Abstract
Objectives
New classes of drugs are needed to treat tuberculosis (TB) in order to combat the emergence of resistance to existing agents and shorten the duration of therapy. Targeting DNA gyrase is a clinically validated therapeutic approach using fluoroquinolone antibiotics to target the gyrase subunit A (GyrA) of the heterotetramer. Increasing resistance to fluoroquinolones has driven interest in targeting the gyrase subunit B (GyrB), which has not been targeted for TB. The biological activities of two potent small-molecule inhibitors of GyrB have been characterized to validate its targeting as a therapeutic strategy for treating TB.
Materials and methods
Novobiocin and aminobenzimidazole 1 (AB-1) were tested for their activity against Mycobacterium tuberculosis (Mtb) H37Rv and other mycobacteria. AB-1 and novobiocin were also evaluated for their interaction with rifampicin and isoniazid as well as their potential for cytotoxicity. Finally, AB-1 was tested for in vivo efficacy in a murine model of TB.
Results
Novobiocin and AB-1 have both been shown to be active against Mtb with MIC values of 4 and 1 mg/L, respectively. Only AB-1 exhibited time-dependent bactericidal activity against drug-susceptible and drug-resistant mycobacteria, including a fluoroquinolone-resistant strain. AB-1 had potent activity in the low oxygen recovery assay model for non-replicating persistent Mtb. Additionally, AB-1 has no interaction with isoniazid and rifampicin, and has no cross-resistance with fluoroquinolones. In a murine model of TB, AB-1 significantly reduced lung cfu counts in a dose-dependent manner.
Conclusions
Aminobenzimidazole inhibitors of GyrB exhibit many of the characteristics required for their consideration as a potential front-line antimycobacterial therapeutic.
Keywords: non-replicating bacteria, topoisomerase, benzimidazole, drug resistance, ciprofloxacin, novobiocin, non-tuberculous mycobacteria
Introduction
Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis (TB), is responsible for ∼2 million deaths each year and remains a major public health hazard throughout the world.1 Although many classes of antibiotics have been approved for the treatment of TB, the long treatment regimens required for cure can result in poor patient compliance and in the rapid emergence of drug-resistant strains. That emergence has made the administration of monotherapy for TB obsolete. The WHO now recommends the Direct Observed Therapy, Short Course (DOTS) programme, a multidrug cocktail consisting of four first-line drugs (isoniazid, rifampicin, pyrazinamide and ethambutol) administered for 2 months, followed by administration of isoniazid and rifampicin for an additional 4 months under direct observation by a healthcare worker. The DOTS programme has proven to be effective for treating TB and for minimizing the emergence of more drug-resistant strains, but it still imposes major burdens on patients and healthcare workers, given the long treatment regimens and the necessity to track drug cocktails.
With the proven effectiveness of DOTS and combination therapy, any new potential antitubercular drugs must be evaluated in the context of a multidrug cocktail regimen and should offer a benefit over the current four-drug cocktail. The most important improvements would entail the creation of a treatment regimen that is shorter and has superior efficacy against the circulating single-drug resistant (SDR) and multidrug-resistant (MDR) TB strains. Additionally, any new treatment regimen should ideally have fewer side effects and a convenient dosing regimen to increase the likelihood of patient compliance.
The long treatments required to cure TB infections are hypothesized to result from the slow-growing nature of Mtb as well as its ability to enter a non-replicating persistent (NRP) state that is much less susceptible or even resistant to many of the first-line antitubercular drugs.1 Potential strategies to shorten the length of treatment would consist of adding or replacing one of the current bacteriostatic drugs (e.g. ethambutol) with an antitubercular drug that is potently bactericidal against NRP Mtb. The clinical benefits of such strategies are still being evaluated, but recent studies with moxifloxacin, which inhibits NRP Mtb, have demonstrated that Mtb can be cleared from the lungs of infected mice in 2 months less time when administered with the standard regimen (isoniazid, rifampicin and pyrazinamide).2 This hypothesis has been further supported by a Phase II clinical trial in which moxifloxacin was used in place of ethambutol (in combination with isoniazid, rifampicin and pyrazinamide) and demonstrated improved culture conversion in the initial phase of TB treatment.3
The results of the studies with moxifloxacin, a potent inhibitor of DNA gyrase subunit A (GyrA), suggest that DNA gyrase may be a good target for reducing the length of TB treatment regimens. DNA gyrase is an essential enzyme that introduces negative supercoils into DNA and regulates the superhelical state of the bacterial chromosomes. The functional DNA gyrase enzyme exists as a heterotetramer with two A subunits and two B subunits (A2B2). Whereas GyrA is targeted by fluoroquinolone antibiotics, the gyrase B subunit (GyrB) is targeted by only one approved antibiotic, the aminocoumarin natural product novobiocin. GyrB has been genetically demonstrated to be a bactericidal drug target in Mtb, but there have not been any effective therapeutics developed against this target for TB.4 Novobiocin was originally approved for the treatment of methicillin-resistant Staphylococcus aureus (MRSA) infections, but has since been withdrawn from the market because it suffered from poor pharmacological properties and raised safety concerns. Recent efforts to identify new classes of compounds targeting the ATP-binding site of GyrB have produced a new class of aminobenzimidazole antibiotics.5,6 These aminobenzimidazole GyrB inhibitors have been developed for the treatment of MRSA and have shown greater antibiotic potency and less toxicity than novobiocin.
