Abstract
The title compound, C16H14O5·2H2O [systematic name: 5-hydroxy-2-(4-hydroxyphenyl)-7-methoxychroman-4-one dihydrate], is a natural phytoalexin flavone isolated from the native chilean species Heliotropium taltalense and crystallizes with an organic molecule and two water molecules in the asymmetric unit. The 5-hydroxy group forms a strong intramolecular hydrogen bond with the carbonyl group, resulting in a six-membered ring. In the crystal, the components are linked by O—H⋯O hydrogen bonds, forming a three-dimensional network. The 4-hydroxyphenyl benzene ring is bonded equatorially to the pyrone ring, which adopts a slightly distorted sofa conformation. The title compound is the hydrated form of a previously reported structure [Shoja (1990 ▶). Acta Cryst. C46, 1969–1971]. There are only slight variations in the molecular geometry between the two compounds.
Related literature
For the first study of the title compound, see: Narasimhachari & Seshadri (1949 ▶); Atkinson & Blakeman (1982 ▶). For its biological properties, see: Plowright et al. (1996 ▶); Atkinson & Blakeman (1982 ▶), Saito et al. (2008 ▶). For its spectroscopic properties, see: Agrawal (1989 ▶); Ogawa et al. (2007 ▶). For the structure of the unsolvated compound, see: Shoja (1990 ▶). For similar compounds, see: Modak et al. (2009 ▶). For graph-set notation, see: Bernstein et al. (1995 ▶). For puckering parameters, see: Cremer & Pople (1975 ▶). For molecular geometry calculations, see: Macrae et al. (2008 ▶). 
Experimental
Crystal data
C16H14O5·2H2O
M r = 322.30
Orthorhombic,
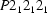
a = 5.0869 (10) Å
b = 9.4622 (19) Å
c = 32.318 (7) Å
V = 1555.6 (5) Å3
Z = 4
Mo Kα radiation
μ = 0.11 mm−1
T = 293 K
0.20 × 0.15 × 0.03 mm
Data collection
Nonius KappaCCD area-detector diffractometer
9743 measured reflections
2021 independent reflections
1623 reflections with I > 2σ(I)
R int = 0.068
Refinement
R[F 2 > 2σ(F 2)] = 0.075
wR(F 2) = 0.180
S = 1.17
2021 reflections
224 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.38 e Å−3
Δρmin = −0.29 e Å−3
Data collection: COLLECT (Nonius, 2000 ▶; cell refinement: DENZO-SMN (Otwinowski & Minor, 1997 ▶); data reduction: DENZO-SMN; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: PLATON (Spek, 2009 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶) and publCIF (Westrip, 2010 ▶).
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536811051221/kj2193sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811051221/kj2193Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811051221/kj2193Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O3—H3⋯O2 | 0.82 | 1.90 | 2.630 (5) | 147 |
| O5—H5⋯O7 | 0.76 (9) | 1.99 (9) | 2.720 (7) | 163 (7) |
| O6—H6A⋯O2i | 0.87 (9) | 2.00 (8) | 2.847 (6) | 166 (7) |
| O6—H6B⋯O3ii | 0.81 (6) | 2.11 (6) | 2.915 (5) | 174 (8) |
| O7—H7A⋯O5i | 0.85 | 2.27 | 3.055 (7) | 153 |
| O7—H7B⋯O6iii | 0.85 | 1.94 | 2.763 (6) | 163 |
Symmetry codes: (i) 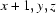 ; (ii)
; (ii) 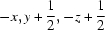 ; (iii)
; (iii) 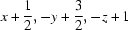 .
.
