Abstract
Tachykinin-related peptide (TRP) refers to a large and structurally diverse family of neuropeptides found in vertebrate and invertebrate nervous systems. These peptides have various important physiological functions, from regulating stress in mammals to exciting the gastric mill (food chewing) and pyloric (food filtering) rhythm in the stomatogastric nervous system (STNS) of decapod crustaceans. Here, a novel TRP, which we named CalsTRP (Callinectes sapidus TRP), YPSGFLGMRamide (m/z 1026.52), was identified and de novo sequenced using a multifaceted mass spectrometry-based platform in both the central nervous system (CNS) and STNS of C. sapidus. We also found, using isotopic formaldehyde labeling, that CalsTRP in the C. sapidus brain and commissural ganglion (CoG) was up-regulated after food intake, suggesting that TRPs in the CNS and STNS are involved in regulating feeding in Callinectes. Using imaging mass spectrometry, we determined that the previously identified CabTRP Ia (APSGFLGMRamide) and CalsTRP were colocalized in the C. sapidus brain. Lastly, our electrophysiological studies show that bath-applied CalsTRP and CabTRP Ia each activate the pyloric and gastric mill rhythms in C. sapidus, as shown previously for pyloric rhythm activation by CabTRP Ia in the crab Cancer borealis. In summary, the newly identified CalsTRP joins CabTRP Ia as a TRP family member in the decapod crustacean nervous system, whose actions include regulating feeding behavior.
Keywords: Neuropeptide, CalsTRP, Callinectes sapidus, tachykinin-related peptide, mass spectrometry, feeding, stomatogastric nervous system
Tachykinins constitute a large and structurally diverse family of neuropeptides in the animal kingdom, with members present in both vertebrates and invertebrates.1−3 In the latter group, these peptides are termed tachykinin-related peptides (TRPs), and they share the conserved C-terminal pentapeptide sequence FX1GX2Ramide.4−10 While TRPs differ in their primary amino acid sequences between vertebrates and invertebrates, the ability to localize TRPs in invertebrates using antisera directed against vertebrate tachykinin1 and to physiologically inhibit the actions of TRPs using vertebrate tachykinin receptor antagonists10 in decapod crustaceans suggests that they have conserved structure and functions in both animal groups.
With their presence in both central and peripheral tissues, tachykinins mediate various physiological and pathological effects including nociception, regulation of stress, pain perception, coordination of gastrointestinal motility, stimulation of contractions of visceral and skeletal muscle, and triggering a motor rhythm in the crab stomatogastric ganglion (STG).10−15 The involvement of tachykinins in various physiological processes makes them very important in drug discovery.16,17 In particular, tachykinins play an important role in feeding, which is critical for energy homeostasis and animal survival. In mammals, the tachykinin peptides substance P and neuropeptide K consistently suppress feeding behavior in rats.2,18,19 In contrast, in crustaceans, TRPs exhibit significant changes in response to food intake20−22 and they have excitatory actions on the motor circuits for chewing (gastric mill motor circuit) and the filtering of chewed food (pyloric motor circuit).10,23,24
The decapod crustacean nervous system is a good invertebrate model system to study the TRP physiological function in feeding-related motor patterns because it is accessible and well-characterized at the level of identified neurons.25−28 In these animals, the TRPs are distributed in CNS interneurons, including some within the stomatogastric nervous system (STNS).1,11,29−31 The STNS is composed of four ganglia, including the STG, esophageal ganglion (OG) and the paired commissural ganglia (CoG). The supraesophageal ganglion (brain) communicates with the STNS via the inferior ventricular nerve (ivn), which connects to the OG, and the paired circumesophageal commissures (cocs), each of which connects to a CoG.32 Our previous work documented a feeding-related change in the amount of CabTRP, including APSGFLGMRa (CabTRP Ia) and TPSGFLGMRa, in the brain,20 suggesting an involvement of these peptides within the CNS in feeding behavior.
In the current study, we discover a novel TRP, YPSGFLGMRamide (CalsTRP), in Callinectes sapidus via a new multifaceted mass spectrometry (MS) based platform combining multiple microseparation techniques with various MS detection strategies. Furthermore, we explore the feeding-related, quantitative changes of CalsTRP in the brain and CoG of C. sapidus through tissue extraction followed by stable isotopic labeling. The results show elevated expression of TRPs, including CabTRP Ia and CalsTRP, after feeding in both tissues. Using imaging mass spectrometry (IMS), we also determined that these two TRP isoforms are colocalized in the brain. Lastly, we show that CalsTRP has biological activity comparable to CabTRP Ia in C. sapidus, insofar as bath applying either peptide (10–6 or 10–7 M) to the STG activated the gastric mill (food chewing) rhythm and the pyloric (food filtering) rhythm. Overall, this study shows that the multifaceted MS approach is a powerful and efficient tool for discovery and functional study of neuropeptides in the crustacean nervous system. The newly identified CalsTRP represents another TRP neuropeptide family member playing a regulatory role in the feeding behavior.
Results and Discussion
Detection of Putative Tachykinins in Different Neural Tissues in C. sapidus
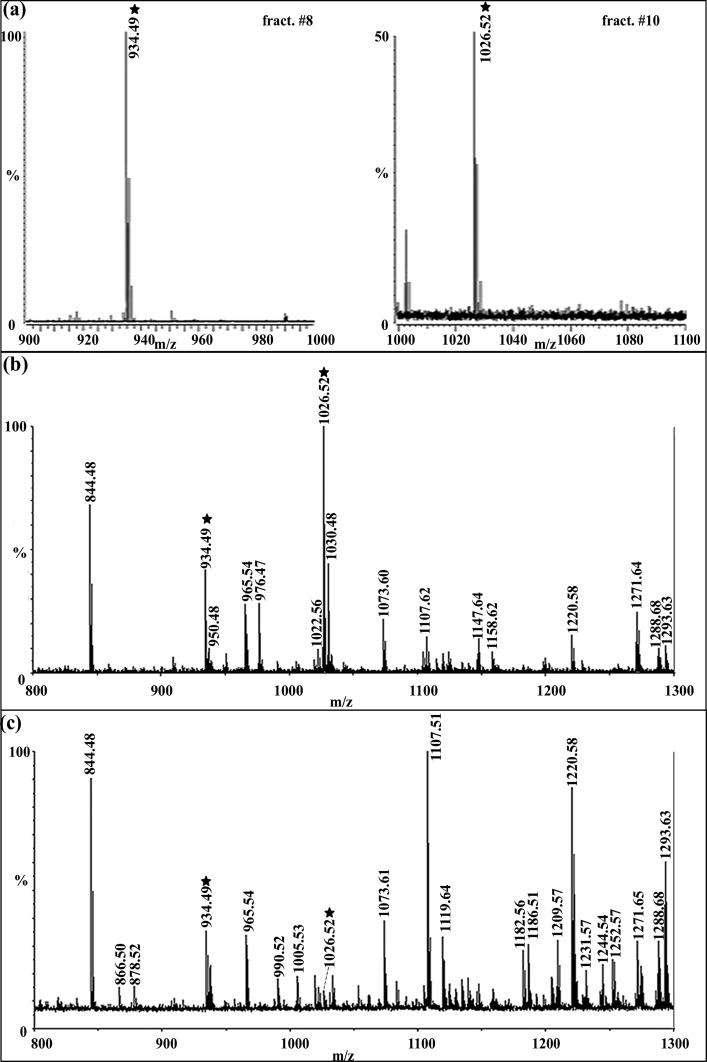
We found that two putative TRPs coexist in the C. sapidus nervous system. Specifically, in this system, we observed two TRPs by accurate mass measurements via different mass spectrometric approaches, including nanoLC-ESI-QTOF, MALDI-FTMS, and MALDI-TOF/TOF, combined with different tissue preparation methods including direct tissue profiling, crude extract, and HPLC fractions. Figure 1a shows the neuropeptide spectra of the brain HPLC fractions #8 and #10 acquired by MALDI-FTMS. In these spectra, CabTRP Ia (m/z 934.49) was observed in fraction #8 and putative C. sapidus TRP (m/z 1026.52) was observed in fraction #10, collected at 16 and 20 min in a 120 min gradient of 5–95% Solution B (see Methods). The similar LC elution profile and simultaneous detection of both peaks in the CoG and STG suggested that the peptide peak at m/z 1026.5 was likely a novel TRP in C. sapidus. Figure 1b shows the neuropeptide profile of CoG extraction by MALDI-TOF/TOF, wherein both m/z 934.49 and m/z 1026.52 peaks were present, with the latter being the most abundant peptide peak. This result agrees with previous findings showing that TRPs are highly abundant in the CoGs.33,34 This high abundance in the CoG may result from the dense collection of TRP-containing nerve terminals of the ∼100 postesophageal commissure neurons that innervate a neurohemal structure within the CoG neuropil.30,33 Figure 1c shows the direct tissue profile for a STG by MALDI-TOF/TOF, in which both putative TRPs were present, albeit in relatively low abundance compared to that in the brain (data not shown) and CoG. If the TRP-containing neuronal organization in C. sapidus is similar to that in C. borealis, then the relatively low TRP abundance in the STG results from these peptides being present only in the axon terminals of a single pair of projection neurons, with no TRP-containing STG neurons.10,11,31 In summary, with high sensitivity MALDI-TOF/TOF, high resolution and high mass measurement accuracy of the MALDI-FT MS, and the de novo sequencing capability of ESI-Q-TOF, we readily detected two TRPs in the brain, CoG and STG of C. sapidus, in direct tissue, crude tissue extracts, and HPLC fractions, providing complementary data to support the presence of TRPs in the blue crab nervous system and enable detailed characterization of these important signaling peptides.
