Abstract
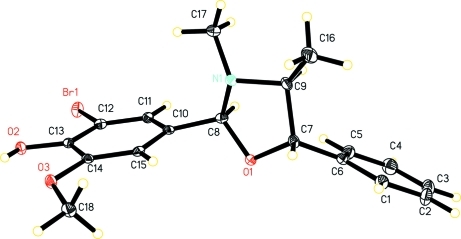
In the title compound, C18H20BrNO3, the oxazolidine ring adopts an envelope conformation with the N atom at the flap position. The mean plane of oxazolidine ring makes dihedral angles of 82.96 (13) and 70.97 (12)°, respectively, with the phenyl and benzene rings. In the crystal, adjacent molecules are connected via O—H⋯O and C—H⋯O hydrogen bonds and C—H⋯π interactions into a zigzag chain along the b axis.
Related literature
For the synthesis and closely related structures, see: Asaruddin et al. (2010 ▶); Diwischeck et al. (2003 ▶); Khruscheva et al. (1997 ▶); Duffy et al. (2004 ▶). For therapeutic properties of oxazolidine derivatives, see: Moloney et al. (1998 ▶); Wang et al. (2010 ▶); Nakano et al. (2010 ▶); Fülöp et al. (2004 ▶); Panneerselvam (2011 ▶). For standard bond lengths, see: Allen et al. (1987 ▶). For the low-temperature device used in the data collection, see: Cosier & Glazer (1986 ▶).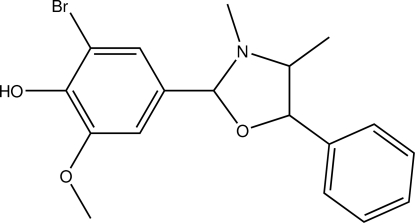
Experimental
Crystal data
C18H20BrNO3
M r = 378.26
Orthorhombic,
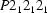
a = 7.8056 (4) Å
b = 11.9034 (6) Å
c = 18.9109 (9) Å
V = 1757.07 (15) Å3
Z = 4
Mo Kα radiation
μ = 2.35 mm−1
T = 100 K
0.50 × 0.36 × 0.23 mm
Data collection
Bruker SMART APEXII CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2009 ▶) T min = 0.383, T max = 0.618
10569 measured reflections
3074 independent reflections
2935 reflections with I > 2σ(I)
R int = 0.037
Refinement
R[F 2 > 2σ(F 2)] = 0.023
wR(F 2) = 0.056
S = 1.08
3074 reflections
215 parameters
1 restraint
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.32 e Å−3
Δρmin = −0.26 e Å−3
Absolute structure: Flack (1983 ▶), 1283 Friedel pairs
Flack parameter: 0.004 (7)
Data collection: APEX2 (Bruker, 2009 ▶); cell refinement: SAINT (Bruker, 2009 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811051269/is5016sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811051269/is5016Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811051269/is5016Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg2 is the centroid of the C1–C6 phenyl ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O2—H2⋯O1i | 0.85 (1) | 2.03 (1) | 2.7853 (19) | 148 (2) |
| C15—H15A⋯O2ii | 0.95 | 2.46 | 3.232 (3) | 138 |
| C18—H18A⋯Cg2i | 0.98 | 2.96 | 3.679 (3) | 131 |
Symmetry codes: (i) 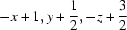 ; (ii)
; (ii) 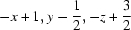 .
.
Acknowledgments
MH, HAW and ASAR acknowledge the Malaysia Ministry of Science, Technology and Innovations (MOSTI) for funding the synthetic chemistry work under the R&D initiative grant Nos. 09-05-lfn-meb-004 and 304/PFARMASI/650544/I121. MH thanks Universiti Sains Malaysia for the award of a postgraduate fellowship.
supplementary crystallographic information
Comment
Oxazolidine compounds are important in understanding drug behaviour in medicinal chemistry (Duffy et al., 2004). Derivatives of oxazolidine have shown inhibitory effects for several diseases or condition such as β-adrenoreseptor antagonist (Moloney et al., 1998), influenza antiviral (Wang et al., 2010; Nakano et al., 2010), antinflammatory agents (Fulop et al., 2004) and antihyperglycemic (Panneerselvam, 2011). In this paper, we report the X-ray crystal structure of the title oxazolidine compound, (I).