Acknowledgments
IB thanks the Spanish Research Council (CSIC) for providing us with a free-of-charge licence for the CSD system. The authors acknowdledge funds from FONDECYT (1110068) and U. Antofagasta (CODEI N° 1383).
supplementary crystallographic information
Comment
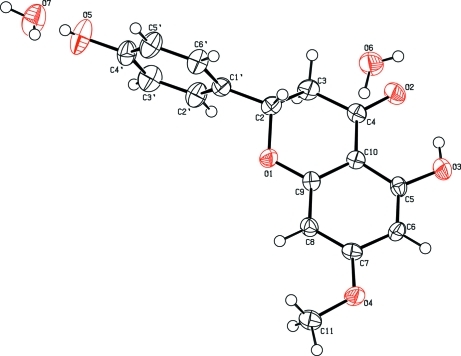
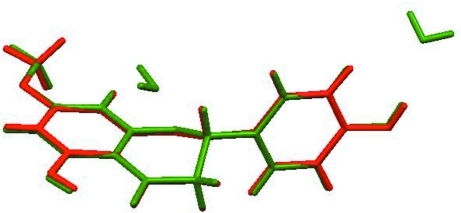
The crystal structure of the flavanone (S)-sakuranetin (5,4'-di-hydroxy-7-methoxyflavanone dihydrate, C16H14O5.2H2O) which was isolated from the native Chilean shrub Heliotropium taltalense (Heliotropiaceae) collected in Quebrada de Paposo II Region of Chile, is presented in this paper. (S)-Sakuranetin is a methylated flavanone obtained from the bark of Prunuspuddum (Narasimhachari & Seshadri, 1949) and later from other Prunus species (Atkinson & Blakeman, 1982). This compound is a phytoalexin produced in response to infection in rice (Plowright et al., 1996) with several beneficial biological properties such as antimicrobial activity (Atkinson & Blakeman, 1982) and the induction of adipogenesis (Saito et al., 2008). H. taltalense, is an endemic shrub or bush that grows in arid regions which obtains survival water from condensation from the Chilean coastal fog. Several Heliotropium species grow freely mainly in the north of Chile and they have glandular secreting trichomes whose principal rol is to produce a gummy exudate aimed to protect the plant from environmental factors and predators. This exudate is composed mainly of waxes and a mixture of phenolic compounds, mainly flavonoids and aromatic geranyl derivatives (Modak et al., 2009). The title compound (Fig. 1) crystallizes with an organic molecule and two water molecules in the asymmetric unit. The hydroxy group at C5 forms a strong intramolecular hydrogen bond with the carbonyl group with graph-set notation S(6) (Bernstein, et al., 1995). This interaction is observed in the unsolvated compound too, (Shoja, 1990). In the crystal, the components are linked by intermolecular O—H···O hydrogen bonds with an average O···O distance of 2.86 (14) Å and O—H···O angles in the range 147–174°, forming a 3-D network (Fig. 2). The 4'-hydroxyphenyl ring is bonded equatorially to the pyrone ring which adopts a slightly distorted sofa conformation with the C2 atom 0.328 (5) Å out of plane defined by the C2/C3/C4/C9/C10/O1 atoms, [puckering amplitude QT= 0.465 (5) Å; θ =124.9 (6)° & φ = 245.3 (7)° (Cremer & Pople, 1975)]. The title compound is the hydrated form of a previously reported structure. There are only slight variations in the molecular geometry of both compounds, so when both compound are superimposed all atoms are fitted within RMSD 0.0715 Å, (with inversion & flexibility), (Macrae et al., 2008), as is shown in Fig. 3.
Experimental
Dried aerial parts of H. taltalense (1.8 kg) were immersed in ethyl acetate (EtOAc) for one minute (2 l) in order to obtain an extract from the exudate. The extract was immediately concentrated in vacuo and the resulting dark brown syrup (47 g) was adsorbed on to silicagel 60 G (50 g) and slurred onto the top of a column containing silica gel 60 H (0.5 kg), partitioned using a medium pressure pump with an isocratic eluent (n-hexane–EtOAc 8:2), to obtain six partitions (fractions A to F: n-hexane, n-hexane–EtOAc 95:5, n-hexane–EtOAc 90:10, n-hexane–EtOAc 80:20, n-hexane–EtOAc 50:50 and pure EtOAc). Further purification by a combination of chromatography on silicagel 60 H and permeation through Sephadex LH-20 (eluting with methanol-water 7:3) of the fraction 20% hexane-ethyl acetate (fraction D, 15 g) afforded the phytoalexin 5,4'-di-hydroxy-7-methoxyflavanone dihydrate for which 1-D and 2-D NMR data are consistent with literature (Agrawal, 1989; Ogawa, et al., 2007). Recrystallization from hexane-ethyl acetate (9:1) at -20 ° C yielded yellow crystals of (S)-sakuranetin dihydrate (0.039 g), suitable for X-ray diffraction analysis.