Figure 1.
Identification of putative tachykinins in C. sapidus brain, CoG and STG prepared with different sample processing methods using multiple mass spectrometers. (a) HPLC fractions # 8 and #10 of brain extract analyzed by MALDI-FTMS reveal the presence of CabTRP Ia (m/z 934.39) and putative CalsTRP (m/z 1026.52), respectively. (b) Mass spectrum of a crude CoG extract acquired on MALDI-TOF/TOF and (c) STG direct tissue profiling on MALDI-TOF/TOF reveal the presence of two putative tachykinins by mass profiling. The abundant peaks in the spectra are labeled with their protonated masses. Mass spectral peaks highlighted by stars are two putative tachykinins, including the previously identified CabTRP Ia (APSGFLGMRa: m/z 934.49) and the novel putative TRP in C. sapidus (m/z 1026.52).
De Novo Sequencing and Sequence Confirmation of Tachykinin Peptides in C. sapidus
To confirm the identity of CabTRP Ia and determine the amino acid sequence of the putative novel TRP in C. sapidus observed by mass profiling, we performed tandem mass spectrometry (MS/MS) experiments. Two TRPs, including CabTRP Ia (m/z 934.49, APSGFLGMRamide)10 and a novel peptide YPSGFLGMRamide (m/z 1026.52), were first sequenced in C. sapidus brain tissue extract and HPLC fractions by nanoLC-ESI-QTOF. Comparison of the two peptide sequences confirmed that the new peptide at m/z 1026.52 is indeed a novel member of the TRP family, and it is named CalsTRP (Callinectes sapidus TRP) in this study. Subsequently, both TRPs were sequenced from CoG and STG tissue extracts.
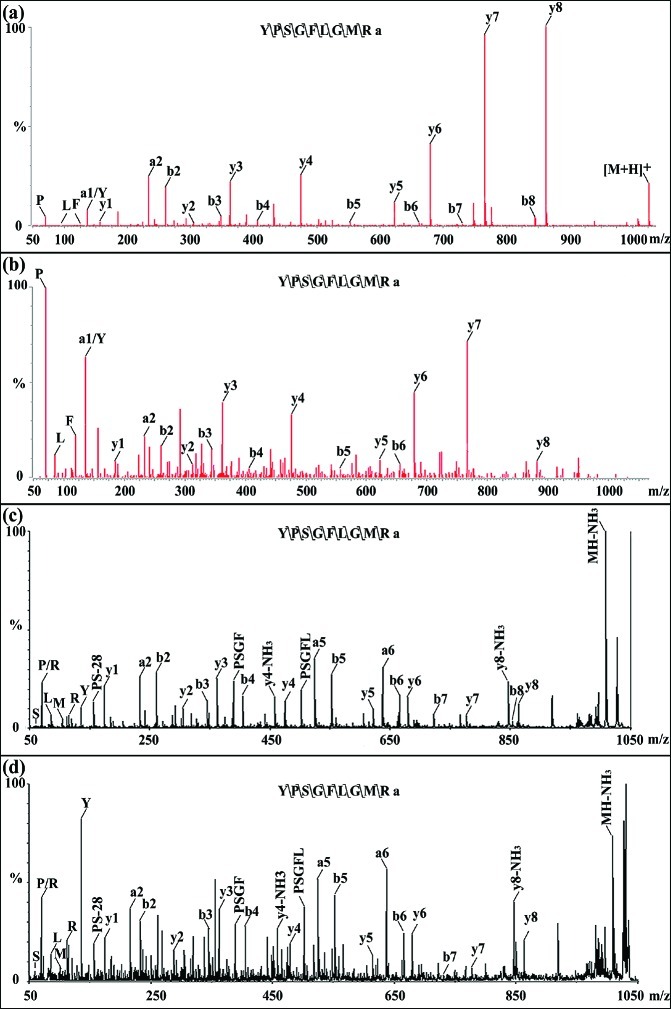
To confirm the sequence of the novel peptide, a peptide standard with the derived sequence YPSGFLGMRamide was synthesized and MS/MS experiments were performed using both nanoLC-ESI-QTOF and MALDI-TOF/TOF. In Figure 2, panels a and b show the MS/MS comparison of the standard peptide to the peptide from brain extract by ESI-QTOF, while panels c and d show the MS/MS comparison of the standard peptide to the CoG extract by MALDI TOF/TOF. In panel a, immonium ions P, L, F, Y, all y-ions (y1–y8) and a1, a2, b2–b8 were observed while b7, b8 were absent in panel b, presumably due to relatively lower abundance of the peptide in the complex brain extract. In panel c, immonium ions for S, P, L, R, M, Y, all y-ions (y1–y8), a2, a5, a6, b2–b8, and some internal fragment ions including PS-28, PSGF, and PSGFL were observed, while in panel d all the above ions were observed except b8. It is evident from both experiments that the fragmentation patterns of the synthesized peptide standard and the endogenous CalsTRP isolated from the brain and CoG tissue extract match very well, further supporting the de novo sequencing result and identity of CalsTRP. In comparison with the crabs Cancer borealis and Carcinus maenas, which contain CabTRP Ia (APSGFLGMRamide; m/z 934.49) and CabTRP II (TPSGFLGMRamide; m/z 964.50),10,35−37 our results show that C. sapidus contains CabTRP Ia and CalsTRP (YPSGFLGMRamide). Thus far, CabTRP Ia is present in all crustacean species studied, whereas one or more additional TRPs are present in different subsets.22,34,37−40 The currently available databases for crustaceans, such as nonredundant protein sequences (nr), only contain several TRP prohormones from limited species such as Homarus americanus, Procambarus clarkii, and Panulirus interruptus that encode APSGFLGMRamide and TPSGFLGMRamide.38,41 The novel TRP described here suggests the existence of a multi-TRP containing preprotachykinin in C. sapidus that is yet to be discovered.
Figure 2.
Tandem mass spectrometry (MS/MS) spectra of synthesized CalsTRP (YPSGFLGMRa, m/z 1026.52) and the novel CalsTRP from real biological samples. (a) MS/MS of synthesized peptide standard YPSGFLGMRa by ESI-QTOF. (b) MS/MS of CalsTRP from brain neuropeptide extract analyzed by nano LC-ESI-QTOF. (c) MS/MS of synthesized peptide standard by MALDI TOF/TOF. (d) MS/MS of CalsTRP from CoG extract by MALDI TOF/TOF. Major sequence-specific fragment ions (immonium ions indicative of amino acid residues, a-, b-, and y-type ions) are indicated in each spectrum. Comparison of synthesized CalsTRP standard and the novel CalsTRP in tissue extract reveals almost identical fragmentation patterns, confirming the tandem MS-derived peptide sequence.
Capillary Electrophoresis (CE) Separation for Improved Quantitation Study
CE is a powerful modern microcolumn separation technique that has gained widespread use in biomolecular analysis because of its compatible buffer system for biological samples, unparalleled resolution, and minimal sample consumption. CE has been successfully employed to enhance neuropeptide profiling coupled to MALDI MS detection in our previous reports.42,43 Here, its coupling to MS detection successfully enabled us to address the issues of the CalsTRP peak overlapping with an RFamide peptide peak (see below) and the limited sample amounts available for the analyses of CoG and brain extracts. Generally, the migration order of peptides in CE is determined by their family specific sequences (amino acid residues and mass) and isoelectric point values (net charge). The TRPs have +2 net charge while the RFamide peptide has a +3 charge under the CE separation conditions (i.e., pH 5.0). Thus, the RFamide migrated faster than CalsTRP which enables them to be separated.
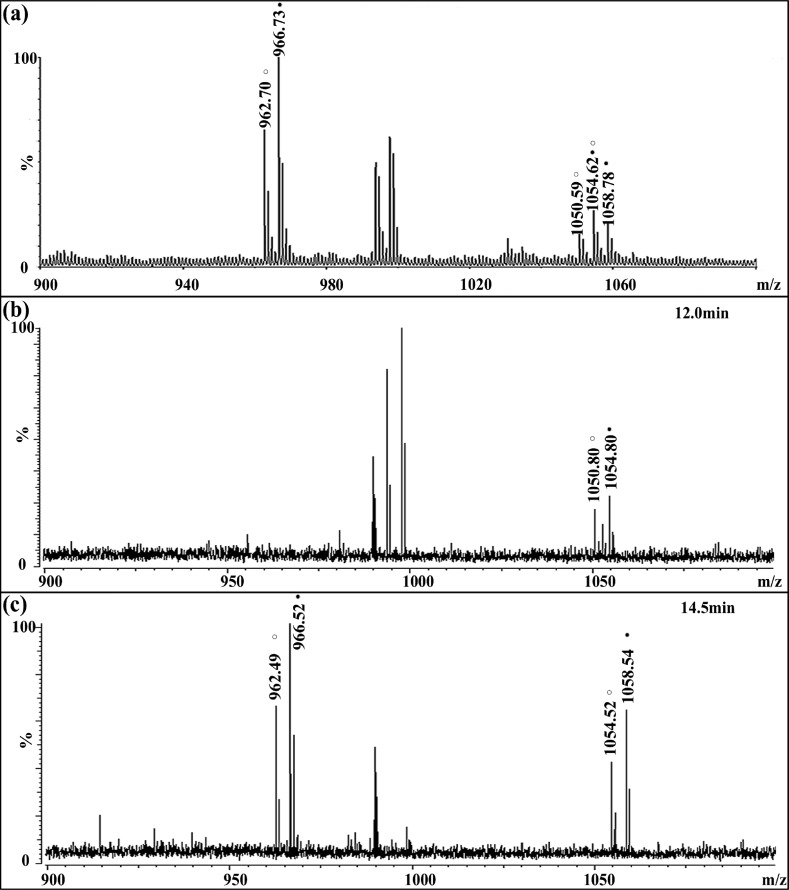
In the C. sapidus CoG, the RFamide peptide GNRNFLRFamide (m/z 1022.56) was detected (Figure 1b) with an appreciable intensity, as was also the case in the brain sample (data not shown). After formaldehyde labeling, CalsTRP peptide YPSGFLGMRamide (m/z 1026.52) was present at m/z 1054.57 with the light label and m/z 1058.59 with the heavy label, as shown in Figure 3a. The differential labeling generated 4 Da of mass difference for the same peptide from control and fed animals. Similarly, GNRNFLRFamide (m/z 1022.56) became the 1050.61/1054.63 pair after labeling. The two 1054 peaks overlapped in the direct MALDI MS spectrum and were highly difficult to resolve, even with the high-resolution FTMS, due to their very close mass values (1054.57 vs 1054.63). To resolve these closely overlapped peptides and thus obtain accurate expression changes of the CalsTRP neuropeptide after feeding, a reliable separation method was needed prior to MS analysis. CE separation was therefore employed to separate these peptide pairs. As shown in Figure 3b, peak pair 1050/1054 eluted at 12.0 min (fraction #24) and two TRP peak pairs (992/996 and 1054/1058) coeluted later, at 14.5 min (fraction #29) (Figure 3c). The separation of the two peptide pairs enabled confident assignment and accurate quantitation of the peak pairs.
