Abstract
Background
The insertion/deletion (I/D) polymorphism in the Angiotensin-converting enzyme (ACE) gene has been implicated in susceptibility to cancer, but a large number of studies have reported inconclusive results. The aim of this study is to assess the association between the I/D polymorphism in the ACE gene and cancer risk by meta-analysis.
Methods
A search was performed in Pubmed database, Embase database, Chinese Biomedical (CBM) database, China National Knowledge Infrastructure (CNKI) database and Weipu database, covering all studies until August 31, 2010. Statistical analysis was performed by using Revman4.2 and STATA 10.0.
Results
A total of 25 case-control studies comprising 3914 cancer patients and 11391 controls were identified. No significant association was found between the I/D polymorphism and over all cancer risks (OR = 0.88, 95%CI = 0.73-1.06, P = 0.17 for DD+DI vs. II). In the subgroup analysis by ethnicity, no significant association was found among Asians and Europeans for the comparison of DD+DI vs. II. In the subgroup analysis by cancer types, no significant associations were found among lung cancer, breast cancer, prostate cancer, colorectal cancer, gastric cancer for the comparison of DD+DI vs. II. Results from other comparative genetic models also indicated the lack of associations between this polymorphism and cancer risks.
Conclusions
This meta-analysis suggested that the ACE D/I polymorphism might not contribute to the risk of cancer.
Keywords: Angiotensin-converting enzyme, cancer, meta-analysis, polymorphism, risk
Background
The angiotensin-converting enzyme (ACE), a key enzyme in the renin-angiotensin system, plays the important roles of regulating of blood pressure and serum electrolytes [1,2]. It has also been implicated in the pathogenesis of several cancers, such as lung cancer, breast cancer, prostate cancer, gastric cancer and oral cancer [2-6]. It is differentially expressed in several carcinomas and may affect tumor cell proliferation, migration, angiogenesis, and metastatic behaviors [7]. Inhibition of ACE activity suppresses tumor growth and angiogenesis in vitro and vivo of animal models; moreover, epidemiologic studies have also indicated that ACE inhibitors might decrease the risk and mortality rate of cancers [2,7].
The human ACE gene is located on chromosome 17q23, and many polymorphisms have been identified [8]. The most widely studied polymorphism Insertion/Deletion (I/D, rs4646994) is located on intron 16[6]. It is characterized by the presence or absence of a 287-bp Alu repetitive sequence, which results in three genotypes: II, DI and DD [6]. The I/D polymorphism accounts for 20% to 50% of the variance in ACE expression or activity in blood and tissues among individuals [7]. Homozygote II may display as low as half of the plasma ACE level compared to the homozygote DD, whereas the heterozygote DI displays an intermediate level[2]. Recently, many studies investigated the role of this polymorphism in the etiology of cancers among various organs, including lung, breast, prostate, gastric, oral and others [3-7,9-18]. However, the observed associations of these studies were inconsistent, and a single study might be insufficient to detect a possible small effect of the polymorphism on cancers. Meta-analysis is a useful method for investigating the associations between diseases and risk factors because it uses a quantitative approach to combine the results of different studies on the same topic, potentially providing more reliable conclusions [19,20]. Considering the extensive role of ACE in the pathogenesis of cancers, a meta-analysis was performed on all eligible case-control studies to estimate the association between this polymorphism and cancer risks.
Methods
Publication search
We searched literatures in Pubmed database, Embase database, Chinese Biomedical database(CBM) database, Chinese National Knowledge Infrastructure(CNKI) database and Weipu database to identify articles that evaluated the associations between polymorphisms in ACE gene and cancer risks(Last search was updated on Aug 31st, 2010). The search terms were used as follows: 'cancer or carcinoma' and 'ACE or angiotensin-converting enzyme' in combination with 'polymorphism or mutation or variant'. The languages were limited to English and Chinese. The following inclusion criteria were used in the meta-analysis: (1) the study should evaluate the I/D polymorphism in ACE gene and cancer risk, (2) the study should be a case-control design, (3) enough information had to be provided to calculate the odds ratio (OR) with 95% confidence interval (CI), (4) the distribution of genotypes in the control groups should be consistent with Hardy-Weinberg equilibrium (HWE). Accordingly, the following exclusion criteria were also used: (1) abstracts and reviews, (2) studies in which the genotype frequencies were not reported, (3) repeated or overlapped publications. For studies with the same case series by the same authors, the most recently published studies or studies with the largest numbers of subjects were included.
Data extraction
Data were independently checked and extracted by two investigators. The following items were collected from each study: first author's name, year of publication, country of origin, ethnicity, genotyping methods, cancer type, total number of cases and controls, genotype distributions in cases and controls.
Statistical analysis
For each case-control study, the HWE of genotypes in the control group was assessed by using Person's X2 test. OR and 95% CI was used to assess the strength of the association between the I/D polymorphism and cancer risk. We calculated the OR and respective 95% CI by comparing the carriers of rare alleles with the wild homozygotes (DI+DD vs. II).
Heterogeneity among studies was assessed by a X2 based Q- and I2- statistic. Heterogeneity was considered significant for P less than 0.10. The fixed-effects model and random-effects model were used to pool the results. When the P value of heterogeneity was greater than 0.10, the fixed-effects model was used, otherwise, the random-effects model was used, as it is more appropriate when heterogeneity is present. The significance of the pooled OR was determined by the Z-test and P less than 0.05 was considered as statistically significant. To evaluate the ethnicity-specific, cancer type-specific effects, subgroup analyses were performed by ethnic group ('European', 'Asian', 'African-American' and 'Latino') and cancer types. Subgroup analyses by ethnicity were preformed if one ethnic population included more than three case-control studies. Subgroup analysis by cancer type were preformed if one cancer type canteined three and more than three individual studies. Comparisons of other genetic models were also performed (DD vs. DI+II, DD vs. II, DI vs. II and D vs. I).
Publication bias was investigated by using several methods. Visual inspection of asymmetry in funnel plots was carried out. The Begg's funnel plots and Egger's test were also used to statistically assess publication bias [21,22]. Sensitivity analysis was performed to assess the stability of the results by sequentially excluding each study [23]. All analyses were performed using the software Revman4.2 and STATA 10.0.
