Abstract
Drought is a major constraint to common bean (Phaseolus vulgaris L.) production, especially in developing countries where irrigation for the crop is infrequent. The Mesoamerican genepool is the most widely grown subdivision of common beans that include small red, small cream and black seeded varieties. The objective of this study was to develop a reliable genetic map for a Mesoamerican × Mesoamerican drought tolerant × susceptible cross and to use this map to analyze the inheritance of yield traits under drought and fully irrigated conditions over 3 years of experiments. The source of drought tolerance used in the cross was the cream-seeded advanced line BAT477 crossed with the small red variety DOR364 and the population was made up of recombinant inbred lines in the F5 generation. Quantitative trait loci were detected by composite interval mapping for the traits of overall seed yield, yield per day, 100 seed weight, days to flowering and days to maturity for each field environment consisting of two treatments (irrigated and rainfed) and lattice design experiments with three repetitions for a total of six environments. The genetic map based on amplified fragment length polymorphism and random amplified polymorphic DNA markers was anchored with 60 simple sequence repeat (SSR) markers and had a total map length of 1,087.5 cM across 11 linkage groups covering the whole common bean genome with saturation of one marker every 5.9 cM. Gaps for the genetic map existed on linkage groups b03, b09 and b11 but overall there were only nine gaps larger than 15 cM. All traits were inherited quantitatively, with the greatest number for seed weight followed by yield per day, yield per se, days to flowering and days to maturity. The relevance of these results for breeding common beans is discussed in particular in the light of crop improvement for drought tolerance in the Mesoamerican genepool.
Keywords: Drought stress, Field tolerance mechanisms, Quantitative trait loci, Seed yield
Introduction
Common bean (Phaseolus vulgaris L.) is the most important legume for direct human consumption, but compared to other grain legumes such as chickpea, cowpea, peanut or pigeonpea it is generally considered to have high water requirements and to be susceptible to drought stress (Broughton et al. 2003; Subbarao et al. 1995). This means that water deficiency can have relatively large effects on common bean production (White and Izquierdo 1991). Drought stress on crops is defined as an inadequacy of water availability which restricts the expression of the full genetic potential of a cultivar (Taiz and Zeiger 2006). Drought can occur throughout the lifecycle of the crop or in a given stage of crop development, having large effects during early plant establishment, vegetative expansion, flowering and grain filling (Rao 2001). Given this, drought stress can be defined as early, intermittent or terminal depending on when it occurs (Frahm et al. 2004). Furthermore, the timing, intensity and duration of drought stress can markedly influence seed yield. It is estimated that 60% of the total production area for common bean is affected by drought, including large regions of Latin America and Africa in any given year (Thung and Rao 1999).
Within Latin America, drought is endemic in northeastern Brazil and the central and northern highlands of Mexico but also occurs with high frequency in Central America and occasionally in the Caribbean and certain parts of the Andes, often associated with El Niño climate events. Yield loss in the region can be up to 80% when severe drought strikes the crop early in crop development (Rao 2001).
Within bean production areas of Africa, intermittent droughts are frequent in large areas of Ethiopia, Kenya, Sudan and Tanzania (Wortmann et al. 1998) while other areas in Malawi, South Africa, Zambia and Zimbabwe are affected by terminal drought. Severe, season-long droughts have occurred in these regions in association with global climate fluctuations (Funk et al. 2008). Irrigation of beans is very infrequent in eastern and southern Africa and it is estimated that 396,000 tons production per year are lost to drought, making this the number one production constraint of any kind in the region. Drought also affects producers in Canada and USA but irrigation is common in some dry areas, although water availability might be limiting in the future (Muñoz Perea et al. 2006).
Breeding and selection for drought tolerance in common bean has shown that there is diversity for the trait. Singh (1995) reported the results of breeding for drought tolerance in a tropical environment and later Singh et al. (2001) and Terán and Singh (2002) identified advanced lines such as BAT477 and SEA5 that are highly tolerant. At higher latitudes in Mexico, cultivars Pinto Villa and Pinto Saltillo from the Durango race were released for drought-stressed environments of the northern highlands (Acosta-Gallegos and White 1995). More recently, Beebe et al. (2008) produced a series of drought-tolerant lines with small red, cream striped and black seed. Certain drought tolerance mechanisms appear to be of greater importance than others in protecting the crop (White and Singh 1991). Both physiological avoidance such as earliness or uptake of greater amounts of water through deep rooting as well as physiological adaptation through moderate stomatal conductance and good grain filling under stress are important as plant responses to drought stress (Rao 2001).
Inheritance of drought tolerance in common beans has been suggested to be quantitative with recurrent selection as an effective breeding strategy (Beebe et al. 2008) and some quantitative trait loci (QTL) were identified previously (Schneider et al. 1997b). However, the genetic components of drought tolerance are not fully analyzed as no map-anchored QTL study has been conducted with current co-dominant markers. Given this, we have developed a recombinant inbred line (RIL) population that is advantageous for genetic and physiological analysis of quantitative drought tolerance traits, based on the BAT477 drought resistance source crossed with the susceptible genotype DOR364. Because the population is made up of stable lines, it can be analyzed over a wide range of drought environments in a consistent and reliable fashion.
The main objective of this research was to analyze the intra-genepool Mesoamerican mapping population from the cross DOR364 × BAT477 for marker polymorphism and for seed yield traits under different levels of drought versus irrigation. The specific objectives were (1) to create an anchored, full-coverage genetic map for the cross using microsatellite, amplified fragment length polymorphism (AFLP) and random amplified polymorphic DNA (RAPD) markers and (2) to identify QTL for yield and yield components under drought versus full irrigation over three dry seasons. We stressed yield-related characteristics since seed yield is the most important economic trait under drought stress and the most widely used method for improving performance. Our medium- to long-term goal is to identify the regions of the genome which affect yield potential under drought as part of a project to develop drought resistant varieties for Latin America and eastern and southern Africa.
Materials and methods
Population development
The recombinant inbred line (RIL) population for DOR364 × BAT477 was developed by artificial hybridization between the two parents to create an initial F1 hybrid followed by single seed descent from the F2 until the F5 generation. Two generations were then used to increase the seed through bulking to the F5:7 generation. BAT477 was selected as the drought-tolerant parent because of its well-characterized adaptation to drought (White et al. 1994a, b; White and Castillo 1989; Sponchiado et al. 1989), and is an advanced line from CIAT derived from the cross (G3834 × G4493) × (G4792 × G5694) which has type III growth habit and small cream-colored seeds. In addition to its adaptation to drought conditions, it has high nitrogen-fixing capacity (Lynch and White 1992). DOR364 was selected as the other parent because it is a commercial type, small red-seeded, improved variety originally from CIAT that is widely grown in Central America and in parts of the Caribbean. DOR364 is susceptible to drought but is advantageous because it is resistant to an important viral disease, bean golden yellow mosaic (Beebe et al. 1995). The pedigree of DOR364 is based on the cross BAT1215 × (RAB166 × DOR125) and this genotype has type II bush growth habit and good yield potential when phosphorus (P) is not limiting, but is susceptible to drought. Both genotypes are from the Mesoamerican genepool but differ not only in drought tolerance but also in plant architecture and seed color, this final characteristic being used as a marker for success of the hybridization. A total of 113 lines were developed for the RIL population and all were used in genetic mapping.
Drought trials and trait measurements
The trials were carried out at the International Center for Tropical Agriculture (CIAT) in Palmira, Valle de Cauca, Colombia (3º29′N latitude, 76º21′W longitude, 1,000 m above sea level, 26°C average yearly temperature) over 3 years (2005, 2006 and 2007) in the June to September dry seasons with two levels of water supply (irrigated and rainfed) as separate experiments. The soil at the site was a Mollisol (Haplustoll) soil type with no major fertility problems (pH = 7.7) and clay-loam texture which is estimated to permit storage of 130 mm of available water (assuming 1.0 m of effective root growth with −0.03 and −1.5 MPa upper and lower limits for soil matrix potential). The experiments included a group of 97 RIL lines from the DOR364 × BAT477 population along with one check (the drought-tolerant advanced line SEA5) and both parents (DOR364, BAT477), giving a total of 100 genotypes. All plantings consisted of 10 × 10 lattice design experiments with three repetitions each so as to control for environmental variability in tropical soils. Experimental units consisted of two-row plots that were 3.72 m in length by 0.6 m wide. For the irrigated treatments, four gravity irrigations (35 mm each) were applied while for the drought stress treatments only two irrigations were applied. Two irrigations were used across both drought and well watered treatments for seed germination (first irrigation) and to ensure early uniform crop establishment (second irrigation) for later terminal or intermittent drought stress during flowering and pod development. The full irrigation treatment continued to receive irrigation during flowering and pod development while drought stress treatment did not.
Trials were managed with recommended disease and pest control using preventative treatments of fungicide seed treatment and foliar application of insecticides as well as two foliar applications of zinc and boron as microelements (300 g ha−1 as chelates) at 14 and 21 days after planting. In all trials, plots were hand harvested and threshed and then mechanically winnowed. The same phenological and yield information was collected for the population in all experiments. In the first category, days to flowering and maturity were evaluated. In the second category, seed yield was measured in kg/ha and in yield per day. Hundred seed weight was also evaluated in grams. Finally, analyses of variance (ANOVA) and Pearson’s correlations were carried out using the program SAS v. 8.2 (SAS Institute, Cary, NC, USA).
