML. Phosphodiester cleavage in ribonuclease H occurs via an associative two-metal-aided catalytic mechanism. Journal of the American Chemical Society. 2008;130:10955–10962. doi: 10.1021/ja8005786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang W. An equivalent metal ion in one- and two-metal-ion catalysis. Nature structural & molecular biology. 2008;15:1228–1231. doi: 10.1038/nsmb.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Batra VK, Beard WA, Shock DD, Krahn JM, Pedersen LC, Wilson SH. Magnesium-induced assembly of a complete DNA polymerase catalytic complex. Structure. 2006;14:757–766. doi: 10.1016/j.str.2006.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horton NC, Perona JJ. DNA cleavage by EcoRV endonuclease: two metal ions in three metal ion binding sites. Biochemistry. 2004;43:6841–6857. doi: 10.1021/bi0499056. [DOI] [PubMed] [Google Scholar]
- 57.Mazur DJ, Perrino FW. Excision of 3' termini by the Trex1 and TREX2 3'-->5' exonucleases. Characterization of the recombinant proteins. J Biol Chem. 2001;276:17022–17029. doi: 10.1074/jbc.M100623200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.