Abstract
Background
Cumulative data has shown that microRNAs (miRNAs) are involved in the etiology and prognosis of colorectal cancer (CRC). Genetic polymorphisms in pre-miRNA genes may influence the biogenesis and functions of their host miRNAs. However, whether these polymorphisms are associated with CRC prognosis remains unknown.
Methods
We analyzed the effects of seven single nucleotide polymorphisms (SNPs) in pre-miRNA genes on the prognosis of a Chinese population with 408 CRC patients with surgically-resected adenocarcinoma.
Results
Two SNPs were identified to be significantly associated with recurrence-free survival and overall survival of the patients. The most significant SNP was rs6505162 in pre-miR-423. Compared to the homozygous wild-type genotype, the variant-containing genotypes of this SNP were significantly associated with both the overall survival (HR=2.12, 95% CI1.34–3.34, P=0.001) and the recurrence-free survival (HR=1.59, 95% CI1.08–2.36, P=0.019). Another SNP, rs4919510 in pre-miR-608, was also associated with altered recurrence-free survival (HR=0.61, 95% CI 0.41–0.92, P=0.017). These effects were evident only in patients receiving chemotherapy but not in those without chemotherapy. In addition, the combined analysis of the two SNPs conferred a 2.84-fold (95% CI 1.50–5.37, P=0.001) increased risk of recurrence and/or death. Similarly, this effect was only prominent in those receiving chemotherapy (P<0.001) but not in those without chemotherapy (P=0.999).
Conclusions
Our data suggest that genetic polymorphisms in pre-miRNA genes may impact CRC prognosis especially in patients receiving chemotherapy, a finding that warrants further independent validation.
Impact
This is one of the first studies showing a prognostic role of pre-miRNA gene SNPs in CRC.
Keywords: Polymorphism, microRNA, colorectal cancer
INTRODUCTION
Worldwide, colorectal cancer (CRC) is the third most commonly diagnosed cancer in males and the second in females, with more than1.2 million new cases and 600,000 deaths annually (1). The highest incidence rates of CRC have been observed in regions of developed countries such as Western Europe, North America, and Oceania. In recent decades, the incidence rates are rapidly increasing in several regions that previously had low CRC risk, including countries within Eastern Asia and Eastern Europe (1, 2). Especially, trends of increased CRC incidence and mortality have been observed in China (2). Moreover, CRC is also ranked as the third most common cause of cancer death among both men and women in the United States (3). Although CRC is a disease that is largely influenced by lifestyle and dietary factors (4), recent studies have suggested that inter-individual genetic variations such as single nucleotide polymorphisms (SNPs) may affect risk for CRC (5–7). In addition, emerging evidence has shown that SNPs may be used as surrogate biomarkers of the genetic background of CRC patients to predict therapeutic response and prognosis (8–11).
MicroRNAs (miRNAs) are a group of endogenous single-stranded small non-coding RNAs that have emerged as key regulators of fundamental biological processes through regulating the expression of more than 30% of human genes (12, 13). MiRNAs are initially transcribed as primary miRNAs (pri-miRNA) with several hundred nucleotides, which are further processed into hairpin-structured precursor miRNAs (pre-miRNA) that have approximately 70 nucleotides (14, 15). Pre-miRNAs are the direct precursors of mature miRNAs that have 18–25 nucleotides in length. It has been reported that SNPs in the pri-miRNAs and pre-miRNAs could affect the processing and subsequent maturation of miRNAs, leading to altered mature miRNA expression levels (16, 17). In addition, Hu et al. screened ~400 human pre-miRNAs and their surrounding regions and found that pre-miRNA genes had a significantly lower number of common SNPs than the surrounding regions, suggesting that pre-miRNAs are highly conserved and may be functionally important (18). Consistently, various subsequent studies have reported that pre-miRNA SNPs might confer altered risk of many solid tumors (18–22). However, the role of these SNPs on cancer clinical outcome remains to be evaluated.
Abnormal expression of miRNAs has been well documented as biomarkers or functional regulators in the tumorigenesis and prognosis of CRC (2, 23–26). Because SNPs in miRNA-related genes may impact mature miRNA expression, it follows that these SNPs may also be implicated in cancer development and clinical outcome. Consistently, several recent studies have suggested that SNPs in miRNA biogenesis genes, pri-miRNAs, and miRNA binding sites were associated with altered CRC risk and outcomes (27–29). Nonetheless, to the best of our knowledge, no studies have been reported on the role of SNPs in pre-miRNA regions in CRC prognosis. In the current study, we sought to assess the association of pre-miRNA SNPs and the overall survival and recurrence-free survival in a homogeneous population of Chinese CRC patients.
MATERIALS AND METHODS
Study population
The population used in this study has been described previously (11). Briefly, newly diagnosed and histologically confirmed CRC patients were enrolled in the Xijing Hospital and Tangdu Hospital affiliated with the Fourth Military Medical University (FMMU) in Xi’an, China. There were no restrictions on age, gender, cancer stage or other demographic variables on enrollment. The patient enrollment began from February 2006. As of April 2010, a total of 496 eligible CRC patients were recruited. The rate of recruitment among all eligible patients is 90%. All patients included in this study had histopathologically CRC tumors only and no other cancers before or at the time of diagnosis. For this study, we excluded 88 of the 496 patients, which comprised of 26 patients who did not undergo surgery or only received palliative operation, 48 patients who had incomplete clinical and/or follow-up data, 6 patients who died within one month of surgery, and 8 patients who had poor quality and/or quantity DNA samples. Finally, we included a total of 408 CRC patients with completely validated demographic, clinical, and follow-up data. All patients were Han Chinese with adenocarcinoma and received surgery after diagnosis. Some patients received adjuvant chemotherapy after surgery. No patients received neo-adjuvant or radiation therapy. Informed consent was obtained from each enrolled patient. This study was approved by the local research ethics committees of the participating institutes.
Demographic and clinical data collection
Demographic data were collected through in-person interview at the time of initial visit or follow-up in the clinics, medical chart review, or consultation with the treating physicians by trained clinical research specialists. For data acquired from multiple sources, the research staff compared and validated that these data were consistent. If discrepancies were identified, the patients, family members, and/or treating physicians were further contacted for verification. Individual who smoked more than 100 cigarettes in their lifetime were defined as ever smokers, otherwise as never smokers. Never drinkers were defined as those who consumed less than or equal to one drink per month. One drink was defined as one bottle or can of beer, one medium glass of wine, or one mixed drink. Detailed clinical information was collected by medical chart review and consultation with treating physicians. The follow-up information on recurrence and death was updated at 6-month intervals through onsite interview, direct calling, or medical chart review by trained clinical specialists. The latest follow-up data in this study were obtained in March 2011. For patients enrolled after August 2008, 5-ml of blood was obtained for genomic DNA extraction. For patients enrolled before August 2008, genomic DNA was extracted from approximately 100 mg of adjacent normal tissues obtained by a pathologist after surgery.
