Abstract
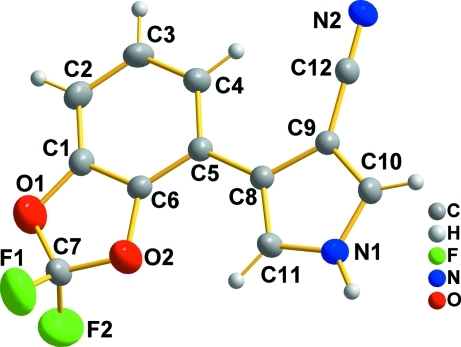
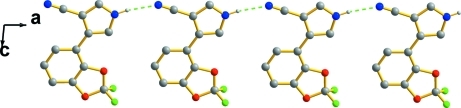
In the title compound, C12H6F2N2O2, the 2,2-difluoro-1,3-benzodioxole ring system is approximately planar [maximum deviation = 0.012 (2) Å] and its mean plane is twisted with respect to the pyrrole ring, making a dihedral angle of 2.51 (9)°. In the crystal, N—H⋯N hydrogen bonds link the molecules into chains running along the a axis. π–π stacking is also observed between parallel benzene rings of adjacent molecules, the centroid–centroid distance being 3.7527 (13) Å.
Related literature
For background to the title compound, see: Li et al. (2009 ▶); Pfluger et al. (1990 ▶). For the synthesis, see: Nyfeler & Ehrenfreund (1986 ▶). 
Experimental
Crystal data
C12H6F2N2O2
M r = 248.19
Triclinic,
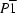
a = 7.5726 (15) Å
b = 7.8114 (16) Å
c = 8.9785 (18) Å
α = 93.58 (3)°
β = 94.65 (3)°
γ = 97.47 (3)°
V = 523.42 (18) Å3
Z = 2
Mo Kα radiation
μ = 0.13 mm−1
T = 293 K
0.39 × 0.32 × 0.15 mm
Data collection
Rigaku R-AXIS RAPID diffractometer
Absorption correction: multi-scan (ABSCOR; Higashi, 1995 ▶) T min = 0.950, T max = 0.980
5120 measured reflections
2359 independent reflections
1485 reflections with I > 2σ(I)
R int = 0.026
Refinement
R[F 2 > 2σ(F 2)] = 0.043
wR(F 2) = 0.118
S = 1.04
2359 reflections
167 parameters
1 restraint
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.19 e Å−3
Δρmin = −0.15 e Å−3
Data collection: RAPID-AUTO (Rigaku, 1998 ▶); cell refinement: RAPID-AUTO; data reduction: CrystalClear (Rigaku/MSC, 2002 ▶); program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536811054523/xu5407sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811054523/xu5407Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811054523/xu5407Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H101⋯N2i | 0.89 (1) | 2.15 (1) | 3.034 (2) | 169 (2) |
Symmetry code: (i) 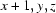 .
.
Acknowledgments
The authors thank the Project of Innovation Service Platform of Heilongjiang Province (PG09J001) and Heilongjiang University, China, for supporting the work.
supplementary crystallographic information
Comment
Fludioxonil also know as Maxim, which is kind of fungicide developed and produced by Novartis (Li et al., 2009; Pfluger et al., 1990). Herein we report its structure.
In the title compound, phenyl and pyrrole ring are almost coplanar with a small dihedral angle of 2.51 (9)° (Figure 1). Intermolecular N—H···N hydrogen bonds link molecules into chains along [100] (Figure 2, Table 1).
Experimental
The title compound was prepared by the reaction of 2-cyano-3-(2,2-difluoro-1,3-benzodioxol-4-yl)-2-propenamide and tosylmethyl isocyanide under alkaline condition (Robert & Josef, 1986). Colorless block crystals suitable for singl crystal X-ray diffraction were obtained by the recrystallization of title compound from a dichloromethane solution.
