Abstract
Active loading of sucrose into phloem companion cells (CCs) is an essential process in apoplastic loaders, such as Arabidopsis or tobacco (Nicotiana sp.), and is even used by symplastic loaders such as melon (Cucumis melo) under certain stress conditions. Reduction of the amount or complete removal of the transporters catalysing this transport step results in severe developmental defects. Here we present analyses of two Arabidopsis lines, suc2-4 and suc2-5, that carry a null allele of the SUC2 gene which encodes the Arabidopsis phloem loader. These lines were complemented with constructs expressing either the Arabidopsis SUC1 or the Ustilago maydis srt1 cDNA from the SUC2 promoter. Both SUC1 and Srt1 are energy-dependent sucrose/H+ symporters and differ in specific kinetic properties from the SUC2 protein. Transgene expression was confirmed by RT-PCRs, the subcellular localization of Srt1 in planta with an Srt1-RFP fusion, and the correct CC-specific localization of the recombinant proteins by immunolocalization with anti-Srt1 and anti-SUC1 antisera. The transport capacity of Srt1 was studied in Srt1-GFP expressing Arabidopsis protoplasts. Although both proteins were found exclusively in CCs, only SUC1 complemented the developmental defects of suc2-4 and suc2-5 mutants. As SUC1 and Srt1 are well characterized, this result provides an insight into the properties that are essential for sucrose transporters to load the phloem successfully.
Keywords: Companion cell, phloem loading, Srt1, SUC1, SUC2, sucrose transport
Introduction
Plants convert a major portion of their photosynthetically fixed CO2 to sucrose, a molecule that is metabolically quite inert and, therefore, ideally suited for long-distance transport and long-term storage. In apoplastic loaders such as Arabidopsis thaliana, sucrose synthesized in the source leaf mesophyll is loaded into the phloem companion cells (CCs) by an energy-dependent H+/sucrose symporter (Stadler et al., 1995; Stadler and Sauer, 1996; Schmitt et al., 2008). Sucrose transporter-mediated phloem loading was also observed in virus-infected melon plants (Cucumis melo; Gil et al., 2011), a species clearly characterized to perform symplastic loading under normal growth conditions (Turgeon and Beebe, 1991). Although all plants analysed to date possess several genes for sucrose transporters (for reviews see Sauer, 2007; Ayre, 2011), studies on transporter mutants demonstrated that only one of these transporters is responsible for this loading step. Arabidopsis mutants with a T-DNA insertion in their SUC2 gene (Gottwald et al., 2000), which encodes the phloem loader of this species (Sauer and Stolz, 1994; Stadler and Sauer, 1996), show compromised carbon partitioning, fail to export sucrose from their source leaves, accumulate anthocyanin, are severely stunted, and only produce very few viable seeds (Gottwald et al., 2000; Srivastava et al., 2008; Srivastava et al., 2009). Similar, although less severe phenotypes were observed in potato (Solanum tuberosum) plants carrying an antisense construct for SUT1, the gene encoding the phloem loader of this species (Kühn et al., 1996).
In addition to this clear characterization of SUC2/SUT1-type sucrose transporters as phloem loaders, other sucrose transporters were reported to act as regulators of SUC2/SUT1-mediated phloem loading (Reinders et al., 2002; Schulze et al., 2003; Chincinska et al., 2008; Kühn and Grof, 2010). However, neither mutants in the genes for these putative interactors nor mutants in any other sucrose transporter gene resulted in phenotypes that came at least close to the strong effects observed with mutants defective in phloem loading (Hackel et al., 2006; Sivitz et al., 2007; Chincinska et al., 2008; S Schneider et al., unpublished data; Payyavula et al., 2011).
