Abstract
Background
Multiple genome-wide and candidate gene association studies have been performed in search of common risk variants for breast cancer. Recent large meta analyses, consolidating evidence from these studies, have been consistent in highlighting the caspase-8 (CASP8) gene as important in this regard. In order to define a risk haplotype and map the CASP8 gene region with respect to underlying susceptibility variant/s, we screened four genes in the CASP8 region on 2q33-q34 for breast cancer risk.
Methods
Two independent data sets from the United Kingdom and the United States, including 3,888 breast cancer cases and controls, were genotyped for 45 tagging single nucleotide polymorphisms (tSNP) in the expanded CASP8 region. SNP and haplotype association tests were carried out using Monte Carlo based methods.
Results
We identified a three-SNP haplotype across rs3834129, rs6723097 and rs3817578 that was significantly associated with breast cancer (p<5×10−6), with a dominant risk ratio and 95% confidence interval of 1.28 (1.21–1.35) and frequency of 0.29 in controls. Evidence for this risk haplotype was extremely consistent across the two study sites and also consistent with previous data.
Conclusions
This three-SNP risk haplotype represents the best characterization so far of the chromosome upon which the susceptibility variant resides.
Impact
Characterization of the risk haplotype provides a strong foundation for re-sequencing efforts to identify the underlying risk variant, which may prove useful for individual-level risk prediction, and provide novel insights into breast carcinogenesis.
Keywords: Single nucleotide polymorphisms, risk haplotype, breast cancer, CASP8, apoptosis pathway
INTRODUCTION
The caspase-8 (CASP8) gene is one of only three genes identified as possessing common variants with strong and noteworthy associations with breast cancer risk based on cumulative evidence from candidate gene and genomewide association studies [1,2]. Similarly, pooled- and meta- analyses focusing on CASP8 specifically have indicated strong associations with breast cancer [3–5]. In particular, these refer to the highly significant association of the minor allele at D302H in exon 12 (rs1045485) and decreased risk. Some data suggests that another variant, a 6-bp deletion in the CASP8 promoter (−652 6N del, rs3834129) is associated with breast cancer, although the evidence for this variant is much less consistent [6,7,8]. There is no known functional effect of rs1045485 [9], and it is very rare in Asian populations. The del allele of rs3834129 has been suggested to remove an Spl transcription binding site, although functional data on the effects of this change in lymphocytes are conflicting [6,9]. Evidence thus far suggests that it is likely that other variant/s in linkage disequilibrium (LD) with rs1045485 and/or rs3834129 will be the critical variant/s.
CASP8 resides at chromosome 2q33. Two other genes, caspase-10 (CASP10) and amyotrophic lateral sclerosis 2 (juvenile) chromosome region candidate 12 (ALS2CR12), lie directly adjacent to CASP8. Like CASP8, CASP10 is an initiator of apoptosis, and the CASP10 V410I variant has been reported to be associated with breast cancer [10]. Another gene, called ‘CASP8 and FADD-like Apoptosis Regulator’ (CFLAR) lies centromeric to CASP10. It is a member of the same gene family as CASP8 and CASP10, but acts as a negative regulator of apoptosis [11]. Given their physical proximity to CASP8 and functional relevance (CASP10/CFLAR), the critical variants could reasonably lie in any of these four genes.
We previously genotyped 14 tagging-SNPs (tSNPs) in CASP8 on 2,450 breast cancer case and control subjects from the Sheffield Breast Cancer Study (SBCS) and identified a four-SNP risk haplotype (1-1-2-1 across SNPs rs7608692, rs1861269, rs6723097, rs3817578; p = 8.0×10−5), with a per allele OR (95% CI) of 1.30 (1.12–1.49) [12]. This haplotype was substantially more significant than any individual SNP, and was consistent with previous findings (i.e. the common (aspartate) allele at rs1045485, and the ins allele of rs3834129, are associated with the increased risk haplotype). The aim of the current study was to consider the broader four gene region and refine the risk haplotype upon which the susceptibility variant/s lie. Here, we have studied 3,888 breast cancer cases and controls from two collaborating sites, genotyped for 45 tSNPs across the four genes in the CASP8 region at chromosome 2q33-q34.