Figure 3.
Representative spectra of isotopic formaldehyde-labeled mixture of extracts from the brain of a fed crab and an unfed crab before and after CE separation. The unfed sample is labeled with FH2, and the fed sample is labeled with FD2. The heavy labeled peaks are indicated with closed circles, and the light labeled peaks are indicated with open circles. (a) Isotopic formaldehyde labeled mixture of brain extract before CE separation showing the overlapping of heavy-labeled RFamde and light-labeled CalsTRP at m/z 1054.62. (b) CE separation of brain extracts mixture at 12.0 min showing the labeled RFamide pair at m/z 1050.80/1054.80. (c) CE separation of brain extraction mixture at 14.5 min showing the labeled CabTRP Ia pair at m/z 962.49/966.52 and CalsTRP pair at m/z 1054.52/1058.54.
Another advantage of the CE separation approach for neuropeptide analysis is that the sample consumption is very low. Each loading requires no more than 50 nL of sample, and thus, it was easy to obtain three to five technical replicates with minimal sample consumption. Subsequently, the ratio of the heavy/light labeled peptide peaks can be averaged from both biological and technical replicates.
Expression Level Change of TRPs in the Brain and CoG between Fed and Unfed Animals Using Formaldehyde Labeling
Isotopic formaldehyde labeling is a well-established, fast, and simple reaction which can be employed for quantitative analysis of differential proteomic and peptidomic studies.44−46 As shown in a previous study from our group,20 formaldehyde labeling is also an efficient and accurate method for measuring the quantitative expression changes involved in behavioral (i.e., feeding) studies. Isotopic formaldehyde labels the N termini of peptides with two methyl groups, resulting in 28 and 32 Da mass shifts for each incorporated light- and heavy-label, respectively, and 4 Da mass differences between light-labeled and heavy-labeled peak pair per incorporated label. This method enables detection of the same peptide in different samples in a single spectrum, and the calculation of the abundance differences based on the signal intensity ratio of each peak pair. Using formaldehyde labeling for the current study, we examined the quantitative changes of neuropeptide expression in crab brain from five fasting and satiated crabs, respectively. Figure 3c shows representative MALDI-FTMS spectra from one group of brain extracts after CE separation. Table 1a lists the five groups of average abundance ratios (crab brain from fed vs unfed crabs) for CalsTRP and CabTRP Ia, calculated from two replicate MS spectra within each group. Table 1b displays four groups of average abundance ratios for the two TRPs in the CoGs from the fed versus unfed crabs.
Table 1. Abundance Ratios of Two Tachykinin Neuropeptides in (a) the Brain from Fed versus Unfed Crabs from Five Groups of Feeding Experiments and (b) the CoG from Fed versus Unfed Crabs from Four Groups of Feeding Experimentsa.
| (a) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Br | 1 | 2 | 3 | 4 | 5 | avg | SEM | p-value |
| 934.39 | 1.56 | 1.52 | 2.24 | 1.68 | 1.20 | 1.64 | 0.17 | <0.01 |
| 1026.52 | 1.4 | 1.52 | 1.27 | 1.70 | 1.24 | 1.43 | 0.08 | <0.01 |
| (b) | |||||||
|---|---|---|---|---|---|---|---|
| CoG | 1 | 2 | 3 | 4 | avg | SEM | p-value |
| 934.39 | 1.43 | 1.22 | 1.40 | 1.20 | 1.31 | 0.06 | <0.01 |
| 1026.52 | 1.02 | 1.24 | 1.45 | 1.08 | 1.20 | 0.11 | <0.05 |
One crab brain (Br) was used to make each extract. Five pairs of crab CoGs were used to make each extract. Each ratio is calculated based on an average of two spectra from MALDI-FTMS analyses. The top row in each table represents values for CabTRP Ia (m/z 934.39), and the bottom row represents values for CalsTRP (m/z 1026.52). Student’s t test was performed to evaluate the differences of each peptide between fed and unfed states, and the p-value of <0.05 was considered as statistically significant.
Significant changes were observed between the fed and unfed conditions for both TRPs (m/z 934.49 and m/z 1026.52). Specifically, CabTRP Ia was consistently elevated in fed crab brain (range: 1.20–2.24-fold increase) and fed crab CoG (1.27–1.70-fold increase), with mean ratios of 1.64 and 1.31, respectively (p < 0.01) (Table 1). Similarly, CalsTRP was up-regulated in the crab brain (1.20–1.43-fold) and CoG (1.02–1.45-fold) after feeding, with a mean ratio of 1.43 (p < 0.01) and 1.20 (p < 0.05), respectively (Table 1).
Members of the tachykinin peptide family have been identified throughout the animal kingdom, from mammals to invertebrates, where they have various physiological and pathological effects on both the CNS and peripheral tissues.18,19 For example, central administration of substance P inhibits feeding behavior in chicks,47 while insect TRPs stimulate the contraction of foregut muscles involved in feeding.48 In a previous feeding study in C. borealis, amounts of two crustacean TRPs were increased in crab brain and CoG after food intake,20 suggesting that this peptide family is related to food intake in crustaceans. The increase in CabTRP Ia levels in the brain observed in this study is consistent with our previous results, as are the parallel results of the novel CalsTRP. This study also presents the first quantitative expression change (increase) of TRPs in fed versus unfed crabs in the CoG, a major component of the STNS which generates feeding-related behaviors.27,28
Tachykinin Localization Study in C. sapidusBrain by MALDI Imaging
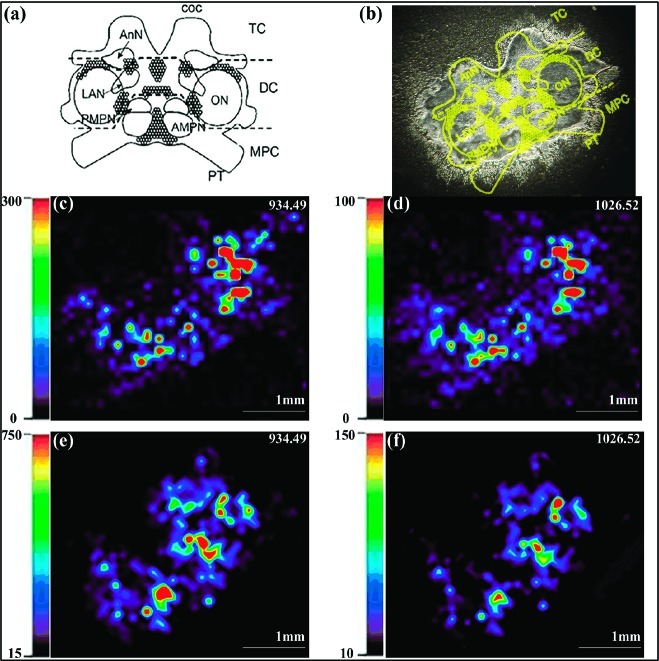
Figure 4a shows a schematic of the crab brain with labeled neuropil regions showing distinctions between the fused ganglia that comprise the brain. Figure 4b shows the optical imaging of a C. sapidus brain tissue slice before application of MALDI matrix. As shown in Figure 4c, the previously identified CabTRP Ia was more concentrated in the anterior (AMPN) and posterior medial protocerebral neuropil (PMPN) and at the boundary of the olfactory lobe (ON) in the C. sapidus brain. CalsTRP exhibited the same localization pattern, as shown in Figure 4d. This observation is consistent with previous results for CabTRP Ia in C. borealis.49,50 Similarly, in fed animals, CabTRP Ia and CalsTRP were present at relatively high amounts in the AMPN and PMPN (Figure 4e, f). The CabTRP Ia signal intensity was consistently higher than that of CalsTRP.
Figure 4.