The title compound, C18H20BrNO3, consists of two aromatic rings which are connected through oxazolidine ring (Fig. 1). The molecule is similar with those reported by Asaruddin et al. (2010), in that only the present of Br atom at β position of 3-hydroxy-4-methoxyphenyl ring is different. The oxazolidine ring (O1/C7–C9/N1) adopts an envelope conformation with puckering parameters of Q = 0.433 (2) Å and φ = 107.3 (3)°. The N1 atom is at the flap position and it deviates from the mean plane through the remaining four atoms by 0.651 (2) Å. The C1–C6 phenyl and C10–C15 benzene rings make dihedral angles of 82.96 (13) and 70.97 (12)°, respectively, with the mean plane of oxazolidine ring. The bond lengths and angles are in normal ranges (Allen et al., 1987) and in agreement with those reported by Asaruddin et al. (2010).
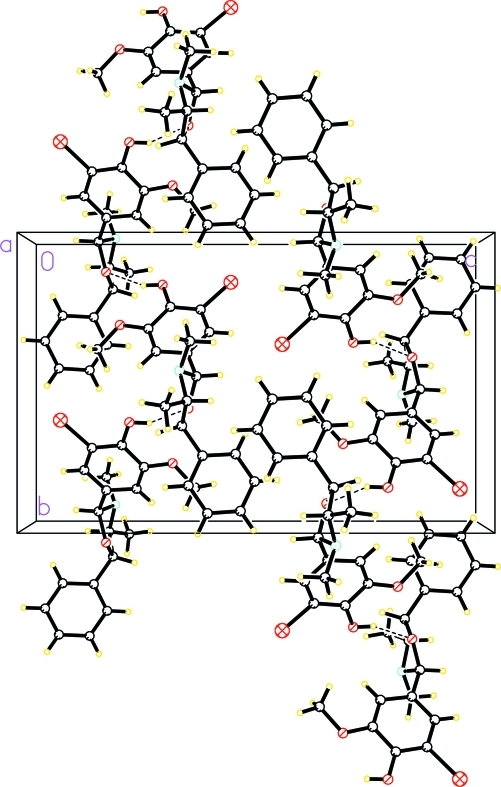
In the crystal structure, adjacent molecules are connected via intermolecular O2—H2···O1 and C15—H15A···O2 hydrogen bonds and C18—H18A···Cg2 interactions (Table 1) to form a chain along the [010] direction; Cg2 is the centroid of the C1–C6 ring.
Experimental
Following a modified method (Asaruddin et al., 2010; Diwischeck et al., 2003; Khruscheva et al., 1997), (1S,2S)-2-methylamino-1-phenylpropan-1-ol (0.17 g, 1 mmol) was mixed with 3-bromo-4-hydroxy-5-methoxybenzaldehyde (0.23 g, 1 mmol) in a two-round neck bottom flask. The mixture was dissolved in methanol (4 ml) and molecular sieve 4Å (0.1 g) was added to the reaction mixture then the solution was refluxed at 333 K for 6 h. The solution was filtered and the solvent was evaporated in vacuo to give a crude product which was then recrystallized three times from methanol to give colourless blocks with a yield 11%. These were washed with n-hexane and dried overnight to afford single crystals suitable for X-ray analysis.
Refinement
X-ray data were collected at 100 K (Cosier & Glazer, 1986). The hydroxyl H atom was located in a difference map and refined freely [O2—H2 = 0.8499 (10) Å]. Other H atoms were positioned geometrically and refined using riding model with C—H = 0.95–1.00 Å and Uiso(H)=1.2 or 1.5Ueq(C). A rotating group model was applied for methyl group.
Figures
Fig. 1.
The molecular structure of the title compound, with displacement ellipsoids are drawn at the 50% probability level.
Fig. 2.
The molecular packing of the title compound viewed down the a axis.