MATERIALS AND METHODS
Case and Control Subjects
A joint resource of 3,888 breast cancer cases and controls were genotyped: SBCS (N=2,049) and Utah Breast Cancer Study (UBCS; N=1,839). The SBCS set consisted of 1,015 histopathologically confirmed breast cancer patients recruited from the surgical outpatient clinics of the Royal Hallamshire Hospital, Sheffield, United Kingdom between 1998 and 2005. Case subjects were a mixture of incident and prevalent cases (median time to diagnosis 2.3 years), with median age at diagnosis (range) 59 (28–92) years. Fifteen percent of SBCS cases had at least one first degree relative with breast cancer. Control SBCS subjects (n=1,034) were healthy women attending the Sheffield Mammography Screening Service between 2000 and 2004. In the UK women are invited for routine mammography screening every 3 years between the ages of 50 and 70 years, and the average uptake in Sheffield is >80%. Women whose mammograms showed no evidence of breast lesions were eligible as controls for this study and median age at recruitment (range) was 57 (45–78) years. Eleven percent of SBCS controls had at least one first degree relative with breast cancer [13]. The UBCS set consisted of 905 breast cancer cases identified using the Utah Population Database (UPDB) and confirmed and ascertained through the Utah Cancer Registry; median age at diagnosis (range) was 56 (21–92) years and 41% of UBCS cases had at least one first degree relative with breast cancer. Controls (n=934) were birth cohort- and sex-matched cancer-free individuals, and 2% of these had at least one first degree relative with breast cancer. Using genealogy from the UPDB, it was established that 208 cases and 564 controls were singletons and the remaining individuals were members of 31 extended pedigrees; although most relationships were distant (average kinship coefficient=0.017, i.e. approximately 6 meioses distant, or second cousins). All cases and controls were of North European ancestry.
We excluded individuals with genotype call rates <80%, resulting in a total sample of 1,882 cases and 1,896 controls included in the genetic analyses.
For replication of significant single SNPs, we used a replication cohort comprising cases with a strong family history of breast cancer and control subjects from Manchester, UK. The cases comprised 713 subjects fulfilling the NICE criteria for BRCA1 and BRCA2 screening (>20% risk of mutation), but negative for BRCA1 or BRCA2 mutations as determined by DNA sequence analysis of coding regions, and multiplex ligation-dependent probe amplification to detect deletions and duplications. The 236 control subjects had no cancer and no immediate family history of breast cancer. All cases and controls were unrelated white British women.
Selection of tSNPs and Genotyping
We identified 60 tSNPs in the four genes for genotyping using LDselect [14], based on an analysis including all known SNPs with data available for the CEPH Utah individuals from HapMap and NIEHS [15]. Genotyping was carried out using the Applied Biosystems SNPlex™ multiplex system (55 SNPs) or 5′ nuclease PCR (TaqMan™) (3 SNPs). Genotype data for the remaining 2 SNPs, rs3834129 and rs6723097, was already available for these subjects [12]. Genotyping quality was assessed by examination of duplicate concordance and call rates for each SNP and a test for compliance with Hardy-Weinberg equilibrium (HWE) in controls. SNPs were removed if, in either SBCS or UBCS, their duplicate concordance rate <98% (n=2), more than one plate failed (n=7), or HWE p<0.005 (n=1). We also removed SNPs that were monomorphic (n=5). This resulted in a final set of 45 tSNPs for analysis (Supplementary Table S1).
Statistical Analyses
In order to account for familial relatedness in the UBCS subjects, all analyses were carried out using the meta-association options in the Genie (single SNPs) and hapConstructor (haplotypes) software packages which use Monte Carlo testing to derive empirical estimates of significance and 95% confidence intervals [16,17]. Odds ratios, confidence intervals and significance tests for individual SNPs were derived based on allele dose, dominant and recessive models. HapConstructor is a data-mining algorithm that builds multi-SNP haplotypes based on association evidence. Starting with evidence from single SNPs, the process adds or removes SNPs using a forward-backwards algorithm. Each step includes tests for dominant, additive, and recessive models for each haplotype. The process continues, provided pre-defined significance thresholds are met with each step. The results from the data-mining are the haplotype, genetic model, risk ratio and p-value. The significance thresholds used for the haplotype construction process were 0.05, 0.005, 0.0005, 0.0001 for haplotypes of one to four SNPs, respectively, and 0.00005 thereafter. Haplotypes were estimated via the estimation maximization algorithm, and any missing genotypes were internally imputed. All p-values were estimated using between 100,000 and 500,000 simulations. Observations more extreme than all simulated data sets were designated p<1/simulations.