Refinement
N-bound H atom was located in a differece Fourier map and positional parameters were refined, Uiso(H) = 1.5Ueq(N). Other H atoms were placed in calculated positions with C—H = 0.93 Å, and refined in riding mode with Uiso(H) = 1.2Ueq(C).
Figures
Fig. 1.
The molecular structure of the title compound, showing displacement ellipsoids at the 50% probability level for non-H atoms.
Fig. 2.
A partial packing view, showing the hydrogen-bonding chain structure along [100].
Crystal data
| C12H6F2N2O2 | Z = 2 |
| Mr = 248.19 | F(000) = 252 |
| Triclinic, P1 | Dx = 1.575 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 7.5726 (15) Å | Cell parameters from 3390 reflections |
| b = 7.8114 (16) Å | θ = 3.4–27.5° |
| c = 8.9785 (18) Å | µ = 0.13 mm−1 |
| α = 93.58 (3)° | T = 293 K |
| β = 94.65 (3)° | Block, colorless |
| γ = 97.47 (3)° | 0.39 × 0.32 × 0.15 mm |
| V = 523.42 (18) Å3 |
Data collection
| Rigaku R-AXIS RAPID diffractometer | 2359 independent reflections |
| Radiation source: fine-focus sealed tube | 1485 reflections with I > 2σ(I) |
| graphite | Rint = 0.026 |
| ω scan | θmax = 27.5°, θmin = 3.4° |
| Absorption correction: multi-scan (ABSCOR; Higashi, 1995) | h = −9→9 |
| Tmin = 0.950, Tmax = 0.980 | k = −10→10 |
| 5120 measured reflections | l = −11→10 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.043 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.118 | w = 1/[σ2(Fo2) + (0.0612P)2 + 0.0143P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.04 | (Δ/σ)max = 0.001 |
| 2359 reflections | Δρmax = 0.19 e Å−3 |
| 167 parameters | Δρmin = −0.15 e Å−3 |
| 1 restraint | Extinction correction: SHELXTL (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.020 (6) |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.0680 (2) | 0.6305 (2) | 0.7663 (2) | 0.0492 (4) | |
| C2 | −0.1137 (2) | 0.6131 (3) | 0.7677 (2) | 0.0580 (5) | |
| H2 | −0.1723 | 0.5712 | 0.8478 | 0.070* | |
| C3 | −0.2036 (2) | 0.6626 (3) | 0.6407 (2) | 0.0604 (5) | |
| H3 | −0.3277 | 0.6534 | 0.6348 | 0.072* | |
| C4 | −0.1158 (2) | 0.7254 (2) | 0.5222 (2) | 0.0533 (5) | |
| H4 | −0.1833 | 0.7566 | 0.4395 | 0.064* | |
| C5 | 0.0715 (2) | 0.7442 (2) | 0.52139 (18) | 0.0412 (4) | |
| C6 | 0.1551 (2) | 0.6918 (2) | 0.64884 (19) | 0.0425 (4) | |
| C7 | 0.3564 (2) | 0.6277 (3) | 0.8215 (2) | 0.0580 (5) | |
| C8 | 0.1679 (2) | 0.8093 (2) | 0.39662 (18) | 0.0406 (4) | |
| C9 | 0.0963 (2) | 0.8705 (2) | 0.25982 (19) | 0.0424 (4) | |