DNA extraction and marker analysis
In preparation for genetic mapping, DNA was extracted from the parents and all the RIL individuals according to methods described in Blair et al. (2003). Briefly, eight seeds per RIL line were germinated on humid paper in a dark growth chamber for 5 days until the first etiolated leaves could be harvested into 1.5 mL Eppendorf tubes where the tissue was ground in liquid nitrogen with a blue pestle. DNA extraction procedure was then carried out according to the protocol of Afanador et al. (1993). DNA concentration was measured in a Hoefer “DNA Quant 200” fluorometer and diluted to a final concentration of 10 ng/μl, with a final volume of 500 μl before use in PCR reactions. For microsatellite (SSR) screening, parental DNAs were evaluated with all the markers from six previous studies (Gaitán et al. 2002; Métais et al. 2002; Blair et al. 2003, 2008, 2009a, b). A total of 50 ng of template DNA was used for PCR amplification in a 20 μL final reaction volume for all primer pairs. PCR conditions, MgCl2 concentration and annealing temperatures were as described in the previous studies with PTC-100 or PTC-200 thermocyclers (MJ Research, Watertown, MA, USA) used for standard amplification. PCR products were run on denaturing 4% polyacrylamide gels in SequiGen electrophoresis units (Bio-Rad, Hercules, CA, USA) and silver stained according to the staining kit from Promega (Madison, WI, USA). Any polymorphic markers were then evaluated on the entire mapping population. Meanwhile, for RAPD analysis a total of 698 Operon primers were evaluated on DOR364 and BAT477 parents in PCR reactions carried out in 96-well plates on the same PTC-100 or PTC-200 thermocyclers. The total reaction volume for RAPDs was 25 μL. Annealing temperatures were 36°C and extension was carried out at 72°C. The PCR products were separated on 1.5% agarose gels that were stained with ethidium bromide and photographed with Polaroid film on a UV light box. The molecular weight standard consisted of PstI digested phage DNA. Finally, for AFLP reactions the methods described in Cichy et al. (2009) were followed with the commercially available AFLP Analysis System I kit, (Invitrogen, Carlsbad, CA, USA) with the modification that the pre-amplification PCR product (primer + 1 base) was run in 1% agarose gel and diluted 1:50. The amplification reactions (primer + 3 bases) were performed with 5 μl of the diluted DNA, 5 μl of Mix I (primers and dNTPs) and 10 μl of Mix II (10 × buffer, MgCl2 and DNA polymerase). PCR conditions were as recommended in Cichy et al. (2009) and amplification products were separated on 4% polyacrylamide gels with silver staining as described above.
Genetic mapping and QTL analysis
Segregation data from the AFLP, RAPD and microsatellite evaluations of the RIL population were used to create a genetic map based on linkage analysis with MapMaker software (v.3.0) for Windows (Lander et al. 1987) and three-point analysis with a minimum LOD of 3.0, which is to say a probability of 1 in 1000. Linkage groups were identified by comparisons to the integrated genetic map for common bean based on microsatellite mapping from Blair et al. (2003). The Kosambi coefficient was used to convert recombination fraction to genetic distance in centiMorgans (cM). QTL were detected with composite interval mapping (CIM) analysis that was carried out using the software program QTL Cartographer v. 1.21 (Basten et al. 2001) and the following parameters: 10 cM window size, 1 cM walkspeed, five significant background markers, analysis by forward and backward multiple linear regression for each chromosomal position with a global significance level of 5% and probability thresholds of 0.05 for the partial F test for both marker inclusion or exclusion. In the CIM analysis, determination coefficients were calculated for each interval separately (R 2) and for each interval given the background markers (TR 2) to determine the phenotypic variance explained by a single QTL (either alone or in conjunction with all other significant intervals). Thresholds for the individual QTL for each trait were determined by the generation of 1000 permutations of the data for that trait (Churchill and Doerge, 1994). Results were displayed using QTL Cartographer and represented graphically with standard drawing software, to designate genomic regions that proved to be significant in the analysis.
Results
Marker polymorphisms and genetic mapping
Three types of markers were evaluated for polymorphisms in the Mesoamerican intra-genepool cross combination, namely AFLP, RAPD and SSR/microsatellites. Among the three marker systems, the AFLP reactions produced only 22 polymorphisms that were useful for genetic mapping, based on seven primer pair combinations (E-AAC/M-CAC, E-AAC/M-CAG, E-AAC/M-CAT, E-AAG/M-CAC, E-AAG/M-CTA, E-ACA/M-CAC, E-ACA/M-CTA) where some of these same combinations were useful in previous studies in our lab (Muñoz et al. 2004; Cichy et al. 2009). RAPD profiling, meanwhile, produced a total of 104 markers that were useful for genetic mapping, based on 59 decamer primers. Apart from the dominant, multi-copy AFLP and RAPD markers, screening of 595 single-copy, co-dominant SSR markers detected 60 polymorphic loci. Therefore, as a result of the marker screening a total of 186 markers were found to be segregating in the RILs and were useful for linkage analysis.
The resulting genetic map covered all 11 linkage groups of the common bean genome and had a total map length of 1087.5 cM. Genetic markers especially the SSR markers were evenly distributed across the majority of linkage groups (Table 1). Linkage groups varied in genetic map distance length ranging from 29.3 (for b11) to 194.8 cM (for b04). Coverage was also low for b09 with only 36.8 cM, while the remaining linkage groups all had genetic map lengths of 72 cM or above.
Table 1.
Summary of markers integrated into the new genetic map for the DOR364 × BAT477 population and average (Avg) distance between SSR or all markers within each linkage group
| LG | AFLP | RAPD | SSR | Total markers | Total linkage group length | Avg distance SSR | Avg distance all markers |
|---|---|---|---|---|---|---|---|
| b01 | 3 | 1 | 6 | 10 | 73.3 | 12.3 | 7.3 |
| b02 | 3 | 18 | 11 | 32 | 159.9 | 14.5 | 5.0 |
| b03 | 0 | 7 | 1 | 8 | 95.6 | NA | 11.9 |
| b04 | 4 | 25 | 8 | 37 | 194.8 | 24.3 | 5.3 |
| b05 | 2 | 10 | 2 | 14 | 72.6 | 36,3 | 5.2 |
| b06 | 1 | 7 | 8 | 16 | 76.2 | 9.5 | 4.8 |
| b07 | 3 | 5 | 4 | 12 | 90.8 | 22.7 | 7.6 |
| b08 | 3 | 10 | 5 | 18 | 112.8 | 22.6 | 6.3 |
| b09 | 0 | 3 | 5 | 8 | 36.8 | 7.4 | 4.6 |
| b10 | 3 | 16 | 7 | 26 | 145.4 | 20.8 | 5.6 |
| b11 | 0 | 2 | 3 | 5 | 29.3 | 9.8 | 5.8 |
| Total | 22 | 104 | 60 | 186 | 1087.5 | 18.1 | 5.85 |
The average distance between all markers on a given linkage group averaged one every 5.9 cM and ranged from one every 4.7 cM to one every 12.0 cM based on each linkage group, showing that the map was at a level of saturation useful for QTL analysis where ideally markers should be distributed one every 10 cM. The microsatellites on the other hand were at an average distance of 18.1 cM with greatest saturation on linkage groups b06, with eight microsatellites at an average distance of 9.5 cM, and b09, with five microsatellites at an average distance of 7.4 cM. Linkage groups b02 and b04 had large numbers of microsatellites (11 and 8, respectively) but were also the first and second largest in genetic map distance. The average distance between SSRs on these linkage groups was 14.5 and 24.4 cM, respectively. When considering all markers, some gaps bigger than 20 cM were found on linkage groups b03, b05, b10 and b11 with some implication for QTL analysis in those specific regions; however, only nine gaps were bigger than 15 cM.
Drought stress conditions
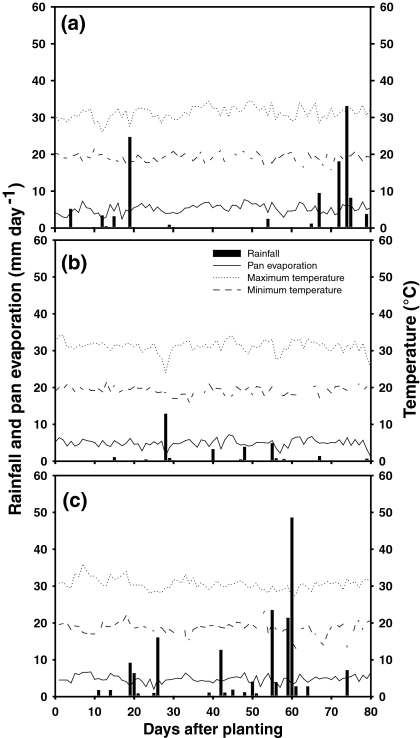
During the crop growing season, average maximum and minimum temperatures (°C), were 34.5 and 15.8 in the first season (2005), 34.2 and 16.0 in the second season (2006) and 30.6 and 18.6 in the third season (2007), respectively. The incident solar radiation (MJ m−2 day−1) range as 10.2–22.8 in the first season, 9.2–23.9 in the second season, and 11.2–25.1 in the third season. Total rainfall was 126, 33.2 and 243 mm in the three seasons, respectively; while the potential pan evaporation was 400, 411 and 431 mm. These data on rainfall and pan evaporation indicated that the crop suffered terminal drought stress in the first and second seasons (2005, 2006) and intermittent drought stress in the third season (2007) during the active crop growth period. Figure 1 shows the distribution of rainfall and the temperature and evaporation regimes in each season.