Results
Studies selection and characteristics in the meta-analysis
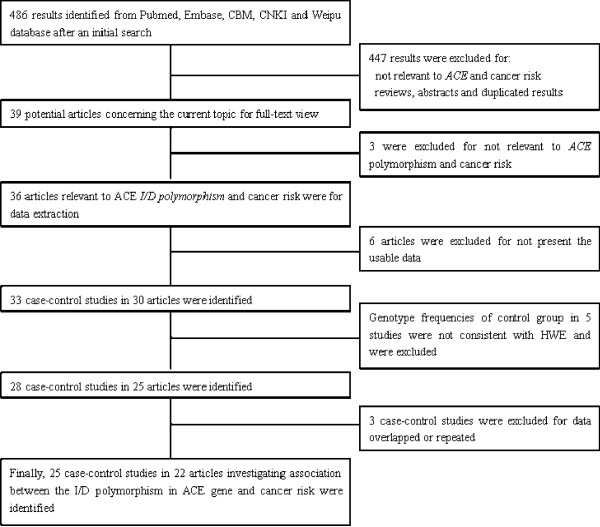
There were 486 results relevant to the search words in the selected databases (Figure 1). After reading the titles and abstracts, 39 potential articles were included for full-text view. Further screening of these articles, three of them were excluded for being not relevant to cancer risk with ACE gene polymorphism. Thus, 36 articles were left for data extraction. Six articles were excluded for not reporting the usable data. One article reported four cohorts each and each cohort was considered as a separate case-control study [6]. Thus, a total of 33 case-control studies in 30 articles were identified. Additionally, 5 case-control studies were excluded for the genotypes in control group not consistent with HWE[24-28], and 3 case-control studies were excluded for data overlapped or duplicated. Thus, a total of 25 case-control studies in 22 articles were finally identified [3-7,9,15-18,29,14,35]. The characteristics of included case-control studies are summarized in Table 1. Genotype and allele distributions for each case-control study are shown in Table 2. There were 8 studies of Asians [4,6,10,11,14,29,30,33], 14 of Europeans [3,5-7,9,12,13,15-17,31,32,34,35], 2 of Latinos [6,18], 1 of African-Americans [6]. In this meta-analysis, the most studied cancers were lung cancer and breast cancer, the genotype methods are a classic PCR assays
Figure 1.
Flow diagram of included/excluded studies.
Table 1.
Characteristics of case-control studies included in meta-analysis
| Author | Year | Country | Ethnicity | Cancer type | No.(Cases/Controls) | Genotyping method |
|---|---|---|---|---|---|---|
| Arima, H[14] | 2006 | Japan | Asian | Cancers | 176/761 | PCR |
| Bardi, E[34] | 2005 | Hungary | European | Cancers | 207/144 | PCR |
| Cheon, K T[29] | 2000 | Korea | Asian | Lung | 218/121 | PCR |
| Ding, × J[30] | 2008 | China | Asian | Lung | 121/33 | PCR |
| Goto, Y[10] | 2005 | Japan | Asian | Gastric | 202/454 | PCR |
| Haiman, C A(AF)[6] | 2003 | USA | African-American | Breast | 257/631 | PCR |
| Haiman, C A(JP)[6] | 2003 | USA | Asian | Breast | 284/357 | PCR |
| Haiman, C A(Latinas)[6] | 2003 | USA | Latino | Breast | 249/652 | PCR |
| Haiman, C A(Whites)[6] | 2003 | USA | European | Breast | 292/402 | PCR |
| Holla, L[35] | 1998 | Czech | European | Leukemia | 25/202 | PCR |
| Nacak, M[3] | 2010 | Turkey | European | Lung | 125/165 | PCR |
| Nikiteas, N[16] | 2007 | Greece | European | colorectal | 92/102 | PCR |
| Rocken, C[7] | 2005 | Germany | European | Gastric | 113/189 | PCR |
| Rocken, C[31] | 2007 | Germany | European | Colorectal | 141/189 | PCR |
| Sierra, Diaz E[18] | 2009 | Mexico | Latino | Prostate | 19/28 | PCR |
| Sugimoto, M[4] | 2006 | Japan | Asian | Gastric | 119/132 | PCR |
| Toma, M[13] | 2009 | Romanian | European | Colorectal | 108/150 | PCR |
| Tunny, T J[17] | 1996 | Australia | European | Aldosterone-producing adenoma |
55/80 | PCR |
| Vairaktaris, E[32] | 2009 | Greece | European | Oral | 160/153 | PCR |
| van der Knaap, R[12] | 2008 | Netherlands | European | Colorectal, lung, breast, prostate |
655/6015 | PCR |
| Vaskù, V[9] | 2004 | Czech | European | T-cell lymphoma | 77/203 | PCR |
| Wang, H W[11] | 2000 | China | Asian | Lung | 34/38 | PCR |
| Yeren, A[15] | 2008 | Turkey | European | Lung | 75/85 | PCR |
| Yigit, B[5] | 2007 | Turkey | European | Prostate | 48/51 | PCR |
| Zhang Q Z[33] | 2005 | China | Asian | Lung | 47/54 | PCR |
Table 2.