Crystal data
| C18H20BrNO3 | F(000) = 776 |
| Mr = 378.26 | Dx = 1.430 Mg m−3 |
| Orthorhombic, P212121 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 8965 reflections |
| a = 7.8056 (4) Å | θ = 2.0–24.9° |
| b = 11.9034 (6) Å | µ = 2.35 mm−1 |
| c = 18.9109 (9) Å | T = 100 K |
| V = 1757.07 (15) Å3 | Block, colourless |
| Z = 4 | 0.50 × 0.36 × 0.23 mm |
Data collection
| Bruker SMART APEXII CCD area-detector diffractometer | 3074 independent reflections |
| Radiation source: fine-focus sealed tube | 2935 reflections with I > 2σ(I) |
| graphite | Rint = 0.037 |
| Detector resolution: 83.66 pixels mm-1 | θmax = 24.9°, θmin = 2.0° |
| φ and ω scan | h = −9→9 |
| Absorption correction: multi-scan (SADABS; Bruker, 2009) | k = −14→14 |
| Tmin = 0.383, Tmax = 0.618 | l = −22→22 |
| 10569 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.023 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.056 | w = 1/[σ2(Fo2) + (0.0197P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 1.08 | (Δ/σ)max = 0.001 |
| 3074 reflections | Δρmax = 0.32 e Å−3 |
| 215 parameters | Δρmin = −0.26 e Å−3 |
| 1 restraint | Absolute structure: Flack (1983), 1283 Friedel pairs |
| Primary atom site location: structure-invariant direct methods | Flack parameter: 0.004 (7) |
Special details
| Experimental. The crystal was placed in the cold stream of an Oxford Cryosystems Cobra open=flow nitrogen cryostat (Cosier & Glazer, 1986) operating at 100.0 (1) K. |
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Br1 | 0.58037 (3) | 0.865565 (17) | 0.942297 (10) | 0.02402 (8) | |
| O1 | 0.2217 (2) | 0.40943 (12) | 0.84900 (7) | 0.0161 (3) | |
| O2 | 0.6507 (2) | 0.85865 (13) | 0.78478 (7) | 0.0187 (3) | |
| O3 | 0.5513 (2) | 0.71024 (11) | 0.68917 (7) | 0.0195 (3) | |
| N1 | 0.0154 (3) | 0.54199 (15) | 0.83295 (9) | 0.0179 (4) | |
| C1 | 0.0051 (3) | 0.1430 (2) | 0.85221 (12) | 0.0259 (5) | |
| H1A | −0.0140 | 0.1381 | 0.8027 | 0.031* | |
| C2 | −0.0108 (4) | 0.0473 (2) | 0.89404 (16) | 0.0362 (7) | |
| H2A | −0.0416 | −0.0224 | 0.8732 | 0.043* | |
| C3 | 0.0182 (4) | 0.0542 (2) | 0.96587 (14) | 0.0365 (7) | |
| H3A | 0.0072 | −0.0110 | 0.9944 | 0.044* | |
| C4 | 0.0633 (4) | 0.1557 (2) | 0.99672 (12) | 0.0341 (6) | |
| H4A | 0.0831 | 0.1600 | 1.0462 | 0.041* | |
| C5 | 0.0792 (4) | 0.25110 (19) | 0.95494 (11) | 0.0257 (5) | |
| H5A | 0.1110 | 0.3205 | 0.9760 | 0.031* | |
| C6 | 0.0490 (3) | 0.24580 (18) | 0.88251 (10) | 0.0191 (5) | |