Fig. 1.
Rainfall distribution, pan evaporation, and maximum and minimum temperatures during crop growing period at Palmira, Colombia across three dry seasons in the years 2005 (a), 2006 (b) and 2007 (c)
Drought intensity index was calculated based on the mean yield of all genotypes under stress versus non-stress conditions for both sets of experiments. In the first season (2005), the mean yield of all the genotypes under the drought stress treatment was 864 kg/ha compared with mean full-irrigation, well-watered yield of 2,133 kg/ha, showing a 59.5% reduction in grain yield due to drought. In the second season (2006), the mean yield under drought conditions was 1,663 kg/ha compared with mean fully irrigated yield of 3,173 kg/ha, showing a 47.6% decrease in mean grain yield due to drought stress. In the last season (2007), the mean yield under drought conditions was 988 kg/ha compared with mean fully irrigated yield of 2,024 kg/ha, resulting in a 51.2% reduction. This showed that drought was more severe in the first season compared to the second two seasons.
Volumetric soil moisture levels were measured in all three seasons at different soil depths (0–5; 5–10; 10–20; 20–40; 40–60 cm). The reductions in volumetric soil moisture content across soil depth in the drought treatment compared to the irrigated treatment were 49.3% versus 21.5% at 50 days after planting (DAP) in 2005, 59.4% versus 23.1% at 46 DAP in 2006, and 64% versus 9.2% at 34 DAP in 2007. This showed that the drought treatment had consistently low soil moisture content in the flowering and reproductive stages compared to the fully-irrigated, well-watered treatment. Volumetric soil moisture content under well-watered, irrigated conditions across soil depth ranged from 20.1 to 37.9%. Table 2 shows the reduction in moisture content across soil depths in the drought conditions at the time points that were evaluated.
Table 2.
Reduction of volumetric soil moisture content (%) under drought conditions compared with the irrigated conditions at Palmira across 3 years
| Year | 2005 | 2006 | 2007 | ||||
|---|---|---|---|---|---|---|---|
| Days after planting | 50 | 33 | 41 | 46 | 10 | 23 | 34 |
| Soil depth (cm) | |||||||
| 0–5 | 49.3 | 8.6 | 25.4 | 59.4 | 21.7 | 0 | 64.0 |
| 5–10 | 50.2 | 28.0 | 35.6 | 48.5 | 13.1 | 0 | 41.8 |
| 10–20 | 39.0 | 18.3 | 27.1 | 36.5 | 6.0 | 0 | 27.0 |
| 20–40 | 18.0 | 5.0 | 19.4 | 26.5 | 0 | 0 | 10.8 |
| 40–60 | 21.5 | 1.8 | 13.1 | 23.1 | 0 | 0 | 9.2 |
Population distributions and parental differences
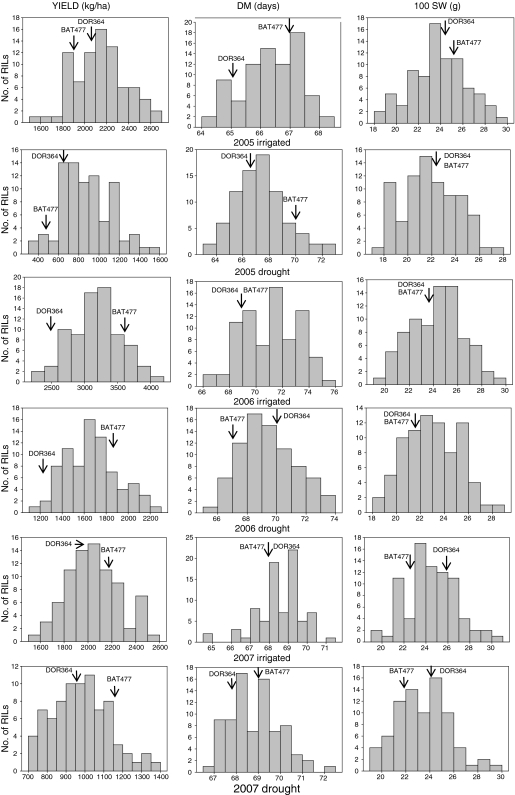
Analyses of variance showed significant differences (P < 0.0001) for yield, yield per day and seed weight in each season and, similarly, differences were significant for days to flowering and maturity in the three seasons (data not shown). Table 3 shows the range in each variable found in the RIL population under drought and irrigated conditions in each season. The population distributions were continuous, showing quantitative inheritance for all the traits measured in each season and in each environment (Fig. 2). Distributions were normal for each trait × season × environment combination except for days to flowering in 2005 irrigation and 2007 drought and days to maturity in 2005 drought and 2007 irrigation.
Table 3.
Descriptive statistics for quantitative traits measured in the DOR364 × BAT477 recombinant inbred line population for each trait × season × environment combination
| Trait | Year | Env.a | RIL average | SE meanb | SDb | Minimum | Maximum | Skew | Kurtosis | Normalityc |
|---|---|---|---|---|---|---|---|---|---|---|
| Yield (kg/ha) | 2005 | Drought | 880.6 | 27.753 | 249.78 | 363.0 | 1543.0 | 0.3889 | −0.1668 | 2.122ns |
| 2005 | Irrig. | 2136.9 | 26.141 | 235.27 | 1512.0 | 2625.0 | −0.0447 | −0.2322 | 0.039ns | |
| 2006 | Drought | 1659.5 | 30.736 | 273.19 | 1064.3 | 2332.0 | 0.1810 | −0.4533 | 3.408ns | |
| 2006 | Irrig. | 3183.5 | 44.119 | 392.14 | 2201.5 | 4002.6 | −0.1158 | −0.0781 | 0.278ns | |
| 2007 | Drought | 977.9 | 16.572 | 148.22 | 701.1 | 1361.6 | 0.3171 | −0.2938 | 1.437ns | |
| 2007 | Irrig. | 2045.2 | 23.708 | 212.05 | 1564.1 | 2527.8 | 0.1747 | −0.3915 | 0.280ns | |
| Yield per day | 2005 | Drought | 13.1 | 0.4240 | 3.8159 | 5.31 | 23.62 | 0.4355 | −0.1888 | 1.967ns |
| 2005 | Irrig. | 32.2 | 0.3871 | 3.4839 | 22.42 | 39.17 | −0.1135 | −0.114 | 0.197ns | |
| 2006 | Drought | 23.9 | 0.4375 | 3.8890 | 16.13 | 34.18 | 0.2347 | −0.3947 | 0.112ns | |
| 2006 | Irrig. | 44.8 | 0.5904 | 5.2477 | 32.63 | 57.72 | 0.0107 | −0.2602 | 0.022ns | |
| 2007 | Drought | 14.2 | 0.2388 | 2.1356 | 10.31 | 20.03 | 0.3428 | −0.1343 | 1.641ns | |
| 2007 | Irrig. | 29.9 | 0.3422 | 3.0604 | 23.40 | 36.95 | 0.1304 | −0.5398 | 0.482ns | |
| Seed weight (g/100s) | 2005 | Drought | 21.9 | 0.2584 | 2.3256 | 17.0 | 28.0 | 0.0991 | −0.6155 | 0.131ns |
| 2005 | Irrig. | 23.9 | 0.2692 | 2.4226 | 18.0 | 29.0 | −0.0951 | −0.2836 | 0.112ns | |
| 2006 | Drought | 22.9 | 0.2504 | 2.2257 | 18.7 | 29.0 | 0.1943 | −0.5422 | 0.989ns | |
| 2006 | Irrig. | 24.1 | 0.2455 | 2.1818 | 19.3 | 29.7 | 0.0289 | −0.5165 | 0.420ns | |
| 2007 | Drought | 23.3 | 0.2479 | 2.2176 | 19.0 | 29.3 | 0.3935 | −0.0001 | 1.137ns | |
| 2007 | Irrig. | 24.3 | 0.2549 | 2.2802 | 19.0 | 30.3 | 0.1605 | −0.0286 | 0.451ns | |
| Days to flowering | 2005 | Drought | 38.5 | 0.1084 | 0.9758 | 36.0 | 41.0 | 0.4932 | 0.4219 | 1.223ns |
| 2005 | Irrig. | 36.6 | 0.1073 | 0.9658 | 34.0 | 38.0 | −0.7420 | 0.4651 | 12.405** | |
| 2006 | Drought | 38.6 | 0.0782 | 0.6954 | 36.7 | 40.0 | −0.3485 | 0.0555 | 1.912ns | |
| 2006 | Irrig. | 36.8 | 0.1308 | 1.1622 | 34.7 | 39.0 | 0.0895 | −1.0072 | 2.629ns | |
| 2007 | Drought | 38.8 | 0.0790 | 0.7070 | 36.0 | 40.7 | −0.6215 | 2.442 | 31.911** | |
| 2007 | Irrig. | 38.3 | 0.1145 | 1.0243 | 35.7 | 40.3 | −0.2651 | −0.189 | 1.868ns | |
| Days to Maturity | 2005 | Drought | 67.3 | 0.2278 | 2.0504 | 64.0 | 73.0 | 0.6827 | 0.4092 | 6.282* |
| 2005 | Irrig. | 66.3 | 0.1071 | 0.9639 | 64.0 | 68.0 | −0.2306 | −0.4739 | 2.463ns | |
| 2006 | Drought | 69.4 | 0.2211 | 1.9656 | 66.0 | 73.3 | 0.1221 | −0.9098 | 0.085ns | |
| 2006 | Irrig. | 71.1 | 0.2382 | 2.1171 | 66.7 | 75.0 | −0.1169 | −0.8896 | 0.579ns | |
| 2007 | Drought | 68.8 | 0.1278 | 1.1432 | 66.7 | 72.0 | 0.4482 | −0.3614 | 0.031ns | |
| 2007 | Irrig. | 68.5 | 0.1354 | 1.2108 | 64.7 | 71.0 | −0.8408 | 1.0964 | 16.052** |
ns not significant
aEnvironments including drought and irrigated treatments as described in text
bStandard error for average in preceding column
cLevel of significance corresponding to * P < 0.05, and ** P < 0.01
Fig. 2.