Distribution of ACE genotype and allele among asthma patients and controls
| Author | Case | Control | Case | Control | HWE for control population | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| II | ID | DD | II | DI | DD | I | D | I | D | X2 | P | |
| Arima, H[14] | 65 | 87 | 24 | 295 | 372 | 94 | 217 | 135 | 962 | 560 | 1.978 | 0.160 |
| Bardi, E[34] | 74 | 89 | 44 | 52 | 71 | 21 | 237 | 177 | 175 | 113 | 0.166 | 0.683 |
| Cheon, K T[29] | 72 | 116 | 30 | 48 | 50 | 23 | 260 | 176 | 146 | 96 | 2.261 | 0.132 |
| Ding, × J[30] | 55 | 56 | 10 | 19 | 10 | 4 | 166 | 76 | 48 | 18 | 1.840 | 0.175 |
| Goto, Y[10] | 76 | 98 | 28 | 209 | 189 | 56 | 250 | 154 | 607 | 301 | 1.674 | 0.196 |
| Haiman, C A(AF)[6] | 62 | 118 | 77 | 100 | 310 | 221 | 242 | 272 | 510 | 752 | 0.254 | 0.614 |
| Haiman, C A(JP)[6] | 119 | 128 | 37 | 154 | 160 | 43 | 366 | 202 | 468 | 246 | 0.021 | 0.884 |
| Haiman, C A(Latinas)[6] | 73 | 127 | 49 | 189 | 301 | 162 | 273 | 225 | 679 | 625 | 3.677 | 0.055 |
| Haiman, C A(Whites)[6] | 79 | 129 | 84 | 91 | 187 | 124 | 287 | 297 | 369 | 435 | 1.613 | 0.204 |
| Holla, L[35] | 25 | 11 | 4 | 40 | 86 | 76 | 61 | 19 | 166 | 238 | 2.937 | 0.087 |
| Nacak, M[3] | 37 | 50 | 38 | 29 | 72 | 64 | 124 | 126 | 130 | 200 | 1.225 | 0.268 |
| Nikiteas, N[16] | 15 | 50 | 27 | 6 | 52 | 44 | 80 | 104 | 64 | 140 | 3.451 | 0.063 |
| Rocken, C[7] | 24 | 57 | 32 | 41 | 95 | 53 | 105 | 121 | 177 | 201 | 0.017 | 0.898 |
| Rocken, C[31] | 37 | 69 | 35 | 41 | 95 | 53 | 143 | 139 | 177 | 201 | 0.017 | 0.898 |
| Sierra, Diaz E[18] | 0 | 7 | 12 | 9 | 12 | 7 | 7 | 31 | 30 | 26 | 0.537 | 0.464 |
| Sugimoto, M[4] | 54 | 53 | 12 | 50 | 60 | 22 | 161 | 77 | 160 | 104 | 0.305 | 0.581 |
| Toma, M[13] | 25 | 50 | 33 | 30 | 73 | 47 | 100 | 116 | 133 | 167 | 0.029 | 0.864 |
| Tunny, T J[17] | 16 | 25 | 14 | 24 | 34 | 22 | 57 | 53 | 82 | 78 | 1.787 | 0.181 |
| Vairaktaris, E[32] | 30 | 70 | 60 | 9 | 66 | 78 | 130 | 190 | 84 | 222 | 1.054 | 0.305 |
| van der Knaap, R[12] | 141 | 329 | 185 | 1332 | 3006 | 1677 | 611 | 699 | 5670 | 6360 | 0.047 | 0.828 |
| Vaskù, V[9] | 19 | 37 | 21 | 43 | 103 | 57 | 75 | 79 | 189 | 217 | 0.078 | 0.780 |
| Wang, H W[11] | 10 | 6 | 18 | 13 | 18 | 7 | 26 | 42 | 44 | 32 | 0.031 | 0.861 |
| Yeren, A[15] | 4 | 39 | 32 | 14 | 37 | 34 | 47 | 103 | 65 | 105 | 0.522 | 0.470 |
| Yigit, B[5] | 4 | 19 | 25 | 12 | 24 | 15 | 27 | 69 | 48 | 54 | 0.157 | 0.692 |
| Zhang Q Z[33] | 21 | 21 | 5 | 20 | 30 | 4 | 63 | 31 | 70 | 38 | 2.567 | 0.109 |
Meta-analysis results
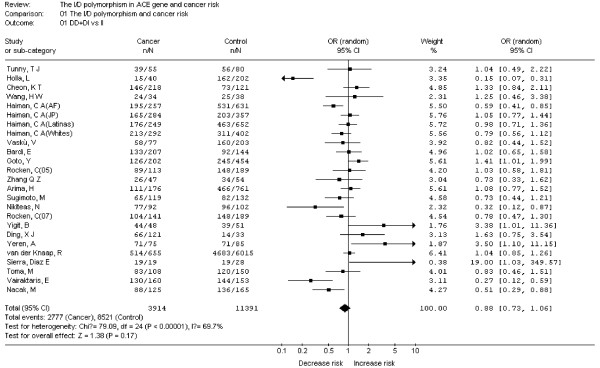
As shown in Figure 2, heterogeneity of DD+DI vs. II for all studies was analyzed and the value of X2 was 79.09 with 24 degrees of freedom and P < 0.00001 in a random-effects model. Additionally, I-square value is another index of the test of heterogeneity. In Figure 2, the I-square was 69.7%, suggesting the presence of heterogeneity. Thus, the random-effects model was chosen to synthesize the data. OR was 0.88(95%CI = 0.73-1.06) and the test for overall effect Z value was 1.38 (P = 0.17). The results suggested that the variant D allele carriers (DI+DD) do not have a significant increased risk of cancer compared with those individuals without D allele (II). Summary of the results of other genetic comparisons are listed in Table 3.
Figure 2.
Meta-analysis with a random-effects model for the association between cancer risk and the ACE I/D polymorphism (DD+DI vs II).
Table 3.
Summary of different comparative results
| Variables | N | Cases/Controls | DD+DI vs II | DD vs DI+II | DD vs II | D vs I | DI vs II | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR(95%CI) | P* | OR(95%CI) | P* | OR(95%CI) | P* | OR(95%CI) | P* | OR(95%CI) | P* | |||
| Total | 25 | 3914/11391 | 0.88(0.73, 1.06) | 0.38 | 0.93(0.80, 1.09) | 0.39 | 0.90(0.73, 1.12) | 0.34 | 0.94(0.83, 1.07) | 0.34 | 0.89(0.75, 1.06) | 0.20 |
| Subgroup by ethnicity | ||||||||||||
| Asian | 8 | 1201/1950 | 1.13(0.96, 1.33) | 0.15 | 1.05(0.74, 1.49) | 0.78 | 1.09(0.83, 1.44) | 0.53 | 1.08(0.93, 1.25) | 0.34 | 0.85(0.68, 1.06) | 0.14 |
| European | 14 | 2188/8130 | 0.89(0.73, 1.09) | 0.26 | 0.89(0.73, 1.09) | 0.26 | 0.76(0.53, 1.08) | 0.12 | 0.86(0.71, 1.03) | 0.10 | 0.78(0.60, 1.01) | 0.06 |
| Subgroup by cancer type | ||||||||||||
| Lung | 7 | 737/6511 | 1.04(0.70, 1.55) | 0.85 | 0.99(0.67, 1.48) | 0.98 | 1.02(0.62, 1.69) | 0.93 | 1.03(0.80, 1.31) | 0.83 | 1.01(0.64, 1.61) | 0.95 |
| Breast | 5 | 1235/8057 | 0.88(0.71, 1.09) | 0.24 | 0.95(0.75, 1.21) | 0.70 | 0.86(0.64, 1.16) | 0.33 | 0.94(0.81, 1.09) | 0.41 | 0.88(0.72, 1.09) | 0.24 |
| Colorectal | 4 | 517/6456 | 0.81(0.53, 1.24) | 0.34 | 0.83(0.66, 1.05) | 0.11 | 0.73(0.45, 1.18) | 0.20 | 0.87(0.71, 1.06) | 0.17 | 0.87(0.58, 1.31) | 0.50 |
| Gastric | 3 | 434/775 | 1.06(0.71, 1.59) | 0.78 | 0.94(0.66, 1.36) | 0.76 | 0.96(0.55, 1.65) | 0.87 | 1.00(0.74, 1.34) | 1.00 | 1.12(0.79, 1.58) | 0.52 |
| Prostate | 3 | 276/6094 | 2.44(0.66, 8.97) | 0.18 | 2.05(0.74, 5.62) | 0.17 | 3.48(0.63, 19.13) | 0.15 | 2.05(0.84, 5.02) | 0.11 | 1.66(0.67, 4.07) | 0.27 |
*: P value for Q-test.