SNP selection and genotyping
The candidate SNPs were selected based on a literature review of pre-miRNA epidemiological studies (20, 21, 30–33). Altogether, we genotyped 10 SNPs, including rs895819 in pre-miR-27a, rs2910164 in pre-miR-146a, rs2292832 in pre-miR-149, rs6505162 in pre-miR-423, rs2289030 in pre-miR-492, rs3746444 in pre-miR-499, rs2368392 in pre-miR-604, rs2043556 in pre-miR-605, rs4919510 in pre-miR-608, and rs17759989 in pre-miR-633. Genotyping was done using the Sequenom iPLEX platform (Sequenom Inc., CA). Laboratory personnel conducting genotyping were blinded to patient information. Strict quality control measures were implemented during genotyping with over 99% concordance between samples genotyped in duplicate.
Statistical and data analysis
Two major endpoints were evaluated in this study: overall survival and recurrence-free survival. Overall survival time was defined as the time from initial surgery to death from any cause. Recurrence-free survival time was defined as the time from initial surgery to local recurrence, distant metastasis, death from any cause, or to the date of last follow-up. Recurrence is defined as, after the treatment of the primary tumor by surgery and/or chemotherapy, the re-growth of tumor in the original organ (local-regional recurrence) or a different organ (distant metastasis). All recurrent tumors in this study were confirmed as having the same histopathological characteristics as the primary tumor. Second primary tumors with different histopathological characteristics were excluded. All patients without recurrence or lost during follow-up were censored for analysis. Recurrence was confirmed through the combined evaluations of imaging findings (ultrasound, computed tomography, positron emission tomography, magnetic resonance imaging) and laboratory results (mainly carcinoembryonic antigen test). The Hardy-Weinberg equilibrium (HWE) of each SNP was tested using a goodness-of-fit chi-square test. Hazard ratios (HR) and 95% confidence interval (CI) were estimated by multivariate Cox proportional hazards model, adjusting for age, gender, education level, body mass index (BMI), smoking status, drinking status, chemotherapy, tumor position, tumor differentiation, and tumor stage, where appropriate. Three genetic models (dominant, recessive, and additive) were tested and the best fitting model was selected for all downstream analyses. The tests for interactions between significant SNPs and demographic and clinical variables were conducted by including a cross-product term into the Cox proportional hazards model. Kaplan-Meier curves and a log-rank test were used to assess the differences in overall survival and recurrence-free survival. STATA software package (version 8, STATA Corp., College Station, TX) was used for these analyses. All P values were two-sided. P ≤ 0.05 was considered the threshold of statistical significance.
RESULTS
Characteristics of the study subjects
The distribution of patients’ demographic and clinicopathologic characteristics are summarized in Table 1. A total of 408 CRC patients were included in this study. The average age at diagnosis was 59.4 (range, 22–90), and average BMI was 22.7 (range, 15.8–32.9). All patients received surgery within two months after diagnosis. All patients were histologically confirmed by pathological examination as having adenocarcinoma. There were 230 (56.4%) male patients and 178 (43.6%) female patients. The majority of patients were never smokers (70.8%) and never drinkers (89.5%). There were approximately equal numbers of patients with colon cancer (47.1%) and rectal cancer (52.9%), which is consistent with previous reports that the incidence rates of colon and rectal cancers are generally of the same magnitude in countries with low CRC risk such as China (34). A total of 192 (47.1%) patients had stage 2 tumor, while stage 0, 1, 3, and 4 tumors were presented in 2.0%, 14.2%, 27.2% and 9.6% of patients, respectively. The majority of patients (66.4%) had moderately differentiated tumors. No patient received neo-adjuvant chemotherapy, radiotherapy or targeted therapy, but most patients (78.2%) received adjuvant chemotherapy after surgery. Among the 319 patients receiving chemotherapy, 266 (83.4%) were treated using the FOLFOX regimen, including folinic acid (FOL), fluorouracil (F) and Oxaliplatin (OX). During follow-up, there were 93 (22.8%) patients who developed recurrences, 94 (23.0%) patients who died, and 134 (32.8%) patients who had at least one event (recurrence and/or death). The median overall survival time was 23.0 months and the median recurrence-free survival time was 19.9 months.
Table 1.
Demographic and clinicopathological characteristics of 408 Chinese CRC patients
| Variables | Number of patients (%), n=408 |
|---|---|
| Age, mean (range) (in years) | 59.4 (22–90) |
| Body mass index, mean (range) | 22.7(15.8–32.9) |
| Gender | |
| Male | 230 (56.4) |
| Female | 178(43.6) |
| Smoking status | |
| Ever | 119 (29.2) |
| Never | 289 (70.8) |
| Drinking status | |
| Ever | 43 (10.5) |
| Never | 365 (89.5) |
| Education | |
| Up to high school | 178(43.6) |
| College degree or higher | 171(41.9) |
| Unknown | 59 (14.5) |
| Tumor position | |
| Colon | 192 (47.1) |
| Rectum | 216 (52.9) |
| Tumor stage | |
| 0 | 8 (2.0) |
| 1 | 58 (14.2) |
| 2 | 192 (47.1) |
| 3 | 111 (27.2) |
| 4 | 39 (9.6) |
| Tumor differentiation | |
| Poor | 37 (9.1) |
| Moderate | 271 (66.4) |
| Well | 100 (24.5) |
| Chemotherapy | |
| Yes | 319 (78.2) |
| No | 89 (21.8) |
| Recurrence | |
| Yes | 93 (22.8) |
| No | 315 (77.2) |
| Death | |
| Yes | 94 (23.0) |
| No | 314 (77.0) |
| Event (recurrence and/or death) | |
| Yes | 134 (32.8) |
| No | 274 (67.2) |
Main effect analyses of individual SNPs
In the 10 genotyped SNPs, three SNPs (rs2292832, rs17759989, and rs3746444) were excluded from downstream analyses because of failing to pass quality control of genotyping. The average call rate of the remaining seven SNPs was 96.8% (range, 90.3%–100%). The detailed genotyping results of the seven SNPs are listed in Table 2. Except for rs2043556 whose HWE P value was 0.001, the HWE P value for all other SNPs were non-significant, ranging from 0.217 to 1.000. The HWE P value was 0.490 for rs6505162 and 0.228 for rs4919510. We did not identify any genotyping error after carefully checking the genotyping results of rs2043556. The significant deviation from HWE for this SNP could potentially result from the differences between our CRC patient population and the general population of cancer-free individuals, and the results related to this SNP need to be interpreted with caution. Overall, two SNPs, rs6505162 in pre-miR-423 and rs4919510 in pre-miR-608, exhibited a significant association with the overall survival and the recurrence-free survival of the CRC patients. For both SNPs, the results of the dominant genetic model analysis were more significant for both the overall and the recurrence-free survivals compared to the recessive and additive genetic models, except for the overall survival analysis for rs4919510, in which the result of the additive model (P=0.063) was slightly more significant than that of the dominant model (P=0.067). We therefore used the dominant model as the best fitting model in this study. Compared to the homozygous wild-type (WW) genotype, the variant-containing (WV + VV) genotypes of this SNP were significantly associated with unfavorable overall and recurrence-free survival with an HR of 2.12 (95% CI, 1.34–3.34; P=0.001) and 1.59 (95% CI, 1.08–2.36; P=0.019), respectively. Another significant SNP was rs4919510. Under a dominant genetic model, the variant-containing genotypes of this SNP were associated with favorable overall and recurrence-free survival with an HR of 0.64 (95% CI, 0.40–1.03; P=0.067) and 0.61 (95% CI, 0.41–0.92; P=0.017). There are 139 (34.1%) subjects whose DNA samples were obtained from tissues and 269 (65.9%) from blood. We conducted a test for interaction between both significant SNPs and DNA source and did not identify any significant interaction (P for interaction for overall survival and recurrence-free survival: 0.371 and 0.646 respectively for rs6505162, and 0.926 and 0.918 respectively for rs4919510).