| C7 | 0.0610 (3) | 0.34994 (17) | 0.83700 (10) | 0.0170 (5) | |
| H7A | 0.0563 | 0.3265 | 0.7862 | 0.020* | |
| C8 | 0.1793 (3) | 0.52295 (17) | 0.86771 (10) | 0.0165 (5) | |
| H8A | 0.1630 | 0.5277 | 0.9201 | 0.020* | |
| C9 | −0.0781 (3) | 0.43775 (18) | 0.84969 (10) | 0.0191 (5) | |
| H9A | −0.1098 | 0.4379 | 0.9009 | 0.023* | |
| C10 | 0.3140 (3) | 0.60567 (17) | 0.84574 (10) | 0.0152 (5) | |
| C11 | 0.3775 (3) | 0.68157 (17) | 0.89478 (11) | 0.0167 (5) | |
| H11A | 0.3434 | 0.6764 | 0.9429 | 0.020* | |
| C12 | 0.4899 (3) | 0.76443 (18) | 0.87389 (10) | 0.0162 (5) | |
| C13 | 0.5436 (3) | 0.77558 (17) | 0.80434 (10) | 0.0153 (4) | |
| C14 | 0.4837 (3) | 0.69524 (17) | 0.75520 (10) | 0.0149 (4) | |
| C15 | 0.3686 (3) | 0.61278 (17) | 0.77486 (10) | 0.0153 (4) | |
| H15A | 0.3264 | 0.5611 | 0.7407 | 0.018* | |
| C16 | −0.2384 (3) | 0.4204 (2) | 0.80546 (13) | 0.0302 (6) | |
| H16A | −0.3266 | 0.4741 | 0.8202 | 0.045* | |
| H16B | −0.2808 | 0.3437 | 0.8122 | 0.045* | |
| H16C | −0.2112 | 0.4324 | 0.7554 | 0.045* | |
| C17 | −0.0712 (3) | 0.64373 (18) | 0.85614 (11) | 0.0258 (5) | |
| H17A | 0.0013 | 0.7090 | 0.8461 | 0.039* | |
| H17B | −0.0931 | 0.6394 | 0.9071 | 0.039* | |
| H17C | −0.1802 | 0.6513 | 0.8309 | 0.039* | |
| C18 | 0.5220 (3) | 0.62188 (19) | 0.63849 (10) | 0.0224 (5) | |
| H18A | 0.5904 | 0.6361 | 0.5960 | 0.034* | |
| H18B | 0.4003 | 0.6199 | 0.6259 | 0.034* | |
| H18C | 0.5554 | 0.5496 | 0.6591 | 0.034* | |
| H2 | 0.659 (3) | 0.856 (2) | 0.74000 (15) | 0.022 (6)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Br1 | 0.03139 (14) | 0.02299 (12) | 0.01767 (10) | −0.00772 (11) | −0.00524 (10) | −0.00103 (8) |
| O1 | 0.0127 (8) | 0.0156 (7) | 0.0200 (7) | −0.0001 (7) | 0.0011 (6) | 0.0004 (5) |
| O2 | 0.0203 (8) | 0.0183 (8) | 0.0176 (7) | −0.0032 (7) | 0.0025 (6) | 0.0005 (6) |
| O3 | 0.0228 (10) | 0.0188 (8) | 0.0171 (6) | −0.0038 (7) | 0.0049 (7) | −0.0016 (5) |
| N1 | 0.0126 (10) | 0.0177 (10) | 0.0234 (8) | −0.0018 (8) | −0.0014 (8) | 0.0019 (7) |
| C1 | 0.0216 (13) | 0.0234 (12) | 0.0328 (11) | −0.0021 (11) | −0.0001 (10) | −0.0009 (10) |
| C2 | 0.0318 (16) | 0.0194 (13) | 0.0574 (16) | −0.0070 (12) | 0.0011 (14) | 0.0022 (11) |
| C3 | 0.0310 (16) | 0.0269 (14) | 0.0517 (15) | −0.0027 (12) | 0.0041 (13) | 0.0184 (11) |
| C4 | 0.0358 (16) | 0.0388 (15) | 0.0278 (11) | 0.0018 (14) | 0.0042 (12) | 0.0111 (10) |
| C5 | 0.0295 (14) | 0.0222 (11) | 0.0252 (10) | 0.0001 (11) | 0.0019 (12) | 0.0020 (8) |
| C6 | 0.0132 (13) | 0.0196 (11) | 0.0245 (10) | 0.0004 (10) | 0.0034 (10) | 0.0023 (8) |
