Abstract
Rationale
The degradation of proteins by the ubiquitin proteasome system (UPS) is required for the maintenance of cellular homeostasis in the heart. An important regulator of metabolic homeostasis is AMP-activated protein kinase (AMPK). AMPK activation inhibits protein synthesis and activates autophagy, but whether AMPK plays a role in regulating protein breakdown through the UPS in the heart is not known.
Objective
To determine whether AMPK enhances UPS-mediated protein degradation by directly regulating the ubiquitin ligases Atrogin-1 and MuRF1 in the heart.
Methods and Results
Nutrient deprivation, pharmacologic or genetic activation of AMPK increased mRNA expression and protein levels of Atrogin-1 and MuRF1, and consequently enhanced protein degradation in neonatal cardiomyocytes. Inhibition of AMPK abrogated these effects. Using gene reporter and chromatin immunoprecipitation assays we found that AMPK regulates MuRF1 expression by acting through the transcription factor MEF2. We further validated these findings in vivo using MEF2-LacZ reporter mice. Furthermore, we demonstrated in adult cardiomoycytes that MuRF1 is necessary for AMPK-mediated proteolysis through the UPS in the heart. Consequently, MuRF1 knockout mice were protected from severe cardiac dysfunction during fasting.
Conclusions
AMPK regulates the transcription of Atrogin-1 and MuRF1 and enhances UPS-mediated protein degradation in heart. Specifically, AMPK regulates MuRF1 through the transcription factor MEF2. The absence of MuRF1 in the heart preserves cardiac function during fasting. The results strengthen the hypothesis that AMPK serves as a modulator of intracellular protein degradation in the heart.
Keywords: AMPK, protein degradation, ubiquitin ligases, transcriptional regulation
Introduction
The heart adapts metabolically, functionally and structurally to changes in its environment. Any stress that reduces the intracellular [ATP]:[AMP] ratio activates the enzyme AMP-activated protein kinase (AMPK), resulting in an increased provision of energy through substrate metabolism and inhibition of energy consuming processes,1 including protein synthesis.2, 3 A role for AMPK in protein degradation in the heart has not been clearly defined, but recent reports have suggested a possible role for AMPK in protein degradation in skeletal muscle.4, 5
Intracellular protein degradation in cardiomyocytes is controlled by independent but interrelated processes: ubiquitin proteasome system (UPS)-mediated proteolysis and autophagy. While macroautophagy can degrade whole organelles,6, 7 individual proteins are degraded through the UPS.8 Ubiquitin ligases confer specificity to the system by the selective ubiquitination of target proteins which are then degraded by the proteasome. Two muscle-specific ubiquitin ligases, muscle atrophy F-box protein (MAFbx) or Atrogin-1, and muscle RING finger protein 1 (MuRF1), are consistently increased in models of skeletal muscle atrophy. Furthermore, mice lacking MAFbx/Atrogin-1 or MuRF1 subjected to atrophic stimuli show reduced levels of skeletal muscle atrophy.9, 10 Both ligases also play a critical role in regulating cardiomyocyte size and heart muscle mass. The overexpression of MAFbx/Atrogin-1 in the heart blunts the development of cardiac hypertrophy in response to both physiologic and pathologic hypertrophic stimuli in vivo.11, 12 Vice versa MuRF1 deficient mice display exaggerated cardiac hypertrophy in response to pressure overload,13 while the overexpression of MuRF1 in cardiomyocytes prevents pharmacologically-induced hypertrophy.14 Collectively these studies suggest that the proteins targeted for degradation by MAFbx/Atrogin-1 and MuRF1 are determinants of cardiomyocyte size.
Early studies in the heart in vitro and in vivo demonstrated that nutrient deprivation decreases protein synthesis and increases fractional rates of protein degradation.15, 16 AMPK is activated during nutrient deprivation in order to provide energy to maintain normal cellular function.1 Although it has recently been reported that nutrient deprivation induces autophagy in cardiomyocytes through AMPK,17 a role of AMPK in the cardiac UPS has never been considered. We have previously proposed that metabolic signals may trigger functional and structural remodeling of the stressed heart,18 therefore we set out to test the hypothesis that AMPK regulates MAFbx/Atrogin-1 and MuRF1 in the heart.
Methods
An expanded methods section is available in the data supplement.
Statistical Analysis
Results are expressed as means ± SEM. Analysis was performed using two-tailed, unpaired Student’s t test or one-way ANOVA with Turkey post hoc test. A value of P<0.05 was considered significant.
Results
Nutrient deprivation upregulates markers of protein degradation in cardiomyocytes
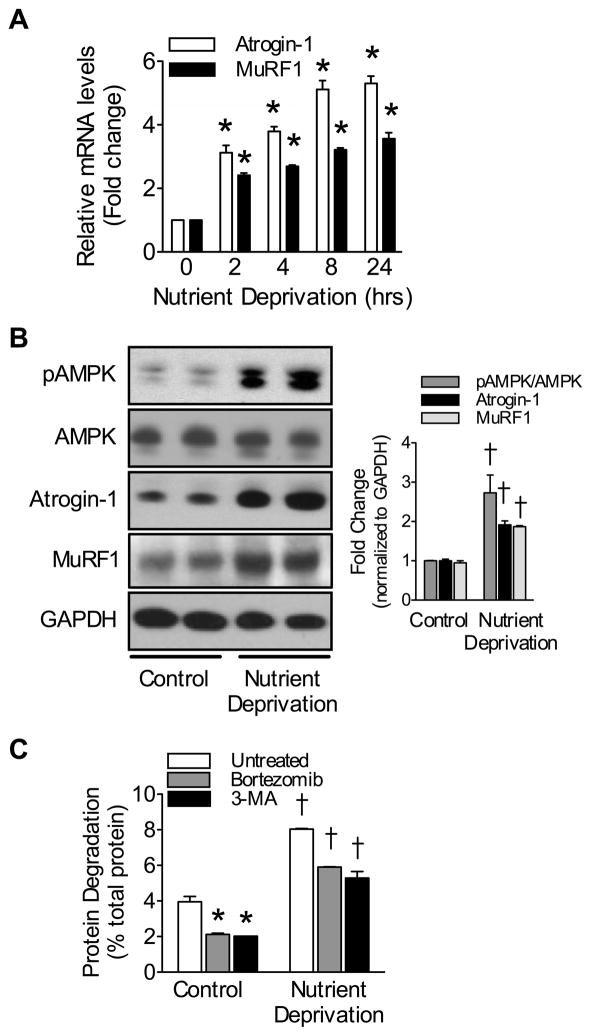
Protein degradation is increased in the heart in response to nutrient deprivation, leading to cardiac atrophy;15, 16 however, the mechanism(s) regulating this process are not entirely known. Molecular mechanisms of skeletal muscle wasting have been investigated before in cultured myoblasts deprived of nutrients.19 Under these conditions, cell size is drastically reduced and molecular markers of atrophy (i.e. atrogenes, including ubiquitin ligases MAFbx/Atrogin-1 and MuRF1) are strongly induced.20 Akin to myoblasts, we found that nutrient deprivation increased the mRNA expression of both MAFbx/Atrogin-1 and MuRF1 in neonatal cardiomyocytes in a time-dependent manner (Figure 1A). The same was the case with glucose deprivation (data not shown). After 24 hours of nutrient deprivation, the protein levels of MAFbx/Atrogin-1 and MuRF1 were similarly increased, which correlated with increased AMPK activation (Figure 1B). Furthermore, in cardiomyocytes protein degradation was significantly increased during nutrient deprivation (Figure 1C). Treatment with either Bortezomib or 3-methyladenine decreased protein degradation in neonatal cardiomyocytes indicating involvement of both the UPS and macroautophagy, respectively. The effect of either inhibitor was incomplete, however, suggesting that both processes regulate protein degradation in the presence and absence of nutrients. Assessment of protein degradation with simultaneous inhibition of the UPS and autophagy was not possible as it resulted in cell death. These data indicate that, akin to autophagy,17 the UPS also plays an important role in regulating protein degradation during nutrient deprivation.