Population distributions for yield, days to maturity and seed weight among the recombinant inbred lines of the DOR364 × BAT477 population grown over three seasons and 3 years in Palmira, Colombia
In each season several RILs were better or worse in yield than the drought-tolerant parent BAT477 or the drought-susceptible parent DOR364. Transgressive segregation for yield was therefore observed. Similarly, transgressive segregation was evident for seed size and yield per day as well as for the phenological traits. BAT477 was higher yielding than DOR364 under drought and irrigated conditions in 2006 and 2007 seasons but not in 2005. The performance of BAT477 in 2005 was not as good as expected, possibly due to higher day temperatures than normal during flowering (up to 34°C).
Differences were not significant (P < 0.05) between the parents in terms of seed weight but were significant for yield; and indeed the seed size was similar for each of the parental genotypes in each of the seasons and environments, except in 2007 where DOR364 produced slightly larger seed than BAT477 under both drought and irrigated conditions. The effect of drought caused a 1–3 g weight decrease per 100 seeds when comparing irrigated and rainfed conditions. Finally, DOR364 was earlier maturing than BAT477 by 2–4 days under both treatments in 2005 and under drought in 2007, but the opposite was true under drought in 2006. Meanwhile, under irrigation in 2006 and 2007 both parental genotypes matured at the same time. In terms of correlations within the same year, the phenological traits tended to be more highly correlated amongst each other compared to correlations between yield and seed weight traits (Table 4). For example, correlations between days to maturity and days to flowering were significant (P < 0.05) in five out of six comparisons made within years and treatments. However, this relationship did not hold for the drought treatment in 2005 although it was found in the other years.
Table 4.
Correlation coefficients for days to flowering (DF), days to maturity (DM), seed weight (SW) and yield (YLD) measured in the DOR364 × BAT477 recombinant inbred line population for each trait × season combination
| DF-irrigated | DM-irrigated | SW-irrigated | YLD-irrigated | |
|---|---|---|---|---|
| 2005 | ||||
| DF-drought | 0.47*** | 0.22* | −0.16ns | 0.00ns |
| DM-drought | −0.13ns | 0.07 ns | −0.12ns | 0.05ns |
| SW-drought | 0.03ns | −0.01ns | 0.65*** | −0.08ns |
| YLD-drought | −0.08ns | 0.19ns | −0.03ns | 0.26* |
| 2006 | ||||
| DF-drought | 0.50*** | 0.36** | 0.06ns | 0.10ns |
| DM-drought | 0.37** | 0.60*** | 0.16ns | −0.08ns |
| SW-drought | −0.10ns | 0.24* | 0.83*** | 0.10ns |
| YLD-drought | 0.01ns | −0.01ns | 0.06ns | 0.12 ns |
| 2007 | ||||
| DF-drought | 0.66*** | 0.47*** | −0.01ns | 0.07ns |
| DM-drought | 0.37** | 0.52*** | 0.00ns | −0.13** |
| SW-drought | 0.02ns | 0.09ns | 0.79*** | 0.06ns |
| YLD-drought | −0.02ns | 0.04ns | 0.05ns | 0.16 ns |
Correlations are all within year, with r-values within the drought treatment shown below the diagonals and r-values within the irrigated treatment for the given year shown above the diagonals. The correlation of each variable for means in drought and irrigated treatments is shown in bold on the diagonals
***, ** and * indicate probabilities of P < 0.001, P < 0.01 and P < 0.05, respectively; ns not significant
Meanwhile, seed weight was positively correlated with days to maturity in the drought treatment in 2006 but not in the other years and yield was negatively correlated with days to maturity in the irrigated treatment in 2007 only. Correlations were highest for the comparisons of the same trait across drought and irrigated treatments and were especially significant (P < 0.001) for seed weight and days to flowering across treatments in all years and for days to maturity across treatments in 2006 and 2007 but not in 2005. Yield was not correlated across treatments in 2006 or 2007 but was correlated (P < 0.05) across treatment in 2005.
QTL identification
A total of 49 QTL were found for yield and yield components under drought or irrigated conditions across the 3 years of the study (Table 5). The majority of these QTL (27) were detected for 100 seed weight rather than for yield or yield per day which had correspondingly fewer QTL (10 and 12, respectively). QTL were also detected for days to flowering and days to maturity in all three seasons (Table 6), with totals of ten and eight loci identified, respectively. Figure 3 shows QTL positions corresponding to the points with maximum LR within the significant intervals.
Table 5.
Quantitative trait loci (QTL) for yield, yield per day and seed weight over 3 years in drought and irrigated treatments in the DOR364 × BAT477 mapping population
| Trait | QTLa | Year | Environmentb | Linkage group | Marker | Additivity | Source | LRc | R 2 | TR 2 | LR threshold |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Yield | Yld3.1 | 2005 | Irrig. | 3 | AE103 | 97.57 | DOR364 | 13.02 | 0.16 | 0.21 | 12.94 |
| Yld8.1 | 2005 | Drought | 8 | O1604 | 93.55 | BAT477 | 13.22 | 0.14 | 0.23 | 13.00 | |
| Yld4.1 | 2006 | Drought | 4 | ×301 | 111.84 | BAT477 | 17.66 | 0.18 | 0.39 | 13.35 | |
| Yld6.1 | 2006 | Drought | 6 | M501 | 155.70 | DOR364 | 19.87 | 0.19 | 0.39 | 13.35 | |
| Yld3.2 | 2006 | Irrig. | 3 | Q1001 | 170.14 | DOR364 | 24.43 | 0.19 | 0.49 | 13.41 | |
| Yld3.3 | 2006 | Irrig. | 3 | BM181 | 155.68 | DOR364 | 15.05 | 0.14 | 0.36 | 13.41 | |
| Yld3.4 | 2006 | Irrig. | 3 | AD1801 | 149.55 | DOR364 | 15.23 | 0.14 | 0.36 | 13.41 | |
| Yld7.1 | 2006 | Irrig. | 7 | CATA06 | 132.32 | BAT477 | 14.13 | 0.11 | 0.53 | 13.41 | |
| Yld8.2 | 2007 | Drought | 8 | AC702 | 78.59 | BAT477 | 27.77 | 0.24 | 0.43 | 13.22 | |
| Yld10.1 | 2007 | Drought | 10 | U1401 | 51.36 | DOR364 | 14.29 | 0.11 | 0.43 | 13.22 | |
| Yield per day | Ypd3.1 | 2005 | Irrig. | 3 | AE103 | 1.47 | DOR364 | 13.63 | 0.17 | 0.22 | 13.25 |
| Ypd8.1 | 2005 | Drought | 8 | O1604 | 1.38 | BAT477 | 12.22 | 0.13 | 0.22 | 10.19 | |
| Ypd4.1 | 2006 | Drought | 4 | ATA288 | 1.39 | BAT477 | 14.60 | 0.14 | 0.30 | 13.40 | |
| Ypd6.1 | 2006 | Drought | 6 | M501 | 2.24 | DOR364 | 20.42 | 0.19 | 0.39 | 13.40 | |