There was speculation as to whether the SUC2/SUT1-type phloem loaders possess specific functional or structural properties that put them in a unique position over all other sucrose transporters, or whether the SUC2/SUT1-type phloem loaders can be replaced by other sucrose transporters normally responsible for different physiological tasks. Therefore, well-characterized sucrose transporters were sought that might be used as substitutes for SUC2 in Arabidopsis plants with a suc2 null allele. Besides SUC2, six additional sucrose transporters were functionally characterized in Arabidopsis, SUC1, SUC3 (synonym SUT2), SUC4 (synonym SUT4), SUC5, SUC8, and SUC9. The Arabidopsis SUC6 and SUC7 genes are pseudogenes and do not encode intact transport proteins (Sauer et al., 2006). SUC3 was excluded from our analyses, as (i) quite divergent Km values have been published for this protein (1.9 mM: Meyer et al., 2000; 11.7 mM: Schulze et al., 2000) and (ii) it was reported to be involved in the regulation of phloem loading (Reinders et al., 2002). SUC4 was also excluded as it is targeted to the tonoplast and not to the plasma membrane (Endler et al., 2006).
Of the four remaining proteins, which all belong to the same phylogenetic group as SUC2 (Sauer, 2007), SUC1 seemed to be the best candidate, as it is perfectly characterized with respect to its plasma membrane localization (Sivitz et al., 2008; Feuerstein et al., 2010), its kinetic properties in baker’s yeast [Saccharomyces cerevisiae (Sauer and Stolz, 1994)] and in Xenopus laevis oocytes (Zhou et al., 1997), and the expression pattern of its gene. SUC1 is expressed primarily in roots and the reproductive organs, but there is no evidence for SUC1 expression in mature leaves or in the vasculature (Stadler et al., 1999; Sivitz et al., 2008; Feuerstein et al., 2010; Hoth et al., 2010). Studies of the Km values for sucrose showed that SUC1 and SUC2 have similar affinities for sucrose (Km SUC1: 0.4–0.5 mM, Km SUC2: 0.8–1.4 mM; Sauer and Stolz, 1994; Zhou et al., 1997; Chandran et al., 2003). Interestingly, however, the two proteins respond quite differently to changes in the extracellular pH values. Whereas SUC2 exhibits a sharp optimum of sucrose transport at pH 4, retains 50% of its activity at pH 5, and has only marginal activities at pH 3 or 6, SUC1 is rather insensitive to changes in the extracellular pH. From its optimum at pH 3 to its minimum at pH 7, the transport rate only decreases by 50% (Sauer and Stolz, 1994; see Supplementary Fig. S1 at JXB online).
The second sucrose transporter chosen for our analyses was the Srt1 protein from Ustilago maydis, a biotrophic fungus that grows during its entire pathogenic development in the apoplast of its host plant (maize, Zea mays), where it feeds on extracellular sucrose. Deletion of the srt1 gene leads to a loss of fungal virulence (Wahl et al., 2010). While Srt1 is highly specific for sucrose, and while the pH-dependence of Srt1 is between that of SUC1 and SUC2, Srt1 has a significantly (20–70-fold) higher affinity for sucrose (Km UmSrt1: 26 μM; Wahl et al., 2010).
Detailed analyses are presented here of two different Arabidopsis suc2 mutant lines, suc2-4 and suc2-5, that express the SUC1 or the srt1 cDNA from the SUC2 promoter and that target these proteins to the plasma membrane of their CCs. Interestingly, only recombinant SUC1 could complement the developmental defects of suc2-4 and suc2-5, whereas srt1-expressing lines looked essentially as the untransformed mutants. The results are discussed against the background of the known kinetic properties of the deleted SUC2 protein and of the recombinant SUC1 and Srt1 proteins.
Materials and methods
Strains and growth conditions
Arabidopsis thaliana plants (Col-0) were used as wild-type (wt) controls. Col-0 plants, suc2 mutant lines [suc2-4 (SALK_038124) heterozygous seeds were provided by Brian Ayre, University of Texas; suc2-5 (SALK_087046) heterozygous seeds were obtained from the Nottingham Arabidopsis Stock Centre] and complemented mutants were germinated and grown on soil under short-day conditions (8/16 h light/dark) at 22 °C and transferred to long day conditions (16/8 h light/dark) after 4 weeks for most applications. The srt1-expressing yeast strain (Saccharomyces cerevisiae) was described by Wahl et al. (2010). Escherichia coli strain DH5α (Hanahan, 1983) was used for all cloning steps. E. coli strain Rosetta™2(DE3) (Merck; Darmstadt, Germany) was used to express fusion protein. Agrobacterium tumefaciens strains C58C1 (Deblaere et al., 1985) and GV3101 (Holsters et al., 1980) were used for plant transformation.