Refinement
The parameters of the three H atoms bonded to atoms O5 (hydroxyl) and O6 (water) were located in difference maps and refined isotropically; all other H atoms were treated using a riding model, with C—H distances are in the range 0.93 – 0.98 Å and O—H distance of 0.82 and 0.85 Å, with Uiso(H) = 1.2Ueq(C,O) or 1.5Ueq(methyl C). The absolute configuration could not be established by the Flack method and the 772 observed Friedel opposites were merged.
Figures
Fig. 1.
: Molecular structure of title compound. Displacement ellipsoids are shown at the 50% probability level.
Fig. 2.
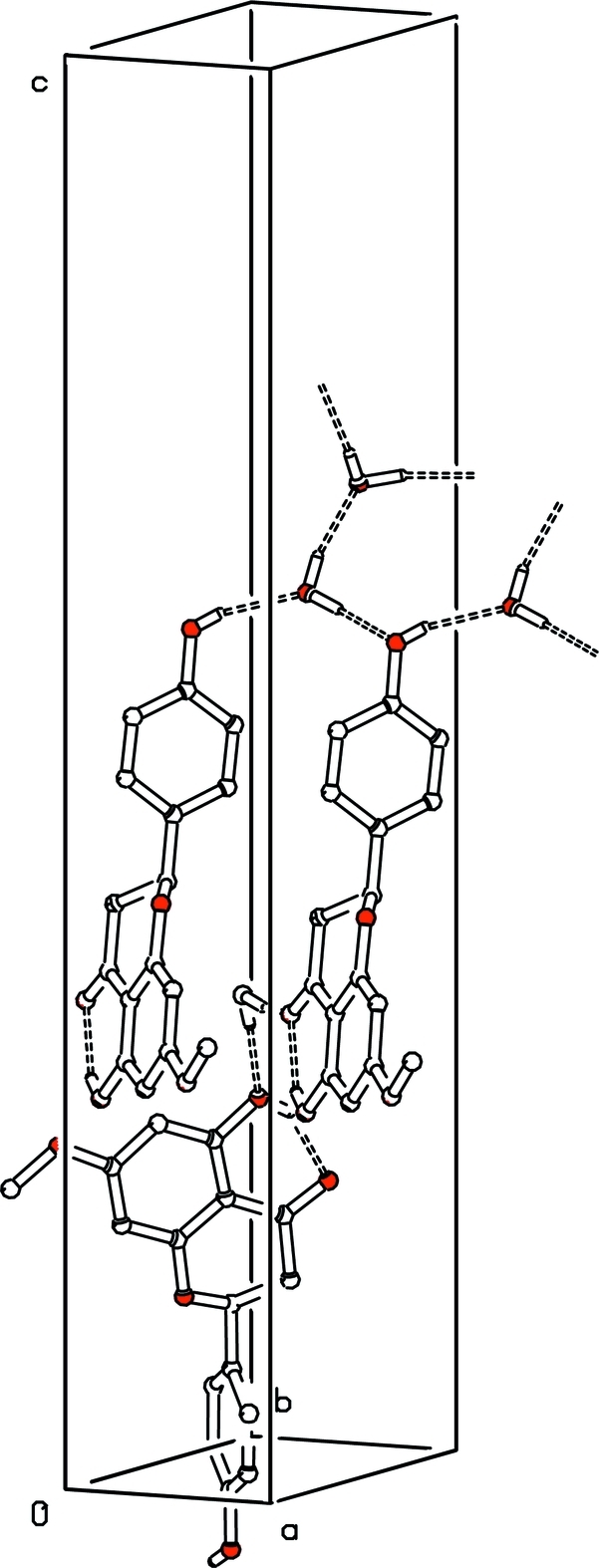
: A view of the crystal packing of the title compound, showing the intermolecular and intramolecular hydrogen bonding.