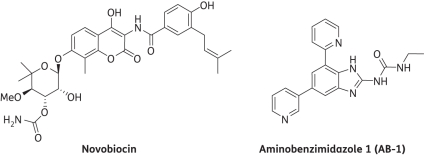
Here, we use novobiocin and aminobenzimidazole 1 (AB-1), a representative aminobenzimidazole GyrB inhibitor, to validate the targeting of GyrB as a therapeutic strategy and the prospect of including GyrB inhibitors in a first-line multidrug cocktail (Figure 1). Both novobiocin and AB-1 have been shown to be potent inhibitors of GyrB, with enzyme inhibition (Ki) and binding (KD) constants in the low nanomolar range (7–15 nM) as well as the ability to inhibit DNA supercoiling in vitro.5,7 Since GyrB is a validated antibiotic target for other pathogenic bacteria, GyrB inhibitors may also prove to be effective in reducing the length of TB treatment regimens and slowing the emergence of drug resistance.8
Figure 1.
Chemical structures of GyrB inhibitors used to validate the target.
Materials and methods
Bacterial strains, growth conditions and chemicals
Tables 1 and 2 list the strains used in this study. Mtb strains were obtained from the ATCC (Manassas, VA, USA) or Colorado State University (CSU, Fort Collins, CO, USA) and cultured in roller bottles (Corning Inc., Corning, NY, USA) using Middlebrook 7H9 broth (Difco Laboratories, Detroit, MI, USA) supplemented with 0.2% glycerol, 0.05% Tween 80 and 10% albumin-dextrose-catalase (Difco Laboratories). Middlebrook 7H10 agar (Difco Laboratories) supplemented with 0.2% glycerol and 10% oleic acid-albumin-dextrose-catalase (Difco Laboratories) was used to visualize colonies. Mtb H37Rv was grown in Middlebrook 7H9 broth at 37°C to the mid-log phase. The culture was diluted with Middlebrook 7H9 to an optical density at 600 nm (OD600) of 0.1 for mouse infection and with Middlebrook 7H9 without Tween 80 to an OD600 of 0.001 for a serum inhibition titration assay. The antibiotics kanamycin, novobiocin, ciprofloxacin, streptomycin, ethambutol, p-aminosalicylic acid, ethionamide, isoniazid, rifampicin and resazurin were purchased from Sigma (St Louis, MO, USA) and resuspended according to the manufacturer's instructions. AB-1 was synthesized using the published procedure and all analytical data matched published values.5,6
Table 1.
Bacterial strains used in the study and their corresponding MICs for AB-1 and novobiocin
| Mycobacterium strain | Resistant to | MIC (mg/L) |
Source | |
|---|---|---|---|---|
| AB-1 | novobiocin | |||
| M. tuberculosis H37Rv | none | 1 | 4 | Colorado State University |
| M. tuberculosis HN878 | none | 2 | 4 | Colorado State University |
| M. tuberculosis ATCC 35827 | kanamycin | 1.25 | 1.25 | ATCC |
| M. tuberculosis ATCC 35820 | streptomycin | 0.31 | 1.25 | ATCC |
| M. tuberculosis ATCC 35822 | isoniazid | 0.31 | 0.62 | ATCC |
| M. tuberculosis ATCC 35837 | ethambutol | 1.25 | 0.62 | ATCC |
| M. tuberculosis ATCC 35821 | p-aminosalicylic acid | 0.15 | 0.62 | ATCC |
| M. tuberculosis ATCC 35830 | ethionamide | 0.62 | 0.62 | ATCC |
| M. tuberculosis H37Rv Cip-Res | ciprofloxacin | 1.25 | 8 | This worka |
| M. abscessus ATCC 19977 | type strain | 128 | ND | ATCC |
| M. chelonae ATCC 35752 | type strain | 32 | ND | ATCC |
| M. avium ATCC 25291 | type strain | 16 | ND | ATCC |
| M. fortuitum ATCC 6841 | type strain | 16 | ND | ATCC |
| M. intracellulare ATCC 13950 | type strain | 4 | ND | ATCC |
ND, not determined.
aThis strains exhibits GAC to GGC (asparagine to lysine) mutation.