T-DNA insertion lines
Seeds of heterozygous mutant lines were germinated on Murashige/Skoog (MS) agar medium (4.4 g MS salts with vitamins l−1, 0.5 g l−1 MES, 8 g l−1 agar) and later transferred to soil. PCR genotyping of homozygous lines was performed using the primers 5′-GAC CGT TGC ACC TCA AGA TTC G-3′ (#1 in Fig. 1A) and 5′-CGA ATA GTT CGT CGA ATG GTC CAC-3′ (#2) for the SUC2 wt allele, 5′-ATT TTG CCG ATT TCG GAA C-3′ (LB) and #1 for the suc2-5 insertion and LB and #2 for the suc2-4 insertion. PCRs were conducted with TaKaRa Ex Taq polymerase (Mobitec, Göttingen, Germany) according to the manufacturer’s instructions. For RT-PCR analyses of transcript abundance, total RNA from mature leaves or (in the case of homozygous suc2 plants) from whole seedlings was reverse transcribed and PCRs were performed with Taq polymerase. A truncated SUC2 RNA fragment from upstream of the insertion site was amplified using the primers #1 and 5′-GAT ACC GAG GAT GGC GAA G-3′ (#3).
Fig. 1.
Characterization of the suc2-5 mutant. (A) Scheme of the SUC2 gene with the confirmed insertion sites of suc2-4 (SALK_038124) and suc2-5 (SALK_087046). Black, exons; white, introns; LB, left border; small black arrows, primer binding sites and primer orientation. (B) Semi-quantitative RT-PCRs on total RNA from wt and suc2-5 plants showing the abundance of a SUC2 mRNA fragment spanning the insertion site (SUC2a, primers #1 and #2) and of an mRNA fragment upstream from the insertion site (SUC2b, primers #1 and #3). ACTIN2 transcript (ACT2) was used as control for amounts of cDNA. (C, D) Phenotype of wt, heterozygous (he) and homozygous (ho) suc2-5 plants at 27 d after germination (dag) (C) and at 41 dag (D). Arrow indicates the tiny homozygous plant. (E) Homozygous suc2-5 plant (48 dag) with anthocyanin accumulation at the leaf margins. Edge length of pots: 6.5 cm.
Constructs for stable and transient transformation
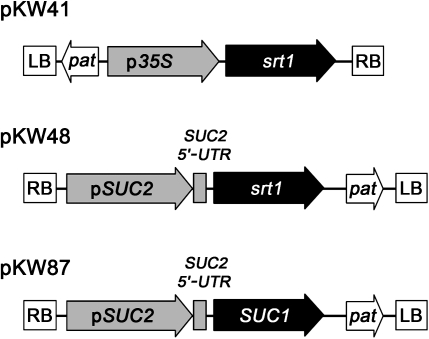
For srt1 over-expression from the 35S promoter or for CC-specific srt1 expression, the srt1 open reading frame (Wahl et al., 2010) was cloned into pENTR™/D/TOPO® (Life Technologies; Darmstadt, Germany) and recombined into the Gateway®-compatible destination vectors pEARLEYGATE100 (p35S; Earley et al., 2006) and pBSUC2 (pSUC2; Thompson and Wolniak, 2008) to obtain pKW41 (p35S::srt1) and pKW48 (pSUC2::srt1), respectively. pKW87 (pSUC2::SUC1) was obtained analogously.
For the investigation of the subcellular localization of Srt1 pENTR™/D/TOPO® containing the srt1 open reading frame was recombined into the Gateway®-compatible destination vector pH7RWG2.0 (Karimi et al., 2002) to obtain pKW45 (p35S::srt1-RFP). For the GFP-INT4 control construct the Arabidopsis INT4 coding sequence was amplified with the primers AtINT4-5-NcoI (5′-CCA TGG TGG AAG GAG GAA TTG-3′) and AtINT4-3-NcoI (5′-CCA TGG CAG CAG CAT CGA CTT CTT TGC-3′), which introduced NcoI sites at both ends and removed the stop codon. The resulting fragment was inserted into pJET1.2 (Fermentas; St Leon-Rot, Germany), sequenced and inserted into the unique NcoI site of pSS87 (S Schneider et al., unpublished data) downstream of the 35S promoter.
