Abstract
Cytochalasins are a group of fungal secondary metabolites with diverse structures and bioactivities, including cytochalasin E produced by Aspergillus clavatus, which is a potent anti-angiogenic agent. Here, we report the identification and characterization of the cytochalasin gene cluster from A. clavatus NRRL 1. As a producer of cytochalasin E and K, the genome of A. clavatus was analyzed and the ~30 kb ccs gene cluster was identified based on the presence of a polyketide synthase-nonribosomal peptide synthetases (PKS-NRPS) and a putative Baeyer-Villiger monooxygenase (BVMO). Deletion of the central PKS-NRPS gene, ccsA, abolished the production of cytochalasin E and K, confirming the association between the natural products and the gene cluster. Based on bioinformatic analysis, a putative biosynthetic pathway is proposed. Furthermore, overexpression of the pathway specific regulator ccsR elevated the titer of cytochalasin E from 25 mg/L to 175 mg/L. Our results not only shed light on the biosynthesis of cytochalasins, but also provided genetic tools for increasing and engineering the production.
Keywords: Cytochalasins, PKS-NRPS, fungal megasynthetases, targeted gene inactivation, pathway-specific regulator, Diels-Alder
1. Introduction
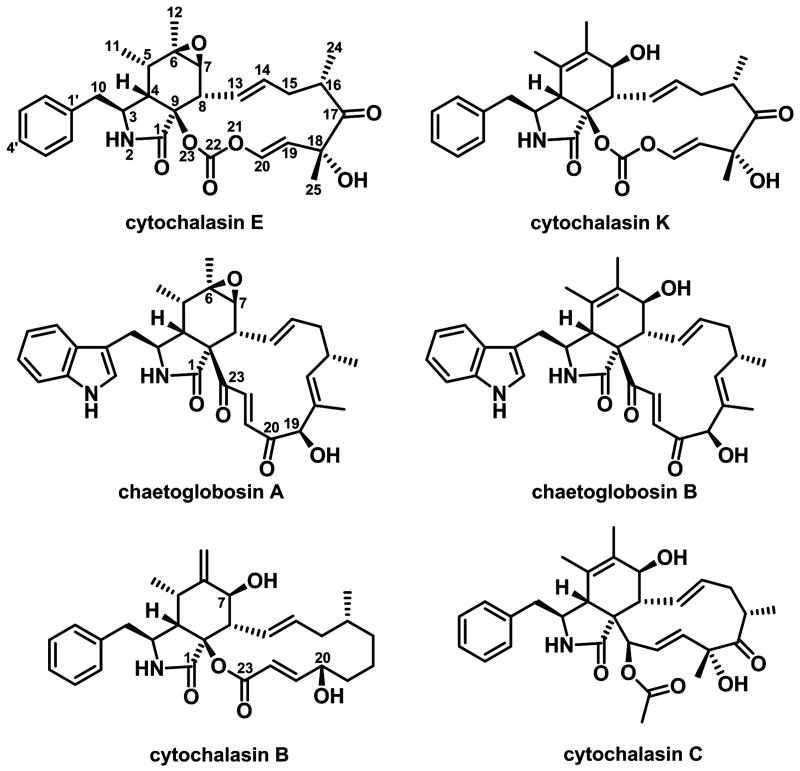
Cytochalasins are a group of polyketide-amino acid hybrid compounds belong to the cytochalasan family of fungal secondary metabolites, which have significant commercial and research values due to their diverse arrays of biological activities and complex molecular structures (Figure 1) (Scherlach et al., 2010). The cytochalasans are characterized structurally by their tricyclic core, which consists of a macrocyclic ring fused to an isoindolone moiety derived from a higly-reduced polyketide backbone and an amino acid (phenylalanine for cytochalasins) (Binder and Tamm, 1973; Scherlach et al., 2010). To date, over 80 different cytochalasans have been isolated from a number of the fungal genera, including Aspergillus, Phomopsis, Penicillium, Zygosporium, Chaetomium, Rosellinia, Metarrhizium, etc (Cole et al., 2003). Many cytochalasins, such as the earliest isolated cytochalasin A and B (Aldridge et al., 1967b), are capable of inhibiting the polymerization of actin and are thus widely used as tools in studying the division and motility of mammalian cells (Cooper, 1987). Besides the well-known actin binding characteristics, cytochalasins are also recognized for their other biological activities. For example, cytochalasin A and B were reported to repress glucose transport in human erythrocytes membrane (Rampal et al., 1980); cytochalasin D was shown to be a reversible inhibitor of protein synthesis in HeLa cells (Ornelles et al., 1986) and derivatives of cytochalasin H can regulate plant growth (Cox et al., 1983). Cytochalasin E, the molecule of interest in this study, was shown to display strong anti-angiogenic activities (Figure 1) (Udagawa et al., 2000). The remarkable structural complexity of cytochalasin E and the potent biological activities make this molecule an interesting target for biosynthetic study.
Figure 1.
Chemical structures of selected cytochalasans.
Previous isotope labeling studies have revealed the mixed malonate and amino acid origin of cytochalasans (Binder et al., 1970; Probst and Tamm, 1981; Vederas and Tamm, 1976), as well as the source of oxygen atoms in these molecules (Oikawa et al., 1992; Vederas et al., 1980). By contrast, the genetic and molecular basis for cytochalasan biosynthesis have only been revealed recently in the context of the (indol-3-yl)methyl-bearing cytochalasan, chaetoglobosin A, from Penicillium expansum (Figure 1) (Schumann and Hertweck, 2007). Using RNA-silencing, the biosynthesis of chaetoglobosin A was shown to involve the cheA gene that encodes for a hybrid iterative type I polyketide synthase-nonribosomal peptide synthetase (PKS-NRPS), and a biosynthetic pathway that could be generalized to other cytochalasans was proposed based on the che gene cluster.
To date, the che gene cluster responsible for chaetoglobosin A biosynthesis remained the only example of a gene cluster that encodes for cytochalasan production (Schumann and Hertweck, 2007). CheA is among the several fungal PKS-NRPS pathways that have been identified and characterized during the last few years, as exemplified by equisetin (Sims et al., 2005), aspyridone A (Bergmann et al., 2007; Xu et al., 2010), pseurotin A (Maiya et al., 2007), cyclopiazonic acid (Liu and Walsh, 2009; Tokuoka et al., 2008), tenellin (Halo et al., 2008) and etc. The PKS module of the PKS-NRPS responsible for the synthesis of a polyketide chain typically consists of several catalytic domains including ketosynthase (KS), malonyl-CoA:ACP transacylase (MAT), dehydratase (DH), methyltransferase (MT), enoylreductase (ER), ketoreductase (KR), and acyl-carrier protein (ACP), arranged in an assembly-line fashion from the N- to C-terminus. A downstream NRPS module, with the canonical set of condensation (C), adenylation (A) and thiolation (T) domains, amidates the carboxyl end of the polyketide with a specific amino acid. Typically, a reductase-like (R) domain is typically found at the C-terminus and can release the PKS-NRPS products via either a Dieckmann cyliczation reaction (Liu and Walsh, 2009; Sims and Schmidt, 2008) or as an aldehyde in a NADPH dependent fashion (Qiao et al., 2011).
Both chaetoglobosin A and cytochalasin E contain a substituted perhydroisoindolone scaffold fused with a macrocyclic ring that is the hallmark of cytochalasans, which is proposed to be derived from an intramolecular Diels-Alder reaction of the PKS-NRPS product following its release and formation of the pyrrolinone dienophile (Schumann and Hertweck, 2007). Unlike chaetoglobosin A however, cytochalasin E is derived from a shorter polyketide chain (octaketide instead of nonaketide), a different amino acid building block (phenylalanine instead of tryptophan), and contains a unique vinyl carbonate moeity, which all warrants further biosynthetic investigation at the molecular genetics levels.
In this work, we report the discovery of the ccs gene cluster involved in the biosynthesis of cytochalasin E and K from A. clavatus NRRL 1 by genome mining. Involvement of the PKS-NRPS (CcsA) was confirmed by gene disruption. Bioinformatic analysis of the genes encoded in the ccs gene cluster revealed insights into the biosynthesis of the unique features present in cytochalasin E and K. With the genetic blueprint in hand, we were able to significantly increase the titer of the cytochalasin products through overexpression of the pathway-specific regulator CcsR.
2. Materials and Methods
2.1. Strains and culture condition
The A. clavatus NRRL1 obtained from the Agriculture Research Service (NRRL) Culture Collection was used as the parental strain in this study. The wild type and mutant strains were maintained on Potato Dextrose Agar (PDA). For sporulation, wild type A. clavatus was grown on malt extract peptone agar (MEPA) (30 g/L malt extract, 3 g/L papaic digest of soybean meal and 15 g/L agar) for 3 days at 25 °C. Escherichia coli strain XL1-Blue (Stratagene) and E. coli TOPO10 (Invitrogen) were used for cloning.