Table 2.
Comparison of antimicrobial activity of AB-1 and novobiocin against Mtb H37Rv under aerobic and low-oxygen conditions
| Assay conditions | MIC (mg/L) |
Source | ||||
|---|---|---|---|---|---|---|
| AB-1 | novobiocin | moxifloxacin | rifampicin | isoniazid | ||
| Aerobic assaya | 0.06 | 15 | 0.18 | 0.09 | 0.24 | ATCC |
| Low oxygen recovery assay | 1 | >32 | >32 | 1.9 | >128 | ATCC |
aVariations in MIC values have been observed historically between different laboratories/assays and strain sources.
Determination of antibiotic susceptibility
To determine the MICs of compounds for Mtb, the resazurin-based microplate assay was performed.9 Briefly, the compounds were resuspended in DMSO and tested in a range from 10 to 0.08 mg/L following a 2-fold dilution scheme. After addition of the bacterial cells with ∼105 cfu/mL, the 96-well plates were incubated at 37°C for 5 days. The addition of 0.05 mL of 0.1% resazurin followed, with additional incubation for 2 days at 37°C. Fluorescence was measured using a Fluoroskan Ascent microplate fluorimeter (Thermo Scientific, Waltham, MA, USA) with an excitation of 530 nm and emission of 590 nm. Wells containing compounds only were used to detect autofluorescence of compounds. The lowest drug concentration that inhibited ≥90% growth was considered to be the MIC. A 2-fold variation in MIC was considered to be within the error range of the assay. The final concentration of DMSO in all wells was 0.625%. These data are presented in Table 1.
Time–kill studies to assess bactericidal action
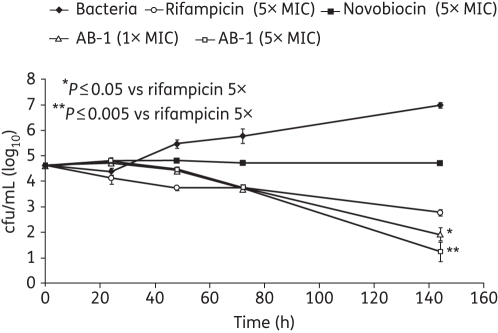
To determine the time–kill kinetics of AB-1, ∼105 cfu of H37Rv were exposed to 1× and 5× MIC concentrations for 5 days. Rifampicin was used as the positive control. Aliquots were removed and bacterial survivors were enumerated on Middlebrook 7H10 agar plates after appropriate dilutions. The plates were incubated for 28 days at 37°C, at which point the colonies were scored. The bactericidal action is plotted in Figure 2. P values were calculated using a two-tailed unpaired Student's t-test.
Figure 2.
Time–kill experiment depicting bactericidal activity of AB-1. Mtb H37Rv was exposed to 1× and 5× MIC of AB-1, rifampicin and novobiocin. The cells were plated at regular intervals and colonies were enumerated 3 weeks later.
Drug interaction studies
To determine whether AB-1 exhibited any in vitro interaction with rifampicin and isoniazid, interaction studies were conducted using the chequerboard microdilution technique.10 Synergy was evaluated using fractional inhibitory concentrations (FICs), which are defined as the sum of the MIC of each drug when used in combination, divided by the MIC of the drug when used alone. The FIC index was calculated as the numerical sum of the two FICs for a given combination. Synergy was defined as an FIC index ≤0.5, no interaction as an FIC index >0.5 to 4.0, and antagonism as an FIC index >4.0.11
Antimicrobial activity against non-replicating Mtb cells
The antimicrobial activity of AB-1 against non-replicating Mtb cells was determined as described previously using the low oxygen recovery assay (LORA) (Table 2).12 Briefly, Mtb H37Rv cells were suspended in Middlebrook 7H12 broth and sonicated for 15 s. Cultures were diluted to obtain an OD570 of 0.03–0.05 and 3000–7000 relative light units (RLUs)/100 μL. Twofold serial dilutions of antimicrobial agents were prepared in a volume of 100 μL in black 96-well microtitre plates and 100 μL of the cell suspension was added. The microplate cultures were placed under anaerobic conditions (oxygen <0.16%) using an Anoxomat Model WS-8080 (MART Microbiology, Drachten, The Netherlands) using three cycles of evacuation and filling with a mixture of 10% H2, 5% CO2, and the balance N2. An anaerobic indicator strip was placed inside the chamber to visually confirm the removal of oxygen. Plates were incubated at 37°C for 10 days and then transferred to an ambient gaseous condition (5% CO2-enriched air) incubator for a 28 h ‘recovery’. On day 11 (after the 28 h aerobic recovery), 100 μL of culture were transferred to white 96-well microtitre plates for determination of luminescence. The MIC was defined as the lowest drug concentration effecting an inhibition of ∼90% relative to drug-free controls.