Figure 1.
Nutrient deprivation increases expression of ubiquitin ligases and enhances protein degradation.
(A) Atrogin-1 and MuRF1 mRNA expression in nutrient deprived NRVM. (B) Relative protein levels and quantification in NRVM after 24 hours of nutrient deprivation. (C) Protein degradation in NRVM after 24 hours of nutrient deprivation with 1μmol/L Bortezomib or 10μmol/L 3-methyladenine treatment. Data are mean ± SEM of 3 independent experiments performed in triplicate. *P< 0.01 vs control or untreated, †P< 0.01 vs control or complete nutrients.
AMPK regulates the expression of ubiquitin ligases in vitro and in vivo
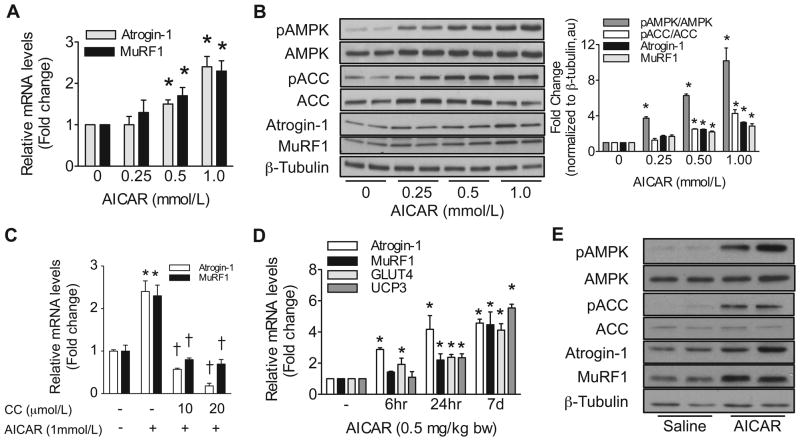
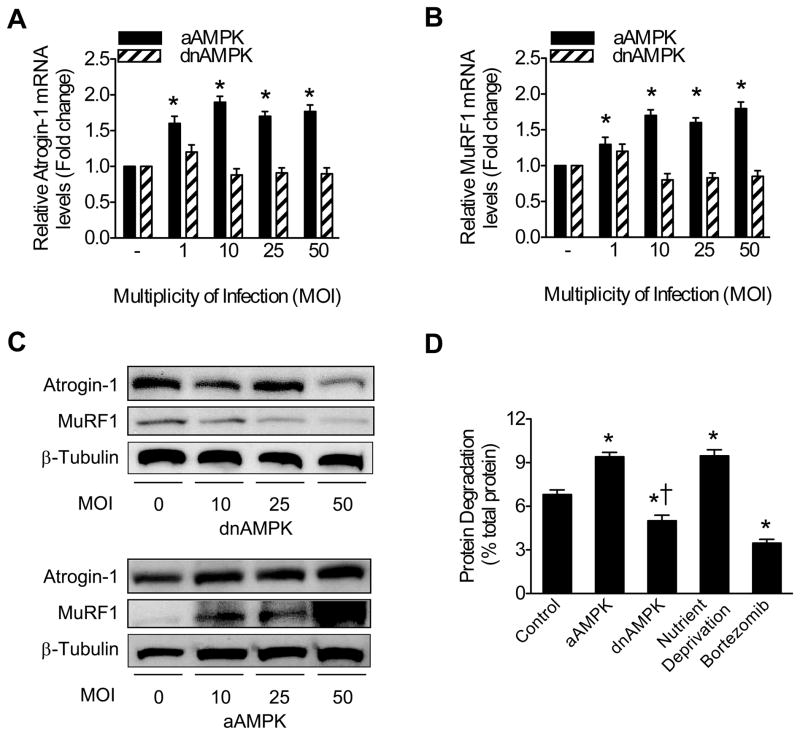
We investigated the effect of direct AMPK activation and inhibition on MAFbx/Atrogin-1 and MuRF1 expression in cardiomyocytes. AICAR (5-Aminoimidazole-4-carboxamide-1-β-D-ribofuranoside) is a known pharmacological activator of AMPK and is readily taken up by cardiomyocytes. AICAR treatment increased MAFbx/Atrogin-1 and MuRF1 mRNA expression and protein levels in a dose-dependent manner (Figure 2A and B) while Compound C, a potent AMPK inhibitor, decreased MAFbx/Atrogin-1 and MuRF1 expression (Figure 2C). The phosphorylation status of acetyl-CoA carboxylase (ACC), a direct target of AMPK, was monitored as a marker of AMPK activity. Intraperitoneal (IP) injection of AICAR in mice is also sufficient to activate AMPK in vivo.21, 22 Either acute or chronic AMPK activation increased MAFbx/Atrogin-1 and MuRF1 mRNA levels in the heart in vivo (Figure 2D). Additionally, two known targets of AMPK in skeletal muscle, GLUT423 and UCP3,24 were also increased in the heart with AMPK activation (Figure 2D). To establish the specificity of the observed effects, we infected neonatal cardiomyocytes with adenoviral constructs to express either active AMPK (aAMPK) or dominant negative AMPK (dnAMPK). Increasing aAMPK MOI resulted in an upregulation of the mRNA expression and protein levels of MAFbx/Atrogin-1 and MuRF1, while dnAMPK had no significant effect on mRNA expression but slightly decreased protein levels at an MOI of 50 (Figure 3A–C).
Figure 2. AMPK activation increases expression of ubiquitin ligases in vitro and in vivo.
(A) Atrogin-1 and MuRF1 mRNA expression and (B) relative protein levels and quantification in NRVM after 24 hours of AMPK activation with AICAR treatment. (C) Atrogin-1 and MuRF1 mRNA expression in NRVM after 24 hours of AMPK activation and inhibition. (D) AMPK activation in vivo with AICAR treatment to increase Atrogin-1, MuRF1, GLUT4, and UCP3 mRNA expression in mouse heart after either acute (6 or 24 hours after injection) or chronic (one injection per day for 7 days) AICAR treatment (n=5–8, 0.5 mg/kg body weight (bw) AICAR). (E) Relative protein levels in mouse heart after 7 days of in vivo AICAR treatment. The in vitro data are mean ± SEM of 3 independent experiments, each performed in triplicate. *P< 0.01 vs untreated or saline, †P< 0.01 vs AICAR.
Figure 3. AMPK regulates expression of ubiquitin ligases and protein degradation in cardiomyocytes.
(A) Atrogin-1 and (B) MuRF1 mRNA expression in NRVM 24 hours after adenoviral transduction of active (aAMPK) or dominant negative (dnAMPK) AMPK. (C) Atrogin-1 and MuRF1 protein levels in response to dnAMPK or aAMPK in NRVM 24 hours after adenoviral transduction. (D) Protein degradation in the presence of aAMPK or dnAMPK (MOI=10, 1μmol/L Bortezomib). Data are mean ± SEM of 3 independent experiments performed in triplicate. *P< 0.01 vs control, †P< 0.01 vs aAMPK.