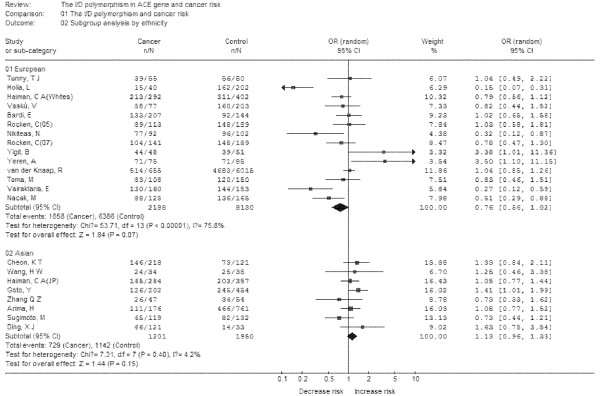
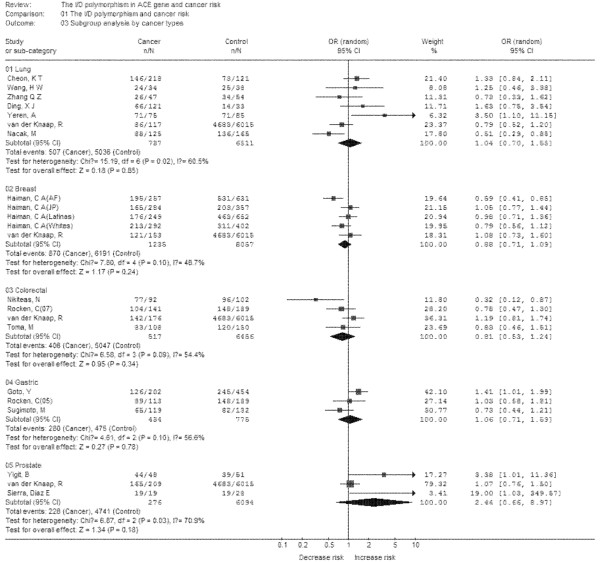
Subgroup analyses were performed after stratifications of the data by ethnicity and cancer types. In the subgroup analysis by ethnicity (Figure 3), no significant increased risks were found in Europeans and Asians. In the subgroup analysis by cancer types (Figure 4), no significant increased risk was found in lung cancer, breast cancer, colorectal cancer, gastric cancer, prostate cancer. Thus, the polymorphism may not increase cancer risk in different population and different cancers.
Figure 3.
Meta-analysis with a random-effects model for the association between cancer risk and the ACE I/D polymorphism (DD+DI vs II): subgroup analysis by ethnicity.
Figure 4.
Meta-analysis with a random-effects model for the association between cancer risk and the ACE I/D polymorphism (DD+DI vs II): subgroup analysis by cancer type.
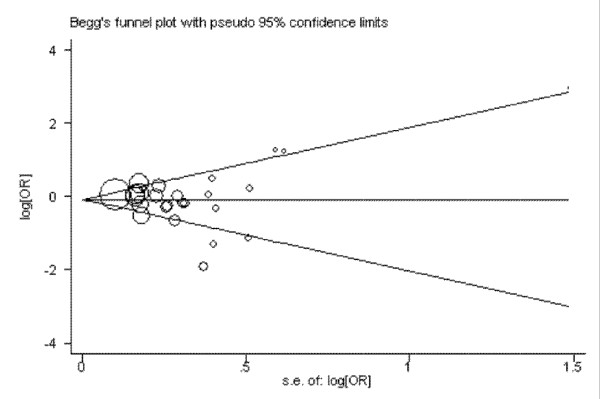
Publication bias
Begg's funnel plot and Egger's test were performed to assess the publication bias of the literatures. The shape of the funnel plots seemed approximately symmetrical (DD+DI vs. II) and the Egger's test did not show any evidence of publication bias (t = 0.45 and P = 0.655 for DD+DI vs. II) (Figure 5).
Figure 5.
Begg's funnel plot for publication bias in selection of studies on the ACE I/D polymorphism (DD+DI vs II).
Sensitivity analysis
Sensitivity analysis was analyzed as previous study [23]. Briefly, after excluding each case-control study for DD+DI vs. II comparative (Table 4), statistically similar results were obtained, suggesting the results of this meta-analysis are stable.
Table 4.