RESULTS
Single SNP analyses
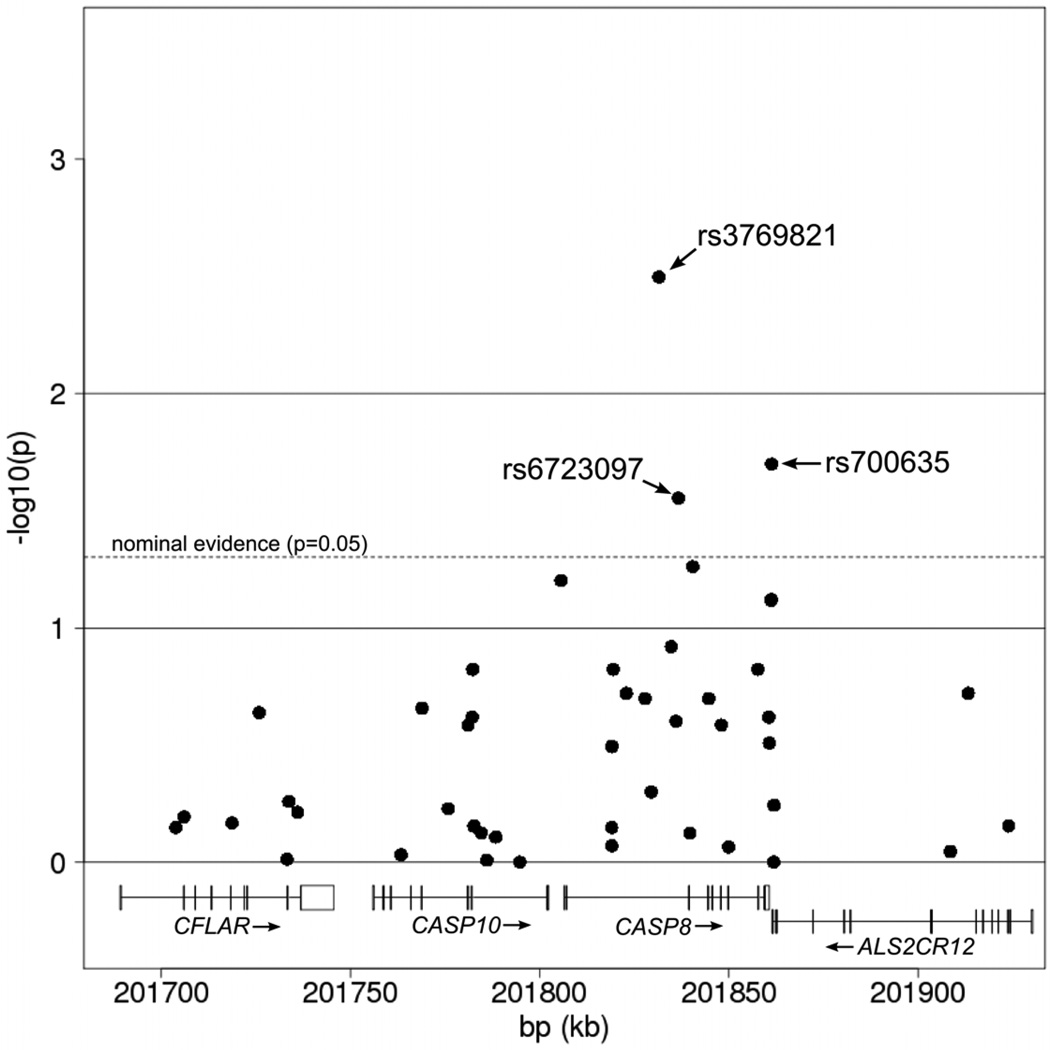
Table 1 and Figure 1 illustrate the individual tSNP results based on the Cochran Mantel Haentzel test for trend (allele dose risk model). For comparison, Supplemental Table S2 and Figure S1 show the most significant evidence for each tSNP (based on dominant or recessive models). Three of the 45 tSNPs showed at least nominally significant association with breast cancer (ptrend<0.05) in single SNP meta-association analyses across the two sites (rs3769821, rs6723097, rs700635 Table 1 and Figure 1). As illustrated in Figure 1, the most significant single SNPs cluster in CASP8 and AL2CR12, with rs3769821 in CASP8 being the most significant, with OR per-allele (95% CI) of 1.17 (1.05–1.30; p=0.0032; Table 1) and ORdom of 1.28 (1.21–1.38; p=5.3×10−4; Table S2). For confirmation, we genotyped the three most significant tSNPs in a cohort of unrelated familial cases negative for mutations in BRCA1 or BRCA2 from Manchester, UK and local controls. Nominally significant results were replicated for rs3769821 (ptrend = 0.042) and rs6723097 (ptrend = 0.014).