Table 2.
Association of pre-miRNA SNPs with overall and recurrence-free survival of CRC patients.
| Gene and SNP | Genotypea | Overall survival
|
Recurrence-free survival
|
||||
|---|---|---|---|---|---|---|---|
| Death/total | HR (95% CI)b | P value | Event/totalc | HR (95% CI) | P value | ||
| pre-miR-27a | WW | 48/213 | 1(reference) | 69/213 | 1(reference) | ||
| rs895819 | WV | 39/167 | 0.88(0.55– 1.41) | 0.599 | 55/167 | 0.75(0.50– 1.13) | 0.168 |
| HWE P 0.375 | VV | 6/25 | 0.66(0.26– 1.70) | 0.393 | 9/25 | 0.76(0.35– 1.64) | 0.478 |
| Dom | 0.85(0.54– 1.34) | 0.476 | 0.75(0.51– 1.11) | 0.153 | |||
| Rec | 0.71(0.28– 1.76) | 0.456 | 0.87(0.41– 1.85) | 0.721 | |||
| Add | 0.85(0.59– 1.22) | 0.374 | 0.81(0.59– 1.11) | 0.195 | |||
| pre-miR-146a | WW | 24/118 | 1(reference) | 34/118 | 1(reference) | ||
| rs2910164 | WV | 50/200 | 1.13(0.68– 1.89) | 0.632 | 69/200 | 1.10(0.72– 1.69) | 0.663 |
| HWE P 1.000 | VV | 19/85 | 0.95(0.49– 1.83) | 0.877 | 30/85 | 0.94(0.54– 1.66) | 0.840 |
| Dom | 1.08(0.66– 1.75) | 0.761 | 1.06(0.70– 1.59) | 0.797 | |||
| Rec | 0.88(0.50– 1.56) | 0.659 | 0.89(0.54– 1.46) | 0.646 | |||
| Add | 0.99(0.73– 1.36) | 0.959 | 0.99(0.76– 1.29) | 0.932 | |||
| pre-miR-423 | WW | 47/242 | 1(reference) | 74/242 | 1(reference) | ||
| rs6505162 | WV | 40/141 | 2.18(1.36– 3.51) | 0.001 | 52/141 | 1.73(1.15– 2.59) | 0.008 |
| HWE P 0.490 | VV | 7/25 | 1.79(0.75– 4.28) | 0.192 | 8/25 | 1.02(0.44– 2.36) | 0.960 |
| Dom | 2.12(1.34– 3.34) | 0.001 | 1.59(1.08– 2.36) | 0.019 | |||
| Rec | 1.29(0.55– 2.99) | 0.557 | 0.82(0.36– 1.87) | 0.642 | |||
| Add | 1.61(1.15– 2.25) | 0.005 | 1.28(0.95– 1.73) | 0.100 | |||
| pre-miR-492 | WW | 54/233 | 1(reference) | 76/233 | 1(reference) | ||
| rs2289030 | WV | 31/136 | 0.72(0.43– 1.18) | 0.194 | 46/136 | 0.83(0.54– 1.26) | 0.379 |
| HWE P 0.217 | VV | 7/28 | 1.05(0.46– 2.40) | 0.910 | 9/28 | 0.95(0.44– 2.03) | 0.885 |
| Dom | 0.77(0.48– 1.23) | 0.274 | 0.85(0.57– 1.26) | 0.411 | |||
| Rec | 1.19(0.53– 2.68) | 0.669 | 1.02(0.48– 2.15) | 0.956 | |||
| Add | 0.88(0.60– 1.28) | 0.498 | 0.90(0.65– 1.24) | 0.530 | |||
| pre-miR-604 | WW | 46/199 | 1(reference) | 67/199 | 1(reference) | ||
| rs2368392 | WV | 35/162 | 1.17(0.73– 1.88) | 0.524 | 50/162 | 0.97(0.64– 1.47) | 0.883 |
| HWE P 0.292 | VV | 11/42 | 1.46(0.69– 3.11) | 0.327 | 15/42 | 1.47(0.79– 2.77) | 0.227 |
| Dom | 1.22(0.78– 1.91) | 0.382 | 1.06(0.72– 1.55) | 0.780 | |||
| Rec | 1.37(0.66– 2.84) | 0.397 | 1.49(0.82– 2.74) | 0.194 | |||
| Add | 1.19(0.85– 1.67) | 0.299 | 1.12(0.84– 1.51) | 0.436 | |||
| pre-miR-605 | WW | 45/184 | 1(reference) | 66/184 | 1(reference) | ||
| rs2043556 | WV | 42/200 | 1.01(0.63– 1.61) | 0.962 | 60/200 | 0.86(0.58– 1.28) | 0.460 |
| HWE P 0.001 | VV | 7/23 | 1.47(0.60– 3.62) | 0.401 | 8/23 | 0.93(0.41– 2.11) | 0.856 |
| Dom | 1.06(0.67– 1.66) | 0.808 | 0.87(0.59– 1.27) | 0.471 | |||
| Rec | 1.46(0.61– 3.50) | 0.392 | 1.00(0.45– 2.23) | 0.999 | |||
| Add | 1.11(0.76– 1.61) | 0.596 | 0.91(0.66– 1.25) | 0.549 | |||
| pre-miR-608 | WW | 30/122 | 1(reference) | 47/122 | 1(reference) | ||
| rs4919510 | WV | 47/213 | 0.67(0.41– 1.10) | 0.112 | 63/213 | 0.62(0.41– 0.95) | 0.027 |
| HWE P 0.228 | VV | 16/72 | 0.54(0.25– 1.16) | 0.112 | 23/72 | 0.58(0.31– 1.08) | 0.086 |
| Dom | 0.64(0.40– 1.03) | 0.067 | 0.61(0.41– 0.92) | 0.017 | |||
| Rec | 0.69(0.34– 1.40) | 0.306 | 0.78(0.43-– 1.38) | 0.389 | |||
| Add | 0.71(0.50– 1.02) | 0.063 | 0.72(0.53– 0.97) | 0.032 | |||
Note: The significant P values (≤0.05) are in bold.
WW, homozygous wild-type genotype; WV heterozygous genotype; VV, homozygous variant genotype; Dom, dominant model; Rec, recessive model; Add, additive model
Adjusted for age, gender, education level, BMI, smoking status, drinking status, chemotherapy, tumor position, tumor differentiation, tumor stage.
Event: recurrence and/or death.