We next investigated the consequence of AMPK activation and inhibition on proteasome-mediated protein degradation in cardiomyocytes. Analogous to the effects of nutrient deprivation, active AMPK increased protein degradation, which was suppressed by dnAMPK or by the proteasome inhibitor Bortezomib (Figure 3D). These data establish that AMPK is directly involved in cardiomyocyte remodeling, both metabolically and structurally, by inhibiting protein synthesis2, 3 and, as shown here, by enhancing protein degradation.
AMPK activation regulates the expression of MuRF1 in vitro through the transcription factor MEF2
To gain an understanding of how AMPK regulates the expression of MAFbx/Atrogin-1 and MuRF1 we performed in silico promoter analyses on both genes. These studies revealed a MEF2 (myocyte enhancer factor 2) consensus binding sequence25, 26 upstream of the MuRF1 transcriptional start site (Figure 4A). Several reports have already shown that AICAR activation of AMPK induces MEF2 transcriptional activity in muscle,27, 28 and we found the same to occur in neonatal cardiomyocytes. To assess MEF2 transcriptional activity in nuclear extracts from neonatal cardiomyocytes, we used the MEF2 TransAM® assay. This ELISA-based method detects the binding of proteins within nuclear extracts to immobilized double stranded oligonucleotides containing MEF2 binding sites.29 We found that pharmacological or genetic activation of AMPK enhanced MEF2 transcriptional activity, while inhibition of AMPK decreased this effect (Figure 4B). To investigate whether MuRF1 transcription is regulated by AMPK through MEF2, we conducted luciferase reporter gene assays. Vectors encoding the MuRF1 promoter, including the endogenous or mutated MEF2 binding site (Figure 4C), were cotransfected in H9c2 cells with a vector encoding β-galactosidase as an internal transfection control. Luciferase activity was increased by nutrient deprivation, AICAR treatment, or by transfection with aAMPK, and conversely decreased by treatment with Compound C or by transfection with dnAMPK. Furthermore, MuRF1 transcription, regulated by AMPK activation, was abolished when the MEF2 binding site on the MuRF1 promoter was mutated (Figure 4D). These results indicate that AMPK promotes MEF2 association with the putative MEF2 binding site in the MuRF1 proximal promoter. To confirm this, we performed chromatin immunoprecipitation assays for endogenous MEF2 on extracts from cardiomyocytes infected with empty adenovirus or adenovirus expressing aAMPK or dnAMPK. In agreement with our other results, we recovered the MuRF1 proximal promoter in MEF2 immunoprecipitates from myocytes expressing aAMPK, but not myocytes expressing the dnAMPK or empty virus control. In contrast, distal promoter sequences could not be amplified in MEF2 immunoprecipitates (Figure 4E). These data demonstrate that MuRF1 is transcriptionally regulated by AMPK through the transcription factor MEF2.
Figure 4. MuRF1 transcription is regulated by an AMPK-MEF2 dependent mechanism in vitro.
(A) MEF2 consensus binding site and potential MEF2 binding site on MuRF1. (B) MEF2 transcriptional activity following activation or inhibition of AMPK in NRVM (1mmol/L AICAR, 20μmol/L Compound C (CC), aAMPK and dnAMPK MOI=10). (C) MuRF1 promoter luciferase reporter vectors. (D) Luciferase activity of the constructs transfected into H9c2 cells. (E) MEF2 chromatin immunoprecipitation. Cardiomyocytes were infected with an empty adenovirus or adenovirus expressing either aAMPK or dnAMPK. PCR assays on input and IP fractions amplified the MuRF1 promoter containing the putative MEF2 site (−191 to −87, top panel) or a distal region of the MuRF1 promoter (−2094 to −1937, bottom panel). Data are mean ± SEM of 3 independent experiments performed in triplicate. *P< 0.01 vs control, †P<0.01 vs AICAR or aAMPK, ‡P< 0.01 vs MuRF1 promoter.
MEF2 transcriptional activity and MuRF1 transcription are regulated by AMPK in the heart
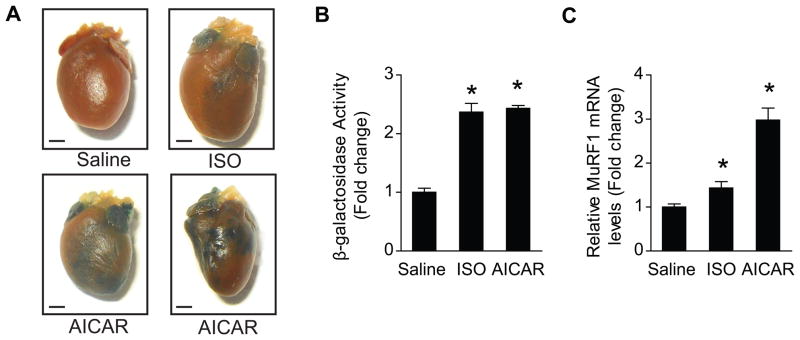
To validate the role of AMPK in MEF2-regulated cardiac transcription of MURF1 in vivo we treated MEF2-lacZ reporter mice30 with AICAR. Consistent with our findings in isolated cardiomyocytes, cardiac MEF2 transcriptional activity was increased in response to AICAR treatment, as evidenced by increased LacZ staining (Figure 5A). We then quantified LacZ staining in cardiac protein extracts using a β-galactosidase activity assay. AICAR treatment significantly increased β-galactosidase activity to the same extent as isoproterenol, used as a positive control31 (Figure 5B). Increased MEF2 transcriptional activity in response to AMPK activation augmented MuRF1 expression in vivo (Figure 5C). Collectively, the results demonstrate that AMPK regulates MEF2-mediated transcription of MuRF1 in vivo.
Figure 5. MEF2 transcriptional activity and MuRF1 transcription are regulated by AMPK in vivo.
(A) MEF2 transcriptional activity, denoted as β-galactosidase staining, after 7 days of AMPK activation in whole hearts and (B) in heart protein extracts. (C) MuRF1 expression in the heart in vivo (15μg/g bw Isoproterenol used as a positive control (ISO), 0.5mg/g bw AICAR, n=6–8). In vitro data are mean ± SEM of 3 independent experiments performed in triplicate. Scale bars=1mm. *P< 0.01 vs saline.