Imaging mass spectrometry of C. sapidus brain shows the colocalization of CabTRP Ia and CalsTRP. (a) Representation of the ventral surface of the isolated brain with labeled neuropil region and neuron clusters. The main body of the decapod crustacean brain includes the median protocerebrum (MPC), deutocerebrum (DC), and tritocerebrum (TC). Each circumesophageal commissure (coc) projects from the tritocerebrum to the thoracic ganglion (TG), part way between which it contains the CoG (not shown). Five major brain neuropil regions include the anterior (AMPN) and posterior medial protocerebral neuropil (PMPN), olfactory lobe (ON), lateral antenna I neuropil (LAN), and antenna II neuropil (AnN). Note: not all neuropil regions are indicated. (b) An optical image of one slice of C. sapidus brain before application of MALDI matrix, with the brain regions highlighted. (c, d) MALDI-MS images of (c) CabTRP Ia and (d) CalsTRP showing their distribution in an unfed C. sapidus brain. (e, f) MALDI-MS images of (e) CabTRP Ia and (f) CalsTRP in a fed C. sapidus brain. The relative abundance is highlighted with a color-scale bar in each respective panel. Triplicate experiments were performed, and same results were obtained.
For each peptide, the signal intensity from the brain and CoG in fed animals was higher than that of unfed ones, as is evident by comparing the magnitude of the intensity bar in Figure 4. Replicate imaging experiments were performed, and the same results were observed. The elevation of both CabTRP Ia and CalsTRP in the nervous system of fed C. sapidus suggests that these two peptides may be encoded by the same preprotachykinin. The increased signal in the brains of fed versus unfed crabs also agreed with the quantitative results of tissue extracts shown in Table 1. Although IMS cannot give accurate quantitative results due to tissue heterogeneity and inherent variation of MALDI signal response from complex tissue samples, IMS provides semiquantitative assessment that is complementary to the quantitative analysis obtained from tissue extracts under different feeding conditions.
Our observation that, in the brains of both unfed and fed crabs, CabTRP Ia had a higher signal intensity than CalsTRP agreed with the tissue extraction profile of brain (data not shown), and suggests that more copies of CabTRP Ia (APSGFLGMRamide) are likely expressed than that of CalsTRP (YPSGFLGMRamide) on the preprohormone. Alternatively, however, CabTRP Ia might be more stable than CalsTRP. For example, it might be less susceptible to peptidase-mediated degradation. Similar observations were made in C. borealis and Homarus americanus, which have a higher abundance of APSGFLGMRamide than TPSGFLGMRamide.34,38
YPSGFLGMRamide Activates the Gastric Mill and Pyloric Motor Patterns in C. sapidus
Because CalsTRP levels in the C. sapidus brain and CoG were consistently elevated after feeding, we determined whether the novel CalsTRP neuropeptide had physiological actions on a feeding-related motor circuit in C. sapidus. Specifically, we focused on the influence of CalsTRP on the C. sapidus gastric mill (chewing) and pyloric (filtering of chewed food) motor patterns, insofar as an identified CabTRP Ia-containing input pathway to the CoG in C. borealis activates a CabTRP Ia-containing CoG projection neuron that drives these motor patterns in the STG.30
The isolated STNS preparation is shown in Figure 5a. The gastric mill and pyloric motor circuits are located in the STG, and selective superfusion of CalsTRP (10–6–10–7 M) to the C. sapidus STG reversibly activated both motor patterns (Figures 5b,c and 6). The gastric mill rhythm is a two-phase (protraction, retraction) motor pattern that is not spontaneously active either in vivo or in vitro.25,27,28 On average, the gastric mill rhythm was activated with a relatively long delay after the start of CalsTRP (10–6 M) superfusion (3.75 ± 1.1 min, n = 5). The activation of the pyloric motor pattern after the start of CalsTRP (10–6 M) superfusion was at a comparable time scale (3.70 ± 1.7 min, n = 3). This prolonged delay was at least partly due to the time required for the superfused peptide to displace the normal saline from the inflow tubing and reach its working concentration in the dish. The CalsTRP-activated gastric mill motor pattern was comparable to that described previously in other decapod crustacean species, including its rhythmic alternating bursting of protractor (e.g., LG) and retractor (e.g., DG) motor neurons (Figure 5b). During these applications, the gastric mill cycle period (13.3 ± 1.7 s, n = 5) was similar to the gastric mill rhythms elicited in C. borealis.25,28,30 This was also the case for several additional parameters commonly used to define the gastric mill rhythm, including the LG neuron burst duration (4.72 ± 0.4 s, n = 5), number of LG neuron spikes per burst (20.65 ± 1.8 spikes, n = 5), and the LG intraburst firing rate (4.15 ± 0.3 Hz, n = 5).
Figure 5.
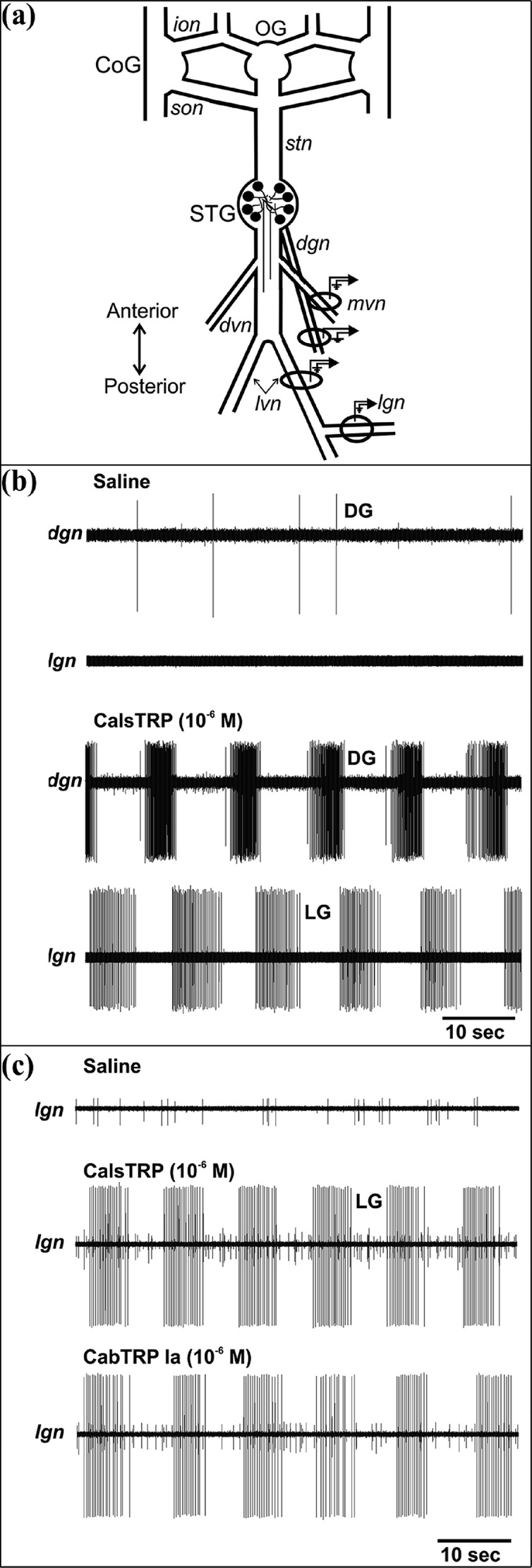
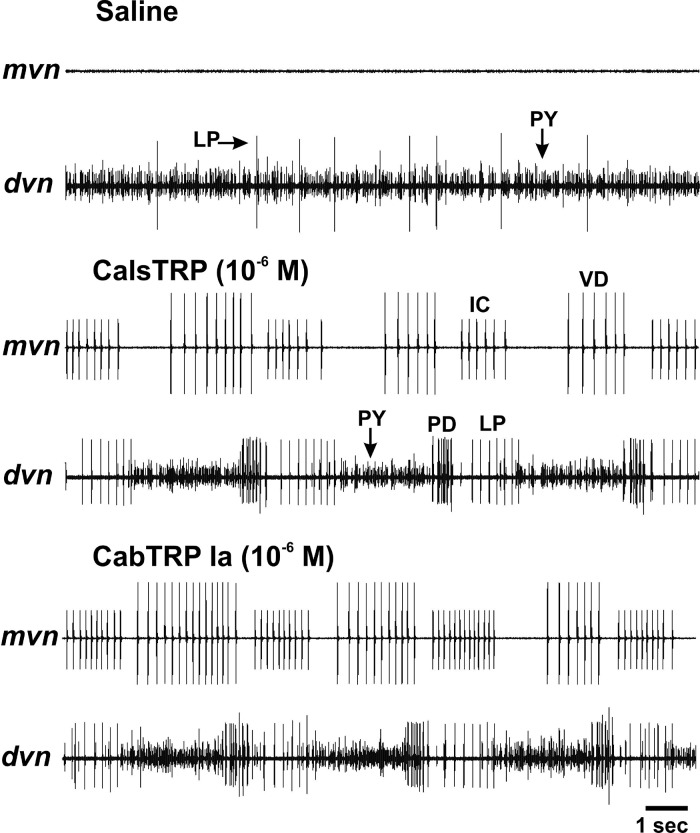
Bath applied CalsTRP and CabTRP each activate the gastric mill rhythm in the C. sapidus STG. (a) Schematic of the isolated C. sapidus STNS. The ovals and associated symbols at the level of the STG nerves represent the sites of extracellular recordings, as performed in panels (b) and (c). Abbreviations: Ganglia: CoG, commissural ganglion; OG, esophageal ganglion; STG, stomatogastric ganglion. Nerves: dgn, dorsal gastric nerve; dvn, dorsal ventricular nerve; ion, inferior esophageal nerve; lgn, lateral gastric nerve; lvn, lateral ventricular nerve; mvn, medial ventricular nerve; son, superior esophageal nerve; stn, stomatogastric nerve. (b) Top: During saline superfusion, there was no ongoing gastric mill rhythm, as indicated by the lack of rhythmic action potential bursts in the gastric mill nerves lgn and dgn. Bottom: Bath-applied CalsTRP activated the gastric mill rhythm. Note the rhythmic, alternating bursting of the LG protractor and DG retractor motor neurons. The time scale bar pertains to both panels. (c) CalsTRP and CabTRP Ia application each activated the gastric mill rhythm in the same preparation. The gastric mill rhythm is evident from the rhythmic action potential bursts of the LG protractor motor neuron. Note the similarity in the LG burst characteristics (cycle period, burst duration, intraburst firing rate) between the two peptide-activated rhythms. There was a 1 h saline wash between the two peptide applications and after the second of the applications. The gastric mill rhythm terminated within 10 min from the onset of each saline wash. The time scale bar pertains to all three panels.