2.2. Analyses of genome sequence of A. clavatus NRRL1
The genome sequence of A. clavatus NRRL1 was obtained from NCBI database (Fedorova et al., 2008). Gene predictions were performed via FGENESH program (www.softberry.com) and manually checked based on homologous gene/protein sequences in the GenBank database. Protein domain functions were deduced using Conserved Domain Search (NCBI).
2.3. DNA manipulation and construction of plasmids
High molecular weight genomic DNA of A. clavatus NRRL1 was prepared according to the protocol described previously (Chooi et al., 2008). DNA restriction enzymes were used as recommended by the manufacturer (New England Biolabs). PCR was performed using Platinum Pfx DNA polymerase (Invitrogen). Sequences of PCR products were confirmed by DNA sequencing (Laragen, CA). The plasmids pBARKS1 and pBARGPE1 (Pall and Brunelli, 1993) were obtained from the Fungal Genetics Stock Center (FGSC). The gene-specific primers in this work are listed in Table S1. The selection marker bar gene with trpC promoter was amplified from the plasmid pBARKS1. Construction of the knock-out cassette for ccsA gene was achieved using the fusion PCR method described previously (Szewczyk et al., 2006), and cloned into pCRblunt (Invitrogen) vector (Table S2). PCR was used to produce up to 10 μg DNA for fungal transformation. For regulator overexpression, the ccsR gene was amplified using PCR and digested with EcoRI/XhoI. The vector was prepared by digesting pBARGPE1 with the same set of restriction enzymes. The digested ccsR gene product was ligated into vector pBARGPE1 to give pKJ150 (Table S2). The other overexpression plasmid for the regulator ACLA_078740 (pKJ151) was cloned using the same strategy as pKJ150.
2.4. DNA transformation of A. clavatus
Preparation and transformation of A. clavatus protoplasts were performed as described previously (Chooi et al., 2010). Transformants were selected on glucose minimal medium agar supplemented with glufosinate (8 mg/mL) and sorbitol (1.2 M) as osmotic stabilizer. Miniprep genomic DNA from A. clavatus transformants was used as templates for PCR screening of gene deletants and was prepared as described for Aspergillus nidulans (Chooi et al., 2008).
2.5. Southern blot hybridization
High molecular weight genomic DNA (10 μg) was digested with restriction enzyme PvuII, separated onto 0.8% agarose gel and blotted onto the positively charged nylon membranes (Roche Applied Science). For Southern blot analysis, the DIG-High Prime DNA Labeling and Detection Starter Kit II (Roche Applied Science) was used. Hybridizations were carried out following the manufacturer’s protocol, except using 0.4% NaOH as transferring buffer instead of 20 × SSC buffer.
2.6. Chemical analysis and characterization of compounds from A. clavatus
For small scale analysis, wild type A. clavatus and transformants were grown in stationary liquid surface culture as a surface mat on 100 × 15 mm Petri dishes with 5 mL malt extract peptone (MEP) medium for 4 days at 25°C. The cultures were extracted with equal volumes of ethyl acetate (EtOAc) and evaporated to dryness. The dried extract was dissolved in methanol and analyzed by liquid chromatography mass spectrometry (LC-MS). LC-MS was conducted with a Shimadzu 2010 EV liquid chromatography mass spectrometer by using both positive and negative electrospray ionization, and a Phenomenex Luna 5 μm 2.0 × 100 mm C18 reverse-phase column. Samples were separated on a linear gradient of 5 to 95% CH3CN (v/v) in H2O supplemented with 0.05% (v/v) formic acid at a flow rate of 0.1 mL/min. The identity of cytochalasin E was confirmed by comparing the UV spectra, retention time and m/z value to the authentic standard (Sigma-Aldrich). To purify cytochalasin K for structural analysis, wild type A. clavatus was grown in stationary liquid surface culture condition in two liters MEP liquid medium divided into 20 large 150 × 15 mm Petri dishes for 7 days at 25 °C. The resulting mycelial mats along with the culture medium were pooled together and extracted three times with equal volumes of EtOAc. The organic extracts were combined and evaporated to dryness, redissolved in methanol, and purified by reverse-phase HPLC (XTerra Prep MS C18 5 μm, 19 mm × 50 mm) on a linear gradient of 50% to 95% CH3CN (v/v) over 20 min and 95% CH3CN (v/v) further for 15 min in H2O at a flow rate of 2.5 mL/min. The eluent was extracted with EtOAc, and dried in vacuo to give cytochalasin K as a pure solid (approximate yield of 18 mg/L). Nuclear magnetic resonance (NMR) spectra were obtained on a Bruker 500 MHz spectrometer using pyridine-d5 as solvent. For comparison of the cytochalasin E production between wild type and ccsR-overexpressing strains, the same stationary liquid surface culture condition as above was employed; while the submerged shaking flask liquid cultures were performed in two liter flasks containing 500 mL MEP medium (250 rpm, 25 °C).
3. Results and Discussion
3.1. Identification of a cytochalasin biosynthetic gene cluster from genome-wide analysis of PKS-NRPS genes from A. clavatus NRRL 1
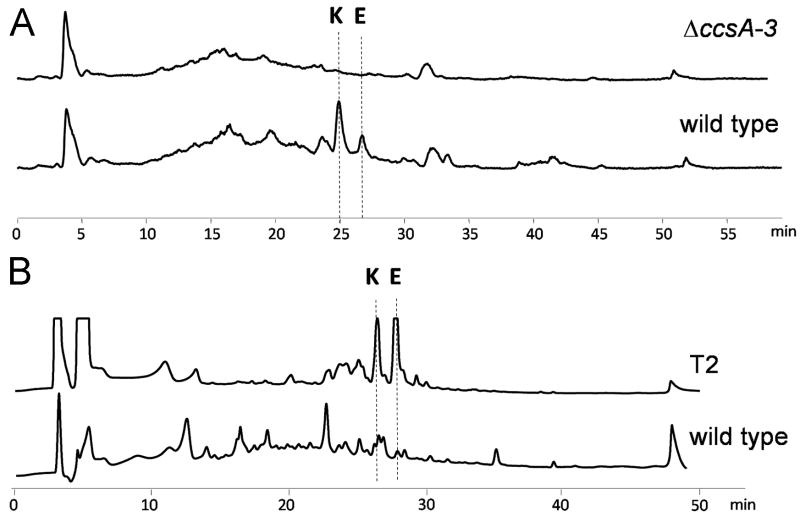
Both cytochalasin E and K have been reportedly isolated from the A. clavatus species (Buchi et al., 1973; Steyn et al., 1982). The production of cytochalasin E in the specific A. clavatus NRRL 1 (CBS 513.65) strain of which the genome has been sequenced (Fedorova et al., 2008), has also been detected in a chemotaxanomic study (Varga et al., 2007). To confirm the production of cytochalasins in A. clavatus NRRL 1, the strain was grown in stationary liquid surface culture in MEP medium for 4 days. LC-MS analysis of the ethyl acetate extract of the culture revealed two primary cytochalasin compounds with identical molecular weight (m/z 518 [M + Na]+) and UV absorption (λmax=235 nm) (Figure 4). The compound that eluted at retention time (RT) of 27 min matched with the authentic standard of cytochalasin E. To confirm the identity of the metabolite with RT = 25 min, the compound was purified from a large scale stationary liquid surface culture of A. clavatus NRRL1 with an approximate yield of 17 mg/L. The 1H NMR spectrum is consistent with that previously reported for cytochalasin K (Liu et al., 2006; Steyn et al., 1982) (Table S5 and Figure S1). Cytochalasin K is an isomer of cytochalasin E in which the 6,7-epoxide is isomerized to a C7 hydroxyl and a C5-C6 double bond, therefore both compounds are expected to be originated from the same gene cluster.
Figure 4.
Metabolic extracts of A. clavatus strains. (A) LC-MS analysis of metabolites produced by wild type and ΔccsA mutant A. clavatus strains. Total ion current chromatogram (m/z range 100-800) is shown. (B) HPLC analysis (210 nm) of metabolites produced by wild type A. clavatus and ccsR-overexpressed strain (T2). E and K stand for cytochalasin E and K, respectively.