Generation and identification of transgenic plants
Transgenic plants were generated via Agrobacterium-mediated transformation of Arabidopsis Col-0 plants with C58C1 carrying pKW41 or GV3101 carrying pKW48 and pSOUP as a co-vector (Hellens et al., 2000). The resulting plant lines were named KW41 (p35S::srt1) and KW48 (pSUC2::srt1). Transgenic plants were identified by Basta® resistance.
KW48 plants were crossed with heterozygous SUC2/suc2 plants. suc2/srt1-transgenic lines were identified by PCR genotyping.
Heterozygous SUC2/suc2 plants were transformed by GV3101 carrying pKW87 and pSOUP. suc2/pSUC2::srt1-transgenic lines were identified by Basta® resistance (T1 generation) and subsequent generations by PCR genotyping. Primers #1 and #2 were used for the SUC2 wt allele, 5′-GAC AAG CAC GGT CAA CTT CC-3′ and 5′-GAA GTC CAG CTG CCA GAA AC-3′ for the Basta® resistance gene, 5′-CAC CAT GGC GTC GTC TTC TCC-3′ and 5′-GCA GAT GTA CGC GTA AAC CG-3′ for the srt1 gene, and 5′-CCT ACG CTA TAG ACA CAG CTC TG-3′ and 5′-GCT ACG TCG AGG ATC CAG AA-3′ for the pSUC2::SUC1 insertion.
RT-PCR analyses of transcript abundance in transgenic plants were performed with the primer 5′-ATT CAG ATG CCC AGA AGT CTT GTT-3′ and 5′-GAA ACA TTT TCT GTG AAC GAT TCC T-3′ for ACTIN2 (ACT2) as control and with 5′-CTC TTC CTC CAC CAC TAC AAC CAC-3′ and 5′-GCT ACG TCG AGG ATC CAG AA-3′ for pSUC2-5′-UTR::SUC1. For the srt1 mRNA levels the same primers were used as for plant genotyping.
Production and purification of antibodies
The sequence encoding the Srt1-C-terminus (69 amino acids) was amplified using the primers 5′-GAG AAT TCA GGA CTT TCT TCG AGA TC-3′ and 5′-CTG AAT TCT CAT TGT GGA CTC GGC-3′ containing EcoRI restriction sites, and cloned into EcoRI sites in the pMAL-c2 polylinker (New England Biolabs; Frankfurt, Germany) yielding a plasmid (pKW80) that encodes a maltose-binding-protein-(MBP)-Srt1-C-terminus fusion. E. coli Rosetta™2(DE3) cells were transformed with this plasmid, induced with 1 mM isopropyl-β-D-thiogalactopyranoside and harvested. The fusion protein was isolated from the cell extract by preparative SDS-PAGE (Laemmli, 1970), extracted from the gel and lyophilized as described by Sauer and Stadler (1993). Immunization of rabbits was done by Pineda Antikörper-Service (Berlin, Germany).
Affinity-purification of the anti-Srt1 antiserum (αSrt1) was carried out as described previously (Sauer and Stadler, 1993; Schmitt et al., 2008). Analogously, purification of available anti-SUC1 raw serum (αSUC1) was done using a synthetic peptide (Feuerstein et al., 2010).
Protein isolation and Western Blot analysis
Soluble and membrane protein fractions from plant leaves or baker’s yeast were isolated as described previously (Sauer and Stolz, 2000; Drechsel et al., 2010). SDS-polyacrylamide gel electrophoresis (Laemmli, 1970) and Western Blot analyses (Burnette, 1981) with affinity-purified αSrt1 (used in a 1:20 dilution) were performed as published.