Figure 6.
Bath applied CalsTRP and CabTRP each activate the pyloric rhythm in the C. sapidus STG. Top: In this preparation, there was no ongoing pyloric rhythm during saline superfusion. As commonly occurs under this condition, there was regular spontaneous activity in the PY motor neurons and intermittent activity in the LP motor neuron (dvn). Middle: CalsTRP application activated the pyloric rhythm. Note the rhythmic, repeating coordinated bursting of the pyloric motor neurons in the dvn (LP, PY, PD) and mvn (IC, VD) nerves. As commonly occurs in other species, there was temporally overlapping activity between the LP and IC neurons and between the VD and PY neurons. Bottom: In the same preparation, CabTRP Ia application also activated the pyloric rhythm. Note the similarity of the rhythms in the presence of each peptide. There was a 1 h saline wash between the two peptide applications and after the second of the applications. Each pyloric rhythm terminated well before the end of the saline wash. The time scale bar pertains to all three panels.
The pyloric motor pattern has three active phases, followed by a brief silent period before the next cycle begins (e.g., Figure 6). The active phases include two successive constriction phases followed by dilation. These movements are driven by identified motor neurons whose activity is readily recorded extracellularly in a single nerve (dvn) that projects posteriorly from the STG (Figure 5a). This motor pattern is commonly spontaneously active in vivo and in vitro, although in some in vitro experiments, when descending modulatory input is removed, there is no spontaneous pyloric rhythm.25,28,30 When there was no ongoing pyloric rhythm in C. sapidus, CalsTRP application routinely activated this motor pattern (n = 4) (Figure 6).
We also applied CabTRP Ia (10–6–10–7 M) to the same preparations as CalsTRP. The gastric mill and pyloric rhythm responses to CabTRP Ia application were equivalent to their above-reported responses to CalsTRP (Figures 5c and 6). For example, with respect to the gastric mill rhythm (n = 5), statistical analysis indicated that each of the studied motor pattern parameters during CalsTRP and CabTRP Ia application were highly likely to derive from the same distribution. These parameters included the gastric mill cycle period (p > 0.05), protractor phase duration (i.e., LG burst duration) (p > 0.05), number of LG spikes per burst (p > 0.05), and LG intraburst firing rate (p > 0.05). Similarly, the pyloric rhythm cycle period resulting from CalsTRP and CabTRP Ia application were statistically equivalent (p > 0.05, n = 3).
Insofar as CalsTRP and CabTRP Ia are identical except for their amino-terminal amino acid, it was not surprising that these two peptides were equally effective on the pyloric rhythm in C. sapidus. In fact, an even more dissimilar TRP, the insect LomTK II peptide (APLSGFYGVRa), can also excite the C. borealis pyloric rhythm, although less strongly than CabTRP Ia.10,11 In contrast, the mammalian tachykinin substance P is ineffective on the C. borealis pyloric rhythm.11
There is a growing list of neuropeptide families with multiple members being present in the same species, and at least sometimes even in the same neurons.40 As in the case of CalsTRP and CabTRP Ia, these individual family members are often nearly identical. This has raised the still unresolved question of what value there is, if any, for colocalizing multiple isoforms from a peptide family for neural signaling. The possible functional consequences of this situation include the different family members activating different receptors, binding to the same receptor yet activating different postsynaptic intracellular signaling systems, and/or being differentially sensitive to extracellular peptidase degradation.51−54 Given their comparable actions on the gastric mill and pyloric rhythms, it seems unlikely that CalsTRP and CabTRP Ia act via different receptors or activate different postsynaptic signaling cascades by binding to the same receptor. With respect to peptidase sensitivity, these two peptides are likely to be equally sensitive to enzymatic cleavage and inactivation by a neprilysin-like endopeptidase, which cleaves many and perhaps all tachykinins.54,55 A neprilysin-like activity regulates the duration of CabTRP Ia actions in C. borealis,23,24,30 and it is likely to have the same action on CalsTRP insofar as these two TRPs have identical amino acid sequences in their internal regions where endopeptidases operate. However, these TRPs might also be regulated by extracellular aminopeptidase activity, which is present in the STG and regulates other neuropeptides therein.56,57 For example, TRPs in at least some insect species are also regulated by extracellular dipeptidyl peptidase activity.58 If the crab TRPs are indeed also sensitive to aminopeptidase activity, then the amino-terminal Ala of CabTRP Ia and the amino-terminal Tyr of CalsTRP are likely to be substrates for different aminopeptidases, because each such peptidase has a strong preference for a specific class of amino acids (e.g., acidic, basic, hydrophobic) at the amino-terminal and/or penultimate amino-terminal position.55
Significance of the Novel Tachykinin
After the discovery of substance P (SP), the first tachykinin in mammals, in 1931,3 three other mammalian tachykinins were sequenced and several other TRPs were detected and sequenced in invertebrates.1 While mammalian tachykinins have the conserved C-terminal motif FXGLMamide, most invertebrate TRPs share the conserved C-terminus GFX1GX2Ramide. In decapod crustaceans specifically, the presence of tachykinin was first identified through immunocytochemical and biological studies,59 and six tachykinin isoforms were subsequently sequenced by mass spectral analysis, including APSGFLGMRamide (CabTRP Ia), SGFLGMRamide (CabTRP Ib),10,37 TPSGFLGMRamide (CabTRP II),37 APAGFLGMRamide (LivTRP I), APSGFNGMRamide (LivTRP II), and APSFGLDMRamide (LivTRP III).39 Among these isoforms, CabTRP Ia has been detected in all decapod crustaceans, whereas other isoforms have only been detected in subsets of decapods. Our identification of CalsTRP reveals the existence of another TRP in decapod crustaceans, with a different amino acid (Tyr) at the N-terminus. While it has only been detected in C. sapidus thus far, its identification suggests the presence of a novel tachykinin prohormone and opens the door for its existence and detection in additional species.
Tachykinin receptors have a conserved sequence from mammals to invertebrates.3 For example, the Drosophila melanogaster tachykinin receptor NKD shares 38% identity in the transmembrane domains with the mammalian NK3 receptor, 35% identity at these domains with mammalian NK1 receptor, and 32% identity with the NK2 receptor.60 Moreover, mammalian substance P produces a dose dependent change in membrane currents by binding with the Drosophila tachykinin receptor DTKR.61 Furthermore, the mammalian tachykinin receptor antagonist Spantide I can suppress CabTRP Ia actions on the C. borealis pyloric and gastric mill rhythms.10,23 Another indication that tachykinins can influence their cognate receptors in a foreign species, presumably due to the structural similarity of receptors and ligands, is that LomTK II (Locusta migratoria tachykinin II) increases inositol triphosphate levels by binding to the NKD receptor.60
Tachykinins are involved in many physiological processes, including the regulation of food intake. For example, administrating the tachykinin peptides neuropeptide K and substance P consistently suppresses feeding behavior in rat and chick.47,62,63 However, in decapod crustaceans, feeding releases TRP in midgut64 and causes the elevation of TRP in brain.20 Tachykinins also stimulate contractions in insects foregut and hindgut1 and can modulate motor rhythms in the crustacean stomatogastric nervous system.10,23,24,30,65 The up-regulation of CalsTRP in our feeding experiments, and its effect on the STG motor rhythms in C. sapidus, suggests that tachykinins are feeding-related neuropeptides from mammals to invertebrates.
Methods
Animals and Feeding Experiment
Blue crabs (C. sapidus) were purchased from a local food market and maintained without food in an artificial seawater tank at 12–13 °C for 1 week before use so that they would be food-deprived for the subsequent experiments. In feeding experiments, the crabs were fed small pieces of seafood until they stopped eating, which usually took 30–45 min. The details of dissection and the feeding experiments were described previously.20,66 Direct tissue preparation and tissue homogenization were also performed as described previously.35 Briefly, small pieces of tissues were dissected, followed by rinsing in acidified methanol and 10 mg/mL of 2,5-dihydroxybenzoic acid (DHB) and prepared for MALDI MS analysis. Acidified methanol was used for homogenization of 10 brains and 30 pairs of CoGs, and prepared for crude extract analysis or HPLC separation as described below.
Instruments
The neuropeptide extracts from brains or CoGs were resuspended in 0.1% formic acid solution, and then were vortexed and briefly (10 min) centrifuged at 16 100g using an Eppendorf 5415D tabletop centrifuge (Eppendorf AG). The resulting supernatants were subsequently fractionated via a Rainin Dynamax high performance liquid chromatography (HPLC) system equipped with a Dynamax UV-D II absorbance detector (Rainin Instrument Inc., Woburn, MA) as described previously.36 The mobile phases included Solution A (deionized water containing 0.1% formic acid) and Solution B (acetonitrile containing 0.1% formic acid). About 20 μL of extract from 10 animals was injected onto a Macrosphere C18 column (2.1 mm i.d. × 250 mm length, 5 μm particle size; Alltech Assoc. Inc., Deerfield, IL). The separations consisted of a 120 min gradient of 5–95% Solution B. Fractions were automatically collected every 2 min using a Rainin Dynamax FC-4 fraction collector.