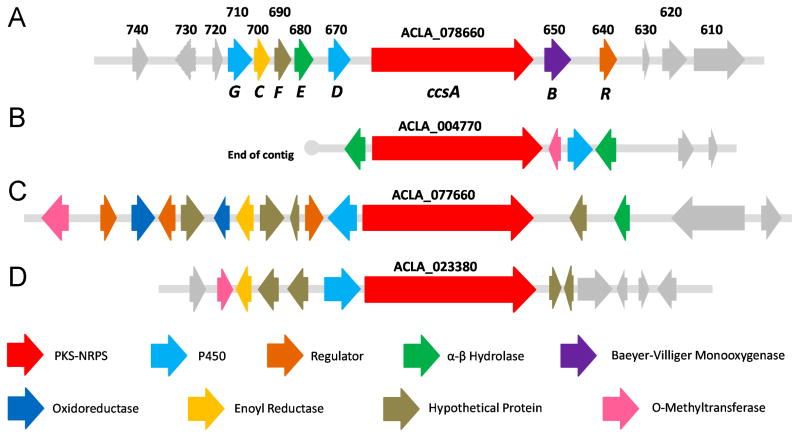
Given that the polyketide-amino acid backbone of cytochalasin E is likely biosynthesized by a PKS-NRPS, we searched the sequenced genome of A. clavatus NRRL 1 for genes encoding PKS-NRPS with the BLASTP program. Using the chaetoglobosin PKS-NRPS CheA as a query sequence (Schumann and Hertweck, 2007), the A. clavatus NRRL 1 genome was found to encode four putative PKS-NRPS genes (ACLA_004770, ACLA_077660, ACLA_023380 and ACLA_078660), none of which have been characterized previously (Figure 2 and Table S4). Among these four hits, ACLA_004770 is orthologous to pseurotin A synthetase from Aspergillus fumigatus (92% similarity, 86% identity) (Maiya et al., 2007), while ACLA_023380 is most closely related to equisetin synthetase from Fusarium heterosporum (64% similarity, 50% identity) (Sims et al., 2005). Due to the significant structural differences of pseurotin A and equisetin to cytochalasin E and K, these two PKS-NRPS loci were considered unlikely to be the desired candidates. Since fungal secondary biosynthetic genes are often clustered together (Keller et al., 2005), adjacent genes of the remaining two PKS-NRPS candidates were scrutinized for additional clues (Figure 2).
Figure 2.
Organizations of the ccs gene cluster and other PKS-NRPS gene loci in the genome of Aspergillus clavatus NRRL 1. (A) The ccs gene cluster. (B) PKS-NRPS gene (ACLA_004770) locus. (C) PKS-NRPS gene (ACLA_077660) locus. (D) PKS-NRPS gene (ACLA_023380) locus.
The vinyl carbonate moiety present in cytochalasins E and K is a unique structural feature not found in other cytochalasans and is rare among known natural products. In a previous feeding study, the successful incorporation of the 13-membered carbocyclic deoxaphomin to yield the 14-membered macrolactone-containing cytochalasin B implied the involvement of an enzymatic Baeyer-Villiger-type oxygen insertion between the C9 bridging carbon and C23 carbonyl (Robert and Tamm, 1975). Accordingly, the 13-membered macrocyclic carbonate in cytochalasin E was proposed to be originated from a corresponding 11-membered carbocyclic cytochalasan, whereby the unusual insertion of two oxygen atoms may occur via two consecutive Baeyer-Villiger oxidations (Robert and Tamm, 1975). In nature, such Baeyer-Villiger oxidations of ketones are known to be mediated by flavin-containing proteins collectively known as Baeyer-Villiger monooxygenases (BVMOs), which can be classified into type I, type II and type “O” with the type I BVMOs being the most commonly found (Leisch et al., 2011). Preliminary analysis of the locus containing the PKS-NRPS gene (ACLA_078660) revealed a gene (ACLA_078650) directly downstream that encodes for a putative flavoprotein (Figure 2A), which exhibits homology to the well-characterized type I BVMOs cyclohexanone monooxygenase (CHMO) from Acinetobacter sp. and cyclopentanone monoxygenase (CPMO) from Comamonas sp. (23 % identity and 24 % identity respectively) (Iwaki et al., 2002; Mirza et al., 2009). Like all type I BVMOs, the flavoprotein encoded by ACLA_078650 contains the conserved FxGxxxHxxxWP fingerprint motif, which serves as a linker that connects the FAD-binding domain to the NADP-binding domain and is important for the catalysis (Fraaije et al., 2002; Malito et al., 2004). The presence of BVMO afforded a strong indication that this gene cluster may encode the targeted cytochalasin E pathway. Furthermore, two cytochrome P450 oxygenases are encoded by genes in close proximity to ACLA_078660, which are consistent with the required enzymatic installation of the C6-C7 epoxide, the C17 keto and the C18 hydroxyl of cytochalasin E (Figure 2A). In contrast, the remaining PKS-NRPS (ACLA_077660) locus lacks a BVMO candidate in the vicinity, but encodes for an O-methyltransferase that is not required in the biosynthesis of cytochalasins E and K (Figure 2C). Therefore, our genome-wide survey of PKS-NRPS concluded that ACLA_078660 (renamed to ccsA) and the enzymes encoded in the corresponding gene cluster (designated as ccs cluster) are most likely to be involved in the biosynthesis of cytochalasins E and K.
3.2. Targeted gene disruption of the ccsA gene in A. clavatus NRRL 1
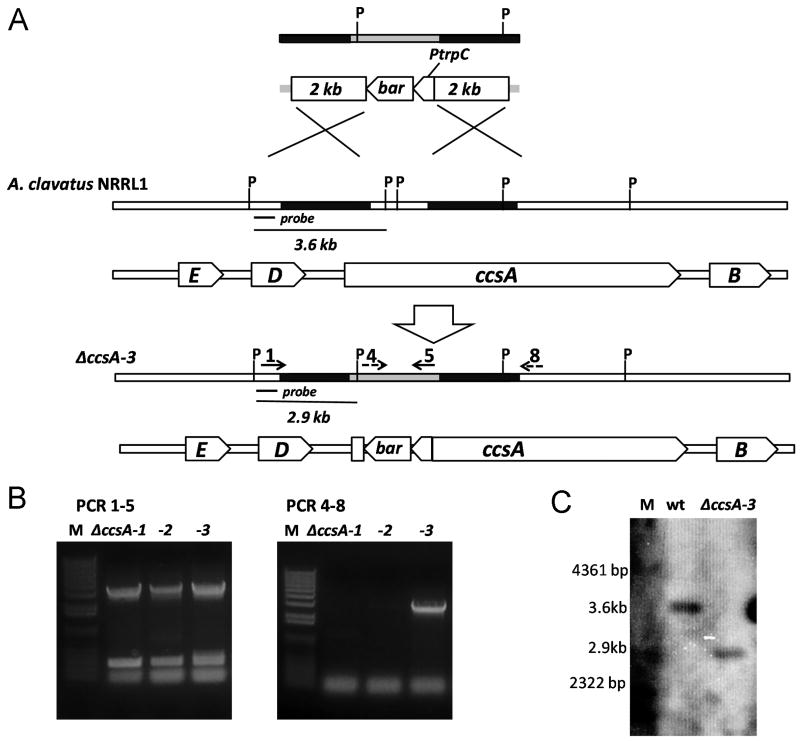
To verify the proposed association between the putative ccs gene cluster and cytochalasin E biosynthesis, a protoplast-based transformation system for A. clavatus and a gene inactivation strategy were developed with the fungal selection marker bar, which confers host resistance towards the selection marker glufosinate (Figure 3A) (Chooi et al., 2010). Successful incorporation of the ccsA deletion cassette by double-crossover recombination expects the replacement of an internal 1 kb region of the KS domain of ccsA gene with the bar resistant marker (Figure 3). Out of 51 glufosinate-resistant transformants, three positive transformants were found to have completely lost the production of cytochalasins E and K by LC-MS screening. The remaining 48 cytochalasin-producing transformants were most likely resulted from the ectopic integration of bar gene cassette in the A. clavatus genome. Further examination by diagnostic PCR showed that only one (ΔccsA-3) out of the three non-producing mutants yielded PCR products with the band sizes expected for a correct double-crossover recombination (Figure 3B). Disruption of cytochalasins production in ΔccsA-1 and ΔccsA-2 may have been a result of single crossover insertion of the deletion cassette as indicated by the diagnostic PCR results (Figure 3B). The disruption of ccsA by double homologous recombination in ΔccsA-3 was further confirmed by southern-blot hybridization of the genomic DNA digested with PvuII with a DIG-labeled probe. As a PvuII site is present in the bar resistant cassette, a 2.9 kb band, instead of the 3.6 kb in wild type, is expected to be detected from the integration of the ccsA deletion cassette into the correct site (Figure 3C). The loss of cytochalasin E and K production upon disruption of ccsA confirmed its essential role in cytochalasin biosynthesis in A. clavatus NRRL 1.
Figure 3.