Isolation and molecular characterization of resistance mutants
Mutants resistant to AB-1 and novobiocin were isolated by plating ∼108–109 H37Rv cells on 7H10OADC agar-containing compounds at a range between 2× and 16× MICs. To reconfirm the resistant phenotype, the selected mutants were plated on the same selection compound concentration and confirmed by MIC.
To isolate genomic DNA, the resistant mutants were grown in 7H9ADC medium, the cells were collected by centrifugation, and genomic DNA was isolated by standard procedures.13 Genomic DNA was used as a template to amplify the ATP-binding domain of GyrB regions from the wild-type and mutants by PCR with the primer pair GyrB-Forward 5′-gcacggcgcggttagatgggtaaaaacgag-3′ and GyrB-Reverse 5′-caccgactccgaatacccggcgttccattg-3′. The PCR was carried out for 35 cycles with denaturation at 95°C for 15 min, annealing at 55°C for 1 min, and extension with Hot Start Taq DNA polymerase (Qiagen, Valencia, CA, USA) at 72°C for 1 min. The PCR products were gel purified using a gel extraction kit (Qiagen) and sequenced using standard automated methods to determine the mutations. The mutations and corresponding MICs are detailed in Table 3.
Table 3.
Molecular characteristics of the Mtb H37Rv resistant mutants generated using novobiocin and AB-1
| Strain | Mutation | Number of mutants isolated | MIC (mg/L) |
|
|---|---|---|---|---|
| AB-1 | novobiocin | |||
| H37Rv | wild-type | none | 1 | 4 |
| Resistant to novobiocin | R180C | 8 | 2 | 15.6 |
| Resistant to AB-1 | A92S | 7 | 7.8 | 4 |
Cytotoxicity determinations for AB-1
To determine the cytotoxic characteristics of AB-1, hepatotoxicity was evaluated using rat hepatocytes. Briefly, 20 000 freshly isolated rat hepatocytes were plated per well in a 96-well bio-coat plate. Following attachment, the cells were exposed to novobiocin and AB-1 in a dose range of 0.35–180 mg/L (0.5% DMSO final) for ∼20 h in a 37°C incubator with 5% CO2. Cytotoxicity was measured using the XTT cell proliferation assay, which measures mitochondrial dehydrogenase activity in viable cells.14 Results were calculated as percentage viability of control cells incubated with 0.5% DMSO alone. A decrease in the percentage viability of the cells indicates that the compounds tested are cytotoxic to the rat hepatocytes under the conditions used. Data were fit to a four-parameter logistic function using GraphPad Prism (GraphPad Software, La Jolla, CA, USA) to calculate a 50% cytotoxicity concentration (CC50).
Serum inhibition titration (SIT) assay
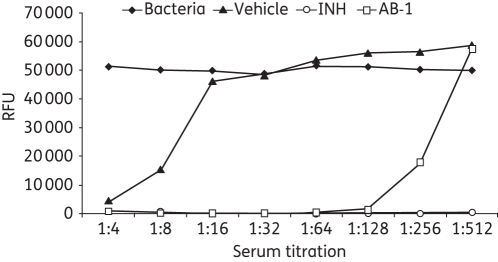
The AB-1 compound was prepared as a homogenous suspension in 0.5% carboxymethyl cellulose (CMC) at 10 mg/mL. Three groups of three mice each were gavaged with one dose of either AB-1 at 100 mg/kg, isoniazid at 10 mg/kg as the positive control, or CMC as the vehicle control. Dosed mice were sacrificed 60 min later and cardiac blood was collected. Serum was separated and collected for the SIT assay. Serum was serially diluted with Middlebrook 7H9 without Tween from 1:2 to 1:64 using a flat-bottomed 96-well plate, with serum background control, CMC vehicle control, isoniazid positive control and growth in Middlebrook 7H9 media control. Equal volumes (0.1 mL) of Middlebrook 7H9 media-diluted H37Rv culture containing ∼104 cfu were added to each well and mixed thoroughly. The final serum titration was brought to 1:4 to 1:128. The plate was sealed and incubated for 7 days at 37°C; 32.5 μL of Alamar Blue in Tween 80 (0.86% and 1% final concentrations) was then added to all wells of the plate. The plate was incubated at 37°C for 16–18 h and fluorescence intensity was measured using a BMG Optima microplate reader (BMG LABTECH, Ortenberg, Germany) at 544 Ex/590 Em. Data are shown as relative fluorescence units (RFUs) in Figure 3.
Figure 3.