Table 1.
Single SNP results with the Cochran Mantel Haentzel test for trend
| Gene | bp | SNP | * ORmeta 95% CI | ORSBCS | ORUBCS | ^ Q phom | # pmeta |
|---|---|---|---|---|---|---|---|
| CFLAR | 201704024 | rs12620435 | 1.03 0.88–1.20 | 1.04 | 1.02 | 0.90 | 0.71 |
| CFLAR | 201706206 | rs11681674 | 1.21 0.54–2.70 | 3.08 | 0.69 | 0.08 | 0.64 |
| CFLAR | 201718871 | rs7573529 | 1.03 0.91–1.16 | 0.97 | 1.10 | 0.31 | 0.68 |
| CFLAR | 201726075 | rs719126 | 1.15 0.91–1.46 | 1.15 | 1.16 | 0.95 | 0.23 |
| CFLAR | 201733376 | rs2518142 | 1.00 0.84–1.19 | 0.91 | 1.14 | 0.22 | 0.97 |
| CFLAR | 201733866 | rs1594 | 1.03 0.93–1.14 | 1.08 | 0.98 | 0.35 | 0.55 |
| CFLAR | 201736187 | rs12721503 | 0.92 0.65–1.29 | 0.78 | 1.06 | 0.38 | 0.61 |
| CASP10 | 201763557 | rs12613347 | 1.01 0.90–1.13 | 1.05 | 0.95 | 0.39 | 0.93 |
| CASP10 | 201769065 | rs3731714 | 0.94 0.84–1.04 | 0.94 | 0.93 | 0.97 | 0.22 |
| CASP10 | 201775958 | rs41343445 | 0.91 0.64–1.30 | 0.70 | 0.95 | 0.52 | 0.59 |
| CASP10 | 201781218 | rs41419046 | 1.34 0.80–2.24 | 1.14 | 1.53 | 0.58 | 0.26 |
| CASP10 | 201782343 | rs13010627 | 1.14 0.92–1.41 | 1.08 | 1.21 | 0.60 | 0.24 |
| CASP10 | 201782452 | rs17860405 | 0.82 0.63–1.07 | 0.95 | 0.65 | 0.18 | 0.15 |
| CASP10 | 201782773 | rs7576306 | 1.02 0.92–1.13 | 1.12 | 0.92 | 0.056 | 0.70 |
| CASP10 | 201784693 | rs6435068 | 1.05 0.80–1.36 | 1.19 | 0.88 | 0.28 | 0.75 |
| CASP10 | 201786202 | rs41514050 | NA | - | - | - | 0.98 |
| CASP10 | 201788555 | rs10931932 | 0.98 0.82–1.16 | 0.92 | 1.06 | 0.41 | 0.78 |
| CASP10 | 201794910 | rs41373151 | 1.00 0.82–1.23 | 1.02 | 0.98 | 0.86 | 1.00 |
| CASP8 | 201805777 | rs3834129 | 0.89 0.79–1.01 | 0.84 | 1.12 | 0.077 | 0.063 |
| CASP8 | 201819185 | SNP50_9898_ct | NA | - | - | - | 0.71 |
| CASP8 | 201819204 | rs7608692 | 0.99 0.87–1.12 | 1.00 | 0.98 | 0.85 | 0.85 |
| CASP8 | 201819265 | rs3820972 | 1.10 0.92–1.32 | 1.13 | 1.06 | 0.72 | 0.32 |
| CASP8 | 201819625 | rs3769825 | 1.08 0.97–1.19 | 1.08 | 1.07 | 0.94 | 0.15 |
| CASP8 | 201823079 | rs7570325 | 1.21 0.91–1.61 | 1.00 | 1.49 | 0.18 | 0.19 |
| CASP8 | 201828012 | rs13402616 | 1.15 0.93–1.41 | 1.08 | 1.21 | 0.58 | 0.20 |
| CASP8 | 201829651 | rs35534358 | 1.73 0.24–12.61 | - | 0.70 | - | 0.50 |