Immunohistochemistry
Leaf tissue was fixed in ethanol:acetic acid (3:1 v/v) and embedded in methacrylate. Microtome sections (4 μm) were prepared as described by Stadler and Sauer (1996). Incubation with primary antibodies was performed overnight at 4 °C. Antibody dilutions in blocking buffer (50 mM TRIS/HCl pH 7.5, 150 mM NaCl, 1% skim milk powder, and 0.1% Triton X-100) were 1:20 for αSrt1, 1:5 for αSUC1, and 1:5 for the αRS6 antibody (Khan et al., 2007). Secondary antibodies (anti-rabbit IgG-Cy2 and anti-mouse IgG-Cy3; Dianova; Hamburg, Germany) were diluted 1:50 in blocking buffer.
Confocal laser scanning microscopy
Confocal laser scanning microscopy for immunolocalizations was done as described by Schmitt et al. (2008). For colocalization of Srt1-RFP and GFP-INT4 in transformed protoplasts, sequential scanning was performed using an excitation wavelength of 488 nm for GFP and 543 nm for RFP. Detection windows were 495–547 nm for GFP, 584–638 nm for RFP, and 675–767 nm for chlorophyll autofluorescence.
Protoplast techniques
Arabidopsis protoplasts were generated and transformed as described by Abel and Theologis (1994). For transport analyses, successful expression of reporter gene constructs was checked microscopically after 24 h. For each transport test, 700 000 protoplasts were harvested by centrifugation for 2 min at 50 g, resuspended in 0.5 ml W5 buffer (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 5 mM glucose, 1.5 mM MES, adjusted to pH 5.2 with KOH), transferred to a 24-well cell culture plate, and 14C-sucrose was added to a final concentration of 0.2 mM. After a 3 h incubation at 22 °C in the light, and after withdrawal of 50 μl of protoplast suspension for scintillation counting (total radioactivity), protoplasts were harvested (2 min, 50 g), washed twice with W5 buffer, and the radioactivity in the protoplast was determined.
Carbohydrate analyses
Plants were grown for 6 weeks under short-day conditions (8/16 h light/dark) at a proton flux density of 100 μmol m−2 s−1. At the end of the light cycle, source leaves from 10 different wt plants (1 leaf per plant) or from 10 plants of a transgenic line were harvested and combined. Soluble carbohydrates were extracted for ion exchange chromatography (IC) as described by Schneider et al. (2008). The eluent was 500 mM NaOH; the run took 80 min. Three biological replicates were analysed.
Results
Characterization of the T-DNA insertion line SALK_087046
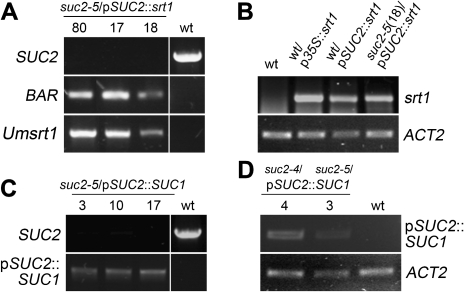
The T-DNA insertion lines SALK_038124 (suc2-4) and SALK_087046 (suc2-5) were used for our analyses. Whereas the position of the T-DNA insertion in the suc2-4 line had previously been determined (Srivastava et al., 2008), the position of the T-DNA insertion in the suc2-5 mutant has only been predicted [SIGnAL: http://signal.salk.edu/cgi-bin/tdnaexpress; predicted insertion site: middle of the 1st exon (Lei et al., 2011)] but not confirmed by sequencing. Therefore, the abundance of different SUC2 mRNA fragments was studied by RT-PCRs on RNA from wt and homozygous suc2-5 plants, and the T-DNA insertion site was sequenced. In contrast to the predicted insertion site (Lei et al., 2011), our analyses identified the insertion in the 1st intron, 1265 base pairs (bp) downstream from the start codon in the genomic sequence (Fig. 1A). Whereas a truncated sequence upstream from this insertion site could be amplified from homozygous suc2-5 mutant plants, no full-length SUC2 transcript could be detected (Fig. 1B). A protein translated from this truncated suc2-5 mRNA would encode 419 of the 512 amino acids of the intact SUC2 protein. A sucrose transport activity of the resulting truncated suc2-5 protein can be excluded, as even the slightly longer but also truncated suc2-4 protein was shown to be functionally inactive (Srivastava et al., 2009).