| C10 | 0.2365 (2) | 0.9151 (2) | 0.1754 (2) | 0.0515 (5) | |
| H10 | 0.2281 | 0.9581 | 0.0812 | 0.062* | |
| C11 | 0.3488 (2) | 0.8223 (3) | 0.3852 (2) | 0.0523 (5) | |
| H11 | 0.4323 | 0.7921 | 0.4572 | 0.063* | |
| C12 | −0.0831 (2) | 0.8924 (2) | 0.2144 (2) | 0.0474 (4) | |
| F1 | 0.46357 (16) | 0.74161 (18) | 0.91577 (13) | 0.0804 (4) | |
| F2 | 0.43750 (16) | 0.48669 (18) | 0.81288 (16) | 0.0803 (4) | |
| N1 | 0.38690 (19) | 0.8860 (2) | 0.25235 (18) | 0.0575 (5) | |
| H101 | 0.4958 (17) | 0.901 (3) | 0.220 (3) | 0.086* | |
| N2 | −0.2273 (2) | 0.9115 (2) | 0.17807 (19) | 0.0631 (5) | |
| O1 | 0.19296 (17) | 0.5902 (2) | 0.87638 (15) | 0.0650 (4) | |
| O2 | 0.33825 (15) | 0.69183 (17) | 0.68299 (13) | 0.0540 (4) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0499 (10) | 0.0565 (11) | 0.0440 (10) | 0.0091 (8) | 0.0105 (8) | 0.0133 (8) |
| C2 | 0.0507 (10) | 0.0722 (13) | 0.0562 (12) | 0.0089 (9) | 0.0222 (9) | 0.0210 (10) |
| C3 | 0.0379 (9) | 0.0835 (14) | 0.0652 (13) | 0.0130 (9) | 0.0160 (8) | 0.0240 (11) |
| C4 | 0.0403 (9) | 0.0718 (12) | 0.0515 (11) | 0.0115 (8) | 0.0092 (8) | 0.0185 (10) |
| C5 | 0.0381 (8) | 0.0465 (9) | 0.0405 (9) | 0.0070 (7) | 0.0077 (7) | 0.0078 (7) |
| C6 | 0.0354 (8) | 0.0514 (9) | 0.0422 (9) | 0.0057 (7) | 0.0092 (7) | 0.0078 (8) |
| C7 | 0.0467 (10) | 0.0838 (14) | 0.0474 (11) | 0.0123 (10) | 0.0066 (8) | 0.0252 (10) |
| C8 | 0.0374 (8) | 0.0470 (9) | 0.0386 (9) | 0.0068 (7) | 0.0065 (7) | 0.0072 (7) |
| C9 | 0.0382 (8) | 0.0512 (10) | 0.0396 (9) | 0.0084 (7) | 0.0061 (7) | 0.0081 (7) |
| C10 | 0.0451 (9) | 0.0727 (12) | 0.0402 (10) | 0.0117 (8) | 0.0082 (7) | 0.0193 (9) |
| C11 | 0.0388 (9) | 0.0759 (12) | 0.0461 (11) | 0.0121 (8) | 0.0066 (7) | 0.0224 (9) |
| C12 | 0.0442 (10) | 0.0602 (11) | 0.0398 (10) | 0.0085 (8) | 0.0064 (7) | 0.0134 (8) |
| F1 | 0.0666 (8) | 0.1180 (11) | 0.0512 (7) | −0.0057 (7) | −0.0045 (6) | 0.0148 (7) |
| F2 | 0.0734 (8) | 0.0941 (9) | 0.0855 (10) | 0.0318 (7) | 0.0215 (6) | 0.0415 (8) |
| N1 | 0.0377 (8) | 0.0870 (12) | 0.0525 (10) | 0.0100 (8) | 0.0134 (7) | 0.0254 (8) |
| N2 | 0.0437 (9) | 0.0914 (13) | 0.0580 (11) | 0.0144 (8) | 0.0042 (7) | 0.0246 (9) |
| O1 | 0.0525 (8) | 0.1003 (11) | 0.0473 (8) | 0.0114 (7) | 0.0113 (6) | 0.0335 (7) |
| O2 | 0.0382 (6) | 0.0824 (9) | 0.0444 (7) | 0.0082 (6) | 0.0065 (5) | 0.0242 (6) |
Geometric parameters (Å, °)
| C1—C2 | 1.366 (3) | C7—F1 | 1.331 (2) |
| C1—C6 | 1.368 (2) | C7—O1 | 1.372 (2) |