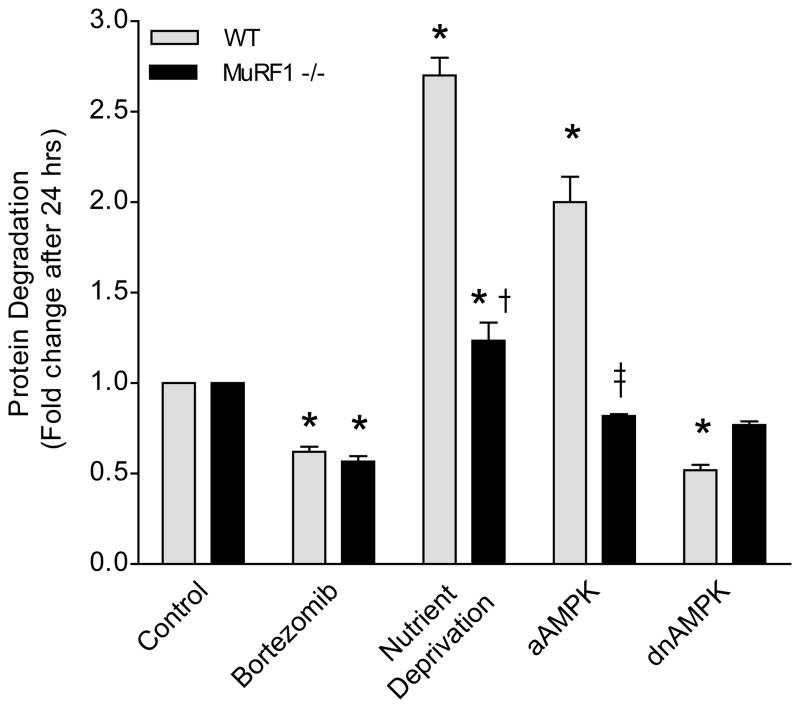
To determine the consequence of AMPK-regulated MuRF1 we investigated whether AMPK-mediated protein degradation through the UPS requires MuRF1. We isolated adult mouse cardiomyocytes from wild type (WT) or MuRF1 knockout hearts and measured rates of protein degradation in vitro (Figure 6). Proteasome-mediated protein degradation was not altered in the absence of MuRF1 under normal conditions, as WT and MuRF1 −/− cardiomyocytes were equally responsive to Bortezomib treatment. Similar to neonatal cardiomyocytes, nutrient deprivation increased protein degradation in WT adult cardiomyocytes. Under the same conditions, increases in protein degradation were less marked in MuRF1 deficient cardiomyocytes, but were still higher than in controls. When active AMPK was expressed, protein degradation was greatly enhanced in WT myocytes, but it was not increased in the absence of MuRF1. When dnAMPK was expressed, protein degradation was decreased in WT and unchanged in MuRF1 deficient myocytes. Collectively, these data show that MuRF1 is required for AMPK-regulated protein degradation in cardiomyocytes.
Figure 6. MuRF1 is required for AMPK-regulated protein degradation in cardiomyocytes.
Protein degradation in adult mouse cardiomyoytes isolated from wild type (WT) or MuRF1 knockout mouse hearts 24 hours after treatment (1mmol/L Bortezomib) or adenoviral transduction (MOI=1). Data are mean ± SEM of 3 independent experiments performed in duplicate. *P< 0.01 vs control, †P< 0.01 vs Nutrient Deprivation, ‡P< 0.01 vs aAMPK.
In order to investigate the physiological importance of the AMPK-MuRF1 axis in vivo we subjected wild type (WT) and MuRF1 knockout (−/−) mice to three days of nutrient deprivation (starvation or fasting). As expected, 3 days of fasting significantly decreased body weight in both WT and MuRF1 −/− mice, however heart weight was decreased only in WT mice (Figure 7A). Interestingly, ejection fraction (EF) was decreased in WT mice in response to fasting, but loss of MuRF1 preserved EF (Figure 7B). Fractional shortening was also decreased after fasting in WT, but not MuRF1 −/− animals (Supplemental Table 1). MuRF1 gene expression was significantly increased in WT starved mice (Figure 7C). We also investigated two known protein targets of MuRF1 in the heart after 3 days of fasting. Indeed the protein levels of both cardiac myosin-binding protein C (cMyBP-C)32 and cardiac troponin I (TnI)33 decreased in WT fasted mice (Figure 7D and E). MyBP-C is also degraded by Atrogin-1, and therefore it is not surprising that protein levels also trended to decrease in hearts from fasted MuRF1 −/− mice. TnI levels, on the other hand were not significantly decreased in fasted MuRF1 −/− hearts, suggesting that degradation of cardiac TnI during fasting requires MuRF1. Together, these data suggest that AMPK-regulated MuRF1 in the heart during fasting is detrimental to structure and function by enhancing the degradation of specific MuRF1 targets in the heart. The proposed MuRF1 transcriptional regulation through the AMPK-MEF2 regulatory axis is summarized in Figure 8.
Figure 7. Absence of MuRF1 preserves cardiovascular function during nutrient deprivation.
(A) Heart weight, body weight, heart weight to body weight ratios, (B) ejection fraction, (C) MuRF1 expression, (D) relative protein levels and (E) protein quantification in wild type (WT) or MuRF1 −/− hearts after 3 days of fasting (n=6–8). *P< 0.01 vs Fed, † P< 0.01 vs WT Fasted, ‡P< 0.01 vs MuRF1 −/− Fed.
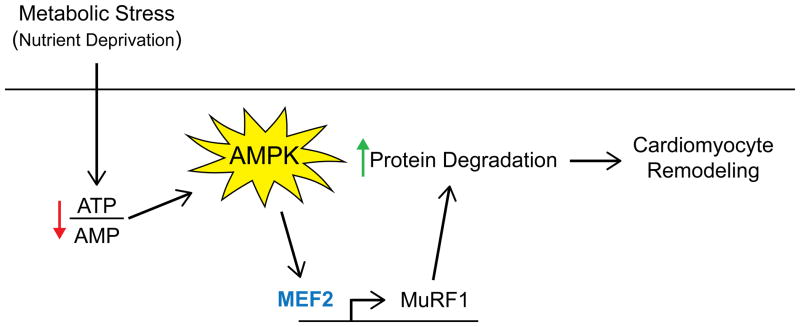
Figure 8. AMPK regulates MuRF1 transcription in a MEF2-dependent manner.
AMPK regulates MuRF1 expression through the transcription factor MEF2. This leads to increased protein degradation in the cardiomyocyte and increased remodeling.
Discussion
We have presented evidence in support of the hypothesis that AMP-activated protein kinase regulates ubiquitin ligases in the rodent heart. The present work extends the long established concept of the “dynamic state of body constituents”34 to a specific situation when the heart adapts to changes in its metabolic environment. Protein turnover (protein synthesis and degradation through the UPS and autophagy) constitutes a major line of defense for protein quality control of the cardiomyocyte35 and is a major mechanism of adaptation in the heart. It is therefore of interest to understand how protein degradation is regulated in the cardiomyocyte under various circumstances. We have previously shown that markers of the UPS are upregrulated in the heart in several settings of cardiac remodeling,36, 37 but how the markers themselves are regulated is not clear. It is known already that AMPK plays a role in cellular homeostasis in part by inhibiting the mTOR pathway2, 3 and thus by decreasing protein synthesis, while at the same time AMPK activates autophagy.17, 38, 39 While AMPK itself has been found to be regulated by the UPS,40 whether AMPK regulates protein degradation through the UPS has not been investigated in the heart until now.
The role of AMPK in fuel homeostasis is well described and studies have predominantly focused on the effects of AMPK activation on energy substrate metabolism.41 AMPKα2, the active subunit highly expressed in the heart,42 is preferentially localized to the nucleus.43 Hence, it is not surprising that AMPK also transcriptionally regulates metabolic gene expression. Although little is known about AMPK-regulated transcription in the heart, earlier reports in liver show that AMPK activation represses transcription by inactivating the transcription factors p300, HNF4-α, ChREBP, and TORC2.44 Consequently, the expression of genes involved in lipogenesis and gluconeogenesis is attenuated. AMPK can also activate transcription by enhancing CREB activity, thus increasing the expression of UCP3 and HKII.24, 45 The activation of PGC1α by AMPK leads to increased mitochondrial gene expression and mitochondrial biogenesis.23 Additionally, the activation of AMPK in muscle increases GLUT4 transcription by increasing both PGC1α23 and MEF2 transcriptional activity, the latter through inactivation of HDAC5.46 The role of AMPK in transcription is only now coming into focus. Akin to its yeast homologue SNF-1,47 AMPK phosphorylates histone 2B in mammalian cells48, 49 suggesting that AMPK regulates entire transcriptional programs, and not only transcription of individual genes. By providing evidence that AMPK regulates the transcription of ubiquitin ligases MAFbx/Atrogin-1 and MuRF1, key regulators of protein degradation in the heart, this study further expands the role of AMPK in both cellular homeostasis and transcriptional regulation in the heart.