Stratified analysis of rs6505162 and rs4919510 by host variables
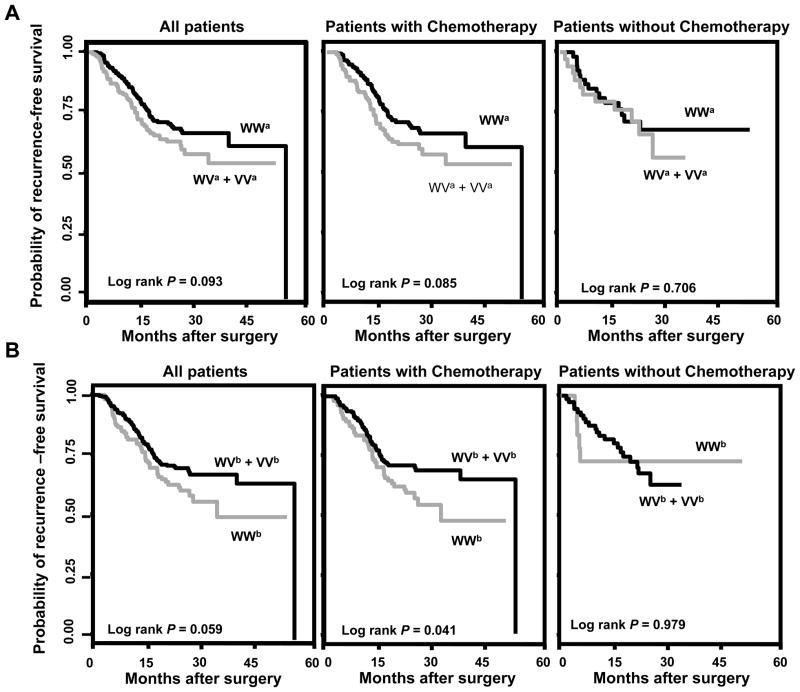
We focused on the recurrence-free survival in all the downstream analyses because this endpoint reflects both recurrence and death. We conducted stratified and interaction analyses of rs6505162 and rs4919510 with the major demographic and clinical variables. As shown in Table 3, four borderline significant (0.05 < P < 0.1) interactions were identified, including the interactions between rs6505162 and BMI (P for interaction = 0.096) and tumor stage (P for interaction = 0.091), as well as the interactions between rs4919510 and gender (P for interaction = 0.092) and chemotherapy (P for interaction = 0.072). The significant association between recurrence-free survival and rs6505162 remained significant in patients with higher BMI (HR=2.60, 95% CI 1.47–4.58, P=0.001) and higher tumor stage (HR=1.87, 95% CI 0.99–3.53, P=0.053), compared to those with lower BMI (HR=0.83, 95% CI 0.43–1.63, P=0.593) and lower tumor stage (HR=1.09, 95% CI 0.62–1.92, P=0.772), respectively. The significant effects conferred by rs4919510 was more evident in male patients (HR=0.47, 95% CI 0.27–0.81, P=0.006) and patients receiving chemotherapy (HR=0.48, 95% CI 0.30–0.75, P=0.001), compared to the female patients (HR=0.94, 95% CI 0.50–1.79, P=0.856) and patients without chemotherapy (HR=1.57, 95% CI 0.48–5.07, P=0.454), respectively. Kaplan-Meier curves indicated at least a borderline significant difference in term of time to recurrence or death, between the wild-type and variant-containing genotypes of both rs6505162 (Log rank P=0.093) (Figure 1A, left panel) and rs4919510 (Log rank P= 0.059) (Figure 1B, left panel). For both SNPs, the significant effect was more evident in patients receiving chemotherapy (Log rank P, 0.085 and 0.041 for rs6505162 and rs4919510, respectively, Figures 1A and 1B, middle panel), but not in those without chemotherapy (Log rank P, 0.706 and 0.979 for rs6505162 and rs4919510, respectively, Figures 1A and 1B, right panel). We also generated recurrence-free survival curves that were adjusted by the demographic and clinical variables listed on Table 1 from the Cox regression, to determine the differences in recurrence-free survival time between the different genotypes of rs6515062 and rs4919510. The adjusted curves were similar to their corresponding Kaplan-Meier curves and consistent with the results of the Cox proportional hazards analyses. That is, for both SNPs, significant differences in recurrence-free survival time were observed in all patients as well as patients receiving chemotherapy, but not in those patients without chemotherapy (Supplementary Figure 1).
Table 3.
Association of rs6505162 and rs4919510 with recurrence-free survival stratified by host characteristics
| Variables | Stratum | Genotypea | rs6505162
|
rs4919510
|
||||
|---|---|---|---|---|---|---|---|---|
| Event/totalc | HR (95% CI)b | P value | Event/total | HR (95% CI) | P value | |||
| Overall | WW | 74/242 | 1(reference) | 47/122 | 1(reference) | |||
| WV + VV | 60/166 | 1.59(1.08–2.36) | 0.019 | 86/285 | 0.61(0.41–0.92) | 0.017 | ||
| Age | ||||||||
| Younger (<61) | WW | 42/117 | 1(reference) | 26/55 | 1(reference) | |||
| WV + VV | 33/83 | 1.33(0.76–2.32) | 0.319 | 49/145 | 0.44(0.25–0.79) | 0.005 | ||
| Older(≥ 61) | WW | 32/125 | 1(reference) | 21/67 | 1(reference) | |||
| WV + VV | 27/83 | 2.05(1.07–3.93) | 0.031 | 37/140 | 0.77(0.41–1.46) | 0.427 | ||
| P for interaction | 0.387 | P for interaction | 0.361 | |||||
| Gender | ||||||||
| Male | WW | 38/131 | 1(reference) | 28/65 | 1(reference) | |||
| WV + VV | 38/99 | 2.17(1.26–3.74) | 0.005 | 48/165 | 0.47(0.27–0.81) | 0.006 | ||
| Female | WW | 36/111 | 1(reference) | 19/57 | 1(reference) | |||
| WV + VV | 22/67 | 1.12(0.60–2.13) | 0.717 | 38/120 | 0.94(0.50–1.79) | 0.856 | ||
| P for interaction | 0.401 | P for interaction | 0.092 | |||||
| Educational level | ||||||||
| Up to high school | WW | 27/114 | 1(reference) | 17/49 | 1(reference) | |||
| WV + VV | 27/64 | 2.06(1.14–3.74) | 0.017 | 36/128 | 0.66(0.35–1.24) | 0.194 | ||
| College or higher | WW | 36/98 | 1(reference) | 25/55 | 1(reference) | |||
| WV + VV | 27/73 | 1.51(0.87–2.60) | 0.144 | 38/116 | 0.52(0.30–0.91) | 0.022 | ||
| P for interaction | 0.469 | P for interaction | 0.302 | |||||
| Body mass index (BMI) | ||||||||
| BMI <22.7 | WW | 36/123 | 1(reference) | 23/64 | 1(reference) | |||
| WV + VV | 24/77 | 0.83(0.43–1.63) | 0.593 | 36/135 | 0.49(0.25–0.97) | 0.041 | ||
| BMI ≥ 22.7 | WW | 38/117 | 1(reference) | 24/56 | 1(reference) | |||
| WV + VV | 34/83 | 2.60(1.47–4.58) | 0.001 | 48/144 | 0.61(0.35–1.06) | 0.078 | ||
| P for interaction | 0.096 | P for interaction | 0.741 | |||||
| Smoking status | ||||||||
| Never smoker | WW | 55/170 | 1(reference) | 32/91 | 1(reference) | |||
| WV + VV | 43/119 | 1.44(0.92–2.27) | 0.113 | 65/197 | 0.69(0.43–1.11) | 0.125 | ||
| Ever smoker | WW | 19/72 | 1(reference) | 15/31 | 1(reference) | |||
| WV + VV | 17/47 | 2.70(1.15–6.31) | 0.022 | 21/88 | 0.34(0.13–0.88) | 0.027 | ||
| P for interaction | 0.401 | P for interaction | 0.206 | |||||
| Drinking status | ||||||||