| Ypd3.2 | 2006 | Irrig. | 3 | AE103 | 2.65 | DOR364 | 26.90 | 0.23 | 0.55 | 13.24 | |
| Ypd3.3 | 2006 | Irrig. | 3 | AD1801 | 2.13 | DOR364 | 20.11 | 0.16 | 0.48 | 13.24 | |
| Ypd4.2 | 2006 | Irrig. | 4 | P1101 | 2.01 | BAT477 | 20.99 | 0.14 | 0.53 | 13.24 | |
| Ypd4.3 | 2006 | Irrig. | 4 | O1602 | 1.95 | BAT477 | 16.83 | 0.14 | 0.52 | 13.24 | |
| Ypd4.4 | 2006 | Irrig. | 4 | AGTA01 | 1.99 | BAT477 | 17.78 | 0.13 | 0.51 | 13.24 | |
| Ypd8.2 | 2007 | Drought | 8 | AC702 | 1.11 | BAT477 | 27.16 | 0.23 | 0.43 | 14.08 | |
| Ypd10.1 | 2007 | Drought | 10 | U1401 | 0.76 | DOR364 | 15.02 | 0.12 | 0.43 | 14.08 | |
| Ypd8.3 | 2007 | Irrig. | 8 | P103 | 1.53 | BAT477 | 16.74 | 0.16 | 0.31 | 13.16 | |
| Seed weight | Sw2.1 | 2005 | Irrig. | 2 | AB1003 | 0.86 | BAT477 | 14.32 | 0.12 | 0.12 | 13.26 |
| Sw6.1 | 2005 | Irrig. | 6 | ACAG02 | 1.37 | DOR364 | 38.22 | 0.31 | 0.31 | 13.26 | |
| Sw6.2 | 2005 | Irrig. | 6 | Y501 | 1.29 | DOR364 | 32.00 | 0.28 | 0.28 | 13.26 | |
| Sw2.2 | 2005 | Drought | 2 | ACAG01 | 1.00 | BAT477 | 15.46 | 0.18 | 0.18 | 13.5 | |
| Sw2.3 | 2005 | Drought | 2 | ATA170 | 0.98 | BAT477 | 14.53 | 0.17 | 0.17 | 13.5 | |
| Sw5.1 | 2006 | Drought | 5 | AGTA02 | 1.39 | DOR364 | 16.14 | 0.12 | 0.54 | 13.27 | |
| Sw6.3 | 2006 | Drought | 6 | AG1301 | 0.99 | DOR364 | 16.65 | 0.18 | 0.41 | 13.27 | |
| Sw6.4 | 2006 | Drought | 6 | U1301 | 0.88 | DOR364 | 17.26 | 0.15 | 0.38 | 13.27 | |
| Sw6.5 | 2006 | Drought | 6 | BM158 | 0.92 | DOR364 | 17.75 | 0.16 | 0.45 | 13.27 | |
| Sw6.6 | 2006 | Drought | 6 | AB1001 | 1.11 | DOR364 | 31.03 | 0.23 | 0.52 | 13.27 | |
| Sw6.7 | 2006 | Drought | 6 | Y501 | 1.08 | DOR364 | 27.15 | 0.22 | 0.51 | 13.27 | |
| Sw6.8 | 2006 | Drought | 6 | BM187 | 1.05 | DOR364 | 25.25 | 0.21 | 0.44 | 13.27 | |
| Sw2.4 | 2006 | Irrig. | 2 | AB1003 | 0.82 | BAT477 | 16.00 | 0.13 | 0.60 | 13.29 | |
| Sw5.2 | 2006 | Irrig. | 5 | U1305 | 0.64 | DOR364 | 13.40 | 0.07 | 0.62 | 13.29 | |
| Sw6.9 | 2006 | Irrig. | 6 | BM158 | 1.02 | DOR364 | 24.25 | 0.22 | 0.54 | 13.29 | |
| Sw6.10 | 2006 | Irrig. | 6 | AB1001 | 1.23 | DOR364 | 44.75 | 0.30 | 0.62 | 13.29 | |
| Sw6.11 | 2006 | Irrig. | 6 | Y501 | 1.13 | DOR364 | 36.02 | 0.25 | 0.57 | 13.29 | |
| Sw2.5 | 2007 | Drought | 2 | AB1003 | 1.09 | BAT477 | 26.33 | 0.22 | 0.70 | 12.71 | |
| Sw2.6 | 2007 | Drought | 2 | ATA16 | 0.90 | BAT477 | 22.67 | 0.14 | 0.63 | 12.71 | |
| Sw5.3 | 2007 | Drought | 5 | U1305 | 0.64 | DOR364 | 12.77 | 0.08 | 0.61 | 12.71 | |
| Sw6.12 | 2007 | Drought | 6 | AB1001 | 0.89 | DOR364 | 24.43 | 0.16 | 0.58 | 12.71 | |
| Sw6.13 | 2007 | Drought | 6 | BM187 | 0.95 | DOR364 | 28.45 | 0.17 | 0.60 | 12.71 | |
| Sw9.1 | 2007 | Drought | 9 | BM114 | 0.87 | BAT477 | 23.76 | 0.15 | 0.61 | 12.71 | |
| Sw9.2 | 2007 | Drought | 9 | N201 | 0.85 | BAT477 | 19.61 | 0.14 | 0.61 | 12.71 | |
| Sw6.14 | 2007 | Irrig. | 6 | AG1301 | 1.09 | DOR364 | 17.53 | 0.21 | 0.41 | 13.18 | |
| Sw6.15 | 2007 | Irrig. | 6 | AB1001 | 1.29 | DOR364 | 37.23 | 0.30 | 0.51 | 13.18 | |
| Sw6.16 | 2007 | Irrig. | 6 | BM187 | 1.28 | DOR364 | 37.11 | 0.30 | 0.50 | 13.18 |
Significant markers that are repeated between the traits yield and yield per day are in bold, while those that are repeated within seed weight between years and treatments are in bold italic
aQTL name based on association with yield (Yld), yield per day (Ypd) or seed weight (Sw). Decimal number represents linkage group and QTL order
bEnvironments including drought and irrigated treatments as described in text
cEmpirical LOD thresholds based on 1000 permutations used for QTL detection as recommended by Churchill and Doerge (1994) are indicated in the final column of the table
Table 6.
QTL for days to flowering and days to maturity over 3 years under drought and irrigated treatments in the DOR364 × BAT477 mapping population
| Trait | QTLa | Year | Environmentb | Linkage group | Marker | Additive | Source | LRc | R 2 d | TR 2 d | LR threshold |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Days to flowering | Df4.1 | 2005 | Irrig. | 4 | X301 | 0.35 | DOR364 | 14.14 | 0.12 | 0.12 | 13.15 |
| Df4.2 | 2005 | Drought | 4 | AGTA03 | 0.38 | DOR364 | 19.60 | 0.17 | 0.45 | 12.87 | |
| Df7.1 | 2005 | Irrig. | 7 | BM185 | 0.44 | DOR364 | 14.63 | 0.19 | 0.19 | 13.15 | |
| Df5.1 | 2006 | Irrig. | 5 | BM138 | 0.42 | DOR364 | 14.22 | 0.12 | 0.41 | 14.06 | |
| Df11.1 | 2006 | Irrig. | 11 | P302 | 0.54 | BAT477 | 16.85 | 0.20 | 0.48 | 14.06 | |
| Df5.2 | 2007 | Drought | 5 | AGTA02 | 0.24 | DOR364 | 13.55 | 0.11 | 0.41 | 12.49 | |
| Df6.1 | 2007 | Drought | 6 | M1601 | 0.27 | BAT477 | 13.18 | 0.14 | 0.45 | 12.49 | |
| Df4.3 | 2007 | Irrig. | 4 | AGTA03 | 0.47 | DOR364 | 21.95 | 0.21 | 0.57 | 13.52 | |
| Df5.3 | 2007 | Irrig. | 5 | V201 | 0.38 | DOR364 | 15.28 | 0.12 | 0.53 | 13.52 | |
| Df6.2 | 2007 | Irrig. | 6 | M1601 | 0.36 | BAT477 | 14.24 | 0.12 | 0.52 | 13.52 | |
| Days to maturity | Dm6.1 | 2005 | Drought | 6 | M1601 | -0.95 | BAT477 | 28.20 | 0.22 | 0.52 | 13.74 |
| Dm6.2 | 2005 | Drought | 6 | BM137 | -0.77 | BAT477 | 15.73 | 0.13 | 0.43 | 13.74 | |
| Dm7.1 | 2005 | Drought | 7 | AGTA07 | -0.82 | BAT477 | 22.06 | 0.17 | 0.52 | 13.74 | |
| Dm5.1 | 2006 | Drought | 5 | U1305 | 0.69 | DOR364 | 14.51 | 0.12 | 0.40 | 13.27 | |
| Dm2.1 | 2006 | Irrig. | 2 | ACAG01 | 1.23 | BAT477 | 24.36 | 0.25 | 0.53 | 13.49 | |
| Dm6.3 | 2006 | Irrig. | 6 | BMc238 | 0.97 | DOR364 | 20.25 | 0.22 | 0.50 | 13.49 | |
| Dm2.2 | 2007 | Drought | 2 | BM221 | 0.67 | BAT477 | 12.90 | 0.16 | 0.33 | 12.77 | |
| Dm2.3 | 2007 | Drought | 2 | P1602 | 0.54 | BAT477 | 14.62 | 0.14 | 0.31 | 12.77 |
Significant markers that are repeated between the traits days to flowering and days to maturity are in bold, while those that are repeated within days to flowering between years and treatments are in bold italic
aQTL name based on association with days to flowering (Df), or days to maturity (Dm). Decimal number represents linkage group and QTL order
bEnvironments including drought and irrigated treatments as described in text
cEmpirical LOD thresholds based on 1000 permutations used for QTL detection as recommended by Churchill and Doerge (1994) are indicated in the final column
dDetermination coefficients based on marker (R 2) and model including background markers (TR 2)
Fig. 3.