Extensive analyses of the MAFbx/Atrogin-1 and MuRF1 promoters have not yet been reported, but independent studies have begun to elucidate the transcriptional regulation of both ligases. The expression of MAFbx/Atrogin-1 and MuRF1 is positively regulated by the transcription factor FoxO3A in the heart, and is negatively regulated through Akt which suppresses FoxO.50–52 TNFα increases the expression of MAFbx/Atrogin-1 and MuRF1 in cardiomyocytes53 and in skeletal muscle54 independent of Akt through Foxo4.55 In C2C12 myotubes56 and in vivo57 MAFbx/Atrogin-1 and MuRF1 expression is increased by glucocorticoid stimulation, and the transcription factor C/EBP1 has been suggested to regulate MAFbx/Atrogin-1 transcription in a glucocorticoid-dependent manner in skeletal muscle.58 The IκKβ/NF-κB pathway also regulates the transcription of MuRF1 in cachexia-induced muscle wasting.59 More recently myogenin has been found to regulate transcription of both MAFbx/Atrogin-1 and MuRF1 in an HDAC-dependent manner.60 Our findings now demonstrate that, in the heart, AMPK contributes to the complex transcriptional regulation of ubiquitin ligases in the setting of nutrient deprivation and fasting.
The duration of fasting may be important, because we observed that prolonged fasting results in impaired contractile function of the heart (Figure 7). Our findings are in agreement with several reports showing that fasting decreased cardiac function in WT mice on the one hand,61 and that the absence of MuRF1 spared muscle and heart from atrophy on the other hand.62 MuRF1 targets EEF1G (a component of the elongation factor complex EF-1) for degradation.63 It also targets key enzymes involved in ATP production (including aldolase a and pyruvate dehydrogenase).64 These studies suggest that MuRF1 not only regulates protein degradation, but also regulates protein synthesis and pathways of energy metabolism. Based on our in vivo findings, we now propose that during starvation, the absence of MuRF1 is cardio-protective through several different mechanisms. First, MuRF1-independent mechanisms regulate protein degradation but protein synthesis is not inhibited by MuRF1. Conversely, in WT hearts of fasted animals the presence of MuRF1 is reflected in an imbalance of protein turnover (enhanced protein degradation and decreased protein synthesis).62 Furthermore, the upregulation of MuRF1 during starvation may degrade metabolic enzymes (like aldolase a and the pyruvate dehydrogenase complex). Others have found that with fasting, intracellular glucose 6-phosphate and fructose 6-phosphate in the heart are increased, and glucose phosphorylation is decreased, most likely by allosteric inhibition of hexokinase.65 Glycogen deposition and citrate levels are also increased in the heart during fasting; the latter inhibits phosphofructokinase activity and therefore puts a brake on glycolysis. 66, 67 It is tempting to speculate that MuRF1 interacts with phosphofructokinase because in muscle atrophy, MuRF1 is upregulated and phosphofructokinase is downregulated,9 and both enzymes can localize to the M-line.68, 69 Therefore, it is possible that upregulation of MuRF1 during starvation leads to the downregulation of metabolic enzymes, decreased ATP production from glycolysis, and decreased cardiac function. Indeed we observed that rates of glucose oxidation in hearts from MuRF1 −/− animals are at least three-fold higher than in the hearts from WT animals (Baskin KK and Taegtmeyer H, unpublished results).
A further point needs to be addressed. Beginning in 2002 a number of clinical studies have reported beneficial outcomes in diabetic heart failure patients treated with the AMPK activator metformin.70–73 Our experimental findings are not inconsistent with the clinical outcomes because in diabetes, in contrast to fasting, the heart is flooded with oxidizable fuel. AMPK-regulated protein degradation may be protective because of enhanced protein quality control.35 We conclude that AMPK is a transcriptional regulator of ubiquitin ligases in heart muscle. Activation of AMPK results in increased rates of protein degradation, and consequently leads to cardiomyocyte remodeling. Whether the remodeling is beneficial or detrimental may be dependent on the immediate cardiometabolic environment. We speculate that the activation of AMPK results in enhanced availability of intracellular amino acids for either ATP production or the synthesis of new proteins as the heart adapts to a new physiologic state.
Supplementary Material
Novelty and Significance.
What Is Known?
AMP-activated protein kinase (AMPK), a key regulator of metabolic homeostasis, inhibits protein synthesis and activates autophagy in the heart.
The ubiquitin proteasome system (UPS) maintains cellular homeostasis by degrading unnecessary and/or damaged proteins through key enzymes, including ubiquitin (E3) ligases.
Two muscle specific E3 ligases, Atrogin-1 and MuRF1, are critical regulators of cardiac size and mass.
What New Information Does This Article Contribute?
Activation of AMPK in vitro and in vivo regulates the transcription of Atrogin-1 and MuRF1 in cardiomyocytes.
AMPK regulates MuRF1 transcription through the transcription factor MEF2.
MuRF1 is necessary for AMPK-mediated proteolysis through the UPS in the heart.
MuRF1 deficient mice are protected from cardiac dysfunction during the metabolic stress of fasting.
The heart adapts both metabolically and structurally to changes in its environment. AMPK is an essential enzyme that regulates many adaptive processes. Not only does AMPK inhibit protein synthesis, but it also activates autophagy, lysosome-mediated protein degradation. Until now, the role of AMPK in proteasome-mediated protein degradation in the heart was not known. We show here that activation of AMPK regulates the transcription of two ubiquitin ligases in the heart: Atrogin-1 and MuRF1. Specifically, AMPK regulates MuRF1 transcription through MEF2 in vitro and in vivo. Consequently, proteasome-mediated protein degradation is increased with AMPK activation. In cardiomyocytes, MuRF1 is necessary for AMPK-mediated proteolysis through the UPS. Excessive proteolysis, which can occur with long-term fasting, induces cardiac dysfunction. However, MuRF1 deficient mice are protected from cardiac dysfunction during fasting. Regulation of protein turnover is especially important in the terminally differentiated cardiomyocyte, where protein quality control is required to maintain normal contractile function. Excessive protein synthesis or degradation has physiological consequences. Therefore it seems important to understand, in detail, how these processes are regulated.
Acknowledgments
We thank Eric N. Olson and Rhonda Bassel-Duby for providing the desMEF2-LacZ reporter mice and Regeneron Pharmaceuticals, INC. for providing the MuRF1 knockout mice. We also thank Meredith Rees for performing the echocardiographic measurements and Rebecca Berdeaux for assistance with the ChIP assays and careful review of the manuscript. Peter Razeghi and Romain Harmancey provided critical comments and suggestions, and Roxy A. Tate helped with the manuscript preparation.
Sources of Funding
These studies were supported in part, by a grant from the National Heart, Lung, and Blood Institute (5R01HL061483-9) of the US Public Health Service. K.K.B. received a predoctoral fellowship from the American Heart Association, National Center (11PRE5200006).
Non-standard Abbreviations and Acronyms
- aAMPK
active AMPK adenovirus
- dnAPMK
dominant negative AMPK adenovirus
- ACC
acetyl-CoA carboxylase
- ISO
isoproterenol
- CC
Compound C
- 3-MA
3-methyladenine
Footnotes
Disclosures
None.