Functional deletion of ccsA. (A) Homologous recombination scheme for deletion of ccsA gene in A. clavatus. P stands for PvuII sites and 1, 4, 5, 8 represent primers ccsA-1, ccsA-4, ccsA-5, ccsA-8 respectively (Table S1). (B) Diagnostic PCR analysis of mutant ΔccsA-1, ΔccsA-2 and ΔccsA-3 using primer pairs described above. (C) Southern blotting analysis of transformant ΔccsA-3 and wild type A. clavatus. Probe positions are shown in (A).
3.3. Overexpression of pathway-specific regulator encoded gene ACLA_078740 and ACLA_078640
Two genes (ACLA_078740 and ACLA_078640) encoding for putative fungal transcriptional factors with Zn(II)2Cys6 motif were found in the ccs gene cluster in the vicinity of the PKS-NRPS gene ccsA. The Zn(II)2Cys6 binuclear cluster proteins have so far been identified solely in fungi and were demonstrated to play crucial roles in transcriptional regulation (MacPherson et al., 2006). Zn(II)2Cys6-type fungal transcriptional regulator genes located inside secondary metabolite gene clusters of filamentous fungi has been shown to regulate the secondary metabolic genes in a pathway-specific manner (Ehrlich et al., 1999; Shimizu et al., 2007) and have been used as tools for genome mining (Bergmann et al., 2007; Chiang et al., 2009).
To functionally examine the two putative fungal transcription factors adjacent to the PKS-NRPS gene ccsA, we cloned both of them into pBARGPE1 vector under the control of A. nidulans gpdA (glyceraldehyde phosphate dehydrogenase gene) promoter. The resultant plasmids, pKJ150 and pKJ151 (containing ACLA_78640 and ACLA78740 respectively; Table S2) were randomly integrated into the genome of A. clavatus NRRL 1. The transformants selected for glufosinate resistance were analyzed by PCR using primer pair gpdA-f/ACLA_078640-r and gpdA-f/ACLA_078740-r, respectively. All transformants with the correct integration of intact overexpression cassettes were grown in stationary liquid surface culture with MEP medium. LC-MS analyses of the culture extracts showed that overexpression of ACLA_078740 did not affect the growth of A. clavatus nor influence the production of cytochalasins, suggesting that ACLA_078740 is not involved in the regulation of cytochalasin biosynthesis. On the other hand, significantly elevated production of both cytochalasin E and K were detected in all four transformants (T2, T6, T10 and T12) carrying the intact ACLA_078640-overexpression cassette, with no significant effect observed on the cell growth. This established that ACLA_078640 (ccsR) is the ccs pathway-specific transcriptional regulator and further confirmed that cytochalasin E and K are the products of the ccs gene cluster.
The titers of cytochalasin E in the four transformants were elevated at least by 3 fold, with T2 as the highest producer of cytochalasin E. The titer of cytochalasin E in a 8-day stationary liquid surface culture of T2 was estimated to be 250 mg/L based on ion current integration of m/z 518 [M + Na]+ in LCMS analysis using cytochalasin E standard curve, and a total of 175 mg/L of cytochalasin E was successfully purified. Therefore, the production of cytochalasin E in ccsR-overexpressing strain T2 had increased by approximately seven fold compared to the titer of wild type A. clavatus NRRL 1 strain (25 mg/L was isolated under the same culture condition and starting inoculation amount). The variation in the production levels of cytochalasin E are likely due to the different copy number of ccsR overexpression cassette present in the individual transformants. We observed during our culture condition testing experiments that cytochalasin E and K were only produced under stationary liquid surface culture condition by wild type A. clavatus NRRL 1, but not in submerged shaking flask condition. This is consistent with previous cytochalasin fermentation optimization experiments that utilized solid substrates without submerging the cells in liquid medium, in which a series of grains were used to culture the fungus in agitated flasks for 2 weeks with barley affording the highest yield (35 mg/kg barley) (Demain et al., 1976). We observed that overexpression of ccsR in A. clavatus T2 strain elevated the titer of cytochalasin E in a 6-day submerged shaking flask culture from undetectable levels in wild type to ~35 mg/L (estimated by MS ion current integration). Although the titer of cytochalasin E for T2 in submerged shaking flask culture is significantly lower than in stationary surface liquid culture, the successful production of cytochalasins in submerged culture condition presents significant advantages for fermentation scale-up in industrial stirred bioreactors.
Regulatory genes often clustered together with the biosynthetic genes in microorganisms. However, not all gene clusters contain a pathway-specific regulator, whereas some gene clusters may contain more than one pathway-specific regulator. The pathway-specific regulator can either play a positive (activator) or a negative (repressor) role in regulating the expression of biosynthetic genes within the cluster. Overexpression of pathway-specific activators is considered a useful metabolic engineering strategy to improve the titer of secondary metabolites. In bacterial systems, this strategy was extensively exploited: overexpression of the Streptomyces antibiotic regulatory protein (SARP) family has been commonly used to improve the production of pharmaceutically important secondary metabolites in actinomycetes (Olano et al., 2008). In fungi, the Zn(II)2Cys6 finger domain-containing proteins are the most common transcriptional activators; it has been demonstrated that overexpression of the corresponding activators AflR and CtnR led to upregulated transcription of the pathway genes and increased production of aflatoxin and citrinin respectively (Flaherty and Payne, 1997; Shimizu et al., 2007). More recently, a novel regulator CefR, which contains the similar “fungal_trans” conserved domain in CtnR but lack the Zn(II)2Cys6 finger domain, was identified in the cephalosporin biosynthetic gene cluster (Teijeira et al., 2011). Overexpression of CefR elevated the cephalosporin C production in the industrial Acremonium chrysogenum strain. Therefore, this approach should prove useful for improving production of pharmaceutically important fungal natural products, provided that such a pathway-specific activator exists in the corresponding biosynthetic gene cluster. In the absence of pathway-specific activator, overexpression of bottleneck enzymes in the pathway or increasing precursor and cofactor supply by engineering of primary metabolic pathways are two alternative strategies that may be considered for increasing fungal secondary metabolite production (Thykaer and Nielsen, 2003; Wattanachaisaereekul et al., 2008).
3.4. Proposed biosynthetic pathway for cytochalasin E/K biosynthesis
In the ccs gene cluster, a total of eight putative genes were predicted to be involved in biosynthesis of cytochalasin E and K (Table 1). At the two boundaries of the ccs gene cluster is a gene encoding for P450 monooxygenase (ccsG) and a gene encoding for the pathway-specific regulator (ccsR) (Figure 2). Immediately upstream of ccsG is a thioredoxin-encoding gene (ACLA_078720) and a mannitol dehydrogenease gene (ACLA_078730), which are both highly conserved in the genomes of other Aspergillus spp. (Table 1). Downstream of ccsR, are ACLA_078630, ACLA_078620 and ACLA_078610, which respectively encode for a hypothetical protein with no significant similarity to any protein sequence in the GenBank database (maybe a misannotation), a glucan 1,4-β-glucosidase (>67% identity to the homologs in A. fumigatus and N. fischeri), and a sensor histidine kinase (60-77% identity to homologs in Aspergillus spp.). As genes further upstream of ccsG and downstream of ccsR are mostly close orthologs shared among Aspergillus spp. and do not appear to involve in secondary metabolic biosynthesis, we reason that these genes are unlikely to participate in cytochalasin biosynthesis and may involve in primary metabolism or housekeeping roles.
Table 1.