| C7 | 0.0159 (12) | 0.0190 (11) | 0.0160 (8) | −0.0046 (10) | −0.0001 (9) | −0.0016 (8) |
| C8 | 0.0167 (13) | 0.0180 (11) | 0.0150 (9) | 0.0035 (10) | −0.0007 (9) | −0.0005 (8) |
| C9 | 0.0143 (11) | 0.0219 (11) | 0.0211 (9) | −0.0030 (11) | 0.0023 (10) | 0.0026 (8) |
| C10 | 0.0110 (11) | 0.0163 (11) | 0.0184 (9) | 0.0043 (9) | −0.0003 (8) | 0.0024 (8) |
| C11 | 0.0165 (12) | 0.0177 (10) | 0.0159 (9) | 0.0022 (9) | −0.0006 (9) | 0.0031 (8) |
| C12 | 0.0146 (12) | 0.0173 (11) | 0.0167 (9) | 0.0016 (9) | −0.0039 (9) | −0.0036 (8) |
| C13 | 0.0105 (12) | 0.0149 (10) | 0.0204 (9) | 0.0012 (9) | −0.0012 (9) | 0.0026 (7) |
| C14 | 0.0118 (11) | 0.0167 (11) | 0.0163 (9) | 0.0040 (9) | 0.0005 (9) | 0.0015 (8) |
| C15 | 0.0149 (11) | 0.0136 (11) | 0.0174 (9) | 0.0034 (9) | −0.0026 (9) | −0.0004 (7) |
| C16 | 0.0171 (13) | 0.0353 (14) | 0.0383 (12) | −0.0027 (12) | −0.0068 (12) | 0.0025 (10) |
| C17 | 0.0178 (12) | 0.0250 (12) | 0.0345 (11) | 0.0039 (14) | 0.0010 (11) | 0.0037 (9) |
| C18 | 0.0256 (12) | 0.0229 (12) | 0.0186 (9) | −0.0033 (11) | 0.0040 (9) | −0.0058 (9) |
Geometric parameters (Å, °)
| Br1—C12 | 1.903 (2) | C7—H7A | 1.0000 |
| O1—C8 | 1.435 (3) | C8—C10 | 1.499 (3) |
| O1—C7 | 1.458 (3) | C8—H8A | 1.0000 |
| O2—C13 | 1.347 (3) | C9—C16 | 1.519 (4) |
| O2—H2 | 0.8499 (10) | C9—H9A | 1.0000 |
| O3—C14 | 1.367 (2) | C10—C11 | 1.386 (3) |
| O3—C18 | 1.441 (2) | C10—C15 | 1.409 (3) |
| N1—C17 | 1.455 (3) | C11—C12 | 1.378 (3) |
| N1—C8 | 1.456 (3) | C11—H11A | 0.9500 |
| N1—C9 | 1.474 (3) | C12—C13 | 1.387 (3) |
| C1—C2 | 1.392 (3) | C13—C14 | 1.413 (3) |
| C1—C6 | 1.394 (3) | C14—C15 | 1.382 (3) |
| C1—H1A | 0.9500 | C15—H15A | 0.9500 |
| C2—C3 | 1.379 (4) | C16—H16A | 0.9800 |
| C2—H2A | 0.9500 | C16—H16B | 0.9800 |
| C3—C4 | 1.387 (4) | C16—H16C | 0.9800 |
| C3—H3A | 0.9500 | C17—H17A | 0.9800 |
| C4—C5 | 1.389 (3) | C17—H17B | 0.9800 |
| C4—H4A | 0.9500 | C17—H17C | 0.9800 |
| C5—C6 | 1.391 (3) | C18—H18A | 0.9800 |
| C5—H5A | 0.9500 | C18—H18B | 0.9800 |
| C6—C7 | 1.512 (3) | C18—H18C | 0.9800 |
| C7—C9 | 1.526 (3) | ||
| C8—O1—C7 | 107.31 (16) | N1—C9—H9A | 109.2 |
| C13—O2—H2 | 107.2 (18) | C16—C9—H9A | 109.2 |
| C14—O3—C18 | 116.80 (16) | C7—C9—H9A | 109.2 |
| C17—N1—C8 | 113.71 (18) | C11—C10—C15 | 119.30 (19) |
| C17—N1—C9 | 113.94 (18) | C11—C10—C8 | 119.56 (18) |
| C8—N1—C9 | 101.95 (17) | C15—C10—C8 | 120.99 (18) |
| C2—C1—C6 | 120.4 (2) | C12—C11—C10 | 120.15 (19) |