Serum titration (bioavailability) of AB-1. Balb/c mice were gavaged with one dose (100 mg/kg), with blood collected and serum separated 60 min later. Growth inhibition of serially diluted serum on H37Rv was tested with Alamar Blue assay. Vehicle, 0.5% CMC; INH, isoniazid at 10 mg/kg; AB-1, AB-1 at 100 mg/kg.
In vivo efficacy/lung cfu burden study
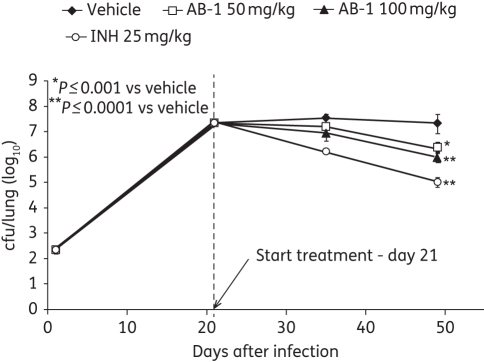
Six-week-old female BALB/c mice were purchased from Charles River Laboratories (Wilmington, MA, USA). Fifty mice were aerosol infected with Mtb H37Rv using the Middlebrook inhalation exposure system (Glas-Col, Terre Haute, IN, USA) with 10 mL of diluted log-phase broth culture at day −21. Implantation of bacterial infection was determined by sacrificing five mice and plating the diluted lung homogenates on selective agar the day after infection and enumerating the cfu 3–4 weeks later. On the day of treatment start (Day 0), five mice were sacrificed and lung cfu burden was determined as mentioned above. Four groups of 10 mice were orally gavaged with either 50 or 100 mg/kg of AB-1, or 25 mg/kg of isoniazid (positive control), or a sham control (5% dextrose). On day 14 and day 28, five mice from each treatment were sacrificed for cfu determinations. Gross lung pathology was also recorded. All animal procedures were approved by the Johns Hopkins University Animal Care and Use Committee. These data are displayed in Figure 4. P values were calculated using a two-tailed unpaired Student's t-test.
Figure 4.
AB-1 reduction of lung cfu after aerosol infection. Mice were aerosol infected with Mtb H37Rv at day −21 and orally gavaged with 50 or 100 mg/kg of AB-1, 25 mg/kg of isoniazid (positive control), or vehicle control (5% dextrose). On day 14 and day 28 of the treatment, five mice from each group were sacrificed for cfu determinations.
Results
Range of antimicrobial activity of AB-1
As shown in Tables 1 and 2, AB-1 exhibited potent activity in the range of 0.312–2 mg/L against all the Mtb strains tested, including single-resistant strains. Most notably, even the presence of ciprofloxacin resistance did not increase the MIC of AB-1, thus demonstrating a lack of cross-resistance between therapeutics targeting the different subunits of the DNA gyrase. On the other hand, novobiocin exhibited less potency against all the Mtb strains tested, with MICs ranging from 0.156 to 8 mg/L. The non-tuberculous mycobacteria (NTM) were appreciably less susceptible to AB-1, with MICs ranging from 4 to 128 mg/L (Table 1). Additionally, testing of AB-1 in the LORA assay, an in vitro assay for antimicrobial activity against non-replicating Mtb, indicated an MIC of 1 mg/L, demonstrating that GyrB may be a potential target for NRP bacterial populations (Table 2).12 The finding is noteworthy because it has been demonstrated that under similar conditions moxifloxacin, which inhibits gyrase A, exhibits a LORA MIC of >32 mg/L, severalfold higher than its MIC under aerobic conditions.12
Isolation of resistant mutants and characterization of the mutation site
Mutants resistant to AB-1 were selected and genetically mapped to determine the key residues involved in the resistance phenotype. The resistance frequency was thus determined to be ∼10−6–10−7 cfu/mL. As can be seen in Table 3, R180C was the only mutation for novobiocin, which is consistent with other in vitro and clinical data indicating that this mutation is most commonly associated with resistance to novobiocin in S. aureus.15 The resistant mutants selected with AB-1 had a conserved A92S mutation. Interestingly, other studies of resistant mutants to the aminobenzimidazole GyrB inhibitors have found that mutations at T206 are most commonly isolated under selection with the aminobenzimidazole class of compounds.6 Although novobiocin and AB-1 both target the ATP-binding site of GyrB, they did not exhibit cross-resistance (Table 3). Also, a genomic analysis of the NTM showed that Mycobacterium abscessus possesses a natural A92S mutation in its gyrB gene, which may partially explain its intrinsic resistance to this class of compounds. None of the other NTMs tested possess this mutation.
Cytotoxicity determinations for AB-1
The hepatotoxicity of AB-1 was determined by rat hepatocyte cell proliferation. AB-1 exhibited minimal toxicity at all concentrations up to 180 mg/L. In comparison, novobiocin exhibited a concentration-dependent toxicity with a CC50 of 25 mg/L against the rat hepatocytes.