QTL for yield, yield per day, seed weight, days to flowering and days to maturity on the genetic map of the DOR364 × BAT477 recombinant inbred line population (refer to Tables 5 and 6 for QTL names). Vertical lines for each QTL represent the range of the QTL that are above the LOD threshold; horizontal marks on the lines indicate the LOD peak for the QTL; and QTL found in drought versus irrigated treatments are coded in different colors
Determination coefficients (R 2 and TR 2) were used to determine the phenotypic variance explained by each QTL (either alone or in conjunction with all other significant intervals). The most important QTL based on R 2 values were found for seed weight with up to 31% of phenotypic variance explained by a single locus. QTL for yield and yield per day explained 11–24% and 12–23% of phenotypic variance, respectively. QTL for days to flowering and maturity had similar R 2 values up to 21 or 25%, respectively. QTL for phenological traits had lower R 2 values when detected under drought compared to those under irrigation, while the opposite was true for yield and yield components. Overlapping QTL were found for yield and yield per day on linkage groups b03, b08 and b10 while different QTL were detected on linkage groups b04 and b07. Similarly, overlapping QTL for days to flowering and days to maturity were found on linkage group b06 but not on other linkage groups.
Positive alleles for the QTL came from each parent, indicating that both contributed to yield and yield components in the drought and irrigated treatments. For the large number of QTL found for seed weight, the majority had a positive allele for larger seed from DOR364 (19) rather than from BAT477 (8). More QTL for seed size were found in the drought stress treatment (16) than in the irrigated treatment (11). All the QTL from BAT477 except one were detected under drought conditions, while for DOR364, ten QTL were detected under drought and nine under irrigation. Meanwhile, the moderate number of QTL for days to flowering and maturity were equally detected under drought conditions and irrigated conditions (nine each) and the allele associated with lateness came evenly from both parents (nine each). However, it was interesting that QTL for days to maturity were more often found in drought while QTL for days to flowering were more often found in irrigated conditions.
Discussion
A major accomplishment of this study was the creation of a genetic map for a Mesoamerican × Mesoamerican intra-genepool cross population that integrates various types of markers into an anchored scaffold of SSRs for full genome coverage. Intra-genepool genetic maps have been rare, especially within the Mesoamerican genepool of common bean, as marker polymorphisms have generally been very low (Frei et al. 2005; Blair et al. 2006a, b). For this reason, we targeted three marker systems for map generation, namely AFLP, RAPD and SSR/microsatellites. The use of various marker systems was envisioned to be complementary among marker types and necessary for map saturation given the small genetic distance between the two parents which both belong to the same race Mesoamerica according to Blair et al. (2006a). A similar strategy was followed by Cichy et al. (2009) who created the first intra-genepool Andean × Andean genetic map.
Of the three marker systems, the AFLP reactions were the least productive, followed by SSR markers and RAPD banding patterns. For example, only 3.1 polymorphic bands were generated per AFLP gel, which is low compared to inter-genepool or inter-specific comparisons (Muñoz et al. 2004), but similar to that found with an Andean intra-genepool population (Cichy et al. 2009). Beebe et al. (2001) also found that AFLPs were more polymorphic for comparisons of Andean and Mesoamerican genotypes and were not very polymorphic for intra-genepool comparisons. RAPD amplifications, on the other hand, were fairly productive as a marker system for uncovering polymorphisms, since 104 polymorphic bands were generated based on 59 random primers. While the average number of bands per primer was 1.76, this did not reflect the large number of RAPD primers that were screened and which were monomorphic. Therefore, the RAPD method, while productive, required a large effort and was also low in polymorphism. Beebe et al. (2000) found a larger amount of polymorphism in a wider screening of Mesoamerican germplasm and races with RAPD markers, but polymorphism was limited to inter-racial combinations and was low within the Mesoamerica race of this genepool.
Microsatellites were of low to intermediate polymorphism in the population and therefore required the screening of a large number of primer pairs (595 in total) to obtain a reasonable number of mappable markers (60). Overall, the polymorphism rate for the microsatellites was 10.1%; however, polymorphism varied according to different microsatellite types. For example, genomic microsatellites from Gaitán et al. (2002), Métais et al. (2002) and Blair et al. (2003, 2008, 2009a) were reasonably polymorphic but genic microsatellites from Blair et al. (2003, 2009b) were of very low polymorphism. Among the genomic microsatellites, the most polymorphic were the AT-rich and GA-derived microsatellites. Genic microsatellites from the EST and Genbank sources were similar in low levels of polymorphism. Similar results of low polymorphism were found for microsatellites screened in an intra-genepool Andean population by Cichy et al. (2009). Blair et al. (2006a) found higher levels of polymorphism for inter-genepool combinations and lower levels of polymorphism for intra-genepool combinations, especially within the Mesoamerican genepool. Polymorphism for DOR364 × BAT477 in that study was 34.1% across 150 microsatellites evaluated and genomic markers were of higher polymorphism than gene-based markers.
The use of multiple marker systems allowed us to create a full-genome map that was 1,087.5 cM long, which compares favorably to previous genetic maps of common bean based on RILs (Blair et al. 2003; Checa and Blair 2008; Ochoa et al. 2006; Cichy et al. 2009). Map saturation was fairly high, given that AFLP and RAPD markers tended to fill in the scaffold created by SSR markers. For example, the majority of linkage groups had similar numbers of markers, with only b03, b09 and b11 standing out for having fewer markers and b02 and b04 for having more markers.
Among the individual marker types, AFLP markers were distributed very evenly on most linkage groups except b03, b09 and b11 where none were mapped and on b06 where only one was mapped. Since AFLP bands are sometimes associated with heterochromatin and centromeres we were not surprised that most of the AFLPs mapped to central locations of the linkage groups. RAPDs were also unevenly distributed with many more on linkage groups b02, b04 and b10 and fewer on linkage groups b01, b09 and b11. This distribution of RAPDs may reflect the distribution of retrotransposons, since many RAPD bands amplify parts of retrotransposons given the inverted repeats present in regions rich in these elements (Blair et al. 2006b). Further support for this was the fact that several RAPD primers detected linked loci which are also typical of retrotransposon loci. The number of RAPD markers was not significantly correlated with the number of AFLP markers or microsatellites on each of the linkage groups.
SSR markers, unlike the other two marker types, were well distributed on all 11 linkage groups, with 4–8 markers on each linkage group except b02 with more and b03 and b05 with fewer. Previous studies have also found a predominance of markers, especially microsatellites, on b02 linkage groups, while some linkage groups such as b03 have also been difficult to saturate before (Blair et al. 2003, 2008). It is likely that for most linkage groups both chromosome arms were represented by the genetic maps, while for b09 and b11 perhaps only one chromosome arm was mapped for each of these linkage groups, although the location of the centromeres in common bean are still being defined in relation to various types of molecular markers.
Overall, the genetic map was enhanced by the inclusion of single-copy microsatellite markers, as these anchored the anonymous and dominant AFLP and RAPD markers to the linkage groups of the common bean core map with the microsatellite markers which were mostly of known map positions (Blair et al. 2003, 2008). This was valuable for comparative mapping and association with known chromosomes. Multiple microsatellite loci on all the linkage groups except b03 allowed us to orient the linkage groups according to Blair et al. (2003), who presented a method for map integration based on this marker type. The only disadvantage of SSRs was that a large number of microsatellites had to be screened based on the low rate of polymorphism for the cross, as discussed in the previous section. However, this makes the present genetic map a clear reference point for genetic mapping within the Mesoamerican genepool. This is especially the case for crosses among race Mesoamerica individuals since it is likely that SSR polymorphisms from the present study are also present in other small seeded beans.
Given the full coverage, the genetic map was found to be useful for QTL discovery in terms of the five traits analyzed for RILs grown under drought and irrigated conditions. QTL mapping was possible since all traits were quantitatively inherited and QTL were found on all linkage groups except b01. Among the traits, QTL for days to flowering and maturity tended to cluster together, with overlapping and co-localizing QTL especially prominent on linkage groups b05 and b06. This suggested pleiotropy for genes controlling both days to flowering and days to maturity, as would be expected based on the correlations found between these traits. Independent QTL for days to flowering on b04 and days to maturity on b02 were also found. Phenological traits are important for drought studies because of the effect of flowering time on detection of QTL for yield under drought conditions and the potential for earlier genotypes to escape drought (Collins et al. 2008). In this study, most QTL for days to flowering or maturity were consistent across years suggesting the higher heritability and lower genotype × environment interaction of the phenological traits compared to the traits discussed below.