| CASP8 | 201831675 | rs3769821 | 1.17 1.05–1.30 | 1.12 | 1.23 | 0.39 | 0.0032 |
| CASP8 | 201834953 | rs7568770 | NA | - | - | - | 0.12 |
| CASP8 | 201836192 | rs6435074 | 1.08 0.95–1.23 | 1.07 | 1.09 | 0.90 | 0.25 |
| CASP8 | 201836863 | rs6723097 | 1.15 1.01–1.30 | 1.15 | 1.15 | 1.00 | 0.028 |
| CASP8 | 201839832 | rs2293554 | 0.97 0.80–1.18 | 0.98 | 0.96 | 0.94 | 0.75 |
| CASP8 | 201840650 | rs34088540 | 0.81 0.65–1.00 | 0.84 | 0.76 | 0.64 | 0.055 |
| CASP8 | 201844840 | rs3817578 | 0.86 0.67–1.09 | 0.90 | 0.81 | 05 | 0.20 |
| CASP8 | 201848183 | rs7571586 | 1.35 0.80–2.28 | 1.20 | 1.57 | 0.62 | 0.26 |
| CASP8 | 201850083 | rs36043647 | 1.02 0.82–1.26 | 0.88 | 1.20 | 0.15 | 0.86 |
| CASP8 | 201857834 | rs1045485 | 0.90 0.77–1.04 | 0.92 | 0.86 | 0.6 | 0.15 |
| CASP8/ALS2CR12 | 201860736 | rs1035140 | 1.06 0.96–1.17 | 1.09 | 1.03 | 0.6 | 0.24 |
| CASP8/ALS2CR12 | 201860874 | rs35010052 | 1.24 0.82–1.90 | 1.26 | 1.23 | 0.96 | 0.31 |
| ALS2CR12 | 201861323 | rs1035142 | 1.10 0.99–1.22 | 1.10 | 1.10 | 0.98 | 0.076 |
| ALS2CR12 | 201861470 | rs700635 | 1.14 1.02–1.28 | 1.16 | 1.12 | 0.78 | 0.020 |
| ALS2CR12 | 201862034 | rs17383714 | 1.00 0.84–1.19 | 1.03 | 0.96 | 0.69 | 1.00 |
| ALS2CR12 | 201862091 | rs11674814 | 0.97 0.88–1.07 | 0.95 | 1.00 | 0.61 | 0.57 |
| ALS2CR12 | 201908695 | rs7579792 | 0.99 0.83–1.17 | 0.94 | 1.05 | 0.56 | 0.90 |
| ALS2CR12 | 201913442 | rs10185177 | 0.89 0.75–1.05 | 0.86 | 0.92 | 0.71 | 0.19 |
| ALS2CR12 | 201924064 | rs3817579 | 0.92 0.59–1.44 | 0.80 | 1.09 | 0.50 | 0.70 |
per allele Odds Ratio for the combined meta-analysis.
Q test p-value for heterogeneity across sites;
p-value for Cochran Mantel Haentzel test;
bold=nominally significant (p≤0.05); bold & italic = borderline interest (0.1<p<0.05). Data for rs6723097, rs3834129 and rs6435074 (UBCS) were taken from previously published data (reference 12). NA; zero cells
Figure 1.
Single SNP results with the Cochran Mantel Haentzel test for trend.
Haplotype analyses
HapConstructor analyses for the genes CASP8 and ALS2CR12 identified highly significant risk haplotypes (p<5×10−6 and p<1×10−5, respectively). No significant haplotypes (p<0.001) were identified in the downstream genes CFLAR or CASP10.