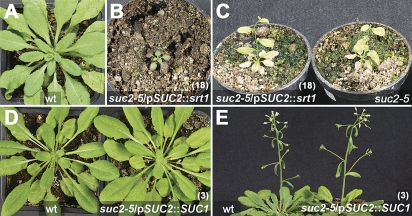
The suc2-5 phenotype matches that of previously described suc2 mutants (Gottwald et al., 2000; Srivastava et al., 2008; Lei et al., 2011). Homozygous plants showed delayed development, stunted growth, and the accumulation of anthocyanin in the leaf margins (Fig. 1C, D, E). Despite these severe defects, suc2-5 plants were able to complete their life cycle and generated viable seeds. Heterozygous plants did not differ phenotypically from the wt.
Generation and genotyping of suc2/pSUC2::srt1 and suc2/pSUC2::SUC1 plants
For complementation of suc2-4 and suc2-5 mutants with an srt1-containing construct, wt plants were transformed with the construct pKW48 (Fig. 2) and these wt/pSUC2::srt1 plants were crossed with heterozygous suc2 mutants. The resulting suc2/pSUC2::srt1 plants should express srt1 from the SUC2 promoter (pSUC2) in the respective mutant background (suc2-4/pSUC2::srt1 or suc2-5/pSUC2::srt1). The absence of a wt SUC2 allele from homozygous suc2/pSUC2::srt1 plants, and the presence of a copy of the srt1 gene and of the BAR gene for Basta® resistance was confirmed by PCR (Fig. 3A). The abundance of the srt1 transcript was checked by RT-PCR on total RNA from transgenic and wt plants. As an additional control, RNA was included from wt plants expressing srt1 from the 35S promoter (wt/p35S::srt1 plants). The srt1 transcript could be detected in all transgenics but not in the wt (Fig. 3B).
Fig. 2.
Constructs used to generate transgenic plants. RB, right border; LB, left border; pSUC2, SUC2 promoter; p35S, 35S promoter; pat, phosphinotricin acetyltransferase (Basta® resistance gene); SUC2 5'-UTR, SUC2 5' untranslated region.
Fig. 3.
Genotyping of suc2/pSUC2::srt1 and suc2/pSUC2::SUC1 plants and determination of transcript abundance. (A) PCR analyses with genomic DNA from suc2/pSUC2::srt1 showing the absence of the SUC2 wt allele and the presence of the BAR gene and of srt1 in three different suc2/pSUC2::srt1 lines (#17, #18, #80). Control PCRs were performed on wt genomic DNA to visualize the SUC2 gene fragment, or on wt/pSUC2::srt1 genomic DNA to show the identity of the amplified BAR and Umsrt1 fragments. (B) Comparative RT-PCR analyses of srt1 transcript abundance on total RNA from wt, wt/p35S::srt1 and wt/pSUC2::srt1 source leaves or from entire suc2/pSUC2::srt1 plants showing srt1 transcripts only in transgenics. ACT2 levels are shown as controls. (C) PCR analyses on genomic DNA from three different suc2/pSUC2::SUC1 lines (#3, #10, #17) showing the absence of the SUC2 wt allele and the presence of the pSUC2::SUC1 insertion. Control PCRs on genomic DNA from wt plants identified the SUC2 gene and failed to amplify the mutant SUC1 allele. (D) RT-PCR analyses on total RNA from source leaves of two different suc2/pSUC2::SUC1 lines (#3, #4) and of wt plants identifying SUC1 mRNA transcribed from the pSUC2::SUC1 insertion only in transgenics. ACT2 transcript levels are shown as controls.
Transgenic lines expressing SUC1 from the SUC2 promoter in the suc2 background (suc2/pSUC2::SUC1) were generated by Agrobacterium-mediated transformation of heterozygous suc2-4 and suc2-5 plants with the construct pKW87 (Fig. 2). Plants carrying the suc2/pSUC2::SUC1 construct were identified by Basta® selection and genotyped (Fig. 3C). Again, homozygous suc2 plants did not contain SUC2 mRNA. However, they contained novel SUC1 mRNA that resulted from the pSUC2::SUC1 insertion as demonstrated by a primer combination specific for this transgene. To avoid amplification of SUC1 transcripts encoded by the wt SUC1 allele present in the suc2 mutants, one of the primers was chosen to bind to the pSUC2-derived 5’-UTR of the pSUC2::SUC1 transgene (Fig. 3D). Of the crosses obtained (srt1) or transformants (SUC1), seven suc2-5/pSUC2::srt1 lines, two suc2-4/pSUC2::SUC1 lines, and three suc2-5/pSUC2::SUC1 lines were used for further analyses).