Synergy/interaction with rifampicin and isoniazid
For any drug to be included in the first line of therapy it has to exhibit either synergy or no interaction with rifampicin and isoniazid. AB-1 manages not to interact with either rifampicin or isoniazid in the chequerboard assay, with FICs of 0.5 and 2.3, respectively. For controls, novobiocin and ciprofloxacin also had no interaction with rifampicin and isoniazid.
Time–kill to determine bactericidal activity
Time–kill studies were performed with H37Rv using novobiocin, AB-1 and rifampicin. As shown in Figure 2, AB-1 exhibits a significant (P values of 0.05 and 0.005) time-dependent bactericidal activity rather than concentration-dependent activity. In comparison, novobiocin at 5× MIC was bacteriostatic, whereas rifampicin at 5× MIC was bactericidal. After 144 h, AB-1 caused an almost 4-log decrease in cfu as compared with the drug-free control, whereas rifampicin caused almost a 2-log decrease. This potent bactericidal activity underscores GyrB as a potential vital target.
SIT assay
Bioavailability was estimated using a SIT assay. As shown in Figure 3, after oral dosing AB-1 was present in the serum at potent concentrations up to a 1:128 dilution. This finding is noteworthy because AB-1 has been shown to have high serum binding, and more optimized analogues have been synthesized to reduce that property.5 In comparison, the positive control drug isoniazid is potently antimicrobial even at 1:512 dilution.
In vivo efficacy/lung cfu burden study
AB-1 demonstrated in vivo efficacy in the murine TB model in a dose-dependent manner. As shown in Figure 4, at day 49, both the 50 and 100 mg/kg doses of AB-1 caused significant (P values of 0.001 and 0.0001, respectively) reductions in cfu in the lung (∼1.5 logs of cfu), whereas, in comparison, isoniazid at 25 mg/kg caused a reduction of ∼2 logs of cfu.
Discussion
Few novel drug targets for Mtb have been identified, in spite of the genomic sequence having been known for more than a decade and the proliferation of new genetic technologies. Abundant information indicates the Mtb genes that are required for growth and pathogenicity, but very few new drug targets have been validated using small molecule inhibitors. The development of novel treatments for TB is further complicated by the requirement that a new drug must provide an improved standard of care over the current multidrug DOTS regimen, which has proven to be quite effective. Thus, it is imperative to validate new targets with potent small molecule inhibitors and demonstrate their potential for therapeutic value in the context of new TB treatment regimens.
Fluoroquinolones target GyrA, one-half of the functional gyrase heterotetramer (A2B2) complex. Resistance to fluoroquinolones is typically generated by a point mutation in the gyrA gene, which gives rise to resistance against the entire class of fluoroquinolones, thus embodying class resistance. However, if GyrB is targeted, it still exerts the same phenotypic effects on bacterial viability as do the fluoroquinolones, thus furthering the life of the DNA gyrase complex as a viable target for TB. As fluoroquinolones become incorporated into clinically used TB drug treatment regimens, alternative drug combinations will be required to combat the emergence of fluoroquinolone-resistant Mtb strains.
As has been shown here, the aminobenzimidazole GyrB inhibitor AB-1 possesses many of the properties required to be part of a first-line anti-TB multidrug cocktail: it is bactericidal; it has excellent activity against drug-resistant Mtb strains, including the fluoroquinolone-resistant strains; and it does not exhibit antagonism against rifampicin and isoniazid. Additionally, the resistance generated against AB-1 does not cross-react with fluoroquinolones. Because DNA gyrase is essential for cell viability and because AB-1 exhibits potent activity against NRP Mtb cells, GyrB is an attractive chemotherapeutic target for incorporation into anti-TB chemotherapy.16,17
GyrB has many of the ideal qualities required for an attractive antibiotic target. It is an essential gene product for bacterial viability, it is present in a single copy, and its inhibition results in significant cell death because there are no viable alternative mechanisms for performing this function. The gyrB genes, among a variety of Mtb isolates that have been sequenced, have an almost 99.9% homology with almost no changes in sequence, further signifying its broad applicability as a drug target.
As evidenced in Table 1, not all mycobacteria are equally susceptible to AB-1. In fact, all of the NTMs that we tested AB-1 against naturally possess a serine residue at position 92 (using Mtb numbering) of their gyrB gene. This same mutation was detected in Mtb strains that were selected to be less susceptible to AB-1 in vitro. It is also possible that additional resistance mechanisms are present in the NTMs, such as permeability barriers or efflux mechanisms, given that the mutation alone cannot account for the extremely high MIC of 128 mg/L found for M. abscessus.