References
- 1.Kim AS, Miller EJ, Young LH. Amp-activated protein kinase: A core signalling pathway in the heart. Acta Physiol (Oxf) 2009;196:37–53. doi: 10.1111/j.1748-1716.2009.01978.x. [DOI] [PubMed] [Google Scholar]
- 2.Chan AY, Soltys CL, Young ME, Proud CG, Dyck JR. Activation of amp-activated protein kinase inhibits protein synthesis associated with hypertrophy in the cardiac myocyte. J Biol Chem. 2004;279:32771–32779. doi: 10.1074/jbc.M403528200. [DOI] [PubMed] [Google Scholar]
- 3.Bolster DR, Crozier SJ, Kimball SR, Jefferson LS. Amp-activated protein kinase suppresses protein synthesis in rat skeletal muscle through down-regulated mammalian target of rapamycin (mtor) signaling. J Biol Chem. 2002;277:23977–23980. doi: 10.1074/jbc.C200171200. [DOI] [PubMed] [Google Scholar]
- 4.Krawiec BJ, Nystrom GJ, Frost RA, Jefferson LS, Lang CH. Amp-activated protein kinase agonists increase mrna content of the muscle-specific ubiquitin ligases mafbx and murf1 in c2c12 cells. Am J Physiol Endocrinol Metab. 2007;292:E1555–1567. doi: 10.1152/ajpendo.00622.2006. [DOI] [PubMed] [Google Scholar]
- 5.Nakashima K, Yakabe Y. Ampk activation stimulates myofibrillar protein degradation and expression of atrophy-related ubiquitin ligases by increasing foxo transcription factors in c2c12 myotubes. Biosci Biotechnol Biochem. 2007;71:1650–1656. doi: 10.1271/bbb.70057. [DOI] [PubMed] [Google Scholar]
- 6.Zheng Q, Li J, Wang X. Interplay between the ubiquitin-proteasome system and autophagy in proteinopathies. Int J Physiol Pathophysiol Pharmacol. 2009;1:127–142. [PMC free article] [PubMed] [Google Scholar]
- 7.Kundu M, Thompson CB. Macroautophagy versus mitochondrial autophagy: A question of fate? Cell Death Differ. 2005;12 (Suppl 2):1484–1489. doi: 10.1038/sj.cdd.4401780. [DOI] [PubMed] [Google Scholar]
- 8.Willis MS, Townley-Tilson WH, Kang EY, Homeister JW, Patterson C. Sent to destroy: The ubiquitin proteasome system regulates cell signaling and protein quality control in cardiovascular development and disease. Circ Res. 2010;106:463–478. doi: 10.1161/CIRCRESAHA.109.208801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 10.Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific f-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci U S A. 2001;98:14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li HH, Kedar V, Zhang C, McDonough H, Arya R, Wang DZ, Patterson C. Atrogin-1/muscle atrophy f-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an scf ubiquitin ligase complex. J Clin Invest. 2004;114:1058–1071. doi: 10.1172/JCI22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li HH, Willis MS, Lockyer P, Miller N, McDonough H, Glass DJ, Patterson C. Atrogin-1 inhibits akt-dependent cardiac hypertrophy in mice via ubiquitin-dependent coactivation of forkhead proteins. J Clin Invest. 2007;117:3211–3223. doi: 10.1172/JCI31757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willis MS, Ike C, Li L, Wang DZ, Glass DJ, Patterson C. Muscle ring finger 1, but not muscle ring finger 2, regulates cardiac hypertrophy in vivo. Circ Res. 2007;100:456–459. doi: 10.1161/01.RES.0000259559.48597.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arya R, Kedar V, Hwang JR, McDonough H, Li HH, Taylor J, Patterson C. Muscle ring finger protein-1 inhibits pkc{epsilon} activation and prevents cardiomyocyte hypertrophy. J Cell Biol. 2004;167:1147–1159. doi: 10.1083/jcb.200402033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Preedy VR, Smith DM, Kearney NF, Sugden PH. Rates of protein turnover in vivo and in vitro in ventricular muscle of hearts from fed and starved rats. Biochem J. 1984;222:395–400. doi: 10.1042/bj2220395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samarel AM, Parmacek MS, Magid NM, Decker RS, Lesch M. Protein synthesis and degradation during starvation-induced cardiac atrophy in rabbits. Circ Res. 1987;60:933–941. doi: 10.1161/01.res.60.6.933. [DOI] [PubMed] [Google Scholar]
- 17.Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J. Distinct roles of autophagy in the heart during ischemia and reperfusion: Roles of amp-activated protein kinase and beclin 1 in mediating autophagy. Circ Res. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 18.Taegtmeyer H, Golfman L, Sharma S, Razeghi P, van Arsdall M. Linking gene expression to function: Metabolic flexibility in the normal and diseased heart. Ann N Y Acad Sci. 2004;1015:202–213. doi: 10.1196/annals.1302.017. [DOI] [PubMed] [Google Scholar]
- 19.Lecker SH, Solomon V, Mitch WE, Goldberg AL. Muscle protein breakdown and the critical role of the ubiquitin-proteasome pathway in normal and disease states. J Nutr. 1999;129:227S–237S. doi: 10.1093/jn/129.1.227S. [DOI] [PubMed] [Google Scholar]
- 20.Jagoe RT, Lecker SH, Gomes M, Goldberg AL. Patterns of gene expression in atrophying skeletal muscles: Response to food deprivation. Faseb J. 2002;16:1697–1712. doi: 10.1096/fj.02-0312com. [DOI] [PubMed] [Google Scholar]
- 21.Aschenbach WG, Hirshman MF, Fujii N, Sakamoto K, Howlett KF, Goodyear LJ. Effect of aicar treatment on glycogen metabolism in skeletal muscle. Diabetes. 2002;51:567–573. doi: 10.2337/diabetes.51.3.567. [DOI] [PubMed] [Google Scholar]
- 22.Holmes BF, Kurth-Kraczek EJ, Winder WW. Chronic activation of 5′-amp-activated protein kinase increases glut-4, hexokinase, and glycogen in muscle. J Appl Physiol. 1999;87:1990–1995. doi: 10.1152/jappl.1999.87.5.1990. [DOI] [PubMed] [Google Scholar]
- 23.Jager S, Handschin C, St-Pierre J, Spiegelman BM. Amp-activated protein kinase (ampk) action in skeletal muscle via direct phosphorylation of pgc-1alpha. Proc Natl Acad Sci U S A. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomson DM, Herway ST, Fillmore N, Kim H, Brown JD, Barrow JR, Winder WW. Amp-activated protein kinase phosphorylates transcription factors of the creb family. J Appl Physiol. 2008;104:429–438. doi: 10.1152/japplphysiol.00900.2007. [DOI] [PubMed] [Google Scholar]
- 25.Andres V, Cervera M, Mahdavi V. Determination of the consensus binding site for mef2 expressed in muscle and brain reveals tissue-specific sequence constraints. J Biol Chem. 1995;270:23246–23249. doi: 10.1074/jbc.270.40.23246. [DOI] [PubMed] [Google Scholar]