| Never drinker | WW | 65/218 | 1(reference) | 40/112 | 1(reference) | |||
| WV + VV | 50/147 | 1.50(0.98–2.30) | 0.064 | 74/252 | 0.64(0.41–1.00) | 0.051 | ||
| Ever drinker | WW | 9/24 | 1(reference) | 7/10 | 1(reference) | |||
| WV + VV | 10/19 | 7.88(1.24–50.07) | 0.029 | 12/33 | 0.46(0.11–1.90) | 0.283 | ||
| P for interaction | 0.591 | P for interaction | 0.636 | |||||
| Chemotherapy | ||||||||
| No | WW | 15/54 | 1(reference) | 5/18 | 1(reference) | |||
| WV + VV | 11/35 | 1.96(0.79–4.83) | 0.144 | 21/71 | 1.57(0.48–5.07) | 0.454 | ||
| Yes | WW | 59/188 | 1(reference) | 42/104 | 1(reference) | |||
| WV + VV | 49/131 | 1.58(1.01–2.49) | 0.046 | 65/214 | 0.48(0.30–0.75) | 0.001 | ||
| P for interaction | 0.852 | P for interaction | 0.072 | |||||
| Tumor position | ||||||||
| Colon | WW | 34/116 | 1(reference) | 24/60 | 1(reference) | |||
| WV + VV | 30/76 | 1.09(0.59–2.00) | 0.785 | 40/132 | 0.60(0.32–1.12) | 0.108 | ||
| Rectum | WW | 40/126 | 1(reference) | 23/62 | 1(reference) | |||
| WV + VV | 30/90 | 1.91(1.06–3.42) | 0.030 | 46/153 | 0.66(0.37–1.19) | 0.169 | ||
| P for interaction | 0.254 | P for interaction | 0.538 | |||||
| Tumor differentiation | ||||||||
| Poor and Moderate | WW | 57/184 | 1(reference) | 41/94 | 1(reference) | |||
| WV + VV | 50/124 | 1.81(1.16–2.82) | 0.009 | 65/213 | 0.54(0.35–0.85) | 0.008 | ||
| Well | WW | 17/58 | 1(reference) | 6/28 | 1(reference) | |||
| WV + VV | 10/42 | 0.88(0.32–2.44) | 0.808 | 21/72 | 1.65(0.46–5.91) | 0.440 | ||
| P for interaction | 0.253 | P for interaction | 0.218 | |||||
| Tumor stage | ||||||||
| Stage 0–2 | WW | 32/148 | 1(reference) | 23/76 | 1(reference) | |||
| WV + VV | 24/110 | 1.09(0.62–1.92) | 0.772 | 32/181 | 0.51(0.28–0.92) | 0.026 | ||
| Stage 3–4 | WW | 42/94 | 1(reference) | 24/46 | 1(reference) | |||
| WV + VV | 36/56 | 1.87(0.99–3.53) | 0.053 | 54/104 | 0.65(0.35–1.22) | 0.183 | ||
| P for interaction | 0.091 | P for interaction | 0.476 | |||||
Note: The significant P values (≤0.05) are in bold.
WW, homozygous wild-type genotype; WV heterozygous genotype; VV, homozygous variant genotype; Dom, dominant model
Adjusted for age, gender, education level, BMI, smoking status, drinking status, chemotherapy, tumor position, tumor differentiation, tumor stage, where appropriate.
Event: recurrence and/or death.
Figure 1.
Kaplan-Meier recurrence-free survival curves of CRC in all patients, in patients with chemotherapy and in patients without chemotherapy. A, rs6505162, dominant model; B, rs4919510, dominant model. WWa, WVa and VVa indicate the homozygous wild-type, heterozygous and homozygous variant genotypes of rs6505162, respectively. WWb, WVb and VVb indicate the homozygous, heterozygous and homozygous variant genotypes of rs4919510, respectively.
Effects of chemotherapy on patient outcome by pre-miRNA SNPs
Because both rs6505162 and rs4919510 conferred significantly altered risk of recurrence-free survival in patients receiving chemotherapy but not in those without chemotherapy, we sought to evaluate whether the effect of chemotherapy on patient outcome was also influenced by these SNPs. As showed in Table 4, patients receiving chemotherapy exhibited a better recurrence-free survival compared to those without chemotherapy (HR=0.61, 95% CI 0.37–1.00, P=0.048). Interestingly, this effect remained at least borderline significant in patients with the low-risk genotypes of either SNPs (WW in rs6505162 or WV + VV in rs4919510). That is, the effect of chemotherapy on recurrence-free survival remained borderline significant in patients with the homozygous wild-type genotype of rs6505162 (HR=0.52, 95% CI 0.27–1.01, P=0.052) and significant in patients with the variant-containing genotypes of rs4919510 (HR=0.41, 95% CI 0.23–0.73, P=0.003). In comparison, no significant effect by chemotherapy on recurrence-free survival was observed in patients with the high-risk genotypes of both SNPs (P value, 0.248 for rs6505162 and 0.329 for rs4919510).
Table 4.
Effects of chemotherapy on CRC recurrence-free survival stratified by rs6505162 or rs4919510
| SNP and Variablesa | Event/totalc | HR (95% CI)b | P value |
|---|---|---|---|
| In all patients | |||
| No chemotherapy | 26/89 | 1(reference) | |
| Chemotherapy | 108/319 | 0.61(0.37–1.00) | 0.048 |
| In patients with WW genotype of rs6505162 | |||
| No chemotherapy | 15/54 | 1(reference) | |
| Chemotherapy | 59/188 | 0.52(0.27–1.01) | 0.052 |
| In patients with WV + VV genotype of rs6505162 | |||
| No chemotherapy | 11/35 | 1(reference) | |
| Chemotherapy | 49/131 | 0.62(0.27–1.40) | 0.248 |
| In patients with WW genotype of rs4919510 | |||
| No chemotherapy | 5/18 | 1(reference) | |
| Chemotherapy | 42/104 | 1.75(0.57–5.41) | 0.329 |
| In patients with WV + VV genotype of rs4919510 | |||
| No chemotherapy | 21/71 | 1(reference) | |
| Chemotherapy | 65/214 | 0.41(0.23–0.73) | 0.003 |
Note: The significant P values (≤0.05) are in bold.
WW, homozygous wild-type genotype; WV heterozygous genotype; VV, homozygous variant genotype; Dom, dominant model
Adjusted for age, gender, education level, BMI, smoking status, drinking status, chemotherapy, tumor position, tumor differentiation, tumor stage, where appropriate.