The most significant risk haplotype with greatest effect size in CASP8 was a six-SNP haplotype 1-2-1-1-1-1 across rs3834129, rs6723097, rs3817578, rs7571586, rs36043647 and rs35010052 (p<5×10−6; ORdom=1.29, 95% CI 1.22–1.33). The first five SNPs reside in CASP8, the last SNP, rs35010052, is contained in the ~700bp region between CASP8 and ALS2CR12. The frequency of this haplotype was 0.27 in controls and 0.30 in cases. Risk estimates from the two separate sites were ORdom = 1.31 95% CI 1.09–1.57 (p=0.004) and 1.27 95% CI 1.22–1.33 (p=0.0002) for SBCS and UBCS, respectively. The association strength of this six-SNP haplotype was driven by a three-SNP sub-haplotype, which gave a slightly lower risk estimate, but attained the same significance in the meta analysis (1-2-1 at rs3834129, rs6723097 and rs3817578; p<5×10−6; ORdom=1.28, 95% CI 1.21–1.35; freqcontrol=0.29, freqcases=0.32). Single site results for this three-SNP sub-haplotype haplotype were ORdom = 1.29 95% CI 1.07–1.55 (p=0.008) and 1.27 95% CI 1.23–1.31 (p=0.0002) for SBCS and UBCS, respectively. These three SNPs are not observed to have substantial levels of LD between them in the population (0.54, 0.03 and 0.05 for rs3834129-rs6723097, rs3834129-rs3817578 and rs6723097- rs3817578, respectively).
The findings for CASP8 are consistent in direction with previous single SNP results for rs3834129 [6] and rs1045485 [3–5], that is, the rs3834129 ins allele and rs1045485 aspartate allele are on or in LD with the risk haplotypes. Similarly, they are consistent with the risk haplotype we defined in our previous single-site SBCS analyses (1-1-2-1 across rs7608692, rs1861269, rs6723097, rs3817578; Table 2) [12].
Table 2.
CASP8 gene haplotype associations
| SNPs | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SOURCE | Sample size | rs3834129 | rs7608692 | rs1861269 | rs6723097 | rs3817578 | rs7571586 | rs36043647 | rs35010052 | alleles | Control freq | Case freq | OR (95% CI) | p-value |
| Current analysis | 3,778 | 1 | 2 | 1 | 1 | 1 | 1 | ins-A-G-G-T-del | 0.27 | 0.30 | 1.29 (1.22,1.33) | 5×10−6 | ||
| Current analysis | 3,778 | 1 | 2 | 1 | ins-A-G | 0.29 | 0.32 | 1.28 (1,21,1.35) | 5×10−6 | |||||
| Shephard et al 2009 | 2,450 | 1 | 1 | 2 | 1 | G-C-A-G | 0.20 | 0.24 | 1.30 (1.12,1.49) | 8×10−5 | ||||
The most significant risk haplotype and highest effect size in ALS2CR12 was a three-SNP haplotype 2-2-1 across rs1035140, rs1035142 and rs10185177 (p<1×10−5; ORdom=1.26, 95% CI 1.17–1.35; freqcontrol=0.27, freqcases=0.30). Single site results were ORdom = 1.29 95% CI 1.08–1.54 (p=0.005) and 1.21 95% CI 1.12–1.31 (p=0.003) for SBCS and UBCS, respectively. The first SNP on this haplotype, rs1035140, lies between CASP8 and ALS2CR12. Due to the strong association between the minor alleles at rs1035142 and rs700635 on this risk haplotype, the four-SNP and three-SNP haplotypes created by either adding allele 2 at rs700635 or substituting rs1035142 with rs700635, gave very similar results (both p<1×10−5; ORdom=1.25, 95% CI 1.20–1.30; freqcontrol=0.27, freqcases=0.30).
We performed a hapConstructor analysis across the CASP8 -ALS2CR12 two-gene region. This analysis of 27 tSNPs converged to the same results identified from the CASP8-only analysis, involving haplotype 1-2-1 at pivotal SNPs rs3834129, rs6723097 and rs3817578. This confirmed the expectation that the risk haplotypes identified in CASP8 and ALS2CR12 are due to the same underlying variant/s and suggests that only one risk haplotype exists at this region.