A model 4800 MALDI TOF/TOF analyzer (Applied Biosystems, Framingham, MA) equipped with a 200 Hz, 355 nm Nd:YAG laser was used for direct tissue analysis, tissue extract analysis, and MALDI imaging as described previously.20 A Varian/IonSpec Fourier transform mass spectrometry (FTMS) instrument (Lake Forest, CA) equipped with MALDI source and a 7.0 T actively shielded superconducting magnet was used for tissue extract analysis and quantitation as reported previously.63 Nanoscale LC-ESI-Q-TOF MS/MS was performed using a Waters nanoAcquity UPLC system coupled to a Q-TOF Micro mass spectrometer (Waters Corp., Milford, MA) to perform de novo sequencing as described previously.35,36
Peptide Prediction and Database Searches
De novo sequencing was performed using a combination of MassLynx 4.1 PepSeq software (Waters) and manual sequencing. Tandem mass spectra acquired on the QTOF were first deconvoluted using MaxEnt 3 software (Waters) to convert multiply charged ions into their singly charged forms. The resulting spectra were pasted into the PepSeq window for sequencing analysis. The candidate sequences generated by the PepSeq software were compared and evaluated for homology with previously identified peptides. The online program blastp (National Center for Biotechnology Information, Bethesda, MD; http://www.ncbi.nlm.nih.gov/BLAST/) was used to search the existing NCBI crustacean protein database, using the candidate peptide sequences as queries. Peptides with partial sequence homology were selected for further examination by comparing theoretical MS/MS fragmentation spectra generated by PepSeq with the raw MS/MS spectra. If the fragmentation patterns did not match well, manual sequencing was performed.
Off-Line CE-MALDI MS Analysis
Off-line CE separation was performed on a home-built CE apparatus equipped with a capillary of 75 cm length (50 μm i.d. × 360 μm o.d.) as described elsewhere.42 The CE runs under 19 kV using ammonium acetate buffer (50 mM, 5% MeOH, pH 5.0) at room temperature (25 °C). The CE fractions were deposited, every 30 s, onto the small matrix spots predeposited onto the hydrophobic Parafilm, as described previously, and analyzed by MALDI FTMS.42
In Solution Formaldehyde Labeling
A 3 μL aliquot of tissue extract from one brain or four pairs of CoG was labeled in solution by adding 0.7 μL sodium cyanoborohydride (NaBH3CN, 120 mM) and then mixed with formaldehyde (FH2, 15% in H2O, 0.5 μL) for unfed samples or deuterium formaldehyde (FD2, 15% in H2O, 0.5 μL) for fed samples. The samples were then placed at room temperature for 10 min for the labeling reaction to be completed. Then 4 μL of each solution was combined and mixed. The resulting mixture was spotted on MALDI plates and analyzed using MALDI-FTMS. The spectra were analyzed manually, and the peak pairs generated from TRPs were selected for quantitative analysis. The relative abundance ratio for each neuropeptide in fed crab versus unfed crab was determined by dividing heavy labeled peak intensity with light labeled peak intensity, followed by averaging the ratios from two replicate spectra. The Student’s t test was performed to evaluate the differences of each peptide between fed and unfed states, and a p-value < 0.05 was considered as statistically significant.
Quantitative Data Analysis
Samples from fed and unfed crabs in each experiment were differentially labeled and mixed in a 1:1 ratio. After CE separation, each spot was analyzed twice. The peak intensity of the monoisotope from FH2 and FD2 labeled peak pairs was extracted from these spectra. The abundance ratio for each neuropeptide in fed crab versus unfed crab was determined by dividing the heavy labeled peak intensity over the light labeled peak intensity. The average ratio and SEM were calculated from five duplicate spectra from brain extracts and four triplicate spectra from CoG extracts. The Student’s t test was also performed to evaluate the differences between the fed and unfed samples, and a p-value < 0.05 was considered as statistically significant.
Electrophysiology
Electrophysiology experiments were performed with male blue crabs (Callinectes sapidus) obtained from local markets. Crabs were housed in commercial tanks containing recirculating, aerated, artificial seawater (10–12 °C). Before dissection, the crabs were cold-anesthetized by packing them in ice for at least 30 min. The foregut was then removed and maintained in chilled physiological saline while the STNS was dissected from it and pinned down in a saline-filled silicone elastomer-lined Petri dish (Sylgard 184, KR Anderson, Santa Clara, CA).
The isolated STNS was maintained in saline containing (in mM) 439 NaCl, 26 MgCl2, 13 CaCl2, 11 KCl, 10 Trizma base, and 5 maleic acid (pH 7.4–7.6). During experiments, the nervous system was superfused continuously (7–12 mL/min, 10–12 °C) with saline via a switching manifold, to enable fast solution changes. A Vaseline well surrounded the STG, enabling this compartment to be superfused separately from the rest of the STNS. CalsTRP and CabTRP Ia were each diluted to their working concentration (10–6–10–7 M) from a stock solution immediately prior to applications. TRP superfusion was limited to the STG compartment.
All electrophysiology experiments in C. sapidus were conducted using the isolated STNS (Figure 5a). In all experiments, one or both CoGs were left connected to the STG. Extracellular recordings of pyloric and gastric mill motor neurons were made using routine methods for this system.30 In brief, each extracellular nerve recording was made using a pair of stainless steel wire electrodes (reference and recording) whose ends were pressed into the Sylgard-coated dish. A differential AC amplifier (model 1700, AM Systems, Carlsborg, WA) amplified the voltage difference between the reference wire, placed in the bath, and the recording wire, placed near an individual nerve and isolated from the bath by petroleum jelly (Vaseline, Lab Safety Supply, Janesville, WI). This signal was further amplified and filtered (model 410 amplifier, Brownlee Precision, Santa Clara, CA).
The pyloric rhythm was monitored by recording activity in the dorsal (dvn) and/or lateral ventricular nerve (lvn) and medial ventricular nerve (mvn). The separate activity of five of six pyloric motor neurons can be readily recorded and analyzed in these two nerves (dvn/lvn: lateral pyloric (LP), pyloric (PY), pyloric dilator (PD) motor neurons; mvn: inferior cardiac (IC), ventricular dilator (VD) motor neurons).27 The gastric mill rhythm was monitored by extracellular recordings of the lateral gastric nerve (lgn) and dorsal gastric nerve (dgn). In these nerves, the activity of the LG protractor motor neuron and dorsal gastric (DG) retractor motor neuron are readily recorded. Individual pyloric and gastric mill motor neurons were identified by their axonal pathways and activity patterns.30,67
Electrophysiology Data Analysis
Data were collected onto a chart recorder (Everest model, Astromed Corp.) and simultaneously onto a PC computer using data acquisition/analysis tools (Spike2; digitized at ∼5 kHz). Data analysis was facilitated by a custom-written program for Spike2 (Cambridge Electronic Design, Cambridge, U.K.) that determines the activity levels and burst relationships of individual neurons (freely available at http://www.neurobiologie.de/spike2). Unless otherwise stated, each data point in a data set was derived by determining the mean of 10 consecutive pyloric or gastric mill cycles. The pyloric cycle period was defined as the duration from the start of two consecutive PD neuron bursts. The gastric mill cycle period was defined as the duration between the onset of two consecutive LG neuron bursts. A gastric mill-timed burst was defined as a cluster of action potentials in a gastric mill neuron during which no interspike interval was longer than 1 s. The burst duration was defined as the duration (s) between the onset of the first and last action potential in an impulse burst. The intraburst spike frequency (Hz) was determined by dividing the number of spikes per burst minus one spike by the burst duration. Figures were made from Spike2 files incorporated into CorelDRAW Graphics Suite X5 (Corel Corporation, Mountain View, CA). Statistical analyses were performed with SigmaStat 3.0 (Systat Software Company, Chicago, IL). All data are expressed as the mean ± standard error (SE).
Acknowledgments
The authors wish to thank the University of Wisconsin-Biotechnology Mass Spectrometry Facility for access to the MALDI TOF/TOF instrument. We also want to thank the University of Wisconsin School of Pharmacy Analytical Instrumentation Center for access to the MALDI FTICR MS instrument.
Glossary
Abbreviations
- AMPN
anterior medial protocerebral neuropil
- AnN
antenna II neuropil
- CabTRP
Cancer borealis tachykinin-related peptide
- CalsTRP
Callinectes sapidus TRP
- CE
capillary electrophoresis
- CNS
central nervous system
- cocs
circumesophageal commissures
- CoG
commissural ganglion
- DC
deutocerebrum
- DG
dorsal gastric
- dgn
dorsal gastric nerve
- dvn
dorsal ventricular nerve
- ESI-QTOF
electrospray ionization-quadrupole/time-of-flight
- HPLC
high-performance liquid chromatography
- IC
inferior cardiac
- IMS
imaging mass spectrometry
- ivn
inferior ventricular nerve
- LAN
lateral antenna I neuropil
- LG
lateral gastric
- lgn
lateral gastric nerve
- LP
lateral pyloric
- MALDI-FTMS
matrix-assisted laser desorption/ionization Fourier transform mass spectrometry
- MPC
median protocerebrum
- mvn
medial ventricular nerve
- OG
esophageal ganglion
- ON
olfactory neuropil
- PD
pyloric dilator
- PMPN
posterior medial protocerebral neuropil
- PY
pyloric
- STG
stomatogastric ganglion
- STNS
stomatogastric nervous system
- TC
tritocerebrum
- TG
thoracic ganglion
- TOF
time-of-flight
- TRP
tachykinin-related peptide
- UPLC
ultraperformance liquid chromatography
- VD
ventricular dilator
Author Contributions
L.H., M.P.N., and L.L. designed research; L.H., Y.Z., J.W., A.C., and H.Y. performed research; L.H. and A.C. analyzed data; L.H., A.C., M.P.N., and L.L. wrote the paper.