Event: recurrence and/or death.
Joint effects of rs6505162 and rs4919510 on CRC risk
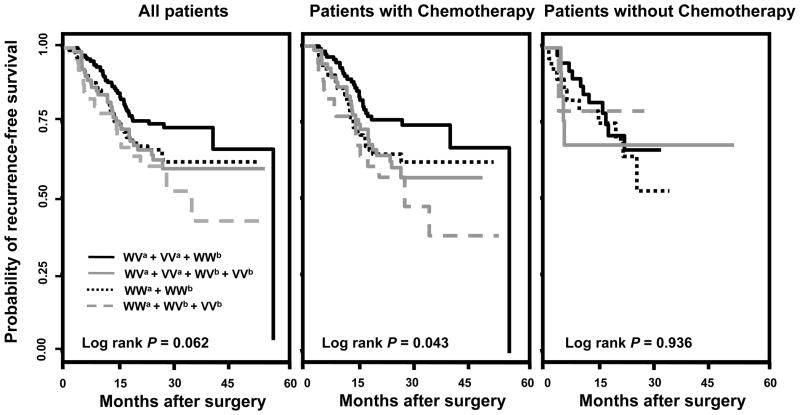
Finally, we conducted joint analysis to assess the cumulative effect of the two significant SNPs on CRC recurrence-free survival. Using the patients with the low-risk genotype in both SNPs as reference group, patients with the high-risk genotype of both SNPs exhibited a 2.84-fold (95% CI, 1.50–5.37, P=0.001) increase in risk of recurrence and/or death (Table 5). Consistently, this effect was more prominent in patients receiving chemotherapy (HR=3.81, 95% CI 1.87–7.78, P<0.001) but not in those without chemotherapy (HR=1.00, 95% CI 0.10–10.24, P=0.999). Kaplan-Meier curves showed a borderline significance in term of recurrence-free survival time in the joint effect analysis (Log rank P=0.062, Figure 2. left panel). Consistent with the Cox analysis, this effect was more evident in those receiving chemotherapy (Log rank P=0.043, Figure 2. middle panel), than those without chemotherapy (Log rank P=0.936, Figure 2. right panel).
Table 5.
Joint effects of rs6505162 and rs4919510 on CRC recurrence-free survival.
| Genotypea | All patients
|
Patients with chemotherapy d
|
Patients without chemotherapy
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Event/totalc | HR (95% CI)b | P value | Event/total | HR (95% CI) | P value | Event/total | HR (95% CI) | P value | ||
| rs6505162 | rs4919510 | |||||||||
| WW | WV + VV | 43/159 | 1(reference) | 32/118 | 1(reference) | 11/41 | 1(reference) | |||
| WV + VV | WV + VV | 43/126 | 1.81(1.11–2.94) | 0.017 | 33/96 | 1.85(1.03–3.31) | 0.040 | 10/30 | 2.09(0.74–5.89) | 0.164 |
| WW | WW | 30/82 | 1.84(1.09–3.13) | 0.024 | 26/69 | 2.34(1.27–4.29) | 0.006 | 4/13 | 0.92(0.21–3.98) | 0.908 |
| WV + VV | WW | 17/40 | 2.84(1.50–5.37) | 0.001 | 16/35 | 3.81(1.87–7.78) | <0.001 | 1/5 | 1.00(0.10–10.24) | 0.999 |
Note: The significant P values (≤0.05) are in bold.
WW, homozygous wild-type genotype; WV heterozygous genotype; VV, homozygous variant genotype.
Adjusted for age, gender, education level, BMI, smoking status, drinking status, chemotherapy, tumor position, tumor differentiation, tumor stage, where appropriate.
Event: recurrence and/or death.
Figure 2.
Kaplan-Meier recurrence-free survival curves of joint effects between rs6505162 and rs4919510 under a dominant model. WWa + WVb + VVb indicate the combination of the low-risk genotypes of both rs6505162 and rs4919510; WVa + VVa + WVb + VVb indicate the combination of the high-risk genotypes of rs6505162 and the low-risk genotypes of rs4919510; WWa + WWb indicate the combination of the low-risk genotypes of rs6505162 and the high-risk genotypes of rs4919510; WVa + VVa + WWb indicate the combination of the high-risk genotypes of both rs6505162 and rs4919510.
DISCUSSION
In this study, we evaluated the effects of SNPs in pre-miRNA on the clinical outcomes of a Chinese CRC population. We found that two SNPs, rs6505162 in pre-miR-423 and rs4919510 in pre-miR-608, were significantly associated with altered overall survival and recurrence-free survival of the patients. In addition, joint effects of the two SNPs on CRC outcome were observed, especially in patients treated with chemotherapy.
Genetic polymorphisms in pre-miRNA genomic regions have been associated with the risk and clinical outcomes of various solid malignancies such as cancers of breast, prostate, lung, bladder, esophagus and kidney (18, 21, 31, 33, 35–37). In addition, a few studies also reported the implications of pre-miRNA SNPs in the risk of CRC (38, 39). However, it remains to be determined whether these SNPs affect the clinical outcome of CRC patients. The only previously reported epidemiological study on miRNA-related SNPs was conducted by Lee et al, who did not identify any significant effect between miRNA-related SNPs and CRC survival on a multivariate basis in a population of 420 Korean CRC patients (40). In another small study with 61 patients, Boni et al. found that SNPs in the primary precursor region (pri-miRNAs) of pri-miR-26a1 and pri-miR-100 were associated with the clinical response of metastatic colon cancer patients treated with 5-fluorouracil and irinotecan (29). In the current study, we found that rs6505162 in pre-miR-423 and rs4919510 in pre-miR-608 were significantly associated with altered overall and recurrence-free survivals of Chinese CRC patients. Both SNPs have been evaluated in previous studies with mixed results. For instance, it has been reported that rs6505162 was associated with a significantly increased risk of ovarian cancer (41), contrary to another three studies showing that it conferred a reduced risk of esophageal cancer, and recurrence or survival of renal cell carcinoma and prostate cancer (37, 42, 43). The expression level of miR-423 has been reported to be significantly increased in breast cancer tissues compared to normal tissues (44). However, whether rs6505162 modulates the expression of miR-423 is not clear. The rs4919510 variant has been associated with an increased risk of recurrence in patients with renal cell carcinoma (42). Different alleles of this SNP has been predicted by in silico algorithms to exhibit differential capacities to bind to the potential target genes of miR-608 such as insulin receptor INSR and tumor suppressor TP53 (45), suggesting that the variant allele of rs4919510 could have different biological functions in relation to its various targets. It has been reported that miR-608 was down-regulated in ovarian cancer (46), but whether rs4919510 has an effect on the expression of miR-608 remains unknown. The conflicting results of these two SNPs among different cancers may be accounted for by differences in study design, sample size, and populations, or biological functions specific to different cancer types. Additional studies with well-powered homogeneous study populations, together with functional characterizations, are warranted to provide additional definitive evidence.