The location of the mutation in the isolated resistant colonies at A92 was unexpected, in that similar studies with aminobenzimidazoles in other species of bacteria resulted in mutations at a conserved threonine/serine that is S208 in TB (Mtb numbering).6 These two residues are far apart in the protein sequence, but spatially are only about 8 Å from each other in the deep part of the ATP-binding site. This region of the binding site is known to form direct interactions with the urea group in AB-1.5 Based on the structure–activity relationship for this class of GyrB inhibitors, the structural interactions made by the urea group are essential for activity, thus it makes sense that mutations to these residues would result in a significant loss of activity. The R180C mutation that resulted in resistance to novobiocin is located on the edge of the ATP-binding pocket and is known to form a strong hydrogen bonding interaction with the coumarin ring of the aminocoumarin GyrB inhibitors. This same arginine has been shown to interact with one of the pyridine rings of AB-1 through a hydrogen bond, but the loss of this residue did not significantly affect the MIC of AB-1 against the novobiocin-resistant mutant (Table 3).
One of the key differentiators of the aminobenzimidazole GyrB inhibitors compared with those of the aminocoumarins is the increased dual-targeting of the topoisomerase IV (topoIV) enzyme in addition to GyrB. Prokaryotic topoIV enzymes are structurally and functionally related to GyrB, but their primary function is thought to be the decatenation of DNA following DNA replication.18 Mechanistically, topoIV enzymes function very similarly to GyrB and can also relax positive supercoils in DNA. Novobiocin is also an inhibitor of the ATP-binding subunit of topoIV in Escherichia coli, ParE, but is an order of magnitude more potent against GyrB than ParE, whereas AB-1 is only 1-fold less potent against ParE.8,15 Both of these enzymes are essential for viability, and the advantage of potently targeting both of them should reduce the development of resistance through target mutation because the occurrence of mutations at two independent targets is statistically less probable. This hypothesis was validated in studies with Enterococcus faecalis: very low spontaneous resistance rates were found for one of the aminobenzimidazole inhibitors compared with mutants when either GyrB or ParE already contained a resistance mutation.6 Because no topoIV protein has been annotated for Mtb, it is unclear whether the dual-targeting activity of the aminobenzimidazoles is pertinent in Mtb.19 Although Mtb must presumably have an enzyme that unlinks DNA following DNA replication, that enzyme may not have been annotated yet because of a sequence or structure different from those for other bacterial topoIV enzymes; alternatively, the GyrB protein may also perform this function.
The functional activities for GyrB inhibitors would be expected to give phenotypes similar to fluoroquinolones because they ultimately inhibit the same DNA gyrase complex (as well as topoIV proteins in other bacteria). In other types of non-replicating and dormancy models, fluoroquinolones have shown bactericidal activity, but are consistently inactive in the LORA model.20 Based on these conflicting data, it is likely that the inactivity of fluoroquinolones in the LORA model is an artefact of the assay and perhaps they interfere with luciferase readout. The fact that the AB-1 GyrB inhibitor is potently active in this assay demonstrates that the Mtb gyrase enzyme is required for survival in this NRP model. The data here demonstrate that, although GyrB does appear to be a good target for developing chemotherapeutics for TB, the nature of the chemical class of inhibitors is important and can affect functional activities in cellular and in vivo models. With recent data suggesting that incorporating fluoroquinolones in first-line anti-TB drug cocktails may shorten the required length of drug treatments, it is important to begin the development of backup drugs to replace fluoroquinolones when resistance for that class of drugs becomes prevalent. The data generated with AB-1 suggest that drugs targeting GyrB may be a viable alternative to include in first-line anti-TB drug cocktails.
Funding
This project was supported by award numbers R56AI090817 and U01AI082070 from the National Institute of Allergy and Infectious Diseases.