Fig. 3.
Superimposed structures for the title compound (green) and the unsolvated compound (red).
Crystal data
| C16H14O5·2H2O | F(000) = 680 |
| Mr = 322.30 | Dx = 1.376 Mg m−3 |
| Orthorhombic, P212121 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 2419 reflections |
| a = 5.0869 (10) Å | θ = 3.3–28.4° |
| b = 9.4622 (19) Å | µ = 0.11 mm−1 |
| c = 32.318 (7) Å | T = 293 K |
| V = 1555.6 (5) Å3 | Block, yellow |
| Z = 4 | 0.20 × 0.15 × 0.03 mm |
Data collection
| Nonius KappaCCD area-detector diffractometer | 1623 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.068 |
| graphite | θmax = 28.4°, θmin = 3.3° |
| φ and ω scans with κ offsets | h = 0→6 |
| 9743 measured reflections | k = 0→12 |
| 2021 independent reflections | l = −40→42 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.075 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.180 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.17 | w = 1/[σ2(Fo2) + (0.0649P)2 + 1.4205P] where P = (Fo2 + 2Fc2)/3 |
| 2021 reflections | (Δ/σ)max < 0.001 |
| 224 parameters | Δρmax = 0.38 e Å−3 |
| 0 restraints | Δρmin = −0.29 e Å−3 |
Special details
| Geometry. All s.u.'s (except the s.u. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell s.u.'s are taken into account individually in the estimation of s.u.'s in distances, angles and torsion angles; correlations between s.u.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell s.u.'s is used for estimating s.u.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.1698 (7) | 0.3272 (3) | 0.39766 (8) | 0.0317 (8) | |
| O2 | −0.3650 (7) | 0.5120 (4) | 0.31855 (10) | 0.0370 (9) | |
| O3 | −0.1585 (7) | 0.3656 (4) | 0.25747 (9) | 0.0350 (8) | |
| H3 | −0.2576 | 0.4224 | 0.2686 | 0.053* | |
| O4 | 0.5380 (7) | 0.0576 (4) | 0.28692 (9) | 0.0341 (8) | |
| O5 | 0.1305 (11) | 0.5341 (6) | 0.58135 (12) | 0.0610 (15) | |
| H5 | 0.247 (18) | 0.564 (8) | 0.593 (2) | 0.07 (3)* | |
| C2 | 0.0708 (9) | 0.4690 (5) | 0.40716 (13) | 0.0266 (10) | |
| H2 | 0.1893 | 0.5381 | 0.3945 | 0.032* | |
| C3 | −0.2023 (9) | 0.4877 (6) | 0.38806 (13) | 0.0345 (12) | |
| H3A | −0.3254 | 0.4250 | 0.4018 | 0.041* | |
| H3B | −0.2613 | 0.5840 | 0.3924 | 0.041* | |
| C4 | −0.2002 (9) | 0.4562 (5) | 0.34223 (13) | 0.0265 (10) | |
| C10 | −0.0071 (9) | 0.3561 (5) | 0.32828 (12) | 0.0247 (9) | |