Discussion
Previously, we defined a four-SNP risk haplotype in CASP8 based on an analysis of 14 tSNPs and data from a single site (SBCS) [12]. Here, we provide a more thorough interrogation of a broader region using 45 tSNPs across four genes on chromosome 2q33 genotyped in two independent data sets (SBCS and UBCS). Two single-SNP results were additionally replicated in a third set of familial cases without BRCA1 or BRCA2 mutations. We have identified a common three-SNP risk haplotype in CASP8 that drives the association evidence at both sites and results in a highly significant meta-association finding (p<5×10−6; ORdom=1.28; freqcontrol=0.29). Evidence from multi-gene analyses indicates haplotypes that are centered in CASP8, and continues to confirm the involvement of CASP8 in risk to breast cancer. The consistency of results across two independent sites lends robustness to the finding and credibility and to the risk haplotype.
A recent genome-wide association study identified that the CASP8 region is associated with melanoma risk, and there are reports that it is also associated with other cancers including chronic lymphocytic leukaemia and pancreatic cancer [18–20]. These observations suggest that this region may be of broader interest for cancer in general. The risk haplotype provides a strong foundation for re-sequencing efforts by refining the haplotype upon which the susceptibility variant likely resides. Identification of the critical underlying risk variant may prove useful for individual-level risk prediction, aid in deciphering the role of CASP8 in risk of other cancers [6,18–20], and provide novel insights into breast carcinogenesis.
Supplementary Material
Acknowledgements
The work reported in this paper would not have been possible without the contribution of many individuals. In particular we thank Steve Backus, Kim Nguyen, Jathine Wong, Thomas Naranjo and Jim Farnham (UBCS) and Sue Higham, Gordon MacPherson, Helen Cramp, Dan Connley and Ian Brock (SBCS). We would also like to thank all the women who took part in these studies.
Grant Support
Data collection for the UBCS was made possible by the Utah Population Database (UPDB) and the Utah Cancer Registry (UCR). Partial support for all datasets within the UPDB was provided by the University of Utah Huntsman Cancer Institute (HCI) and the HCI Cancer Center Support grant, P30 CA42014 from the NCI. The UCR is funded by contract HHSN261201000026C from the NCI SEER program with additional support from the Utah State Department of Health and the University of Utah. The genotyping and analysis was supported by funding from the Susan G. Komen Foundation (BCTR0706911) and the Avon Foundation (02-2009-080), the Breast Cancer Campaign (2004Nov 49), and Yorkshire Cancer Research (S295, S299 and core funding). L-C-A acknowledges support from the Huntsman Cancer Foundation. HMcB, AL, WGN and DGE were all supported by the Manchester NIHR Biomedical Research Centre.
Footnotes
Conflict of Interest: No conflicts of interest to declare.
References
- 1.Zhang B, Beeghly-Fadiel A, Long J, Zheng W. Genetic variants associated with breast-cancer risk: comprehensive research synopsis, meta-analysis, and epidemiological evidence. Lancet Oncol. 2011;12:477–488. doi: 10.1016/S1470-2045(11)70076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peng S, Lü B, Ruan W, Zhu Y, Sheng H, Lai M. Genetic polymorphisms and breast cancer risk: evidence from meta-analyses, pooled analyses, and genome-wide association studies. Breast Cancer Res Treat. 2011;127:309–324. doi: 10.1007/s10549-011-1459-5. [DOI] [PubMed] [Google Scholar]
- 3.Cox A, Dunning AM, Garcia-Closas M, Balasubramanian S, Reed MW, Pooley KA, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007;39:352–358. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 4.Sergentanis TN, Economopoulos KP. Association of two CASP8 polymorphisms with breast cancer risk: a meta-analysis. Breast Cancer Res Treat. 120:229–234. doi: 10.1007/s10549-009-0471-5. 201. [DOI] [PubMed] [Google Scholar]