Genes within and flanking the ccs gene cluster.
| Assigned gene name | Gene locus (ACLA_) | Gene size (bp) | Closest characterized homolog | Identity/Similarity (%) | Conserved domains | Deduced function |
|---|---|---|---|---|---|---|
| - | 078610 | 3980 | Aspergillus fumigates XP_746424.2* | 77/86 | HATPase_c[cd00075], Histidine kinase-like ATPases; | Sensor histidine kinase |
| - | 078620 | 1970 | Neosartorya fischeri XP_001258671.1* | 70/83 | Glyco_hydro_15 [pfam00723], Glycosyl hydrolases family 15. | Glucan 1,4- alpha- glucosidase |
| - | 078630 | 585 | n/a | n/a | n/a | n/a |
| ccsR | 078640 | 1593 | Aspergillus fumigatus XP_747734.1 * | 27/43 | GAL4-like Zn(II)2Cys6 binuclear cluster DNA- binding domain. | Transcription regulator |
| ccsB | 078650 | 1967 | Pseudomonas sp. HI-70 BAE93346 | 42/57 | PLN02172, flavin-containing monooxygenase FMO GS-OX. | Baeyer-Villiger monooxygenase |
| ccsA | 078660 | 12374 | Fusarium heterosporum AAV66106.2 | 42/61 | KS-AT-DH-MT-ER0-KR- ACP-C-A-T-R | PKS-NRPS hybrid |
| ccsD | 078670 | 1598 | Aspergillus fumigatus AAW03300.1 (Afu8g00560) | 32/52 | P450 super family [cl12078], Cytochrome P450. | P450 epoxidase |
| ccsE | 078680 | 1381 | Metarhizium anisopliae EFY94436.1* | 43/61 | Abhydrolase_1[pfam00561], alpha/beta hydrolase fold. | Esterase |
| ccsF | 078690 | 1190 | Metarhizium anisopliae EFZ03366.1* | 73/83 | n/a | Unknown |
| ccsC | 078700 | 1154 | Aspergillus terreus 3B6Z_A (LovC) | 42/59 | Enoyl_reductase_like [cd08249], enoyl reductase of the MDR family | Enoyl reductase |
| ccsG | 078710 | 1797 | Gibberella moniliformis CAQ16961.1 (GA-P450-4) | 39/55 | P450 [pfam00067], Cytochrome P450. | P450 monooxygenase |
| - | 078720 | 730 | Aspergillus oryzae XP_001827639.2* | 72/88 | DsbA_FrnE [cd03024], DsbA family, FrnE subfamily. | Thioredoxin |
| - | 078730 | 1509 | Neosartorya fischeri XP_001266605.1* | 88/94 | Mannitol_dh [pfam01232], Mannitol dehydrogenase Rossmann domain; | Mannitol dehydrogenase |
| - | 078740 | 1132 | Metarhizium anisopliae EFY97644.1* | 34/49 | n/a | Transcription regulator |
No close characterized homologs. Uncharacterized closest homologs based on BLASTP search are shown.
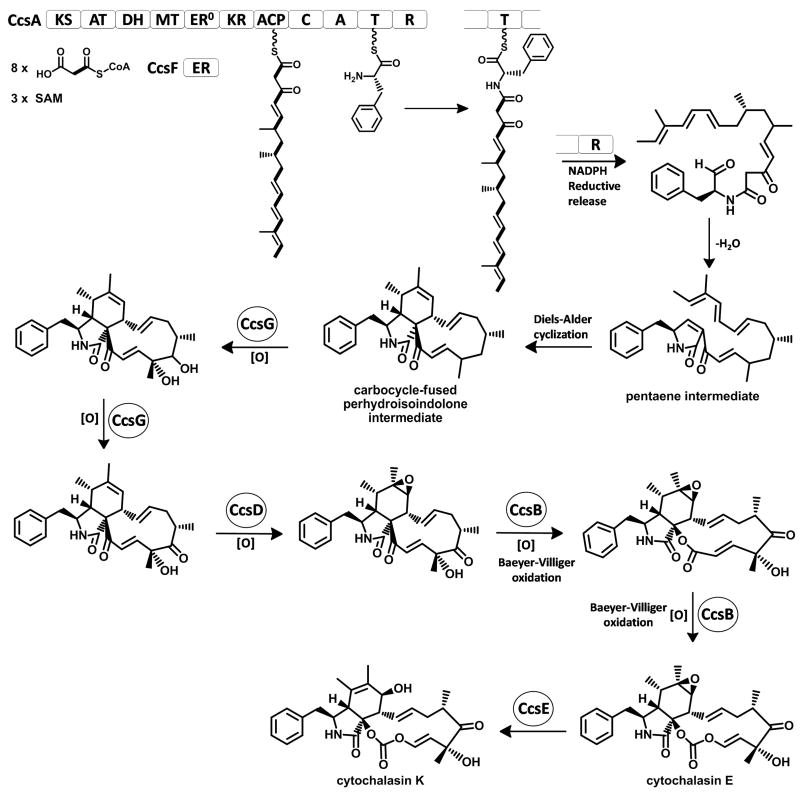
Based on the previous isotope labeling studies and deduced gene functions of ccs gene cluster, the biosynthetic pathway for cytochalasin E and K can be proposed (Figure 5) (Probst and Tamm, 1981; Scherlach et al., 2010). The domain organization of the PKS-NRPS encoded by ccsA is similar to that of other reported PKS-NRPSs genes, including CheA in P. expansum (35% identity to CcsA), TenS in Beauveria bassiana (33 % identity to CcsA) and ApdA in A. nidulans (36 % identity to CcsA) (Bergmann et al., 2007; Halo et al., 2008; Schumann and Hertweck, 2007). Due to the lack of the key NADPH binding motif (LXHXG(A)XGGVG), the ER domain of CcsA is proposed to be inactive. Correspondingly, a dissociated ER (encoded by gene ccsC) downstream of ccsA is proposed to function in trans with the PKS module of CcsA to synthesize a reduced β-keto octaketide backbone, analogous to the roles played by TenC and ApdC during the biosynthesis of tenillin and aspyridone respectively (Halo et al., 2008; Xu et al., 2010). Subsequently, one molecule of phenylalanine is selectively activated by the A domain of the NRPS module and transferred to the phosphopantetheinyl (pPant) arm of T domain. C domain then catalyzes the condensation between the nucleophilic amino group of phenylalanine and the electrophilic carbonyl of the upstream nascent octaketide chain to yield the T domain-tethered amide intermediate (Figure 5).
Figure 5.
Proposed biosynthetic pathway for cytochalasin E and K.
The R domain at the end of the NRPS module is proposed to catalyze the reductive release of the nascent aminoacyl-thioester to give an aminoaldehyde intermediate, which can readily undergo an intramolecular Knoevenagel condensation to yield the 3,5-disubstituted 3-pyrrolin-2-one (Figure 5). The terminal R domains of some PKS-NRPSs was demonstrate to offload the polyketide-amino acid intermediates via a Dieckmann condensation to afford a tetramic acid moiety (Liu and Walsh, 2009; Sims and Schmidt, 2008). Under this scenario, the released tetramic acid intermediate from CcsA would have to undergo further keto reduction and dehydration to form the proposed 3-pyrrolin-2-one dienophile. The lack of the required reductase encoded in ccs gene cluster is inconsistent with this possibility, while the reductive release catalyzed by CcsA R domain is supported by the protein sequence analysis. The CcsA R domain harbors an intact conserved NADPH binding motif GXSXXG and the catalytic triad Ser-Tyr-Lys shared among the short chain dehydrogenase/reductase SDR superfamily proteins, whereas the characterized Dieckmann R* domains contain a leucine or phenylalanine in place of the tyrosine in the catalytic triad (Liu and Walsh, 2009).
The intramolecular [4+2] Diels-Alder endo cycloaddition of the pentaene intermediate was proposed to occur between the terminal diene of the polyketide chain and the 3-pyrrolin-2-one dienophile to afford the 11-membered carbocycle-fused perhydroisoindolone scaffold (Figure 5). This proposed enzymatic reaction step, which may involve a so-called “Diels-Alderase”, has been shown to be feasible as demonstrated by previous biomimetic synthesis of cytochalasins (Oikawa et al., 1992; Vedejs et al., 1982), although the identity and enzymatic mechanism of such a “Diels-Alderase” remains enigmatic. In biosynthesis of lovastatin, the Diels-Alder reaction was proposed to occur at the hexaketide stage where the polyketide intermediate is still tethered to the ACP of the PKS LovB (Auclair et al., 2000). In contrast, the Diels-Alder cycloaddition in cytochalasan biosynthesis is predicted to occur after the release of the hybrid polyketide-amino acid molecule from the PKS-NRPS. Therefore, a discrete post PKS-NRPS tailoring enzyme, such as in solonapyrone and spinosyn A biosyntheses (Kasahara et al., 2010; Kim et al., 2011), may be required for the post PKS-NRPS Diels-Alder reaction in cytochalasin biosynthesis.
Upon formation of the 11-membered carbocycle-fused perhydroisoindolone intermediate, a number of oxidative steps are required to afford the final cytochalasin E and K, including two hydroxylations at C17 and C18, one alcohol oxidation at C17, one epoxidation at C6 and C7 and two Baeyer-Villiger oxidations. Based on the previous 13C-labeled acetate feeding experiments of cytochalasin B pathway in Phoma exigua (Vederas et al., 1980), the carbonyl (C-22) on the vinyl carbonate and the lactam carbonyl (C-1) of cytochalasin E and K are likely to be retained from the acetate units. Previous cytochrome P450 inhibitory experiment in chaetoglobosin-producing fungus Chaetomium subaffine showed that the oxygen atoms on the C20 carbonyl, C19 hydroxyl and the 6,7-epoxide are all added by cytochrome P450 enzymes (Oikawa et al., 1992). Similarly, we propose that the oxidative modification at C17, C18 and the C6-C7 epoxidation are likely to be catalyzed by the two cytochrome P450 oxygenases (CcsD and CcsG) found in the ccs gene cluster. CcsD exhibits similarity to GliF from A. fumigatus (42 % protein identity) that possibly catalyzes the formation of an arene oxide in gliotoxin biosynthesis (Gardiner and Howlett, 2005); hence, CcsD may be responsible for the epoxidation of the C6-C7 double bond. The closest characterized homolog of CcsG is GA-P450-4 (39% identity) from Gibberella moniliformis, which is an ent-kaurene oxidase that catalyzes the multiple oxidations of ent-kaurene to ent-kaurenoic acid (Bomke et al., 2008). This hints that CcsG may be responsible for the successive oxidative modifications at C17 and C18.