- 26.Cserjesi P, Olson EN. Myogenin induces the myocyte-specific enhancer binding factor mef-2 independently of other muscle-specific gene products. Mol Cell Biol. 1991;11:4854–4862. doi: 10.1128/mcb.11.10.4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al-Khalili L, Chibalin AV, Yu M, Sjodin B, Nylen C, Zierath JR, Krook A. Mef2 activation in differentiated primary human skeletal muscle cultures requires coordinated involvement of parallel pathways. Am J Physiol Cell Physiol. 2004;286:C1410–1416. doi: 10.1152/ajpcell.00444.2003. [DOI] [PubMed] [Google Scholar]
- 28.Holmes BF, Sparling DP, Olson AL, Winder WW, Dohm GL. Regulation of muscle glut4 enhancer factor and myocyte enhancer factor 2 by amp-activated protein kinase. Am J Physiol Endocrinol Metab. 2005;289:E1071–1076. doi: 10.1152/ajpendo.00606.2004. [DOI] [PubMed] [Google Scholar]
- 29.Yamaguchi N, Takahashi N, Xu L, Smithies O, Meissner G. Early cardiac hypertrophy in mice with impaired calmodulin regulation of cardiac muscle ca release channel. J Clin Invest. 2007;117:1344–1353. doi: 10.1172/JCI29515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naya FJ, Wu C, Richardson JA, Overbeek P, Olson EN. Transcriptional activity of mef2 during mouse embryogenesis monitored with a mef2-dependent transgene. Development. 1999;126:2045–2052. doi: 10.1242/dev.126.10.2045. [DOI] [PubMed] [Google Scholar]
- 31.Zhao XS, Gallardo TD, Lin L, Schageman JJ, Shohet RV. Transcriptional mapping and genomic analysis of the cardiac atria and ventricles. Physiol Genomics. 2002;12:53–60. doi: 10.1152/physiolgenomics.00086.2002. [DOI] [PubMed] [Google Scholar]
- 32.Mearini G, Gedicke C, Schlossarek S, Witt CC, Kramer E, Cao P, Gomes MD, Lecker SH, Labeit S, Willis MS, Eschenhagen T, Carrier L. Atrogin-1 and murf1 regulate cardiac mybp-c levels via different mechanisms. Cardiovasc Res. 2010;85:357–366. doi: 10.1093/cvr/cvp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kedar V, McDonough H, Arya R, Li HH, Rockman HA, Patterson C. Muscle-specific ring finger 1 is a bona fide ubiquitin ligase that degrades cardiac troponin i. Proc Natl Acad Sci U S A. 2004;101:18135–18140. doi: 10.1073/pnas.0404341102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schoenheimer R. The dynamic state of body constituents. Cambridge, MA: Harvard University Press; 1942. [Google Scholar]
- 35.Wang X, Robbins J. Heart failure and protein quality control. Circ Res. 2006;99:1315–1328. doi: 10.1161/01.RES.0000252342.61447.a2. [DOI] [PubMed] [Google Scholar]
- 36.Razeghi P, Baskin KK, Sharma S, Young ME, Stepkowski S, Faadiel Essop M, Taegtmeyer H. Atrophy, hypertrophy, and hypoxemia induce transcriptional regulators of the ubiquitin proteasome system in the rat heart. Biochem Biophys Res Commun. 2006;342:361–364. doi: 10.1016/j.bbrc.2006.01.163. [DOI] [PubMed] [Google Scholar]
- 37.Baskin KK, Taegtmeyer H. Taking pressure off the heart: The ins and outs of atrophic remodelling. Cardiovasc Res. 2011;90:243–250. doi: 10.1093/cvr/cvr060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, Shaw RJ. Phosphorylation of ulk1 (hatg1) by amp-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J, Kundu M, Viollet B, Guan KL. Ampk and mtor regulate autophagy through direct phosphorylation of ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zungu M, Schisler JC, Essop MF, McCudden C, Patterson C, Willis MS. Regulation of ampk by the ubiquitin proteasome system. Am J Pathol. 2011;178:4–11. doi: 10.1016/j.ajpath.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hardie DG. Role of amp-activated protein kinase in the metabolic syndrome and in heart disease. FEBS Lett. 2008;582:81–89. doi: 10.1016/j.febslet.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 42.Li J, Coven DL, Miller EJ, Hu X, Young ME, Carling D, Sinusas AJ, Young LH. Activation of ampk alpha- and gamma-isoform complexes in the intact ischemic rat heart. Am J Physiol Heart Circ Physiol. 2006;291:H1927–1934. doi: 10.1152/ajpheart.00251.2006. [DOI] [PubMed] [Google Scholar]
- 43.Salt I, Celler JW, Hawley SA, Prescott A, Woods A, Carling D, Hardie DG. Amp-activated protein kinase: Greater amp dependence, and preferential nuclear localization, of complexes containing the alpha2 isoform. Biochem J. 1998;334 ( Pt 1):177–187. doi: 10.1042/bj3340177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Towler MC, Hardie DG. Amp-activated protein kinase in metabolic control and insulin signaling. Circ Res. 2007;100:328–341. doi: 10.1161/01.RES.0000256090.42690.05. [DOI] [PubMed] [Google Scholar]
- 45.Stoppani J, Hildebrandt AL, Sakamoto K, Cameron-Smith D, Goodyear LJ, Neufer PD. Amp-activated protein kinase activates transcription of the ucp3 and hkii genes in rat skeletal muscle. Am J Physiol Endocrinol Metab. 2002;283:E1239–1248. doi: 10.1152/ajpendo.00278.2002. [DOI] [PubMed] [Google Scholar]
- 46.McGee SL, van Denderen BJ, Howlett KF, Mollica J, Schertzer JD, Kemp BE, Hargreaves M. Amp-activated protein kinase regulates glut4 transcription by phosphorylating histone deacetylase 5. Diabetes. 2008;57:860–867. doi: 10.2337/db07-0843. [DOI] [PubMed] [Google Scholar]
- 47.Lo WS, Duggan L, Emre NC, Belotserkovskya R, Lane WS, Shiekhattar R, Berger SL. Snf1--a histone kinase that works in concert with the histone acetyltransferase gcn5 to regulate transcription. Science. 2001;293:1142–1146. doi: 10.1126/science.1062322. [DOI] [PubMed] [Google Scholar]
- 48.Bungard D, Fuerth BJ, Zeng PY, Faubert B, Maas NL, Viollet B, Carling D, Thompson CB, Jones RG, Berger SL. Signaling kinase ampk activates stress-promoted transcription via histone h2b phosphorylation. Science. 2010;329:1201–1205. doi: 10.1126/science.1191241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hardie DG. Transcription. Targeting the core of transcription. Science. 2010;329:1158–1159. doi: 10.1126/science.1195447. [DOI] [PubMed] [Google Scholar]
- 50.Skurk C, Izumiya Y, Maatz H, Razeghi P, Shiojima I, Sandri M, Sato K, Zeng L, Schiekofer S, Pimentel D, Lecker S, Taegtmeyer H, Goldberg AL, Walsh K. The foxo3a transcription factor regulates cardiac myocyte size downstream of akt signaling. J Biol Chem. 2005;280:20814–20823. doi: 10.1074/jbc.M500528200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO, Gonzalez M, Yancopoulos GD, Glass DJ. The igf-1/pi3k/akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting foxo transcription factors. Mol Cell. 2004;14:395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- 52.Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adams V, Linke A, Wisloff U, Doring C, Erbs S, Krankel N, Witt CC, Labeit S, Muller-Werdan U, Schuler G, Hambrecht R. Myocardial expression of murf-1 and mafbx after induction of chronic heart failure: Effect on myocardial contractility. Cardiovasc Res. 2007;73:120–129. doi: 10.1016/j.cardiores.2006.10.026. [DOI] [PubMed] [Google Scholar]