Transparency declarations
None to declare.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
References
- 1.Dye C, Williams BG. Slow elimination of multidrug-resistant tuberculosis. Sci Transl Med. 2009;1:3ra8. doi: 10.1126/scitranslmed.3000346. doi:10.1126/scitranslmed.3000346. [DOI] [PubMed] [Google Scholar]
- 2.Nuermberger EL, Yoshimatsu T, Tyagi S, et al. Moxifloxacin-containing regimen greatly reduces time to culture conversion in murine tuberculosis. Am J Respir Crit Care Med. 2004;169:421–6. doi: 10.1164/rccm.200310-1380OC. doi:10.1164/rccm.200310-1380OC. [DOI] [PubMed] [Google Scholar]
- 3.Conde MB, Efron A, Loredo C, et al. Moxifloxacin versus ethambutol in the initial treatment of tuberculosis: a double-blind, randomised, controlled phase II trial. Lancet. 2009;373:1183–9. doi: 10.1016/S0140-6736(09)60333-0. doi:10.1016/S0140-6736(09)60333-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaur P, Agarwal S, Datta S. Delineating bacteriostatic and bactericidal targets in mycobacteria using IPTG inducible antisense expression. PLoS One. 2009;4:e5923. doi: 10.1371/journal.pone.0005923. doi:10.1371/journal.pone.0005923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charifson PS, Grillot A-L, Grossman TH, et al. Novel dual-targeting benzimidazole urea inhibitors of DNA gyrase and topoisomerase IV possessing potent antibacterial activity: intelligent design and evolution through the judicious use of structure-guided design and structure-activity relationships. J Med Chem. 2008;51:5243–63. doi: 10.1021/jm800318d. doi:10.1021/jm800318d. [DOI] [PubMed] [Google Scholar]
- 6.Grossman TH, Bartels DJ, Mullin S, et al. Dual targeting of GyrB and ParE by a novel aminobenzimidazole class of antibacterial compounds. Antimicrob Agents Chemother. 2007;51:657–66. doi: 10.1128/AAC.00596-06. doi:10.1128/AAC.00596-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glaser BT, Malerich JP, Duellman SJ, et al. A high-throughput fluorescence polarization assay for inhibitors of gyrase B. J Biomol Screen. 2011;16:230–8. doi: 10.1177/1087057110392038. doi:10.1177/1087057110392038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mdluli K, Ma Z. Mycobacterium tuberculosis DNA gyrase as a target for drug discovery. Infect Disord Drug Targets. 2007;7:159–68. doi: 10.2174/187152607781001763. doi:10.2174/187152607781001763. [DOI] [PubMed] [Google Scholar]
- 9.Collins L, Franzblau SG. Microplate alamar blue assay versus BACTEC 460 system for high-throughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob Agents Chemother. 1997;41:1004–9. doi: 10.1128/aac.41.5.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hindler J. Antimicrobial susceptibility testing. In: Henry DI, editor. Clinical Microbiology Procedures Handbook. Washington, DC: American Society for Microbiology; 1992. [Google Scholar]
- 11.Odds FC. Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother. 2003;52:1. doi: 10.1093/jac/dkg301. doi:10.1093/jac/dkg301. [DOI] [PubMed] [Google Scholar]
- 12.Cho SH, Warit S, Wan B, et al. Low-oxygen-recovery assay for high-throughput screening of compounds against nonreplicating Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2007;51:1380–5. doi: 10.1128/AAC.00055-06. doi:10.1128/AAC.00055-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Helden PD, Victor TC, Warren RM, et al. Isolation of DNA from Mycobacterium tuberculosis. In: Parish T, Stoker NG, editors. Mycobacterium tuberculosis protocols. New York: Humana Press; 2001. pp. 19–30. [DOI] [PubMed] [Google Scholar]
- 14.Scudiero DA, Shoemaker RH, Paull KD, et al. Evaluation of a soluble tetrazolium/formazan assay for cell growth and drug sensitivity in culture using human and other tumor cell lines. Cancer Res. 1988;48:4827–33. [PubMed] [Google Scholar]
- 15.Maxwell A. DNA gyrase as a drug target. Trends Microbiol. 1997;5:102–9. doi: 10.1016/S0966-842X(96)10085-8. doi:10.1016/S0966-842X(96)10085-8. [DOI] [PubMed] [Google Scholar]
- 16.Piton J, Petrella S, Delarue M, et al. Structural insights into the quinolone resistance mechanism of Mycobacterium tuberculosis DNA gyrase. PLoS One. 2010;5:e12245. doi: 10.1371/journal.pone.0012245. doi:10.1371/journal.pone.0012245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sassetti CM, Boyd DH, Rubin EJ. Comprehensive identification of conditionally essential genes in mycobacteria. Proc Natl Acad Sci USA. 2001;98:12712–7. doi: 10.1073/pnas.231275498. doi:10.1073/pnas.231275498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson VE, Gootz TD, Osheroff N. Topoisomerase IV catalysis and the mechanism of quinolone action. J Biol Chem. 1998;273:17879–85. doi: 10.1074/jbc.273.28.17879. doi:10.1074/jbc.273.28.17879. [DOI] [PubMed] [Google Scholar]
- 19.Reddy TB, Riley R, Wymore F, et al. TB database: an integrated platform for tuberculosis research. Nucleic Acids Res. 2009;37:D499–508. doi: 10.1093/nar/gkn652. doi:10.1093/nar/gkn652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu Y, Coates AR, Mitchison DA. Sterilizing activities of fluoroquinolones against rifampin-tolerant populations of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2003;47:653–7. doi: 10.1128/AAC.47.2.653-657.2003. doi:10.1128/AAC.47.2.653-657.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]