| C5 | 0.0125 (9) | 0.3128 (5) | 0.28607 (12) | 0.0248 (10) | |
| C6 | 0.1952 (10) | 0.2153 (5) | 0.27344 (13) | 0.0278 (10) | |
| H6 | 0.2063 | 0.1899 | 0.2457 | 0.033* | |
| C7 | 0.3658 (9) | 0.1537 (5) | 0.30263 (13) | 0.0251 (9) | |
| C8 | 0.3505 (9) | 0.1902 (5) | 0.34455 (13) | 0.0267 (9) | |
| H8 | 0.4586 | 0.1465 | 0.3639 | 0.032* | |
| C9 | 0.1720 (8) | 0.2925 (5) | 0.35667 (12) | 0.0221 (9) | |
| C1' | 0.0839 (9) | 0.4870 (5) | 0.45324 (13) | 0.0265 (10) | |
| C2' | −0.0739 (11) | 0.4059 (6) | 0.48002 (14) | 0.0379 (12) | |
| H2' | −0.1923 | 0.3410 | 0.4691 | 0.046* | |
| C3' | −0.0542 (12) | 0.4220 (6) | 0.52256 (15) | 0.0439 (14) | |
| H3' | −0.1581 | 0.3673 | 0.5400 | 0.053* | |
| C4' | 0.1203 (10) | 0.5197 (6) | 0.53925 (13) | 0.0367 (12) | |
| C5' | 0.2769 (12) | 0.6004 (6) | 0.51347 (15) | 0.0441 (14) | |
| H5' | 0.3940 | 0.6656 | 0.5246 | 0.053* | |
| C6' | 0.2585 (11) | 0.5837 (6) | 0.47061 (14) | 0.0361 (12) | |
| H6' | 0.3645 | 0.6381 | 0.4534 | 0.043* | |
| C11 | 0.7076 (11) | −0.0153 (6) | 0.31527 (15) | 0.0364 (12) | |
| H11A | 0.6039 | −0.0734 | 0.3334 | 0.055* | |
| H11B | 0.8043 | 0.0523 | 0.3314 | 0.055* | |
| H11C | 0.8280 | −0.0737 | 0.3001 | 0.055* | |
| O6 | 0.2193 (9) | 0.7134 (5) | 0.32036 (13) | 0.0500 (11) | |
| H6A | 0.354 (18) | 0.659 (8) | 0.316 (2) | 0.07 (2)* | |
| H6B | 0.209 (15) | 0.761 (6) | 0.2996 (19) | 0.06 (2)* | |
| O7 | 0.5931 (9) | 0.6486 (6) | 0.60698 (12) | 0.0586 (13) | |
| H7A | 0.7450 | 0.6441 | 0.5961 | 0.088* | |
| H7B | 0.6306 | 0.6736 | 0.6315 | 0.088* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0332 (19) | 0.0370 (19) | 0.0248 (15) | 0.0105 (18) | −0.0035 (13) | −0.0066 (13) |
| O2 | 0.0244 (17) | 0.046 (2) | 0.0406 (18) | 0.0133 (18) | −0.0064 (14) | 0.0006 (16) |
| O3 | 0.0334 (19) | 0.039 (2) | 0.0324 (16) | 0.0081 (19) | −0.0069 (14) | 0.0014 (14) |
| O4 | 0.0292 (18) | 0.041 (2) | 0.0324 (16) | 0.0126 (17) | −0.0009 (14) | −0.0085 (14) |
| O5 | 0.061 (3) | 0.096 (4) | 0.0258 (18) | −0.028 (3) | 0.0028 (19) | −0.006 (2) |
| C2 | 0.018 (2) | 0.032 (2) | 0.030 (2) | 0.001 (2) | 0.0035 (17) | −0.0057 (18) |
| C3 | 0.022 (2) | 0.044 (3) | 0.038 (2) | 0.012 (3) | 0.0049 (19) | −0.006 (2) |
| C4 | 0.018 (2) | 0.029 (2) | 0.032 (2) | 0.000 (2) | −0.0033 (18) | 0.0001 (18) |