In stratified analyses, we found that the effects of both rs6505162 and rs4919510 on recurrence-free survival were only evident in patients receiving chemotherapy (Table 3). A test for interaction revealed a borderline significant interaction effect between chemotherapy and rs4919510. The finding of this potential interaction is in accordance with the observation that the hazard ratios of this SNP in the stratified analysis were opposite in patients with chemotherapy (HR=0.48) and those without chemotherapy (HR=1.57) (Table 3). We did not identify a significant interaction between chemotherapy and rs6505162, which is consistent with the observation that for this SNP, both of the patients with and without chemotherapy had elevated risk of recurrence and/or death (HR, 1.58 and 1.96, respectively). Nonetheless, given the modest samples size of our study population, the interaction analysis may not be adequately powered. Therefore, the results are not definitive at this point and future studies with larger patient populations are needed to validate these findings. When we conducted the joint effect analysis by combining the low-risk genotypes of both SNPs as the reference group, we found those patients with the high-risk genotypes of both SNPs had a significantly increased risk of death and/or recurrence (P=0.001). In accordance, this effect was more prominent in patients receiving chemotherapy (HR=3.81, P<0.001) than those without chemotherapy (HR=1.00, P=0.999) (Table 5). MicroRNAs have been extensively implicated in affecting the response to chemotherapy in patients with various cancers including CRC (23, 47–49). However, it remains to be assessed whether SNPs in miRNA gene regions influence CRC chemo-response by affecting the expression and function of the host miRNAs. Our data in this study suggest that the two significant SNPs identified in this study may possibly modulate the effects of chemotherapy on CRC outcome individually and jointly. If validated, they have the potential to be incorporated with other prognostic factors to select high-risk patients that are more likely to benefit from the therapeutic benefits of chemotherapy.
Our study has several strengths. First, the patients were enrolled from Xi’an and adjacent area. This region is highly attractive in conducting population-based research because of the geographical stability with low mobility rate. Second, the patients analyzed in this study were highly homogenous, in that all the patients had adenocarcinoma and were surgically treated to remove the primary tumor. Additionally, all chemotherapy treatments were started within two months of surgery and 83.4% of the 319 chemotherapy-treated patients received the same first-line adjuvant FOLFOX chemotherapy. The highly homogenous patient characteristics and treatments, as well as low rate of patient loss to follow-up, greatly reduced the confounding effects of the heterogeneous therapeutics modalities identified in most biomarker studies of cancer clinical outcome. Our study also has limitations. First, our study was restricted to Han Chinese and whether the findings can be generalized to other ethnic groups needs further evaluations. In addition, we could not rule out the possibility of chance findings in our study due to the relatively short follow-up time (median follow-up time, 23.7 months) and modest sample size. The enrollment of this study is still ongoing with a low rate of patient loss, which will further enable us to obtain higher statistical power for in-depth analyses. Another interesting observation in this study was that the HR was stronger for overall survival than recurrence-free survival for rs6505162. This could be explained by the possibility that the variant allele of this SNP conferred a higher risk of dying of primary cancer or its sequelae. However, the lack of complete data on sequelae in this patient population prevented us from further analyzing the patient outcome based on different causes of death.
In conclusion, our data showed for the first time that polymorphisms in pre-miRNAs were significantly associated with CRC clinical outcomes in a Chinese population, especially in patients receiving chemotherapy. Further validations of these findings are needed using independent populations and functional characterizations.
Supplementary Material
Acknowledgments
The work reported here is supported by a start-up grant from Thomas Jefferson University, grant 2009CB521704 from The National Basic Research Program of China, and grant 30872927 from National Natural Science Foundation of China. We thank Dr. Marie Dennis and Ms. Heidi Swan (Division of Population Science, Department of Medical Oncology, Thomas Jefferson University) for scientific editing.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Center MM, Jemal A, Smith RA, Ward E. Worldwide variations in colorectal cancer. CA Cancer J Clin. 2009;59:366–78. doi: 10.3322/caac.20038. [DOI] [PubMed] [Google Scholar]
- 3.Gellad ZF, Provenzale D. Colorectal Cancer: National and International Perspective on the Burden of Disease and Public Health Impact. Gastroenterology. 2010;138:2177–90. doi: 10.1053/j.gastro.2010.01.056. [DOI] [PubMed] [Google Scholar]
- 4.Ferrari P, Jenab M, Norat T, Moskal A, Slimani N, Olsen A, et al. Lifetime and baseline alcohol intake and risk of colon and rectal cancers in the European prospective investigation into cancer and nutrition (EPIC) Int J Cancer. 2007;121:2065–72. doi: 10.1002/ijc.22966. [DOI] [PubMed] [Google Scholar]
- 5.Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41:882–4. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, Kivioja T, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41:885–90. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- 7.Goel A, Boland CR. Recent insights into the pathogenesis of colorectal cancer. Curr Opin Gastroenterol. 2010;26:47–52. doi: 10.1097/MOG.0b013e328332b850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, Xie F, Chen K, Wang D, Jiang H, Li J, et al. ERCC5 promoter polymorphisms at −763 and +25 predict the response to oxaliplatin-based chemotherapy in patients with advanced colorectal cancer. Cancer Biol Ther. 2009;8:1424–30. doi: 10.4161/cbt.8.14.8889. [DOI] [PubMed] [Google Scholar]
- 9.Ulrich CM, Robien K, McLeod HL. Cancer pharmacogenetics: polymorphisms, pathways and beyond. Nat Rev Cancer. 2003;3:912–20. doi: 10.1038/nrc1233. [DOI] [PubMed] [Google Scholar]
- 10.Castro FA, Forsti A, Buch S, Kalthoff H, Krauss C, Bauer M, et al. TLR-3 polymorphism is an independent prognostic marker for stage II colorectal cancer. Eur J Cancer. 47:1203–10. doi: 10.1016/j.ejca.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 11.Xing J, Myers RE, He X, Qu F, Zhou F, Ma X, et al. GWAS-identified colorectal cancer susceptibility locus associates with disease prognosis. Eur J Cancer. 2011 doi: 10.1016/j.ejca.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–70. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 16.Mishra PJ, Bertino JR. MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics. 2009;10:399–416. doi: 10.2217/14622416.10.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;118:2600–8. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Z, Liang J, Wang Z, Tian T, Zhou X, Chen J, et al. Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum Mutat. 2009;30:79–84. doi: 10.1002/humu.20837. [DOI] [PubMed] [Google Scholar]