This work was supported by National Institute of Digestive and Kidney Disease Grant R01DK071801 (L.L.) and National Institute of Neurological Disorders and Stroke Grants R37-NS 29436 and R37-NS 29436-S1 (M.P.N.). L.L. acknowledges a UW-Madison Vilas Associate Award and an H. I. Romnes Faculty Fellowship.
Funding Statement
National Institutes of Health, United States
References
- Nässel D. R. (1999) Tachykinin-related peptides in invertebrates: a review. Peptides 20, 141–158. [DOI] [PubMed] [Google Scholar]
- Severini C.; Improta G.; Falconieri-Erspamer G.; Salvadori S.; Erspamer V. (2002) The tachykinin peptide family. Pharmacol. Rev. 54, 285–322. [DOI] [PubMed] [Google Scholar]
- Vanden Broeck J.; Torfs H.; Poels J.; Van Poyer W.; Swinnen E.; Ferket K.; De Loof A. (1999) Tachykinin-like peptides and their receptors - A review. Ann. N.Y. Acad. Sci. 897, 374–387. [DOI] [PubMed] [Google Scholar]
- Johansson K. U. I.; Lundquist C. T.; Hallberg E.; Nässel D. R. (1999) Tachykinin-related neuropeptide in the crayfish olfactory midbrain. Cell Tissue Res 296, 405–415. [Google Scholar]
- Lundquist C. T.; Clottens F. L.; Holman G. M.; Nichols R.; Nachman R. J.; Nässel D. R. (1994) Callitachykinin-I and Callitachykinin-Iii, 2 Novel Myotropic Peptides Isolated from the Blowfly, Calliphora-Vomitoria, That Have Resemblances to Tachykinins. Peptides 15, 761–768. [DOI] [PubMed] [Google Scholar]
- Lundquist C. T.; Clottens F. L.; Holman G. M.; Riehm J. P.; Bonkale W.; Nässel D. R. (1994) Locustatachykinin immunoreactivity in the blowfly central nervous system and intestine. J. Comp. Neurol. 341, 225–240. [DOI] [PubMed] [Google Scholar]
- Muren J. E.; Nässel D. R. (1996) Isolation of five tachykinin-related peptides from the midgut of the cockroach Leucophaea maderae: Existence of N-terminally extended isoforms. Regul. Pept. 65, 185–196. [DOI] [PubMed] [Google Scholar]
- Muren J. E.; Nässel D. R. (1997) Seven tachykinin-related peptides isolated from the brain of the madeira cockroach: Evidence for tissue-specific expression of isoforms. Peptides 18, 7–15. [DOI] [PubMed] [Google Scholar]
- Meola S. M.; Clottens F. L.; Holman G. M.; Nachman R. J.; Nichols R.; Schoofs L.; Wright M. S.; Olson J. K.; Hayes T. K.; Pendleton M. W. (1998) Isolation and immunocytochemical characterization of three tachykinin-related peptides from the mosquito, Culex salinarius. Neurochem. Res. 23, 189–202. [DOI] [PubMed] [Google Scholar]
- Christie A. E.; Lundquist C. T.; Nässel D. R.; Nusbaum M. P. (1997) Two novel tachykinin-related peptides from the nervous system of the crab Cancer borealis. J. Exp. Biol. 200, 2279–2294. [DOI] [PubMed] [Google Scholar]
- Blitz D. M.; Christie A. E.; Marder E.; Nusbaum M. P. (1995) Distribution and effects of tachykinin-like peptides in the stomatogastric nervous system of the crab, Cancer borealis. J. Comp. Neurol. 354, 282–294. [DOI] [PubMed] [Google Scholar]
- Evans P. D. (1994) The effects of myomodulin and structurally related neuropeptides on skeletal neuromuscular-transmission in the locust. J. Exp. Biol. 190, 253–264. [DOI] [PubMed] [Google Scholar]
- Nässel D. R.; Eckert M.; Muren J. E.; Penzlin H. (1998) Species-specific action and distribution of tachykinin-related peptides in the foregut of the cockroaches Leucophaea maderae and Periplaneta americana. J. Exp. Biol. 201, 1615–1626. [DOI] [PubMed] [Google Scholar]
- Bartho L.; Holzer P. (1985) Search for a physiological-role of substance-P in gastrointestinal motility. Neuroscience 16, 1–32. [DOI] [PubMed] [Google Scholar]
- Pernow B. (1983) Substance-P. Pharmacol. Rev. 35, 85–141. [PubMed] [Google Scholar]
- Kulkarni S. K.; Dhir A. (2009) Current investigational drugs for major depression. Expert Opin. Invest. Drugs 18, 767–788. [DOI] [PubMed] [Google Scholar]
- Poulsen A.; Liljefors T.; Gundertofte K.; Bjornholm B. (2002) A pharmacophore model for NK2 antagonist comprising compounds from several structurally diverse classes. J. Comput-Aided. Mol. Des. 16, 273–286. [DOI] [PubMed] [Google Scholar]
- Satake H.; Kawada T.; Nomoto K.; Minakata H. (2003) Insight into tachykinin-related peptides, their receptors, and invertebrate tachykinins: A review. Zool. Sci. 20, 533–549. [DOI] [PubMed] [Google Scholar]
- Otsuka M.; Yoshioka K. (1993) Neurotransmitter functions of mammalian tachykinins. Physiol. Rev. 73, 229–308. [DOI] [PubMed] [Google Scholar]
- Chen R. B.; Hui L. M.; Cape S. S.; Wang J. H.; Li L. J. (2010) Comparative neuropeptidomic analysis of food intake via a multifaceted mass spectrometric approach. ACS Chem. Neurosci. 1, 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R. B.; Ma M. M.; Hui L. M.; Zhang J.; Li L. J. (2009) Measurement of neuropeptides in crustacean hemolymph via MALDI mass spectrometry. J. Am. Soc. Mass Spectrom. 20, 708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson P. S.; Stemmler E. A.; Christie A. E. (2008) The pyloric neural circuit of the herbivorous crab Pugettia producta shows limited sensitivity to several neuromodulators that elicit robust effects in more opportunistically feeding decapods. J. Exp. Biol. 211, 1434–1447. [DOI] [PubMed] [Google Scholar]
- Wood D. E.; Stein W.; Nusbaum M. P. (2000) Projection neurons with shared cotransmitters elicit different motor patterns from the same neural circuit. J. Neurosci. 20, 8943–8953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein W.; DeLong N. D.; Wood D. E.; Nusbaum M. P. (2007) Divergent co-transmitter actions underlie motor pattern activation by a modulatory projection neuron. Eur. J. Neurosci. 26, 1148–1165. [DOI] [PubMed] [Google Scholar]
- Nusbaum M. P.; Beenhakker M. P. (2002) A small-systems approach to motor pattern generation. Nature 417, 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusbaum M. P. (2002) Regulating peptidergic modulation of rhythmically active neural circuits. Brain Behav. Evol. 60, 378–387. [DOI] [PubMed] [Google Scholar]
- Marder E.; Bucher D. (2007) Understanding circuit dynamics using the stomatogastric nervous system of lobsters and crabs. Annu. Rev. Physiol. 69, 291–316. [DOI] [PubMed] [Google Scholar]
- Stein W. (2009) Modulation of stomatogastric rhythms. J. Comp. Physiol., A 195, 989–1009. [DOI] [PubMed] [Google Scholar]
- Sousa G. L.; Lenz P. H.; Hartline D. K.; Christie A. E. (2008) Distribution of pigment dispersing hormone- and tachykinin-related peptides in the central nervous system of the copepod crustacean Calanus finmarchicus. Gen. Comp. Endrocrinol. 156, 454–459. [DOI] [PubMed] [Google Scholar]
- Blitz D. M.; White R. S.; Saideman S. R.; Cook A.; Christie A. E.; Nadim F.; Nusbaum M. P. (2008) A newly identified extrinsic input triggers a distinct gastric mill rhythm via activation of modulatory projection neurons. J. Exp. Biol. 211, 1000–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitz D. M.; Christie A. E.; Coleman M. J.; Norris B. J.; Marder E.; Nusbaum M. P. (1999) Different proctolin neurons elicit distinct motor patterns from a multifunctional neuronal network. J. Neurosci. 19, 5449–5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby M. S.; Nusbaum M. P. (2007) Central nervous system projections to and from the commissural ganglion of the crab Cancer borealis. Cell Tissue Res. 328, 625–637. [DOI] [PubMed] [Google Scholar]
- Messinger D. I.; Kutz K. K.; Le T.; Verley D. R.; Hsu Y. W. A.; Ngo C. T.; Cain S. D.; Birmingham J. T.; Li L. J.; Christie A. E. (2005) Identification and characterization of a tachykinin-containing neuroendocrine organ in the commissural ganglion of the crab Cancer productus. J. Exp. Biol. 208, 3303–3319. [DOI] [PubMed] [Google Scholar]
- Stemmler E. A.; Cashman C. R.; Messinger D. I.; Gardner N. P.; Dickinson P. S.; Christie A. E. (2007) High-mass-resolution direct-tissue MALDI-FTMS reveals broad conservation of three neuropeptides (APSGFLGMRamide, GYRKPPFNGSIFamide and pQDLDHVFLRFamide) across members of seven decapod crustaean infraorders. Peptides 28, 2104–2115. [DOI] [PubMed] [Google Scholar]