| C10 | 0.018 (2) | 0.028 (2) | 0.028 (2) | −0.001 (2) | 0.0005 (17) | −0.0006 (17) |
| C5 | 0.023 (2) | 0.027 (2) | 0.025 (2) | −0.005 (2) | −0.0066 (17) | 0.0041 (17) |
| C6 | 0.032 (2) | 0.031 (2) | 0.0204 (19) | 0.002 (2) | 0.0008 (18) | −0.0029 (17) |
| C7 | 0.017 (2) | 0.026 (2) | 0.032 (2) | 0.002 (2) | 0.0021 (17) | −0.0008 (18) |
| C8 | 0.023 (2) | 0.031 (2) | 0.026 (2) | 0.002 (2) | −0.0012 (17) | 0.0009 (18) |
| C9 | 0.0115 (18) | 0.029 (2) | 0.0259 (19) | −0.0042 (19) | −0.0017 (15) | −0.0002 (17) |
| C1' | 0.022 (2) | 0.029 (2) | 0.028 (2) | −0.001 (2) | −0.0004 (17) | −0.0030 (18) |
| C2' | 0.032 (3) | 0.048 (3) | 0.033 (2) | −0.008 (3) | −0.002 (2) | −0.006 (2) |
| C3' | 0.039 (3) | 0.054 (4) | 0.039 (3) | −0.012 (3) | 0.012 (2) | 0.003 (2) |
| C4' | 0.034 (3) | 0.050 (3) | 0.027 (2) | −0.001 (3) | 0.005 (2) | 0.000 (2) |
| C5' | 0.039 (3) | 0.057 (4) | 0.036 (3) | −0.008 (3) | −0.003 (2) | −0.009 (2) |
| C6' | 0.029 (3) | 0.047 (3) | 0.031 (2) | −0.012 (2) | 0.0050 (19) | −0.001 (2) |
| C11 | 0.027 (3) | 0.040 (3) | 0.042 (3) | 0.014 (3) | 0.000 (2) | −0.001 (2) |
| O6 | 0.048 (3) | 0.058 (3) | 0.044 (2) | 0.020 (2) | 0.007 (2) | 0.001 (2) |
| O7 | 0.049 (3) | 0.084 (3) | 0.042 (2) | −0.008 (3) | −0.0053 (19) | −0.007 (2) |
Geometric parameters (Å, °)
| O1—C9 | 1.365 (5) | C7—C8 | 1.400 (6) |
| O1—C2 | 1.465 (5) | C8—C9 | 1.383 (6) |
| O2—C4 | 1.252 (5) | C8—H8 | 0.9300 |
| O3—C5 | 1.364 (5) | C1'—C6' | 1.393 (6) |
| O3—H3 | 0.8200 | C1'—C2' | 1.408 (7) |
| O4—C7 | 1.361 (5) | C2'—C3' | 1.387 (7) |
| O4—C11 | 1.435 (6) | C2'—H2' | 0.9300 |
| O5—C4' | 1.369 (6) | C3'—C4' | 1.391 (7) |
| O5—H5 | 0.75 (8) | C3'—H3' | 0.9300 |
| C2—C1' | 1.501 (6) | C4'—C5' | 1.382 (7) |
| C2—C3 | 1.530 (6) | C5'—C6' | 1.397 (6) |
| C2—H2 | 0.9800 | C5'—H5' | 0.9300 |
| C3—C4 | 1.511 (6) | C6'—H6' | 0.9300 |
| C3—H3A | 0.9700 | C11—H11A | 0.9600 |
| C3—H3B | 0.9700 | C11—H11B | 0.9600 |
| C4—C10 | 1.437 (6) | C11—H11C | 0.9600 |
| C10—C9 | 1.426 (6) | O6—H6A | 0.87 (9) |
| C10—C5 | 1.427 (6) | O6—H6B | 0.81 (6) |
| C5—C6 | 1.372 (6) | O7—H7A | 0.8500 |
| C6—C7 | 1.408 (6) | O7—H7B | 0.8501 |
| C6—H6 | 0.9300 | ||
| C9—O1—C2 | 115.3 (3) | C9—C8—H8 | 120.6 |
| C5—O3—H3 | 109.5 | C7—C8—H8 | 120.6 |
| C7—O4—C11 | 118.0 (4) | O1—C9—C8 | 116.7 (4) |
| C4'—O5—H5 | 123 (6) | O1—C9—C10 | 121.2 (4) |