- 20.Liu Z, Li G, Wei S, Niu J, El-Naggar AK, Sturgis EM, et al. Genetic variants in selected pre-microRNA genes and the risk of squamous cell carcinoma of the head and neck. Cancer. 2010;116:4753–60. doi: 10.1002/cncr.25323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang H, Dinney CP, Ye Y, Zhu Y, Grossman HB, Wu X. Evaluation of genetic variants in microRNA-related genes and risk of bladder cancer. Cancer Res. 2008;68:2530–7. doi: 10.1158/0008-5472.CAN-07-5991. [DOI] [PubMed] [Google Scholar]
- 22.Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2008;105:7269–74. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. Jama. 2008;299:425–36. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slaby O, Svoboda M, Michalek J, Vyzula R. MicroRNAs in colorectal cancer: translation of molecular biology into clinical application. Mol Cancer. 2009;8:102. doi: 10.1186/1476-4598-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nana-Sinkam SP, Croce CM. MicroRNAs as therapeutic targets in cancer. Transl Res. 2011;157:216–25. doi: 10.1016/j.trsl.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 26.Wu WKK, Law PTY, Lee CW, Cho CH, Fan D, Wu K, et al. MicroRNA in colorectal cancer: from benchtop to bedside. Carcinogenesis. 2011;32:247–53. doi: 10.1093/carcin/bgq243. [DOI] [PubMed] [Google Scholar]
- 27.Landi D, Gemignani F, Naccarati A, Pardini B, Vodicka P, Vodickova L, et al. Polymorphisms within micro-RNA-binding sites and risk of sporadic colorectal cancer. Carcinogenesis. 2008;29:579–84. doi: 10.1093/carcin/bgm304. [DOI] [PubMed] [Google Scholar]
- 28.Zhang W, Winder T, Ning Y, Pohl A, Yang D, Kahn M, et al. A let-7 microRNA-binding site polymorphism in 3′-untranslated region of KRAS gene predicts response in wild-type KRAS patients with metastatic colorectal cancer treated with cetuximab monotherapy. Ann Oncol. 2011;22:104–9. doi: 10.1093/annonc/mdq315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boni V, Zarate R, Villa JC, Bandres E, Gomez MA, Maiello E, et al. Role of primary miRNA polymorphic variants in metastatic colon cancer patients treated with 5-fluorouracil and irinotecan. Pharmacogenomics J. 2010 doi: 10.1038/tpj.2010.58. [DOI] [PubMed] [Google Scholar]
- 30.Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L, et al. Genetic variants of miRNA sequences and non†“small cell lung cancer survival. The Journal of Clinical Investigation. 2008;118:2600–8. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffman AE, Zheng T, Yi C, Leaderer D, Weidhaas J, Slack F, et al. microRNA miR-196a-2 and breast cancer: a genetic and epigenetic association study and functional analysis. Cancer Res. 2009;69:5970–7. doi: 10.1158/0008-5472.CAN-09-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang R, Schlehe B, Hemminki K, Sutter C, Bugert P, Wappenschmidt B, et al. A genetic variant in the pre-miR-27a oncogene is associated with a reduced familial breast cancer risk. Breast Cancer Research and Treatment. 2010;121:693–702. doi: 10.1007/s10549-009-0633-5. [DOI] [PubMed] [Google Scholar]
- 33.Bao B-Y, Pao J-B, Huang C-N, Pu Y-S, Chang T-Y, Lan Y-H, et al. Polymorphisms inside MicroRNAs and MicroRNA Target Sites Predict Clinical Outcomes in Prostate Cancer Patients Receiving Androgen-Deprivation Therapy. Clinical Cancer Research. 2011;17:928–36. doi: 10.1158/1078-0432.CCR-10-2648. [DOI] [PubMed] [Google Scholar]
- 34.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 35.Liang D, Meyer L, Chang DW, Lin J, Pu X, Ye Y, et al. Genetic variants in MicroRNA biosynthesis pathways and binding sites modify ovarian cancer risk, survival, and treatment response. Cancer Res. 2010;70:9765–76. doi: 10.1158/0008-5472.CAN-10-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horikawa Y, Wood CG, Yang H, Zhao H, Ye Y, Gu J, et al. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin Cancer Res. 2008;14:7956–62. doi: 10.1158/1078-0432.CCR-08-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye Y, Wang KK, Gu J, Yang H, Lin J, Ajani JA, et al. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev Res (Phila Pa) 2008;1:460–9. doi: 10.1158/1940-6207.CAPR-08-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhan JF, Chen LH, Chen ZX, Yuan YW, Xie GZ, Sun AM, et al. A Functional Variant in MicroRNA-196a2 Is Associated with Susceptibility of Colorectal Cancer in a Chinese Population. Arch Med Res. 42:144–8. doi: 10.1016/j.arcmed.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 39.Chen H, Sun LY, Chen LL, Zheng HQ, Zhang QF. A variant in microRNA-196a2 is not associated with susceptibility to and progression of colorectal cancer in Chinese. Intern Med J. doi: 10.1111/j.1445-5994.2011.02434.x. [DOI] [PubMed] [Google Scholar]
- 40.Lee HC, Kim JG, Chae YS, Sohn SK, Kang BW, Moon JH, et al. Prognostic impact of microRNA-related gene polymorphisms on survival of patients with colorectal cancer. J Cancer Res Clin Oncol. 2010;136:1073–8. doi: 10.1007/s00432-009-0754-6. [DOI] [PubMed] [Google Scholar]
- 41.Kontorovich T, Levy A, Korostishevsky M, Nir U, Friedman E. Single nucleotide polymorphisms in miRNA binding sites and miRNA genes as breast/ovarian cancer risk modifiers in Jewish high-risk women. Int J Cancer. 2010;127:589–97. doi: 10.1002/ijc.25065. [DOI] [PubMed] [Google Scholar]
- 42.Lin J, Horikawa Y, Tamboli P, Clague J, Wood CG, Wu X. Genetic variations in microRNA-related genes are associated with survival and recurrence in patients with renal cell carcinoma. Carcinogenesis. 2010;31:1805–12. doi: 10.1093/carcin/bgq168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bao BY, Pao JB, Huang CN, Pu YS, Chang TY, Lan YH, et al. Polymorphisms inside microRNAs and microRNA target sites predict clinical outcomes in prostate cancer patients receiving androgen-deprivation therapy. Clin Cancer Res. 17:928–36. doi: 10.1158/1078-0432.CCR-10-2648. [DOI] [PubMed] [Google Scholar]
- 44.Arriola E, Marchio C, Tan DS, Drury SC, Lambros MB, Natrajan R, et al. Genomic analysis of the HER2/TOP2A amplicon in breast cancer and breast cancer cell lines. Lab Invest. 2008;88:491–503. doi: 10.1038/labinvest.2008.19. [DOI] [PubMed] [Google Scholar]
- 45.Landi D, Gemignani F, Barale R, Landi S. A catalog of polymorphisms falling in microRNA-binding regions of cancer genes. DNA Cell Biol. 2008;27:35–43. doi: 10.1089/dna.2007.0650. [DOI] [PubMed] [Google Scholar]
- 46.Dahiya N, Sherman-Baust CA, Wang T-L, Davidson B, Shih I-M, Zhang Y, et al. MicroRNA Expression and Identification of Putative miRNA Targets in Ovarian Cancer. PLoS ONE. 2008;3:e2436. doi: 10.1371/journal.pone.0002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hamano R, Miyata H, Yamasaki M, Kurokawa Y, Hara J, Moon JH, et al. Overexpression of miR-200c induces chemoresistance in esophageal cancers mediated through activation of the Akt signaling pathway. Clin Cancer Res. 17:3029–38. doi: 10.1158/1078-0432.CCR-10-2532. [DOI] [PubMed] [Google Scholar]
- 48.Yang H, Kong W, He L, Zhao JJ, O’Donnell JD, Wang J, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–33. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 49.Xi Y, Formentini A, Chien M, Weir DB, Russo JJ, Ju J, et al. Prognostic Values of microRNAs in Colorectal Cancer. Biomark Insights. 2006;2:113–21. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.