Sensitivity analysis for the current meta-analysis: ORs with 95%CIs and P value were results after excluding each case-control study for DD+DI vs.II.
| Author | OR | 95%CT | P |
|---|---|---|---|
| Arima, H[14] | 0.87 | 0.71-1.06 | 0.16 |
| Bardi, E[34] | 0.87 | 0.72-1.06 | 0.16 |
| Cheon, K T[29] | 0.86 | 0.71-1.04 | 0.12 |
| Ding, × J[30] | 0.86 | 0.71-1.04 | 0.12 |
| Goto, Y[10] | 0.85 | 0.71-1.03 | 0.10 |
| Haiman, C A(AF)[6] | 0.90 | 0.74-1.09 | 0.27 |
| Haiman, C A(JP)[6] | 0.87 | 0.71-1.06 | 0.16 |
| Haiman, C A(Latinas)[6] | 0.87 | 0.71-1.06 | 0.17 |
| Haiman, C A(Whites)[6] | 0.88 | 0.78-1.07 | 0.21 |
| Holla, L[35] | 0.93 | 0.79-1.09 | 0.38 |
| Nacak, M[3] | 0.90 | 0.75-1.09 | 0.27 |
| Nikiteas, N[16] | 0.90 | 0.75-1.08 | 0.26 |
| Rocken, C[7] | 0.87 | 0.72-1.06 | 0.16 |
| Rocken, C[31] | 0.88 | 0.73-1.07 | 0.21 |
| Sierra, Diaz E[18] | 0.87 | 0.72-1.04 | 0.13 |
| Sugimoto, M[4] | 0.89 | 0.73-1.07 | 0.22 |
| Toma, M[13] | 0.88 | 0.73-1.07 | 0.19 |
| Tunny, T J[17] | 0.87 | 0.72-1.06 | 0.16 |
| Vairaktaris, E[32] | 0.91 | 0.76-1.09 | 0.31 |
| van der Knaap, R[12] | 0.87 | 0.71-1.07 | 0.18 |
| Vaskù, V[9] | 0.88 | 0.73-1.07 | 0.19 |
| Wang, H W[11] | 0.87 | 0.72-1.05 | 0.15 |
| Yeren, A[15] | 0.86 | 0.71-1.03 | 0.10 |
| Yigit, B[5] | 0.86 | 0.71-1.03 | 0.10 |
| Zhang Q Z[33] | 0.88 | 0.73-1.07 | 0.20 |
Discussion
ACE is a key enzyme in the renin-angiotensin system, which is involved in the regulation of blood pressure and serum electrolytes. In recent years, more evidences indicated that the enzyme was associated with the pathogenesis of cancers. It may influence tumor cell proliferation, migration, angiogenesis and metastatic behaviors. Given the important roles of ACE in cancer etiology, it is possible that genetic variations of the ACE gene may modulate the risk of cancer. The I/D polymorphism in intron 16 of the ACE gene is the most extensively studied polymorphism. This polymorphism is based on insertion or deletion of a 287-bp Alu sequence, leading to a change in the plasma ACE level. Growing number of studies have been published to investigate the associations between this polymorphism with cancer risk; however, the results were inconsistent and conflict. In order to resolve this issue, we conducted a meta-analysis of 25 case-control studies, including 3914 cases and 11391 controls, to evaluate the associations between the ACE I/D polymorphism and cancer risks.
Our results showed that the ACE I/D polymorphism was not associated with cancer risks. Moreover, in other comparative genetic models, no significant associations were found in any genetic models. These results indicated that this polymorphism may not contribute to cancer risks. Although previous studies revealed possible roles of ACE in cancer etiology, our result suggested that these roles may not account by the variant of ACE gene. Although the exact mechanism of this enzyme in cancer etiology is not so clear, our results may indicate that ACE I/D polymorphism may not influence cancer risk. In addition, considering the possible role of this polymorphism in serum ACE level, it is possible that the cancer risk may be modified by ACE level, but not by the variant. Thus, future studies are warranted to identify the associations between ACE polymorphism, ACE levels and cancer risk.
Considering the property of genetic background may affect the results of genetic association studies, we performed subgroup analysis by ethnicity. Two subgroups were included in this meta-analysis: 'European' and 'Asian'. In this meta-analysis, we didn't find a significant association between this polymorphism and cancer risk in any sub-populations. Moreover, no significant associations were found in any other genetic models. Interestingly, this polymorphism and cancer risk in Asians and Europeans were all inversely associated, although they were not statistically significant. These results may suggest that this polymorphism may exert varying effects in different populations. However, all included studies were from European, Asian, African-American and Latino populations, further studies are necessary to validate these findings for other ethnic populations, especially in Africans.
In another subgroup analysis by cancer types, we found that this polymorphism is not associated with increased risks in all sub-cancer types (lung, breast, prostate, gastric and colorectal). Considering the limitation of studies in each subgroup, the weak associations between this polymorphism and different cancers should be discussed. It is possible that these null associations may be due to chance because studies with small sample size may have insufficient statistical power to detect a slight effect. Considering the limited studies in each cancer type, our results should be explained with caution.
Heterogeneity is one of the important issues when performing meta-analysis. We found that heterogeneity between studies existed in overall comparisons. After subgroup analysis by ethnicity and cancer types, the heterogeneity was effectively decreased or removed in Asians and some cancer types, suggesting that certain effects of genetic variants are cancer specific and ethnic specific. The stability of this meta-analysis was analyzed by sequentially excluding individual studies; our results indicated stability of results. Publication bias is another important issue which should also be discussed in meta-analysis. After evaluating the publication bias by Egger's funnel plots and Begg's test, we did not detect a publication bias, indicating the strength of the results.
Cancers have been considered as genetic diseases, and many genetic variants are contributed to cancer risks [36-40]. It is worthy to mention the recently studies of genetic analysis by genome wide-association studies (GWAS). Up to now, a large number of GWAS for cancers have been published, and a large number of cancer susceptible loci were found [36-38,41-46]. However, none of them indicated the ACE D/I polymorphism, which can be partly explained by the chips which were used in the original studies. There is probably a lack of adequate coverage on chips which also identify insertion or deletion a 287-bp sequence. It is noted worthy, the SNP rs4343, which is considered as a good proxy in Caucasians for I/D variant (r2 > 0.80), have been studied by Illumina genotyping arrays [47,48]. Thus, further studies may investigate rs4343 for cancer risk by GWAS to help to resolve whether the I/D polymorphism is associated with cancer risk.
We have to mention a recently published study by Ruiter in 2011[49]. They also investigated the ACE I/D polymorphism with the risk of cancers. There were some differences between these two studies. First, the current meta-analysis included more case-control studies than Ruiter's study. Second, some issues which may affect the results of meta-analysis were addressed in our study, such as publication bias, sensitivity analysis and HWE analysis. Third, the current study is a meta-analysis, and Ruiter's study is more like a review. Despite of these differences, our study also indicated the ACE I/D polymorphism might not contribute to the risk of cancer, which is consistent with Ruiter's study.