| O1—C2—C1' | 107.3 (4) | C8—C9—C10 | 122.2 (4) |
| O1—C2—C3 | 109.5 (4) | C6'—C1'—C2' | 118.3 (4) |
| C1'—C2—C3 | 115.3 (4) | C6'—C1'—C2 | 120.2 (4) |
| O1—C2—H2 | 108.2 | C2'—C1'—C2 | 121.5 (4) |
| C1'—C2—H2 | 108.2 | C3'—C2'—C1' | 120.6 (5) |
| C3—C2—H2 | 108.2 | C3'—C2'—H2' | 119.7 |
| C4—C3—C2 | 111.5 (4) | C1'—C2'—H2' | 119.7 |
| C4—C3—H3A | 109.3 | C2'—C3'—C4' | 120.2 (5) |
| C2—C3—H3A | 109.3 | C2'—C3'—H3' | 119.9 |
| C4—C3—H3B | 109.3 | C4'—C3'—H3' | 119.9 |
| C2—C3—H3B | 109.3 | O5—C4'—C5' | 121.5 (5) |
| H3A—C3—H3B | 108.0 | O5—C4'—C3' | 118.4 (5) |
| O2—C4—C10 | 123.0 (4) | C5'—C4'—C3' | 120.1 (5) |
| O2—C4—C3 | 120.7 (4) | C4'—C5'—C6' | 119.8 (5) |
| C10—C4—C3 | 116.3 (4) | C4'—C5'—H5' | 120.1 |
| C9—C10—C5 | 116.7 (4) | C6'—C5'—H5' | 120.1 |
| C9—C10—C4 | 120.9 (4) | C1'—C6'—C5' | 121.1 (4) |
| C5—C10—C4 | 122.4 (4) | C1'—C6'—H6' | 119.5 |
| O3—C5—C6 | 118.5 (4) | C5'—C6'—H6' | 119.5 |
| O3—C5—C10 | 119.9 (4) | O4—C11—H11A | 109.5 |
| C6—C5—C10 | 121.6 (4) | O4—C11—H11B | 109.5 |
| C5—C6—C7 | 119.8 (4) | H11A—C11—H11B | 109.5 |
| C5—C6—H6 | 120.1 | O4—C11—H11C | 109.5 |
| C7—C6—H6 | 120.1 | H11A—C11—H11C | 109.5 |
| O4—C7—C8 | 124.2 (4) | H11B—C11—H11C | 109.5 |
| O4—C7—C6 | 115.0 (4) | H6A—O6—H6B | 105 (6) |
| C8—C7—C6 | 120.8 (4) | H7A—O7—H7B | 101.2 |
| C9—C8—C7 | 118.9 (4) | ||
| C9—O1—C2—C1' | −180.0 (4) | C2—O1—C9—C8 | 155.2 (4) |
| C9—O1—C2—C3 | 54.2 (5) | C2—O1—C9—C10 | −26.6 (6) |
| O1—C2—C3—C4 | −54.4 (5) | C7—C8—C9—O1 | −178.2 (4) |
| C1'—C2—C3—C4 | −175.4 (4) | C7—C8—C9—C10 | 3.6 (6) |
| C2—C3—C4—O2 | −153.5 (5) | C5—C10—C9—O1 | 179.7 (4) |
| C2—C3—C4—C10 | 28.6 (6) | C4—C10—C9—O1 | −1.7 (6) |
| O2—C4—C10—C9 | −178.3 (4) | C5—C10—C9—C8 | −2.2 (6) |
| C3—C4—C10—C9 | −0.5 (6) | C4—C10—C9—C8 | 176.4 (4) |
| O2—C4—C10—C5 | 0.3 (7) | O1—C2—C1'—C6' | 112.6 (5) |
| C3—C4—C10—C5 | 178.0 (4) | C3—C2—C1'—C6' | −125.2 (5) |
| C9—C10—C5—O3 | 177.7 (4) | O1—C2—C1'—C2' | −66.1 (6) |
| C4—C10—C5—O3 | −0.9 (7) | C3—C2—C1'—C2' | 56.1 (6) |
| C9—C10—C5—C6 | −0.2 (6) | C6'—C1'—C2'—C3' | −0.3 (8) |
| C4—C10—C5—C6 | −178.8 (4) | C2—C1'—C2'—C3' | 178.5 (5) |
| O3—C5—C6—C7 | −176.9 (4) | C1'—C2'—C3'—C4' | 0.6 (8) |
| C10—C5—C6—C7 | 1.1 (7) | C2'—C3'—C4'—O5 | 178.9 (5) |
| C11—O4—C7—C8 | 2.6 (7) | C2'—C3'—C4'—C5' | −0.6 (9) |