- Ma M. M.; Bors E. K.; Dickinson E. S.; Kwiatkowski M. A.; Sousa G. L.; Henry R. P.; Smith C. M.; Towle D. W.; Christie A. E.; Li L. J. (2009) Characterization of the Carcinus maenas neuropeptidome by mass spectrometry and functional genomics. Gen. Comp. Endrocrinol. 161, 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M. M.; Wang J. H.; Chen R. B.; Li L. J. (2009) Expanding the crustacean neuropeptidome using a multifaceted mass spectrometric approach. J. Proteome Res. 8, 2426–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmler E. A.; Peguero B.; Bruns E. A.; Dickinson P. S.; Christie A. E. (2007) Identification, physiological actions, and distribution of TPSGFLGMRamide: a novel tachykinin-related peptide from the midgut and stomatogastric nervous system of Cancer crabs. J. Neurochem. 101, 1351–1366. [DOI] [PubMed] [Google Scholar]
- Christie A. E.; Cashman C. R.; Stevens J. S.; Smith C. M.; Beale K. M.; Stemmler E. A.; Greenwood S. J.; Towle D. W.; Dickinson P. S. (2008) Identification and cardiotropic actions of brain/gut-derived tachykinin-related peptides (TRPs) from the American lobster Homarus americanus. Peptides 29, 1909–1918. [DOI] [PubMed] [Google Scholar]
- Ma M. M.; Gard A. L.; Xiang F.; Wang J. H.; Davoodian N.; Lenz P. H.; Malecha S. R.; Christie A. E.; Li L. J. (2010) Combining in silico transcriptome mining and biological mass spectrometry for neuropeptide discovery in the Pacific white shrimp Litopenaeus vannamei. Peptides 31, 27–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie A. E.; Stemmler E. A.; Dickinson P. S. (2010) Crustacean neuropeptides. Cell. Mol. Life Sci. 67, 4135–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda-Kamatani Y.; Yasuda A. (2004) APSGFLGMRamide is a unique tachykinin-related peptide in crustaceans. Eur. J. Biochem. 271, 1546–1556. [DOI] [PubMed] [Google Scholar]
- Wang J. H.; Ma M.; Chen R.; Li L. (2008) Enhanced neuropeptide profiling via capillary electrophoresis off-line coupled with MALDI FTMS. Anal. Chem. 80, 6168–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. H.; Zhang Y. Z.; Xiang F.; Zhang Z. C.; Li L. J. (2010) Combining capillary electrophoresis matrix-assisted laser desorption/ionization mass spectrometry and stable isotopic labeling techniques for comparative crustacean peptidomics. J. Chromatogr., A 1217, 4463–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J. L.; Huang S. Y.; Chow N. H.; Chen S. H. (2003) Stable-isotope dimethyl labeling for quantitative proteomics. Anal. Chem. 75, 6843–6852. [DOI] [PubMed] [Google Scholar]
- DeKeyser S. S.; Li L. J. (2006) Matrix-assisted laser desorption/ionization Fourier transform mass spectrometry quantitation via in cell combination. Analyst 131, 281–290. [DOI] [PubMed] [Google Scholar]
- Ji C. J.; Li L. (2005) Quantitative proteome analysis using differential stable isotopic labeling and microbore LC-MALDI MS and MS/MS. J. Proteome Res. 4, 734–742. [DOI] [PubMed] [Google Scholar]
- Tachibana T.; Khan M. S. I.; Matsuda K.; Ueda H.; Cline M. A. (2010) Central administration of substance P inhibits feeding behavior in chicks. Horm. Behav. 57, 203–208. [DOI] [PubMed] [Google Scholar]
- Audsley N.; Weaver R. J. (2009) Neuropeptides associated with the regulation of feeding in insects. Gen. Comp. Endocr. 162, 93–104. [DOI] [PubMed] [Google Scholar]
- Chen R. B.; Hui L. M.; Sturm R. M.; Li L. J. (2009) Three dimensional mapping of neuropeptides and lipids in crustacean brain by mass spectral imaging. J. Am. Soc. Mass Spectrom. 20, 1068–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeKeyser S. S.; Kutz-Naber K. K.; Schmidt J. J.; Barrett-Wilt G. A.; Li L. J. (2007) Imaging mass spectrometry of neuropeptides in decapod crustacean neuronal tissues. J. Proteome Res. 6, 1782–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poels J.; Birse R. T.; Nachman R. J.; Fichna J.; Janecka A.; Broeck J. V.; Nassel D. R. (2009) Characterization and distribution of NKD, a receptor for Drosophila tachykinin-related peptide 6. Peptides 30, 545–556. [DOI] [PubMed] [Google Scholar]
- Poels J.; Nachman R. J.; Akerman K. E.; Oonk H. B.; Guerrero F.; De Loof A.; Janecka A. E.; Torfs H.; Vanden Broeck J. (2005) Pharmacology of stomoxytachykinin receptor depends on second messenger system. Peptides 26, 109–114. [DOI] [PubMed] [Google Scholar]
- Poels J.; Verlinden H.; Fichna J.; Van Loy T.; Franssens V.; Studzian K.; Janecka A.; Nachman R. J.; Broeck J. V. (2007) Functional comparison of two evolutionary conserved insect neurokinin-like receptors. Peptides 28, 103–108. [DOI] [PubMed] [Google Scholar]
- Turner A. J.; Isaac R. E.; Coates D. (2001) The neprilysin (NEP) family of zinc metalloendopeptidases: genomics and function. Bioessays 23, 261–269. [DOI] [PubMed] [Google Scholar]
- Isaac R. E.; Bland N. D.; Shirras A. D. (2009) Neuropeptidases and the metabolic inactivation of insect neuropeptides. Gen. Comp. Endrocrinol. 162, 8–17. [DOI] [PubMed] [Google Scholar]
- Coleman M. J.; Konstant P. H.; Rothman B. S.; Nusbaum M. P. (1994) Neuropeptide degradation produces functional inactivation in the crustacean nervous system. J. Neurosci. 14, 6205–6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood D. E.; Nusbaum M. P. (2002) Extracellular peptidase activity tunes motor pattern modulation. J. Neurosci. 22, 4185–4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac R. E.; Parkin E. T.; Keen J. N.; Nässel D. R.; Siviter R. J.; Shirras A. D. (2002) Inactivation of a tachykinin-related peptide: identification of four neuropeptide-degrading enzymes in neuronal membranes of insects from four different orders. Peptides 23, 725–733. [DOI] [PubMed] [Google Scholar]
- Goldberg D.; Nusbaum M. P.; Marder E. (1988) Substance-P-like immunoreactivity in the stomatogastric nervous systems of the crab Cancer borealis and the lobsters Panulirus interruptus and Homarus americanus. Cell Tissue Res. 252, 515–522. [DOI] [PubMed] [Google Scholar]
- Monnier D.; Colas J. F.; Rosay P.; Hen R.; Borrelli E.; Maroteaux L. (1992) Nkd, a developmentally regulated tachykinin receptor in Drosophila. J. Biol. Chem. 267, 1298–1302. [PubMed] [Google Scholar]
- Li X. J.; Wolfgang W.; Wu Y. N.; North R. A.; Forte M. (1991) Cloning, heterologous expression and developmental regulation of a Drosophila receptor for tachykinin-like peptides. EMBO J. 10, 3221–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra S. P.; Dube M. G.; Kalra P. S. (1991) Neuropeptide-K (Npk) suppresses copulatory behavior in male rats. Physiol. Behav. 49, 1297–1300. [DOI] [PubMed] [Google Scholar]
- Kalra S. P.; Sahu A.; Dube M. G.; Kalra P. S. (1991) Effects of various tachykinins on pituitary LH-Secretion, feeding, and sexual-behavior in the rat. Ann. N.Y. Acad. Sci. 632, 332–338. [DOI] [PubMed] [Google Scholar]
- Christie A. E.; Kutz-Naber K. K.; Stemmler E. A.; Klein A.; Messinger D. I.; Goiney C. C.; Conterato A. J.; Bruns E. A.; Hsu Y. W. A.; Dickinson P. S. (2007) Midgut epithelial endocrine cells are a rich source of the neuropeptides APSGFLGMRamide (Cancer borealis tachykinin-related peptide Ia) and GYRKPPFNGSIFamide (Gly(1)-SIFamide) in the crabs Cancer borealis, Cancer magister and Cancer productus. J. Exp. Biol. 210, 699–714. [DOI] [PubMed] [Google Scholar]
- Thirumalai V.; Marder E. (2002) Colocalized neuropeptides activate a central pattern generator by acting on different circuit targets. J. Neurosci. 22, 1874–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutz K. K.; Schmidt J. J.; Li L. J. (2004) In situ tissue analysis of neuropeptides by MALDI FTMS in-cell accumulation. Anal. Chem. 76, 5630–5640. [DOI] [PubMed] [Google Scholar]
- Weimann J. M.; Meyrand P.; Marder E. (1991) Neurons that form multiple pattern generators - identification and multiple activity patterns of gastric pyloric neurons in the crab stomatogastric system. J. Neurophysiol. 65, 111–122. [DOI] [PubMed] [Google Scholar]