- 54.Li YP, Chen Y, John J, Moylan J, Jin B, Mann DL, Reid MB. Tnf-alpha acts via p38 mapk to stimulate expression of the ubiquitin ligase atrogin1/mafbx in skeletal muscle. Faseb J. 2005;19:362–370. doi: 10.1096/fj.04-2364com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moylan JS, Smith JD, Chambers MA, McLoughlin TJ, Reid MB. Tnf induction of atrogin-1/mafbx mrna depends on foxo4 expression but not akt-foxo1/3 signaling. Am J Physiol Cell Physiol. 2008;295:C986–993. doi: 10.1152/ajpcell.00041.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sacheck JM, Ohtsuka A, McLary SC, Goldberg AL. Igf-i stimulates muscle growth by suppressing protein breakdown and expression of atrophy-related ubiquitin ligases, atrogin-1 and murf1. Am J Physiol Endocrinol Metab. 2004;287:E591–601. doi: 10.1152/ajpendo.00073.2004. [DOI] [PubMed] [Google Scholar]
- 57.Clarke BA, Drujan D, Willis MS, Murphy LO, Corpina RA, Burova E, Rakhilin SV, Stitt TN, Patterson C, Latres E, Glass DJ. The e3 ligase murf1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab. 2007;6:376–385. doi: 10.1016/j.cmet.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 58.Yang H, Mammen J, Wei W, Menconi M, Evenson A, Fareed M, Petkova V, Hasselgren PO. Expression and activity of c/ebpbeta and delta are upregulated by dexamethasone in skeletal muscle. J Cell Physiol. 2005;204:219–226. doi: 10.1002/jcp.20278. [DOI] [PubMed] [Google Scholar]
- 59.Cai D, Frantz JD, Tawa NE, Jr, Melendez PA, Oh BC, Lidov HG, Hasselgren PO, Frontera WR, Lee J, Glass DJ, Shoelson SE. Ikkbeta/nf-kappab activation causes severe muscle wasting in mice. Cell. 2004;119:285–298. doi: 10.1016/j.cell.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 60.Moresi V, Williams AH, Meadows E, Flynn JM, Potthoff MJ, McAnally J, Shelton JM, Backs J, Klein WH, Richardson JA, Bassel-Duby R, Olson EN. Myogenin and class ii hdacs control neurogenic muscle atrophy by inducing e3 ubiquitin ligases. Cell. 2010;143:35–45. doi: 10.1016/j.cell.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suzuki J, Ueno M, Uno M, Hirose Y, Zenimaru Y, Takahashi S, Osuga JI, Ishibashi S, Takahashi M, Hirose M, Yamada M, Kraemer FB, Miyamori I. Effects of hormone-sensitive lipase-disruption on cardiac energy metabolism in response to fasting and refeeding. Am J Physiol Endocrinol Metab. 2009 doi: 10.1152/ajpendo.91031.2008. [DOI] [PubMed] [Google Scholar]
- 62.Koyama S, Hata S, Witt CC, Ono Y, Lerche S, Ojima K, Chiba T, Doi N, Kitamura F, Tanaka K, Abe K, Witt SH, Rybin V, Gasch A, Franz T, Labeit S, Sorimachi H. Muscle ring-finger protein-1 (murf1) as a connector of muscle energy metabolism and protein synthesis. J Mol Biol. 2008;376:1224–1236. doi: 10.1016/j.jmb.2007.11.049. [DOI] [PubMed] [Google Scholar]
- 63.Witt CC, Witt SH, Lerche S, Labeit D, Back W, Labeit S. Cooperative control of striated muscle mass and metabolism by murf1 and murf2. EMBO J. 2008;27:350–360. doi: 10.1038/sj.emboj.7601952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hirner S, Krohne C, Schuster A, Hoffmann S, Witt S, Erber R, Sticht C, Gasch A, Labeit S, Labeit D. Murf1-dependent regulation of systemic carbohydrate metabolism as revealed from transgenic mouse studies. J Mol Biol. 2008;379:666–677. doi: 10.1016/j.jmb.2008.03.049. [DOI] [PubMed] [Google Scholar]
- 65.Newsholme EA, Randle PJ. Regulation of glucose uptake by muscle. 5. Effects of anoxia, insulin, adrenaline and prolonged starving on concentrations of hexose phosphates in isolated rat diaphragm and perfused isolated rat heart. Biochem J. 1961;80:655–662. doi: 10.1042/bj0800655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adrouny GA. Differential patterns of glycogen metabolism in cardiac and skeletal muscles. Am J Physiol. 1969;217:686–693. doi: 10.1152/ajplegacy.1969.217.3.686. [DOI] [PubMed] [Google Scholar]
- 67.Schneider CA, Taegtmeyer H. Fasting in vivo delays myocardial cell damage after brief periods of ischemia in the isolated working rat heart. Circ Res. 1991;68:1045–1050. doi: 10.1161/01.res.68.4.1045. [DOI] [PubMed] [Google Scholar]
- 68.Centner T, Yano J, Kimura E, McElhinny AS, Pelin K, Witt CC, Bang ML, Trombitas K, Granzier H, Gregorio CC, Sorimachi H, Labeit S. Identification of muscle specific ring finger proteins as potential regulators of the titin kinase domain. J Mol Biol. 2001;306:717–726. doi: 10.1006/jmbi.2001.4448. [DOI] [PubMed] [Google Scholar]
- 69.Lange S, Auerbach D, McLoughlin P, Perriard E, Schafer BW, Perriard JC, Ehler E. Subcellular targeting of metabolic enzymes to titin in heart muscle may be mediated by dral/fhl-2. J Cell Sci. 2002;115:4925–4936. doi: 10.1242/jcs.00181. [DOI] [PubMed] [Google Scholar]
- 70.Johnson JA, Majumdar SR, Simpson SH, Toth EL. Decreased mortality associated with the use of metformin compared with sulfonylurea monotherapy in type 2 diabetes. Diabetes Care. 2002;25:2244–2248. doi: 10.2337/diacare.25.12.2244. [DOI] [PubMed] [Google Scholar]
- 71.Masoudi FA, Inzucchi SE, Wang Y, Havranek EP, Foody JM, Krumholz HM. Thiazolidinediones, metformin, and outcomes in older patients with diabetes and heart failure: An observational study. Circulation. 2005;111:583–590. doi: 10.1161/01.CIR.0000154542.13412.B1. [DOI] [PubMed] [Google Scholar]
- 72.Shah DD, Fonarow GC, Horwich TB. Metformin therapy and outcomes in patients with advanced systolic heart failure and diabetes. J Card Fail. 2010;16:200–206. doi: 10.1016/j.cardfail.2009.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aguilar D, Chan W, Bozkurt B, Ramasubbu K, Deswal A. Metformin use and mortality in ambulatory patients with diabetes and heart failure. Circ Heart Fail. 2011;4:53–58. doi: 10.1161/CIRCHEARTFAILURE.110.952556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Johnson JA, Simpson SH, Toth EL, Majumdar SR. Reduced cardiovascular morbidity and mortality associated with metformin use in subjects with type 2 diabetes. Diabet Med. 2005;22:497–502. doi: 10.1111/j.1464-5491.2005.01448.x. [DOI] [PubMed] [Google Scholar]
- 75.Giamouzis G, Triposkiadis F, Butler J. Metformin use in patients with diabetes mellitus and heart failure: Friend or foe? J Card Fail. 2010;16:207–210. doi: 10.1016/j.cardfail.2009.12.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.