| C1—O1 | 1.391 (2) | C7—O2 | 1.373 (2) |
| C2—C3 | 1.383 (3) | C8—C11 | 1.373 (2) |
| C2—H2 | 0.9300 | C8—C9 | 1.437 (2) |
| C3—C4 | 1.382 (2) | C9—C10 | 1.375 (2) |
| C3—H3 | 0.9300 | C9—C12 | 1.421 (2) |
| C4—C5 | 1.407 (2) | C10—N1 | 1.336 (2) |
| C4—H4 | 0.9300 | C10—H10 | 0.9300 |
| C5—C6 | 1.373 (2) | C11—N1 | 1.358 (2) |
| C5—C8 | 1.468 (2) | C11—H11 | 0.9300 |
| C6—O2 | 1.3960 (19) | C12—N2 | 1.145 (2) |
| C7—F2 | 1.330 (2) | N1—H101 | 0.891 (10) |
| C2—C1—C6 | 122.93 (17) | F2—C7—O2 | 109.90 (18) |
| C2—C1—O1 | 127.94 (16) | F1—C7—O2 | 109.89 (16) |
| C6—C1—O1 | 109.12 (15) | O1—C7—O2 | 110.86 (15) |
| C1—C2—C3 | 114.73 (17) | C11—C8—C9 | 104.79 (14) |
| C1—C2—H2 | 122.6 | C11—C8—C5 | 126.80 (15) |
| C3—C2—H2 | 122.6 | C9—C8—C5 | 128.40 (14) |
| C4—C3—C2 | 122.42 (16) | C10—C9—C12 | 123.08 (16) |
| C4—C3—H3 | 118.8 | C10—C9—C8 | 107.74 (15) |
| C2—C3—H3 | 118.8 | C12—C9—C8 | 129.12 (15) |
| C3—C4—C5 | 122.74 (17) | N1—C10—C9 | 108.11 (15) |
| C3—C4—H4 | 118.6 | N1—C10—H10 | 125.9 |
| C5—C4—H4 | 118.6 | C9—C10—H10 | 125.9 |
| C6—C5—C4 | 112.87 (15) | N1—C11—C8 | 109.45 (15) |
| C6—C5—C8 | 123.29 (14) | N1—C11—H11 | 125.3 |
| C4—C5—C8 | 123.83 (15) | C8—C11—H11 | 125.3 |
| C1—C6—C5 | 124.29 (15) | N2—C12—C9 | 179.4 (2) |
| C1—C6—O2 | 108.24 (15) | C10—N1—C11 | 109.90 (14) |
| C5—C6—O2 | 127.46 (14) | C10—N1—H101 | 125.5 (15) |
| F2—C7—F1 | 105.63 (16) | C11—N1—H101 | 124.5 (15) |
| F2—C7—O1 | 110.18 (16) | C7—O1—C1 | 105.74 (14) |
| F1—C7—O1 | 110.26 (18) | C7—O2—C6 | 106.02 (13) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H101···N2i | 0.89 (1) | 2.15 (1) | 3.034 (2) | 169 (2) |
Symmetry codes: (i) x+1, y, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: XU5407).
References
- Higashi, T. (1995). ABSCOR Rigaku Corporation, Tokyo, Japan.
- Li, C., Miu, H.-D., Zeng, Z.-W., Wang, M.-J., Wu, Z.-X., Yang, F. & Shi, W.-J. (2009). Modern Agrochem. 8, 19–24.
- Nyfeler, R. & Ehrenfreund, J. (1986). Switzerland Patent No. EP0206999.
- Pfluger, R. W., Indermühle, J. & Felix, F. (1990). Switzerland Patent No. EP0378046.
- Rigaku (1998). RAPID-AUTO Rigaku Corporation, Tokyo, Japan.
- Rigaku/MSC (2002). CrystalClear Rigaku/MSC Inc., The Woodlands, Texas, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536811054523/xu5407sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811054523/xu5407Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811054523/xu5407Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report