The double Baeyer-Villiger oxidations are among the final steps leading to cytochalasin E and K (Figure 5). Multiple sequence alignment of the primary protein sequences of CcsB and previously characterized BVMOs indicated that CcsB contains the two intact conserved Rossmann fold motifs GxGxxG and GxGxxA, as well as the so-called BVMO signature motif FxGxxxHxxxW (Iwaki et al., 2006; Leisch et al., 2011). Compared to the well-known BVMOs that function on smaller cyclic compounds, CcsB exhibits a significantly higher similarity toward the more recently characterized Pseudomonas sp. HI-70 cyclopentadecanone monooxygenase (CPDMO) CpdB (41% identity to CcsB) and Rhodococcus ruber SC1 cyclododecanone monooxygenase (CDMO) CddA (38% identity to CcsB), which are capable of lactonizing the C15 and C12 cyclic ketones respectively (Iwaki et al., 2006). This coincides with the proposed role of CcsB being involved in the oxidations of the 11-membered carbocyclic intermediate (Figure 5). Given that there are no additional gene that encodes for BVMO-like enzyme in the ccs gene cluster beside ccsB, CcsB may be responsible for the consecutive Baeyer-Villiger insertion of two oxygen atoms into the 11-membered carbocyclic intermediate to generate the unique vinyl carbonate moiety in cytochalasin E and K. A two-step oxidation is proposed: the first oxidation step is consistent with the mechanism in the biosynthesis of cytochalasin B (Robert and Tamm, 1975), while the second Baeyer oxidation is likely occurred on an acrylate moiety (Figure 5). Examples of such oxygen insertion between a ketone and an α-β double bond are rare but was reportedly observed in a microbial steroid degradation study where the A-ring of cholest-4-en-3-one was cleaved presumably via an enzymatic Baeyer-Villiger oxidation (Turfitt, 1948). If proven, CcsB may represent the first example of a BVMO that can catalyze such a double Baeyer-Villiger oxidation.
The epoxide-containing cytochalasin E is likely to be the compound that precedes cytochalasin K in the pathway. CcsE, which belongs to the large family of α/β hydrolase, may catalyze hydrolysis of epoxide bond to afford cytochalasin K. The similarity of CcsE to the Afu8g00540 (46% identity) in the pseurotin A pathway, in which the α/β hydrolase was proposed to hydrolyze the epoxide bond on the pyrrolidinone ring, supports the proposed function of CcsE. ccsF encodes for a protein that has no significant similarity to characterized proteins and no conserved domains are detected, thus its function has not been assigned. Interestingly, most CcsF homologs seems to be in the vicinity of PKS-NRPS genes in several sequenced fungal genomes, including Metarhizium anisopliae ARSEF 23, Magnaporthe oryzae 70-15, Phaeosphaeria nodorum SN15 and Chaetomium globosum CBS 148.51. Therefore, CcsF and its homologs may play a role in post-PKS-NRPS biosynthetic step (possibly Diels-Alder cyclization), resistance or transport of cytochalasins and related PKS-NRPS products.
3.5. ccs-like gene clusters in other fungal genomes
Closer examination of the structures of cytochalasin E and chaetoglobosin A suggests that the reduction, dehydration and methylation steps occur during the initial four polyketide iterations of both CcsA and CheA are identical. Therefore, it is surprising that CcsA shares the lowest head-to-tail protein similarity with CheA (39% identity between the PKS modules) among all the four PKS-NRPSs encoded in the A. clavatus NRRL 1 genome. In fact, CheA shares a slightly higher similarity (43% identity) to TenS and DmbS that produce pretenellin A and predesmethylbassianin A, both of which are structurally distinctive from cytochalasans. Furthermore, the tailoring enzymes in the che and ccs cluster do not appear to share close homology either (Table S6). Therefore, it is reasonable to speculate that the pathway for (indol-3-yl)methyl-containing cytochalasans may have diverged significantly from the benzyl-containing cytochalasins, or alternatively the two pathways may have arisen convergently.
The discovery and identification of ccs gene cluster opened up the possibility to use the PKS-NRPS gene ccsA and its associated tailoring genes for genome mining of gene clusters of both known and novel cytochalasins. Cytochalasin C and D have been isolated from the entomopathogenic fungus M. anisopliae (Aldridge et al., 1967a; Aldridge and Turner, 1969). Like cytochalasin E and K, both cytochalasin C and D are derived from octaketide chain and contain a benzyl group originated from phenylalanine but lack the vinyl carbonate moeity (Figure 1). Cytochalasin D in particular, has been used extensively to study cellular processes and is known to impair maintenance of long term potentiation (LTP) of actin filaments (Krucker et al., 2000). Since the genome of M. anisopliae ARSEF 23 has been sequenced recently (Gao et al., 2011), we examined the genome sequence for candidate cytochalasin gene cluster using CcsA as a query sequence in BLASTP search. A PKS-NRPS gene MAA_00428 was found to share the highest similarity to CcsA among all the PKS-NRPS genes in the GenBank database at the time of query. The corresponding homologs of the post PKS-NRPS tailoring genes (ccsCDEFG) in the ccs gene cluster can also be found in vicinity of MAA_00428 PKS-NRPS gene (Table S6). As expected, the BVMO ccsB homolog is missing in the gene cluster, while a putative acetyltransferase is present. However, the MAA_00428 PKS-NRPS appear to be truncated after the C domain of the NRPS module. Therefore, it is likely that this proposed cytochalasin-producing gene cluster from M. anisopliae ARSEF 23 strain may not have the ability to produce cytochalasin C and D due to truncation of the A-T-C domain in a relatively recent mutation event. Although this speculation remains to be proven, it is possible that such a homologous gene cluster with the complete CcsA homolog maybe present in cytochalasin-producing M. anisopliae strains. Besides M. anisopliae, ccs homologs (ccsACDEFG) are also found clustered in other fungal genomes, including the chaetoglobosin-producing Chaetomium globosum (CHGG_01239, 49% identity) (Table S6) and several plant pathogenic fungi, such as Magnaporthe grisea (syn2, 55% identity) and Phaeosphaeria nodurum (SNOG_00308, 50% identity), which may responsible for production of cytochalasin-related PKS-NRPS products.
4. Conclusion
In summary, we have identified the gene cluster for biosynthesis of cytochalasin E and K from the genome of A. clavatus NRRL 1. Disruption of ccsA provided the direct evidence that the PKS-NRPS gene ccsA is essential for the biosynthesis of cytochalasin E and K. Overexpression of the cytochalasin pathway-specific regulator ccsR led to a significantly increased cytochalasin production. The detail mechanistic steps of Diels-Alder reaction and the double Baeyer-Villiger oxidations of ketone to carbonate are currently under investigation. The identification of ccs gene cluster not only allows for continuous investigation of the molecular basis of the structural diversity generated by fungal PKS-NRPSs, but also opens the door for genome mining of cytochalasin gene cluster and application of molecular engineering to generate novel cytochalasan derivatives.
Supplementary Material
Highlights.
-
➢
We identified the gene cluster for cytochalasin E/K in Aspergillus clavatus genome
-
➢
The involvement of a PKS-NRPS gene (ccsA) was confirmed by targeted gene disruption
-
➢
A biosynthetic route for cytochalasin E/K was proposed based on gene cluster analyses
-
➢
Overexpression of the ccsR regulator gene improved cytochalasin production
-
➢
Genome mining revealed ccs-like gene clusters among other sequenced fungal genomes
Acknowledgments
This work was supported by NIH Grants 1R01GM085128 and 1R01GM092217, and the David and Lucile Packard Fellowship to YT. We thank the anonymous reviewers for their valuable comments and suggestions.