Some limitations of this study should be addressed. First, only published studies in Chinese and English which were included by the selected databases were included for data analysis, some potential studies which were included by other databases or published with other languages or unpublished could be missed. Second, due to lack of original data, we could not evaluate the potential interactions of gene-gene and gene-environment. Third, this meta-analysis included data from Europeans, Asians, African-American and Latino populations, so that, the results are applicable to only these ethnic groups. Fourth, it is reported that PCR amplification of ACE I/D polymorphism using only flanking primer pairs would misclassify 4-5% of the ID genotype as the DD genotype and a second PCR should performed to confirm the DD genotype[50]. However, only a small portion of included studies performed a second PCR, indicating the possibility of imprecise results of the meta-analysis. Fifth, some other important factors may also bias our results, such as smoking status, the heterogeneity of cancer patients (pre- or post -menopausal breast cancer patients); genotyping technique changes over time and other unknown function of RAS system et al. However, this meta-analysis also has some advantages. First, the comprehensive meta-analysis included more than 15,000 individuals; it is statistically more powerful than any single study. Second, this is a comprehensive meta-analysis concerning cancer risk and ACE D/I polymorphism and the result also indicated a gene in RAS may not contribute to cancer risk. Third, the result of publication bias is not significant; indicating the conclusion of this study may be unbiased.
In summary, this meta-analysis suggests that the I/D polymorphism in the ACE gene may not contribute to susceptibility to cancer. However, larger well-designed studies are warranted to validate these findings. Moreover, future studies should also investigate gene-gene and gene-environment interactions to better display the association between the polymorphisms in ACE gene and cancer risk.
Conclusion
These results suggest that the D/I polymorphism in ACE gene may not contribute to susceptibility to cancer.
Conflict of interests
The authors declare that they have no competing interests.
Authors' contributions
HF designed the research. YGZ and JZ searched the publications, extracted the data and wrote the materials and methods, results, discussion. YD checked all data. ZPX, HLH and CT was responsible for data synthesis and helped designed the study's analytic strategy. JH wrote the introduction. JH and XBL edited the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Yonggang Zhang, Email: yonggang-zhang@sohu.com.
Jie He, Email: jieh321@sohu.com.
Yao Deng, Email: chagallday@gmail.com.
Jie Zhang, Email: jiezhangwzyx@sohu.com.
Xiaobo Li, Email: lxb166@yeah.net.
Zhangpeng Xiang, Email: zpxwscu@sohu.com.
Honglang Huang, Email: hlhwscu@sohu.com.
Can Tian, Email: doudouniao24@163.com.
Jin Huang, Email: huang.jin2010@sohu.com.
Hong Fan, Email: z85445417@yahoo.cn.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (30470761 and 30871117).
References
- Eriksson U, Danilczyk U, Penninger JM. Just the beginning: novel functions for angiotensin-converting enzymes. Curr Biol. 2002;12:R745–R752. doi: 10.1016/S0960-9822(02)01255-1. [DOI] [PubMed] [Google Scholar]
- Vairaktaris E, Yapijakis C, Tsigris C, Vassiliou S, Derka S, Nkenke E, Spyridonidou S, Vylliotis A, Vorris E, Ragos V. et al. Association of angiotensin-converting enzyme gene insertion/deletion polymorphism with increased risk for oral cancer. Acta Oncol. 2007;46:1097–1102. doi: 10.1080/02841860701373579. [DOI] [PubMed] [Google Scholar]
- Nacak M, Nacak I, Sanli M, Ozkur M, Pektas M, Aynacioglu AS. Association of angiotensin converting enzyme gene insertion/deletion polymorphism with lung cancer in Turkey. Cancer Genet Cytogenet. 2010;198:22–26. doi: 10.1016/j.cancergencyto.2009.11.019. [DOI] [PubMed] [Google Scholar]
- Sugimoto M, Furuta T, Shirai N, Ikuma M, Sugimura H, Hishida A. Influences of chymase and angiotensin I-converting enzyme gene polymorphisms on gastric cancer risks in Japan. Cancer Epidemiol Biomarkers Prev. 2006;15:1929–1934. doi: 10.1158/1055-9965.EPI-06-0339. [DOI] [PubMed] [Google Scholar]
- Yigit B, Bozkurt N, Narter F, Yilmaz H, Yucebas E, Isbir T. Effects of ACE I/D polymorphism on prostate cancer risk, tumor grade and metastatis. Anticancer Res. 2007;27:933–936. [PubMed] [Google Scholar]
- Haiman CA, Henderson SO, Bretsky P, Kolonel LN, Henderson BE. Genetic variation in angiotensin I-converting enzyme (ACE) and breast cancer risk: the multiethnic cohort. Cancer Res. 2003;63:6984–6987. [PubMed] [Google Scholar]
- Rocken C, Lendeckel U, Dierkes J, Westphal S, Carl-McGrath S, Peters B, Kruger S, Malfertheiner P, Roessner A, Ebert MP. The number of lymph node metastases in gastric cancer correlates with the angiotensin I-converting enzyme gene insertion/deletion polymorphism. Clin Cancer Res. 2005;11:2526–2530. doi: 10.1158/1078-0432.CCR-04-1922. [DOI] [PubMed] [Google Scholar]
- Kitsios G, Zintzaras E. ACE (I/D) polymorphism and response to treatment in coronary artery disease: a comprehensive database and meta-analysis involving study quality evaluation. BMC Med Genet. 2009;10:50. doi: 10.1186/1471-2350-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasku V, Vasku JA, Goldbergova MP, Vasku A. Association of variants in angiotensin-converting enzyme and endothelin-1 genes with phototherapy in cutaneous T-cell lymphoma. Acta Dermatovenerol Alp Panonica Adriat. 2004;13:111–116. 118. [PubMed] [Google Scholar]