- 5.Yin M, Yan J, Wei S, Wei Q. CASP8 polymorphisms contribute to cancer susceptibility: evidence from a meta-analysis of 23 publications with 55 individual studies. Carcinogenesis. 2010;31:850–857. doi: 10.1093/carcin/bgq047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun T, Gao Y, Tan W, Ma S, Shi Y, Yao J, et al. A six-nucleotide insertion-deletion polymorphism in the CASP8 promoter is associated with susceptibility to multiple cancers. Nat Genet. 2007;39:605–613. doi: 10.1038/ng2030. [DOI] [PubMed] [Google Scholar]
- 7.Frank B, Rigas SH, Bermejo JL, Wiestler M, Wagner K, Hemminki K, et al. The CASP8 -652 6N del promoter polymorphism and breast cancer risk: a multicenter study. Breast Cancer Res Treat. 2008;111:139–144. doi: 10.1007/s10549-007-9752-z. [DOI] [PubMed] [Google Scholar]
- 8.Haiman CA, Garcia RR, Kolonel LN, Henderson BE, Wu AH, Le Marchand L. A promoter polymorphism in the CASP8 gene is not associated with cancer risk. Nat Genet. 2008;40:259–260. doi: 10.1038/ng0308-259. [DOI] [PubMed] [Google Scholar]
- 9.Li C, Lu J, Liu Z, Wang L-E, Zhao H, El-Naggar AK, et al. The six-nucleotide deletion/insertion variant in theCASP8 promoter region is inversely associated with risk of squamous cell carcinoma of the head and neck. Cancer Prev Res. 2010;3:246–252. doi: 10.1158/1940-6207.CAPR-08-0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank B, Hemminki K, Wappenschmidt B, Meindl A, Klaes R, Schmutzler RK, et al. Association of the CASP10 V410I variant with reduced familial breast cancer risk and interaction with the CASP8 D302H variant. Carcinogenesis. 2006;27:606–609. doi: 10.1093/carcin/bgi248. [DOI] [PubMed] [Google Scholar]
- 11.Rogers KM, Thomas M, Galligan L, Wilson TR, Allen WL, Sakai H, et al. Cellular FLICE-inhibitory protein regulates chemotherapy-induced apoptosis in breast cancer cells. Mol Cancer Ther. 2007;6:1544–1551. doi: 10.1158/1535-7163.MCT-06-0673. [DOI] [PubMed] [Google Scholar]
- 12.Shephard ND*, Abo R*, Rigas SH, Frank B, Lin WY, Brock IW, et al. A breast cancer risk haplotype in the caspase-8 gene. Cancer Res. 2009;69:2724–2728. doi: 10.1158/0008-5472.CAN-08-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacPherson G, Healey CS, Teare MD, Balasubramanian SP, Reed MW, Pharoah PD, et al. Association of a common variant of the CASP8 gene with reduced risk of breast cancer. J Natl Cancer Inst. 2004;96:1866–1869. doi: 10.1093/jnci/dji001. [DOI] [PubMed] [Google Scholar]
- 14.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. http://hapmap.ncbi.nlm.nih.gov/; http://egp.gs.washington.edu.
- 16.Curtin K, Wong J, Allen-Brady K, Camp NJ. PedGenie: meta genetic association testing in mixed family and case-control designs. BMC Bioinformatics. 2007;8:448. doi: 10.1186/1471-2105-8-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abo R, Knight S, Wong J, Cox A, Camp NJ. hapConstructor: automatic construction and testing of haplotypes in a Monte Carlo framework. Bioinformatics. 2008;24:2105–2107. doi: 10.1093/bioinformatics/btn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrett JH, Iles MM, Harland M, Taylor JC, Aitken JF, Andresen PA, et al. Genome-wide association study identifies three new melanoma susceptibility loci. Nat Genet. 2011 doi: 10.1038/ng.959. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enjuanes A, Benavente Y, Bosch F, Martín-Guerrero I, Colomer D, Pérez-Alvarez S, et al. Genetic variants in apoptosis and immunoregulation-related genes are associated with risk of chronic lymphocytic leukemia. Cancer Res. 2008;68:10178–10186. doi: 10.1158/0008-5472.CAN-08-2221. [DOI] [PubMed] [Google Scholar]
- 20.Couch FJ, Wang X, McWilliams RR, Bamlet WR, de Andrade M, Petersen GM. Association of breast cancer susceptibility variants with risk of pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:3044–3048. doi: 10.1158/1055-9965.EPI-09-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.