For yield and yield per day, a similar number of QTL were found for the drought and irrigated treatments, although there were differences in QTL detection per year. Among the individual QTL for these traits, the most promising for yield were Yld3.1 and Yld8.1 which were detected in two seasons each at approximately the same location. Similarly, the same QTL were found for yield per day at these locations and at the locus Ypd4.1 over both drought and irrigated conditions in 2006. From summarizing the QTL found, we noticed that the number of QTL for yield per day was slightly higher than for yield alone, indicating the influence of days to maturity on detecting QTL for yield per day and that the duration of overall reproductive period or pod filling affected yield potential in the population; however, this may have been a result of population size, which affects the power to detect QTL. Most QTL for yield per day matched locations of QTL for days to maturity or yield and together the traits would be reliable measures of drought tolerance.
For seed weight, it was notable that more QTL were found under drought than under irrigated conditions across the years and within each year. For example, in 2007 no seed weight QTL was found under irrigation but four were found under drought. Overall there was a high number of QTL of greater R 2 values found for seed weight than for yield or other traits, perhaps due to seed weight being more highly heritable under drought and non-drought conditions, as was found by Schneider et al. (1997a, b). The seed weight trait was important given that seed filling is inhibited under drought stress, so large seed may indicate tolerance to drought and lead to higher yields (Ramírez-Vallejo and Kelly 1998). Interestingly, the positive allele for seed size under drought came from both BAT477, the drought-tolerant parent, and from DOR364, the drought-susceptible parent, and there was substantial transgressive segregation for this trait, indicating that seed filling may be one mechanism by which DOR364 contributed to yield potential in the RILs. Most seed weight QTL were stable across years and this would make them useful for molecular breeding (Collins et al. 2008). Overall there were fewer QTL detected in 2005 than under 2006 and 2007, perhaps because of the more severe drought stress in this first season or because of differences in experimental conditions across the years, with intermittent rather than terminal drought found in the last of the seasons.
The results from this study agree with a previous analysis of recombinant inbred line populations under drought from Schneider et al. (1997a) who also found a small number of QTL for yield and a larger number of QTL for seed weight. Having said this, the number of QTL can be overestimated since many QTL were near the same marker. Schneider et al. (1997a) also studied the possibility of molecular-marker-assisted selection to improve drought tolerance (Schneider et al. 1997b). In one-way analysis of variance and multiple regressions, they identified four RAPD markers in one population and five in another that were significantly associated with yield under drought, yield without drought, and/or geometric mean yield. QTL positions, however, were on an un-anchored genetic map so comparisons between the RIL populations cannot be made.
Some seed weight and phenological QTL aligned with previously identified QTL from other QTL studies in bean (http://www.css.msu.edu/bic/PDF/Bean_Core_map_2009.pdf). These included a photoperiod induced delay in flowering QTL on linkage group b11 near the Dl1 gene (Hannah et al. 2007) and the seed weight QTL found on linkage group b06 which aligned with a QTL from the inter-genepool population of Park et al. (2000). These QTL are likely to be general for various conditions, as was found for Sw6.1 and Sw6.2, which were expressed in various years and under both drought stress and high irrigation treatments. The cluster of seed weight QTL on linkage group b02 may be homologous with those found by Tar’an et al. (2002). The fact that we did not detect QTL for seed weight or phenological traits on linkage group b01 may be associated with our use of an intra-genepool population of Mesoamerican beans compared to previous studies of inter-genepool populations (reviewed in Broughton et al. 2003).
In conclusion, microsatellites proved to be very useful for anchoring the genetic map and assuring that all chromosomes were being assayed in the present study. The genetic map was also one of the first based on a Mesoamerican × Mesoamerican cross and was an important achievement, especially given that both parents are from the same Mesoamerica race (Blair et al. 2006a). We should note that monomorphism has been a large problem in this population due to its inter-genepool, within-race origin and it might be that some sections of the genome are identical by descent. We plan, therefore, to fill gaps in the map that are larger than 15 cM with new markers in the future. In terms of physiology, we found yield under drought stress as well as seed weight or grain filling to be good general measures of drought tolerance in the evaluation of the Mesoamerican RIL population. This is important since grain filling can be considered a measure of photosynthate mobilized to seed formation. QTL were located for yield, seed weight and phenology traits and showed the relationships between these traits.
Genetic improvement strategies could use these QTL once they are validated by additional lines, although the use of lattice design experiments for populations of this size appears to be a way to minimize overestimation of QTL effects (Vales et al. 2005). Meanwhile, in terms of breeding for drought tolerance, BAT477 has been used to improve various classes of common beans with a range of genotypes partially derived from this parental source (Terán and Singh 2002, Beebe et al. 2008). Therefore, other advanced lines derived from BAT477 can provide a background to confirm QTL from this study. In terms of gene transfer within the Mesoamerica race and the Mesoamerican genepool, the results of this study were very important given that small seeded beans like DOR364 are among the most widely grown beans around the world and are found in areas where drought stress is on the increase (Beebe et al. 2008; Wortmann et al. 1998).
Acknowledgments
We are grateful to the full field crews of the bean breeding and physiology projects for their diligence in trial management and data collection. Mariela Rivera and José Polania collected soil moisture data which was especially helpful. This work was funded in part by the Tropical Legumes I project and by the German government through the BMZ project No. (2002.7860.6–001.00) and by CIAT core funds.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Acosta-Gallegos JA, White JW. Phenological plasticity as an adaptation by common bean to rainfed environments. Crop Sci. 1995;35:199–204. doi: 10.2135/cropsci1995.0011183X003500010037x. [DOI] [Google Scholar]
- Afanador L, Haley S, Kelly JD. Adoption of a “mini-prep” DNA extraction method for RAPD’s marker analysis in common bean Phaseolus vulgaris. Bean Improv Coop. 1993;36:10–11. [Google Scholar]
- Basten CJ, Weir BS, Zeng ZB. QTL cartographer: a reference manual and tutorial for QTL mapping. Raleigh. NC: Department of Statistics, North Carolina State University; 2001. [Google Scholar]
- Beebe SE, Ochoa I, Skroch P, Nienhhuis J, Tivang J. Genetic diversity among common bean breeding lines developed for Central America. Crop Sci. 1995;35:1178–1183. doi: 10.2135/cropsci1995.0011183X003500040045x. [DOI] [Google Scholar]
- Beebe S, Skroch P, Tohme J, Duque MC, Pedraza F, Nienhhuis J. Structure of genetic diversity among common bean landraces of middle American origin based on correspondence analysis of RAPD. Crop Sci. 2000;40:264–273. doi: 10.2135/cropsci2000.401264x. [DOI] [Google Scholar]
- Beebe S, Rengifo J, Gaitán-Solís E, Duque MC, Tohme J. Diversity and origin of Andean landraces of common bean. Crop Sci. 2001;41:854–862. doi: 10.2135/cropsci2001.413854x. [DOI] [Google Scholar]
- Beebe SE, Rao IM, Cajiao C, Grajales M. Selection for drought resistance in common bean also improves yield in phosphorus limited and favorable environments. Crop Sci. 2008;48:582–592. doi: 10.2135/cropsci2007.07.0404. [DOI] [Google Scholar]
- Blair MW, Pedraza F, Buendia HF, Gaitán-Solís E, Beebe SE, Gepts P, Tohme J. Development of a genome-wide anchored microsatellite map for common bean (Phaseolus vulgaris L.) Theor Appl Genet. 2003;107:1362–1374. doi: 10.1007/s00122-003-1398-6. [DOI] [PubMed] [Google Scholar]
- Blair MW, Giraldo MC, Buendía HF, Tovar E, Duque MC, Beebe SE. Microsatellite marker diversity in common bean (Phaseolus vulgaris L.) Theor Appl Genet. 2006;113:100–109. doi: 10.1007/s00122-006-0276-4. [DOI] [PubMed] [Google Scholar]