- Goto Y, Ando T, Nishio K, Ishida Y, Kawai S, Goto H, Hamajima N. The ACE gene polymorphism is associated with the incidence of gastric cancer among H. pylori seropositive subjects with atrophic gastritis. Asian Pac J Cancer Prev. 2005;6:464–467. [PubMed] [Google Scholar]
- Wang HW, Nie ZS, Duan YZ, Liu DD, Han ZH, Zhang XH, Hua B. Association between polymorphism in ACE gene with lung cancer and COPD patients(Chinese) Bei Jing Yi Xue. 2000;22:364–365. [Google Scholar]
- van der Knaap R, Siemes C, Coebergh JW, van Duijn CM, Hofman A, Stricker BH. Renin-angiotensin system inhibitors, angiotensin I-converting enzyme gene insertion/deletion polymorphism, and cancer: the Rotterdam Study. Cancer. 2008;112:748–757. doi: 10.1002/cncr.23215. [DOI] [PubMed] [Google Scholar]
- Toma M, Cimponeriu D, Apostol P, Stavarachi M, Cojocaru M, Belusica L, Craciun AM, Radu I, Gavrila L. Lack of association between ACE ID polymorphism and colorectal cancer in Romanian patients. Chirurgia (Bucur) 2009;104:553–556. [PubMed] [Google Scholar]
- Arima H, Kiyohara Y, Tanizaki Y, Nakabeppu Y, Kubo M, Kato I, Sueishi K, Tsuneyoshi M, Fujishima M, Iida M. Angiotensin I-converting enzyme gene polymorphism modifies the smoking-cancer association: the Hisayama Study. Eur J Cancer Prev. 2006;15:196–201. doi: 10.1097/01.cej.0000199506.15571.37. [DOI] [PubMed] [Google Scholar]
- Yaren A, Oztop I, Turgut S, Turgut G, Degirmencioglu S, Demirpence M. Angiotensin-converting enzyme gene polymorphism is associated with anemia in non small-cell lung cancer. Exp Biol Med (Maywood) 2008;233:32–37. doi: 10.3181/0705-RM-141. [DOI] [PubMed] [Google Scholar]
- Nikiteas N, Tsigris C, Chatzitheofylaktou A, Yannopoulos A. No association with risk for colorectal cancer of the insertion/deletion polymorphism which affects levels of angiotensin-converting enzyme. In Vivo. 2007;21:1065–1068. [PubMed] [Google Scholar]
- Tunny TJ, Xu L, Richardson KA, Stowasser M, Gartside M, Gordon RD. Insertion/deletion polymorphism of the angiotensin-converting enzyme gene and loss of the insertion allele in aldosterone-producing adenoma. J Hum Hypertens. 1996;10:827–830. [PubMed] [Google Scholar]
- Sierra DE, Sanchez CJ, Rosales GR, Gutierrez RS, Vazquez CJ, Solano MH, Moran MM. Angiotensin-converting enzyme insertion/deletion and angiotensin type 1 receptor A1166C polymorphisms as genetic risk factors in benign prostatic hyperplasia and prostate cancer. J Renin Angiotensin Aldosterone Syst. 2009;10:241–246. doi: 10.1177/1470320309352800. [DOI] [PubMed] [Google Scholar]
- Tong Y, Lin Y, Zhang Y, Yang JY, Zhang YW, Liu HC, Zhang B. Association between TCF7L2 gene polymorphisms and susceptibility to Type 2 Diabetes Mellitus: a large Human Genome Epidemiology (HuGE) review and meta-analysis. BMC Med Genet. 2009;10:15. doi: 10.1186/1471-2350-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao TF, Zhao JP. Association of the apolipoprotein A5 gene -1131 T > C polymorphism with fasting blood lipids: a meta-analysis in 37859 subjects. BMC Med Genet. 2010;10:120. doi: 10.1186/1471-2350-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang J, Huang J, Li X, He C, Tian C, Peng C, Guo L, Xiao Y, Fan H. Polymorphisms in the transforming growth factor-beta1 gene and the risk of asthma: A meta-analysis. Respirology. 2010;15:643–650. doi: 10.1111/j.1440-1843.2010.01748.x. [DOI] [PubMed] [Google Scholar]
- Liu L, Zhuang W, Wang C, Chen Z, Wu XT, Zhou Y. Interleukin-8 -251 A/T gene polymorphism and gastric cancer susceptibility: A meta-analysis of epidemiological studies. Cytokine. 2010;50:328–334. doi: 10.1016/j.cyto.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Zhang YG, Huang J, Zhang J, Li XB, He C, Xiao YL, Tian C, Wan H, Zhao YL, Tsewang YG. et al. RANTES gene polymorphisms and asthma risk: A meta-analysis. Arch Med Res. 2010;41:50–58. doi: 10.1016/j.arcmed.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Freitas-Silva M, Pereira D, Coelho C, Bicho M, Lopes C, Medeiros R. Angiotensin I-converting enzyme gene insertion/deletion polymorphism and endometrial human cancer in normotensive and hypertensive women. Cancer Genet Cytogenet. 2004;155:42–46. doi: 10.1016/j.cancergencyto.2004.03.020. [DOI] [PubMed] [Google Scholar]
- Koh WP, Yuan JM, Sun CL, van den Berg D, Seow A, Lee HP, Yu MC. Angiotensin I-converting enzyme (ACE) gene polymorphism and breast cancer risk among Chinese women in Singapore. Cancer Res. 2003;63:573–578. [PubMed] [Google Scholar]
- Srivastava K, Srivastava A, Mittal B. Angiotensin I-converting enzyme insertion/deletion polymorphism and increased risk of gall bladder cancer in women. Dna Cell Biol. 2010;29:417–422. doi: 10.1089/dna.2010.1033. [DOI] [PubMed] [Google Scholar]
- Alves CS, Ribeiro DNS, Nogueira-de-Souza NC, Valleta DCC, Massad CA, Juvenal LJ, Vieira GM, Guerreiro DSI. Association between the angiotensin-converting enzyme (insertion/deletion) and angiotensin II type 1 receptor (A1166C) polymorphisms and breast cancer among Brazilian women. J Renin Angiotensin Aldosterone Syst. 2009;10:51–58. doi: 10.1177/1470320309102317. [DOI] [PubMed] [Google Scholar]
- Yaren A, Turgut S, Kursunluoglu R, Oztop I, Turgut G, Kelten C, Erdem E. Association between the polymorphism of the angiotensin-converting enzyme gene and tumor size of breast cancer in premenopausal patients. Tohoku J Exp Med. 2006;210:109–116. doi: 10.1620/tjem.210.109. [DOI] [PubMed] [Google Scholar]