Abbreviations
- PKS-NRPS
polyketide synthase-nonribosomal peptide synthetase
- PDA
potato dextrose agar
- MEPA
malt extract peptone medium agar
- CDD
Conserved Domain Database
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aldridge DC, Armstron JJ, Speake RN, Turner WB. Cytochalasins, a new class of biologically active mould metabolites. Chem Commun (London) 1967a:26–27. [Google Scholar]
- Aldridge DC, Armstrong JJ, Speake RN, Turner WB. The structures of cytochalasins A and B. J Chem Soc C. 1967b:1667–1676. [Google Scholar]
- Aldridge DC, Turner WB. Structures of cytochalasins C and D. J Chem Soc C. 1969:923–928. [Google Scholar]
- Auclair K, Sutherland A, Kennedy J, Witter DJ, Van den Heever JP, Hutchinson CR, Vederas JC. Lovastatin nonaketide synthase catalyzes an intramolecular Diels-Alder reaction of a substrate analogue. J Am Chem Soc. 2000;122:11519–11520. [Google Scholar]
- Bergmann S, Schumann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat Chem Biol. 2007;3:213–217. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- Binder M, Kiechel JR, Tamm C. Zur biogenese des antibioticums phomin. 1 Teil: die grundbausteine Helv Chim Acta. 1970;53:1797–1812. doi: 10.1002/hlca.19700530728. [DOI] [PubMed] [Google Scholar]
- Binder M, Tamm C. The cytochalasans: a new class of biologically active microbial metabolites. Angew Chem Int Ed. 1973;12:370–380. doi: 10.1002/anie.197303701. [DOI] [PubMed] [Google Scholar]
- Bomke C, Rojas MC, Hedden P, Tudzynski B. Loss of gibberellin production in Fusarium verticillioides (Gibberella fujikuroi MP-A) is due to a deletion in the gibberellic acid gene cluster. Appl Environ Microbiol. 2008;74:7790–7801. doi: 10.1128/AEM.01819-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchi G, Kitaura Y, Yuan SS, Wright HE, Clardy J, Demain AL, Ginsukon T, Hunt N, Wogan GN. Structure of cytochalasin E, a toxic metabolite of Aspergillus clavatus. J Am Chem Soc. 1973;95:5423–5425. doi: 10.1021/ja00797a060. [DOI] [PubMed] [Google Scholar]
- Chiang YM, Szewczyk E, Davidson AD, Keller N, Oakley BR, Wang CC. A gene cluster containing two fungal polyketide synthases encodes the biosynthetic pathway for a polyketide, asperfuranone, in Aspergillus nidulans. J Am Chem Soc. 2009;131:2965–2970. doi: 10.1021/ja8088185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chooi YH, Cacho R, Tang Y. Identification of the viridicatumtoxin and griseofulvin gene clusters from Penicillium aethiopicum. Chem Biol. 2010;17:483–494. doi: 10.1016/j.chembiol.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chooi YH, Stalker DM, Davis MA, Fujii I, Elix JA, Louwhoff SH, Lawrie AC. Cloning and sequence characterization of a non-reducing polyketide synthase gene from the lichen Xanthoparmelia semiviridis. Mycol Res. 2008;112:147–161. doi: 10.1016/j.mycres.2007.08.022. [DOI] [PubMed] [Google Scholar]
- Cole RJ, Schweikert MA, Jarvis BB. Handbook of secondary fungal metabolites. Academic, Amsterdam; Boston: 2003. [Google Scholar]
- Cooper JA. Effects of cytochalasin and phalloidin on actin. J Cell Biol. 1987;105:1473–1478. doi: 10.1083/jcb.105.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RH, Cutler HG, Hurd RE, Cole RJ. Proton and C-13 nuclear magnetic resonance studies of the conformation of cytochalasin-H derivatives and plant growth regulating effects of cytochalasins. J Agr Food Chem. 1983;31:405–408. [Google Scholar]
- Demain AL, Hunt NA, Malik V, Kobbe B, Hawkins H, Matsuo K, Wogan GN. Improved procedure for production of cytochalasin E and tremorgenic mycotoxins by Aspergillus clavatus. Appl Environ Microbiol. 1976;31:138–140. doi: 10.1128/aem.31.1.138-140.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich KC, Montalbano BG, Cary JW. Binding of the C6-zinc cluster protein, AFLR, to the promoters of aflatoxin pathway biosynthesis genes in Aspergillus parasiticus. Gene. 1999;230:249–257. doi: 10.1016/s0378-1119(99)00075-x. [DOI] [PubMed] [Google Scholar]
- Fedorova ND, Khaldi N, Joardar VS, Maiti R, Amedeo P, Anderson MJ, Crabtree J, Silva JC, Badger JH, Albarraq A, Angiuoli S, Bussey H, Bowyer P, Cotty PJ, Dyer PS, Egan A, Galens K, Fraser-Liggett CM, Haas BJ, Inman JM, Kent R, Lemieux S, Malavazi I, Orvis J, Roemer T, Ronning CM, Sundaram JP, Sutton G, Turner G, Venter JC, White OR, Whitty BR, Youngman P, Wolfe KH, Goldman GH, Wortman JR, Jiang B, Denning DW, Nierman WC. Genomic islands in the pathogenic filamentous fungus Aspergillus fumigatus. PLoS Genet. 2008;4:e1000046. doi: 10.1371/journal.pgen.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty JE, Payne GA. Overexpression of aflR leads to upregulation of pathway gene transcription and increased aflatoxin production in Aspergillus flavus. Appl Environ Microbiol. 1997;63:3995–4000. doi: 10.1128/aem.63.10.3995-4000.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraaije MW, Kamerbeek NM, van Berkel WJ, Janssen DB. Identification of a Baeyer-Villiger monooxygenase sequence motif. FEBS Lett. 2002;518:43–47. doi: 10.1016/s0014-5793(02)02623-6. [DOI] [PubMed] [Google Scholar]
- Gao Q, Jin K, Ying SH, Zhang Y, Xiao G, Shang Y, Duan Z, Hu X, Xie XQ, Zhou G, Peng G, Luo Z, Huang W, Wang B, Fang W, Wang S, Zhong Y, Ma LJ, St Leger RJ, Zhao GP, Pei Y, Feng MG, Xia Y, Wang C. Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLoS Genet. 2011;7:e1001264. doi: 10.1371/journal.pgen.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner DM, Howlett BJ. Bioinformatic and expression analysis of the putative gliotoxin biosynthetic gene cluster of Aspergillus fumigatus. FEMS Microbial Lett. 2005;248:241–248. doi: 10.1016/j.femsle.2005.05.046. [DOI] [PubMed] [Google Scholar]
- Halo LM, Marshall JW, Yakasai AA, Song Z, Butts CP, Crump MP, Heneghan M, Bailey AM, Simpson TJ, Lazarus CM, Cox RJ. Authentic heterologous expression of the tenellin iterative polyketide synthase nonribosomal peptide synthetase requires coexpression with an enoyl reductase. Chembiochem. 2008;9:585–594. doi: 10.1002/cbic.200700390. [DOI] [PubMed] [Google Scholar]
- Iwaki H, Hasegawa Y, Wang S, Kayser MM, Lau PC. Cloning and characterization of a gene cluster involved in cyclopentanol metabolism in Comamonas sp. strain NCIMB 9872 and biotransformations effected by Escherichia coli-expressed cyclopentanone 1,2-monooxygenase. Appl Environ Microbiol. 2002;68:5671–5684. doi: 10.1128/AEM.68.11.5671-5684.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaki H, Wang S, Grosse S, Bergeron H, Nagahashi A, Lertvorachon J, Yang J, Konishi Y, Hasegawa Y, Lau PC. Pseudomonad cyclopentadecanone monooxygenase displaying an uncommon spectrum of Baeyer-Villiger oxidations of cyclic ketones. Appl Environ Microbiol. 2006;72:2707–2720. doi: 10.1128/AEM.72.4.2707-2720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara K, Miyamoto T, Fujimoto T, Oguri H, Tokiwano T, Oikawa H, Ebizuka Y, Fujii I. Solanapyrone synthase, a possible Diels-Alderase and iterative type I polyketide synthase encoded in a biosynthetic gene cluster from Alternaria solani. Chembiochem. 2010;11:1245–1252. doi: 10.1002/cbic.201000173. [DOI] [PubMed] [Google Scholar]
- Keller NP, Turner G, Bennett JW. Fungal secondary metabolism - from biochemistry to genomics. Nat Rev Microbiol. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Ruszczycky MW, Choi S-h, Liu Y-n, Liu H-w. Enzyme-catalysed [4+2] cycloaddition is a key step in the biosynthesis of spinosyn A. Nature. 2011;473:109–112. doi: 10.1038/nature09981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krucker T, Siggins GR, Halpain S. Dynamic actin filaments are required for stable long-term potentiation (LTP) in area CA1 of the hippocampus. Proc Natl Acad Sci USA. 2000;97:6856–6861. doi: 10.1073/pnas.100139797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisch H, Morley K, Lau PCK. Baeyer–Villiger Monooxygenases: More Than Just Green Chemistry. Chem Rev. 2011;111:4165–4222. doi: 10.1021/cr1003437. [DOI] [PubMed] [Google Scholar]