| C11—O4—C7—C6 | −176.5 (4) | O5—C4'—C5'—C6' | −179.3 (5) |
| C5—C6—C7—O4 | 179.5 (4) | C3'—C4'—C5'—C6' | 0.2 (9) |
| C5—C6—C7—C8 | 0.4 (7) | C2'—C1'—C6'—C5' | −0.1 (8) |
| O4—C7—C8—C9 | 178.2 (4) | C2—C1'—C6'—C5' | −178.9 (5) |
| C6—C7—C8—C9 | −2.7 (7) | C4'—C5'—C6'—C1' | 0.2 (9) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O3—H3···O2 | 0.82 | 1.90 | 2.630 (5) | 147 |
| O5—H5···O7 | 0.76 (9) | 1.99 (9) | 2.720 (7) | 163 (7) |
| O6—H6A···O2i | 0.87 (9) | 2.00 (8) | 2.847 (6) | 166 (7) |
| O6—H6B···O3ii | 0.81 (6) | 2.11 (6) | 2.915 (5) | 174 (8) |
| O7—H7A···O5i | 0.85 | 2.27 | 3.055 (7) | 153 |
| O7—H7B···O6iii | 0.85 | 1.94 | 2.763 (6) | 163 |
Symmetry codes: (i) x+1, y, z; (ii) −x, y+1/2, −z+1/2; (iii) x+1/2, −y+3/2, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: KJ2193).
References
- Agrawal, P. K. (1989). In Carbon-13 NMR of Flavonoids Amsterdam: Elsevier.
- Atkinson, P. & Blakeman, J. P. (1982). New Phytol. 92, 63–74.
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl. 34, 1555–1573.
- Cremer, D. & Pople, J. A. (1975). J. Am. Chem. Soc. 97, 1354–1358.
- Farrugia, L. J. (1999). J. Appl. Cryst. 32, 837–838.
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Modak, B., Rojas, M. & Torres, R. (2009). Molecules, 14, 1980–1989. [DOI] [PMC free article] [PubMed]
- Narasimhachari, N. & Seshadri, T. R. (1949). Proc. Indian Acad. Sci. A 29, 265–268.
- Nonius (2000). COLLECT Nonius BV, Delft, The Netherlands.
- Ogawa, Y., Oku, H., Iwaoka, E., Iinuma, M. & Ishiguro, K. (2007). Chem. Pharm. Bull. 55, 675–678. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods in Enzymology, Vol. 276, Macromolecular Crystallography, Part A, edited by C. W. Carter Jr & R. M. Sweet, pp. 307–326. New York: Academic Press.
- Plowright, R. A., Grayer, R. J., Gill, J. R., Rahman, M. L. & Harborne, J. B. (1996). Nematologica, 42, 564–578.
- Saito, T., Abe, D. & Sekiya, K. (2008). Biochem. Biophys. Res. Commun. 372, 835–839. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Shoja, M. (1990). Acta Cryst. C46, 1969–1971.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536811051221/kj2193sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811051221/kj2193Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811051221/kj2193Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report