- Cheon KT, Choi KH, Lee HB, Park SK, Rhee YK, Lee YC. Gene polymorphisms of endothelial nitric oxide synthase and angiotensin-converting enzyme in patients with lung cancer. Lung. 2000;178:351–360. doi: 10.1007/s004080000039. [DOI] [PubMed] [Google Scholar]
- Ding XJ. Analysis of the relationship between the polymorphism of angiotensin-converting enzyme gene and lung cancer (Chinese) 2008. [DOI] [PubMed]
- Rocken C, Neumann K, Carl-McGrath S, Lage H, Ebert MP, Dierkes J, Jacobi CA, Kalmuk S, Neuhaus P, Neumann U. The gene polymorphism of the angiotensin I-converting enzyme correlates with tumor size and patient survival in colorectal cancer patients. Neoplasia. 2007;9:716–722. doi: 10.1593/neo.07418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vairaktaris E, Serefoglou Z, Avgoustidis D, Yapijakis C, Critselis E, Vylliotis A, Spyridonidou S, Derka S, Vassiliou S, Nkenke E. et al. Gene polymorphisms related to angiogenesis, inflammation and thrombosis that influence risk for oral cancer. Oral Oncol. 2009;45:247–253. doi: 10.1016/j.oraloncology.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Zhang QZ, Liu XM, Zhang ZG, Wang LH. Analysis of the relationship between polymorphism of angiotensin-converting enzyme gene and lung cancer(Chinese) Zhong Guo Fei Ai Za Zhi. 2005;8:211–214. doi: 10.3779/j.issn.1009-3419.2005.03.11. [DOI] [PubMed] [Google Scholar]
- Bardi E, Jenei C, Kiss C. Polymorphism of angiotensin converting enzyme is associated with severe circulatory compromise in febrile neutropenic children with cancer. Pediatr Blood Cancer. 2005;45:217–221. doi: 10.1002/pbc.20400. [DOI] [PubMed] [Google Scholar]
- Holla L, Vasku A, Soucek M, Krahulcova E, Hajek D, Znojil V. Angiotensin-converting enzyme gene polymorphism and blood groups of the ABO system in Leukaemias compared with controls and hypertensives. Scripta Medica(Brno) 1998;71:471–477. [Google Scholar]
- Gold B, Kirchhoff T, Stefanov S, Lautenberger J, Viale A, Garber J, Friedman E, Narod S, Olshen AB, Gregersen P. et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Natl Acad Sci USA. 2008;105:4340–4345. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landa I, Ruiz-Llorente S, Montero-Conde C, Inglada-Perez L, Schiavi F, Leskela S, Pita G, Milne R, Maravall J, Ramos I. et al. The variant rs1867277 in FOXE1 gene confers thyroid cancer susceptibility through the recruitment of USF1/USF2 transcription factors. PLoS Genet. 2009;5:e1000637. doi: 10.1371/journal.pgen.1000637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Marchand L. Genome-wide association studies and colorectal cancer. Surg Oncol Clin N Am. 2009;18:663–668. doi: 10.1016/j.soc.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner AJ, Hooper NM. The angiotensin-converting enzyme gene family: genomics and pharmacology. Trends Pharmacol Sci. 2002;23:177–183. doi: 10.1016/S0165-6147(00)01994-5. [DOI] [PubMed] [Google Scholar]
- Campa D, Vodicka P, Pardini B, Naccarati A, Carrai M, Vodickova L, Novotny J, Hemminki K, Försti A, Barale R. et al. A gene-wide investigation on polymorphisms in the taste receptor 2R14 (TAS2R14) and susceptibility to colorectal cancer. BMC Med Genet. 2010;11:88. doi: 10.1186/1471-2350-11-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Eeles RA. Genome-wide association studies in cancer. Hum Mol Genet. 2008;17:R109–R115. doi: 10.1093/hmg/ddn287. [DOI] [PubMed] [Google Scholar]
- Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, Wacholder S, Wang Z, Welch R, Hutchinson A. et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiemeney LA, Grotenhuis AJ, Vermeulen SH, Wu X. Genome-wide association studies in bladder cancer: first results and potential relevance. Curr Opin Urol. 2009;19:540–546. doi: 10.1097/MOU.0b013e32832eb336. [DOI] [PubMed] [Google Scholar]
- Song H, Ramus SJ, Tyrer J, Bolton KL, Gentry-Maharaj A, Wozniak E, Anton-Culver H, Chang-Claude J, Cramer DW, DiCioccio R. et al. A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nat Genet. 2009;41:996–1000. doi: 10.1038/ng.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, Yu K, Chatterjee N, Welch R, Hutchinson A. et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- Petersen GM, Amundadottir L, Fuchs CS, Kraft P, Stolzenberg-Solomon RZ, Jacobs KB, Arslan AA, Bueno-de-Mesquita HB, Gallinger S, Gross M. et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet. 2010;42:224–228. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CM, Wang RY, Chen JW, Fann CS, Leu HB, Ho HY, Ting CT, Lin TH, Sheu SH, Tsai WC. et al. A genome-wide association study identifies new loci for ACE activity: potential implications for response to ACE inhibitor. Pharmacogenomics J. 2010;10:537–544. doi: 10.1038/tpj.2009.70. [DOI] [PubMed] [Google Scholar]
- Feulner TM, Laws SM, Friedrich P, Wagenpfeil S, Wurst SH, Riehle C, Kuhn KA, Krawczak M, Schreiber S, Nikolaus S. et al. Examination of the current top candidate genes for AD in a genome-wide association study. Mol Psychiatry. 2010;15:756–766. doi: 10.1038/mp.2008.141. [DOI] [PubMed] [Google Scholar]
- Ruiter R, Visser E, Van Duijn M, HCh Stricker B. The ACE Insertion/Deletion Polymorphism and Risk of Cancer, a Review and Meta-Analysis of the Literature. Current Cancer Drug Targets. 2011;11:421–430. doi: 10.2174/156800911795538147. [DOI] [PubMed] [Google Scholar]
- Meyer BR, Vashishtha A, Hillis G, Al-Mohammad A, Jennings K, Ohishi M, Rakugi H, Higaki J, Miki T, Ogihara T. Angiotensin-Converting-Enzyme Genotype and Ischemic Heart Disease. New Engl J Med. 1995;333:458–460. [PubMed] [Google Scholar]