- Liu R, Gu Q, Zhu W, Cui C, Fan G, Fang Y, Zhu T, Liu H. 10-Phenyl-[12]-cytochalasins Z7, Z8, and Z9 from the marine-derived fungus Spicaria elegans. J Nat Prod. 2006;69:871–875. doi: 10.1021/np050201m. [DOI] [PubMed] [Google Scholar]
- Liu X, Walsh CT. Cyclopiazonic acid biosynthesis in Aspergillus sp.: characterization of a reductase-like R* domain in cyclopiazonate synthetase that forms and releases cyclo-acetoacetyl-L-tryptophan. Biochemistry. 2009;48:8746–8757. doi: 10.1021/bi901123r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson S, Larochelle M, Turcotte B. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol. Mol Biol Rev. 2006;70:583–604. doi: 10.1128/MMBR.00015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiya S, Grundmann A, Li X, Li SM, Turner G. Identification of a hybrid PKS/NRPS required for pseurotin A biosynthesis in the human pathogen Aspergillus fumigatus. Chembiochem. 2007;8:1736–1743. doi: 10.1002/cbic.200700202. [DOI] [PubMed] [Google Scholar]
- Malito E, Alfieri A, Fraaije MW, Mattevi A. Crystal structure of a Baeyer-Villiger monooxygenase. Proc. Natl Acad Sci USA. 2004;101:13157–13162. doi: 10.1073/pnas.0404538101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza IA, Yachnin BJ, Wang S, Grosse S, Bergeron H, Imura A, Iwaki H, Hasegawa Y, Lau PC, Berghuis AM. Crystal structures of cyclohexanone monooxygenase reveal complex domain movements and a sliding cofactor. J Am Chem Soc. 2009;131:8848–8854. doi: 10.1021/ja9010578. [DOI] [PubMed] [Google Scholar]
- Oikawa H, Murakami Y, Ichihara A. Biosynthetic study of chaetoglobosin A - origins of the oxygen and hydrogenatoms, and indirect evidence for biological Diels-Alder reaction. J Chem Soc Perk Trans. 1992;1:2955–2959. [Google Scholar]
- Olano C, Lombo F, Mendez C, Salas JA. Improving production of bioactive secondary metabolites in actinomycetes by metabolic engineering. Metab Eng. 2008;10:281–292. doi: 10.1016/j.ymben.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Ornelles DA, Fey EG, Penman S. Cytochalasin releases mRNA from the cytoskeletal framework and inhibits protein synthesis. Mol Cell Biol. 1986;6:1650–1662. doi: 10.1128/mcb.6.5.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall M, Brunelli J. A series of six compect fungal transformation vectors containing polylinkers with multiple unique restriction sites. Fungal Genet Newsl. 1993;40:59. [Google Scholar]
- Probst A, Tamm C. Biosynthesis of the cytochalasans. Biosynthetic studies on chaetoglobosin A and 19-O-acetylchaetoglobosin A. Helv Chim Acta. 1981;64:2065–2077. [Google Scholar]
- Qiao KJ, Zhou H, Xu W, Zhang WJ, Garg N, Tang Y. A fungal nonribosomal peptide synthetase module that can synthesize thiopyrazines. Org Lett. 2011;13:1758–1761. doi: 10.1021/ol200288w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampal AL, Pinkofsky HB, Jung CY. Structure of cytochalasins and cytochalasin B binding sites in human erythrocyte membranes. Biochemistry. 1980;19:679–683. doi: 10.1021/bi00545a011. [DOI] [PubMed] [Google Scholar]
- Robert JL, Tamm C. Biosynthesis of cytochalasans. Part 5. The incorporation of deoxaphomin into cytochalasin B (phomin) Helv Chim Acta. 1975;58:2501–2504. doi: 10.1002/hlca.19750580830. [DOI] [PubMed] [Google Scholar]
- Scherlach K, Boettger D, Remme N, Hertweck C. The chemistry and biology of cytochalasans. Nat Prod Rep. 2010;27:869–886. doi: 10.1039/b903913a. [DOI] [PubMed] [Google Scholar]
- Schumann J, Hertweck C. Molecular basis of cytochalasan biosynthesis in fungi: gene cluster analysis and evidence for the involvement of a PKS-NRPS hybrid synthase by RNA silencing. J Am Chem Soc. 2007;129:9564–9565. doi: 10.1021/ja072884t. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Kinoshita H, Nihira T. Identification and in vivo functional analysis by gene disruption of ctnA, an activator gene involved in citrinin biosynthesis in Monascus purpureus. Appl Environ Microbiol. 2007;73:5097–5103. doi: 10.1128/AEM.01979-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JW, Fillmore JP, Warner DD, Schmidt EW. Equisetin biosynthesis in Fusarium heterosporum. Chem Commun. 2005:186–188. doi: 10.1039/b413523g. [DOI] [PubMed] [Google Scholar]
- Sims JW, Schmidt EW. Thioesterase-like role for fungal PKS-NRPS hybrid reductive domains. J Am Chem Soc. 2008;130:11149–11155. doi: 10.1021/ja803078z. [DOI] [PubMed] [Google Scholar]
- Steyn PS, Vanheerden FR, Rabie CJ. Cytochalasin E and cytochalasin K, toxic metabolites from Aspergillus clavatus. J Chem Soc Perk Trans. 1982;1:541–544. [Google Scholar]
- Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani SA, Oakley BR. Fusion PCR and gene targeting in Aspergillus nidulans. Nat Protoc. 2006;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- Teijeira F, Ullan RV, Fernandez-Aguado M, Martin JF. CefR modulates transporters of beta-lactam intermediates preventing the loss of penicillins to the broth and increases cephalosporin production in Acremonium chrysogenum. Metab Eng. 2011;13:532–543. doi: 10.1016/j.ymben.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Thykaer J, Nielsen J. Metabolic engineering of beta-lactam production. Metab Eng. 2003;5:56–69. doi: 10.1016/s1096-7176(03)00003-x. [DOI] [PubMed] [Google Scholar]
- Tokuoka M, Seshime Y, Fujii I, Kitamoto K, Takahashi T, Koyama Y. Identification of a novel polyketide synthase-nonribosomal peptide synthetase (PKS-NRPS) gene required for the biosynthesis of cyclopiazonic acid in Aspergillus oryzae. Fungal Genet Biol. 2008;45:1608–1615. doi: 10.1016/j.fgb.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Turfitt GE. The microbiological degradation of steroids: 4. Fission of the steroid molecule. Biochem. J. 1948;42:376–383. [PMC free article] [PubMed] [Google Scholar]
- Udagawa T, Yuan J, Panigrahy D, Chang YH, Shah J, D’Amato RJ. Cytochalasin E, an epoxide containing Aspergillus-derived fungal metabolite, inhibits angiogenesis and tumor growth. J Pharmacol Exp Ther. 2000;294:421–427. [PubMed] [Google Scholar]
- Varga J, Due M, Frisvad JC, Samson RA. Taxonomic revision of Aspergillus section Clavati based on molecular, morphological and physiological data. Stud Mycol. 2007;59:89–106. doi: 10.3114/sim.2007.59.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedejs E, Campbell JB, Gadwood RC, Rodgers JD, Spear KL, Watanabe Y. Synthesis of the cytochalasin D isoindolone unit - solutions to the problem of regiochemistry in N-benzoylpyrrolinone Diels-Alder reactions. J Org Chem. 1982;47:1534–1546. [Google Scholar]
- Vederas JC, Nakashima TT, Diakur J. Detetion of 18O-label in Cytochalasin B by 13C-NMR. J Med Plants Res. 1980;39:201–202. [Google Scholar]
- Vederas JC, Tamm C. Biosynthesis of cytochalasans. Part 6. The mode of incorporation of phenylalanine into cytochalasin D1. Helv Chim Acta. 1976;59:558–566. doi: 10.1002/hlca.19760590221. [DOI] [PubMed] [Google Scholar]
- Wattanachaisaereekul S, Lantz AE, Nielsen ML, Nielsen J. Production of the polyketide 6-MSA in yeast engineered for increased malonyl-CoA supply. Metab Eng. 2008;10:246–254. doi: 10.1016/j.ymben.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Xu W, Cai X, Jung ME, Tang Y. Analysis of intact and dissected fungal polyketide synthase-nonribosomal peptide synthetase in vitro and in Saccharomyces cerevisiae. J Am Chem Soc. 2010;132:13604–13607. doi: 10.1021/ja107084d. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.