| C2—C1—H1A | 119.8 | C12—C11—H11A | 119.9 |
| C6—C1—H1A | 119.8 | C10—C11—H11A | 119.9 |
| C3—C2—C1 | 119.8 (2) | C11—C12—C13 | 122.20 (19) |
| C3—C2—H2A | 120.1 | C11—C12—Br1 | 119.62 (15) |
| C1—C2—H2A | 120.1 | C13—C12—Br1 | 118.16 (16) |
| C2—C3—C4 | 120.5 (2) | O2—C13—C12 | 121.22 (18) |
| C2—C3—H3A | 119.7 | O2—C13—C14 | 121.44 (18) |
| C4—C3—H3A | 119.7 | C12—C13—C14 | 117.32 (19) |
| C3—C4—C5 | 119.7 (2) | O3—C14—C15 | 126.11 (18) |
| C3—C4—H4A | 120.2 | O3—C14—C13 | 112.63 (18) |
| C5—C4—H4A | 120.2 | C15—C14—C13 | 121.26 (18) |
| C4—C5—C6 | 120.5 (2) | C14—C15—C10 | 119.68 (19) |
| C4—C5—H5A | 119.7 | C14—C15—H15A | 120.2 |
| C6—C5—H5A | 119.7 | C10—C15—H15A | 120.2 |
| C5—C6—C1 | 119.1 (2) | C9—C16—H16A | 109.5 |
| C5—C6—C7 | 120.85 (18) | C9—C16—H16B | 109.5 |
| C1—C6—C7 | 120.07 (18) | H16A—C16—H16B | 109.5 |
| O1—C7—C6 | 111.27 (17) | C9—C16—H16C | 109.5 |
| O1—C7—C9 | 104.75 (16) | H16A—C16—H16C | 109.5 |
| C6—C7—C9 | 115.34 (19) | H16B—C16—H16C | 109.5 |
| O1—C7—H7A | 108.4 | N1—C17—H17A | 109.5 |
| C6—C7—H7A | 108.4 | N1—C17—H17B | 109.5 |
| C9—C7—H7A | 108.4 | H17A—C17—H17B | 109.5 |
| O1—C8—N1 | 103.74 (17) | N1—C17—H17C | 109.5 |
| O1—C8—C10 | 112.86 (18) | H17A—C17—H17C | 109.5 |
| N1—C8—C10 | 112.88 (17) | H17B—C17—H17C | 109.5 |
| O1—C8—H8A | 109.1 | O3—C18—H18A | 109.5 |
| N1—C8—H8A | 109.1 | O3—C18—H18B | 109.5 |
| C10—C8—H8A | 109.1 | H18A—C18—H18B | 109.5 |
| N1—C9—C16 | 113.81 (18) | O3—C18—H18C | 109.5 |
| N1—C9—C7 | 101.0 (2) | H18A—C18—H18C | 109.5 |
| C16—C9—C7 | 113.98 (18) | H18B—C18—H18C | 109.5 |
| C6—C1—C2—C3 | −0.5 (5) | C6—C7—C9—N1 | 149.13 (17) |
| C1—C2—C3—C4 | 0.0 (5) | O1—C7—C9—C16 | 148.90 (18) |
| C2—C3—C4—C5 | 0.0 (5) | C6—C7—C9—C16 | −88.4 (2) |
| C3—C4—C5—C6 | 0.5 (5) | O1—C8—C10—C11 | −129.0 (2) |
| C4—C5—C6—C1 | −1.0 (4) | N1—C8—C10—C11 | 113.7 (2) |
| C4—C5—C6—C7 | 178.2 (3) | O1—C8—C10—C15 | 55.5 (3) |
| C2—C1—C6—C5 | 1.0 (4) | N1—C8—C10—C15 | −61.8 (3) |
| C2—C1—C6—C7 | −178.2 (2) | C15—C10—C11—C12 | 1.4 (3) |
| C8—O1—C7—C6 | −124.92 (17) | C8—C10—C11—C12 | −174.2 (2) |
| C8—O1—C7—C9 | 0.36 (18) | C10—C11—C12—C13 | 0.0 (3) |
| C5—C6—C7—O1 | 50.7 (3) | C10—C11—C12—Br1 | −178.39 (17) |
| C1—C6—C7—O1 | −130.1 (2) | C11—C12—C13—O2 | 178.6 (2) |
| C5—C6—C7—C9 | −68.4 (3) | Br1—C12—C13—O2 | −3.0 (3) |
| C1—C6—C7—C9 | 110.7 (2) | C11—C12—C13—C14 | −2.5 (3) |
| C7—O1—C8—N1 | −27.71 (18) | Br1—C12—C13—C14 | 175.97 (16) |
| C7—O1—C8—C10 | −150.23 (16) | C18—O3—C14—C15 | −10.8 (3) |
| C17—N1—C8—O1 | 167.85 (16) | C18—O3—C14—C13 | 169.59 (19) |