- Blair MW, Muñoz C, Garza R, Cardona C. Molecular mapping of genes for resistance to the bean pod weevil (Apion godmani Wagner) in common bean. Theor Appl Genet. 2006;112:913–923. doi: 10.1007/s00122-005-0195-9. [DOI] [PubMed] [Google Scholar]
- Blair MW, Buendía HF, Giraldo MC, Métais I, Peltier D. Characterization of AT-rich microsatellites in common bean (Phaseolus vulgaris L.) Theor Appl Genet. 2008;118:91–103. doi: 10.1007/s00122-008-0879-z. [DOI] [PubMed] [Google Scholar]
- Blair MW, Muñoz M, Pedraza F, Giraldo MC, Buendía HF, Hurtado N. Development of microsatellite markers for common bean (Phaseolus vulgaris L.) based on screening of non-enriched small insert genomic libraries. Genome. 2009;52:772–782. doi: 10.1139/G09-053. [DOI] [PubMed] [Google Scholar]
- Blair MW, Muñoz-Torres M, Giraldo MC, Pedraza F. Development and diversity assessment of Andean-derived, gene-based microsatellites for common bean (Phaseolus vulgaris L.) BMC Plant Biol. 2009;9:100. doi: 10.1186/1471-2229-9-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton WJ, Hernandez G, Blair M, Beebe S, Gepts P, Vanderleyden J. Beans (Phaseolus spp.)–model food legumes. Plant Soil. 2003;252:55–128. doi: 10.1023/A:1024146710611. [DOI] [Google Scholar]
- Checa OE, Blair MW. Mapping QTL for climbing ability and component traits in common bean (Phaseolus vulgaris L.) Mol Breeding. 2008;22:201–215. doi: 10.1007/s11032-008-9167-5. [DOI] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichy KA, Caldas GV, Snapp SS, Blair MW. QTL analysis of seed iron, zinc, and phosphorus levels in an Andean bean population. Crop Sci. 2009;49:1742–1750. doi: 10.2135/cropsci2008.10.0605. [DOI] [Google Scholar]
- Collins NC, Tardieu F, Tuberosa R. Quantitative trait loci and crop performance under abiotics stress: where do we stand? Plant Physiol. 2008;147:469–486. doi: 10.1104/pp.108.118117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frahm MA, Rosas JC, Mayek-Perez N, Lopez-Salinas E, Acosta-Gallegos JA, Kelly JD. Breeding beans for resistance to terminal drought in the lowland tropics. Euphytica. 2004;136:223–232. doi: 10.1023/B:euph.0000030671.03694.bb. [DOI] [Google Scholar]
- Frei A, Blair MW, Cardona C, Beebe SE, Gu H, Dorn S. QTL mapping of resistance to Thrips palmi Karny in Common Bean (Phaseolus vulgaris L.) Crop Sci. 2005;45:379–387. doi: 10.2135/cropsci2005.0379. [DOI] [Google Scholar]
- Funk F, Dettinger MD, Michaelsen JC, Verdin JP, Brown ME, Barlow M, Hoell A. Warming of the Indian Ocean threatens eastern and southern African food security but could be mitigated by agricultural development. Proc Natl Acad Sci USA. 2008;105:11081–11086. doi: 10.1073/pnas.0708196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitán E, Duque MC, Edwards KJ, Thome J. Microsatellite repeats in common bean (Phaseolus vulgaris): isolation, characterization, and cross-species amplification in Phaseolus ssp. Crop Sci. 2002;42:2128–2136. doi: 10.2135/cropsci2002.2128. [DOI] [Google Scholar]
- Hannah MA, Krämer KM, Geffroy V, Kopka J, Blair MW, Erban A, Vallejos CE, Heyer AG, Sanders FET, Millner PA, Pilbeam DJ. The DL gene system in common bean (Phaseolus vulgaris L.) causes programmed root death due to a shoot-derived inhibitory signal. New Phytol. 2007;176:537–549. doi: 10.1111/j.1469-8137.2007.02215.x. [DOI] [PubMed] [Google Scholar]
- Lander E, Green P, Abrahamson J, Barlow A, Daly M, Lincoln D, Newburg L. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Lynch J, White JW. Shoot nitrogren dynamics in tropical common bean. Crop Sci. 1992;32:392–397. doi: 10.2135/cropsci1992.0011183X003200020023x. [DOI] [Google Scholar]
- Métais I, Hamon B, Jalouzot R, Peltier D. Structure and level of genetic diversity in various bean types evidenced with microsatellite markers isolated from a genomic enriched library. Theor Appl Genet. 2002;104:1346–1352. doi: 10.1007/s00122-002-0901-9. [DOI] [PubMed] [Google Scholar]
- Muñoz Perea CG, Terán H, Allen RG, Wright JL, Westermann DT, Singh SP. Selection for drought resistance in dry bean landraces and cultivars. Crop Sci. 2006;46:2111–2120. doi: 10.2135/cropsci2006.01.0029. [DOI] [Google Scholar]
- Muñoz LC, Blair MW, Duque MC, Tohme J, Roca W. Introgression in common bean × tepary bean interspecific congruity-backcross lines as measured by AFLP markers. Crop Sci. 2004;44:637–645. doi: 10.2135/cropsci2004.0637. [DOI] [Google Scholar]
- Ochoa IE, Blair MW, Lynch JP. QTL analysis of adventitious root formation in common bean (Phaseolus vulgaris L.) under contrasting phosphorus availability. Crop Sci. 2006;46:1609–1621. doi: 10.2135/cropsci2005.12-0446. [DOI] [Google Scholar]
- Park SO, Coyne DP, Jung G, Skroch PW, Arnaud-Santana E, Steadman JR, Ariyarathne HM, Nienhuis J. Mapping of QTL for seed size and shape traits in common bean. J Am Soc Hort Sci. 2000;125:466–475. [Google Scholar]
- Ramírez-Vallejo P, Kelly JD. Traits related to drought resistance in common bean. Euphytica. 1998;99:127–136. doi: 10.1023/A:1018353200015. [DOI] [Google Scholar]
- Rao IM. Role of physiology in improving crop adaptation to abiotic stresses in the tropics: the case of common bean and tropical forages. In: Pessarakli M, editor. Handbook of plant and crop physiology. New York, USA: Marcel Dekker, Inc.; 2001. pp. 583–613. [Google Scholar]
- Schneider KA, Rosales-Serna R, Ibarra-Perez F, Cazares-Enriquez B, Acosta-Gallego JA, Ramirez-Vallejo P, Wassimi N, Kelly JD. Improving common bean performance under drought stress. Crop Sci. 1997;37:43–50. doi: 10.2135/cropsci1997.0011183X003700010007x. [DOI] [Google Scholar]
- Schneider KA, Brothers ME, Kelly JF. Marker-assisted selection to improve drought resistance in common bean. Crop Sci. 1997;37:51–60. doi: 10.2135/cropsci1997.0011183X003700010008x. [DOI] [Google Scholar]
- Singh SP. Selection for water-stress tolerance in interracial populations of common bean. Crop Sci. 1995;35:118–124. doi: 10.2135/cropsci1995.0011183X003500010022x. [DOI] [Google Scholar]
- Singh SP, Terán H, Gutierrez JA. Registration of SEA5 and SEA13 drought tolerant dry bean germplasm. Crop Sci. 2001;41:276–277. doi: 10.2135/cropsci2001.411276x. [DOI] [Google Scholar]
- Sponchiado BN, White JW, Castillo JA, Jones PG. Root growth of four common bean cultivars in relation to drought tolerance in environments with contrasting soil types. Exp Agric. 1989;25:249–257. doi: 10.1017/S0014479700016756. [DOI] [Google Scholar]
- Subbarao GV, Johansen C, Sinkard AE, Rao RCN, Saxena NP, Chauhan YS. Strategies for improving drought resistance in grain legumes. Crit Rev Plant Sci. 1995;14:469–523. [Google Scholar]
- Taiz L, Zeiger E (2006) Water and Plant Cells, Plant Physiology, 4th edn. Sinauer Associates Inc, p 672
- Tar’an B, Michaels TE, Pauls KP. Genetic mapping of agronomic traits in common bean. Crop Sci. 2002;42:544–556. doi: 10.2135/cropsci2002.0544. [DOI] [Google Scholar]
- Terán H, Singh SP. Comparison of sources and lines selected for drought resistance in common bean. Crop Sci. 2002;42:64–70. doi: 10.2135/cropsci2002.0064. [DOI] [PubMed] [Google Scholar]
- Thung M, Rao IM. Integrated management of abiotic stresses. In: Singh SP, editor. Common bean improvement in the twenty-first century. Dordrecht: Kluwer Academic Publishers; 1999. pp. 331–370. [Google Scholar]
- Vales MI, Schön CC, Capettini F, Chen XM, Corey AE, Mather DE, Mundt CC, Richardson KL, Sandoval-Islas JS, Utz HF, Hayes PM. Effect of population size on the estimation of QTL. Theor Appl Genet. 2005;111:1260–1270. doi: 10.1007/s00122-005-0043-y. [DOI] [PubMed] [Google Scholar]
- White JW, Castillo JA. Relative effect of root and shoot genotypes on yield of common bean under drought stress. Crop Sci. 1989;29:360–362. doi: 10.2135/cropsci1989.0011183X002900020026x. [DOI] [Google Scholar]
- White JW, Izquierdo J. Physiology of yield potential and stress tolerance. In: van Schoonhoven A, Voysest O, editors. Common beans: research for crop improvement. Cali: C.A.B. International/CIAT; 1991. pp. 287–382. [Google Scholar]
- White JW, Singh SP. Breeding for adaptation to drought. In: van Schoonhoven A, Voysest O, editors. Common beans: research for crop improvement. Wallingford, Cali: CAB International, CIAT; 1991. pp. 501–560. [Google Scholar]
- White JW, Castillo JA, Ehleringer JR, Garcia-C JA, Singh SP. Relations of carbon isotope discrimination and other physiological traits to yield in common bean (Phaseolus vulgaris) under rainfed conditions. J Agric Sci (Camb) 1994;122:275–284. doi: 10.1017/S0021859600087463. [DOI] [Google Scholar]
- White JW, Ochoa R, Ibarra F, Singh SP. Inheritance of seed yield, maturity and seed weight of common bean (Phaseolus vulgaris) under semi-arid rainfed conditions. J Agric Sci (Cambridge) 1994;122:265–273. doi: 10.1017/S0021859600087451. [DOI] [Google Scholar]
- Wortmann CA, Kirkby RA, Eledu CA, Allen DJ. Atlas of common bean (Phaseolus vulgaris L.) production in Africa. Cali: CIAT; 1998. [Google Scholar]