| C9—N1—C8—O1 | 44.77 (18) | O2—C13—C14—O3 | 2.2 (3) |
| C17—N1—C8—C10 | −69.6 (2) | C12—C13—C14—O3 | −176.71 (19) |
| C9—N1—C8—C10 | 167.27 (17) | O2—C13—C14—C15 | −177.4 (2) |
| C17—N1—C9—C16 | 71.3 (2) | C12—C13—C14—C15 | 3.6 (3) |
| C8—N1—C9—C16 | −165.77 (19) | O3—C14—C15—C10 | 178.0 (2) |
| C17—N1—C9—C7 | −166.13 (17) | C13—C14—C15—C10 | −2.3 (3) |
| C8—N1—C9—C7 | −43.20 (18) | C11—C10—C15—C14 | −0.2 (3) |
| O1—C7—C9—N1 | 26.45 (18) | C8—C10—C15—C14 | 175.3 (2) |
Hydrogen-bond geometry (Å, °)
| Cg2 is the centroid of the C1–C6 phenyl ring. |
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2—H2···O1i | 0.85 (1) | 2.03 (1) | 2.7853 (19) | 148 (2) |
| C15—H15A···O2ii | 0.95 | 2.46 | 3.232 (3) | 138 |
| C18—H18A···Cg2i | 0.98 | 2.96 | 3.679 (3) | 131 |
Symmetry codes: (i) −x+1, y+1/2, −z+3/2; (ii) −x+1, y−1/2, −z+3/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: IS5016).
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Asaruddin, M. R., Wahab, H. A., Mohamed, N., Goh, J. H. & Fun, H.-K. (2010). Acta Cryst. E66, o1452–o1453. [DOI] [PMC free article] [PubMed]
- Bruker (2009). APEX2, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Cosier, J. & Glazer, A. M. (1986). J. Appl. Cryst. 19, 105–107.
- Diwischeck, F., Heller, E. & Holzgrabe, U. (2003). Monatsh. Chem. 134, 1105–1111.
- Duffy, M., Gallagher, J. F. & Lough, A. J. (2004). Acta Cryst. E60, o234–o236.
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Fülöp, F., Lázár, L., Szakonyi, Z., Pihlavisto, M., Alaranta, S., Vainio, P. J., Juhakoski, A., Marjamäki, M. & Smith, D. J. (2004). Pure Appl. Chem. 76, 965–972.
- Khruscheva, N. S., Loim, N. M., Sokolov, V. I. & Makhaev, V. D. (1997). J. Chem. Soc. Perkin Trans. 1, pp. 2425–2427.
- Moloney, G. P., Craik, D. J., Iskander, M. N. & Nero, T. L. (1998). J. Chem. Soc. Perkin Trans. 2, pp. 199–206.
- Nakano, H., Osone, K., Takeshita, M., Kwon, E., Seki, C., Matsuyama, H., Takano, N. & Kohari, Y. (2010). Chem. Commun. 46, 4827–4829. [DOI] [PubMed]
- Panneerselvam, T. (2011). J. Adv. Pharm. Tech. Res. 2, 43–46. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Wang, Y., Chan, C., Su, Z. & Chen, C. (2010). Biophys. Chem. 147, 74–80. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811051269/is5016sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811051269/is5016Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811051269/is5016Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report