Phenotypes of suc2/pSUC2::srt1 and suc2/pSUC2::SUC1 plants
Although both sucrose transporter genes, SUC1 and srt1, were expressed in suc2/pSUC2::SUC1 and suc2/pSUC2::srt1 plants, respectively (Fig. 3B, D), they showed strong morphological differences. Whereas suc2 mutants expressing srt1 from the SUC2 promoter looked essentially like un-transformed suc2 mutants (Fig. 4A, B, C), suc2/pSUC2::SUC1 plants looked like wt plants and did not show any recognizable developmental defect (Fig. 4D, E). This suggested that transformation with the pKW87 construct (pSUC2::SUC1) but not with the pKW48 (pSUC2::srt1) construct (Fig. 2) leads to successful complementation of the suc2 phenotype.
Fig. 4.
Phenotype of suc2/pSUC2::srt1 and suc2/pSUC2::SUC1 plants. (A) wt and (B) suc2-5/pSUC2::srt1 (line 18) plant at 46 dag. (C) suc2-5/pSUC2::srt1 (line 18) and suc2-5 flowering plants at 80 dag. (D) and (E) suc2-5/pSUC2:SUC1 (line 3) and wt plant at 88 dag (D) and flowering at 99 dag (E). Edge length of squared pots: 6.5 cm, diameter of round pots: 6 cm.
An Srt1-RFP fusion localizes to the plasma membrane and is a functionally active transporter in planta
The observed lack of complementation in suc2/pSUC2::srt1 plants might indicate (i) that the identified srt1 mRNA (Fig. 3B) is not translated, (ii) that the Srt1 protein is not targeted to the plasma membrane, (iii) that the Srt1 protein is not functional in the plant plasma membrane, or (iv) that the pSUC2::srt1 construct is not expressed in the correct cell type, i.e. in the CCs. To exclude the first three options, the subcellular localization of an Srt1-RFP fusion in plant cells was checked, the presence of Srt1 protein was tested on Western blots, and the sucrose transport capacity was studied in Srt1-RFP-expressing Arabidopsis protoplasts.
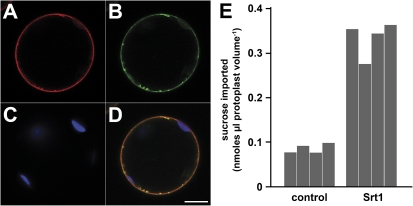
To determine the subcellular localization of Srt1, co-localization analyses were performed in Arabidopsis protoplasts co-transformed with constructs for Srt1-RFP and GFP-INT4 fusions. INT4 is an inositol transporter of the Arabidopsis plasma membrane and a GFP fusion was previously shown to be targeted to the plasma membrane (Schneider et al., 2006). In confocal sections from Srt1-RFP and GFP-INT4 co-expressing protoplasts, both the red Srt1-RFP fluorescence (Fig. 5A) and the green GFP-INT4 fluorescence (Fig. 5B) labelled the plasma membrane, which is most obvious in a merge of these images (Fig. 5D).
Fig. 5.
Srt1-RFP localizes to the plasma membrane of Arabidopsis mesophyll protoplasts where it catalyses the uptake of radiolabelled sucrose. (A–C) Optical sections of an Arabidopsis protoplast cotransformed with an Srt1-RFP and an GFP-INT4 construct. (A) Localization of Srt1-RFP. (B) Localization of GFP-INT4. (C) Detection of chloroplasts by chlorophyll autofluorescence. (D) Merge of (A–C). Bar=10 μm. (E) Uptake of 14C-sucrose into Arabidopsis protoplasts expressing GFP-INT4 (control) or Srt1-RFP (Srt1). Srt1 and control data show results from two different protoplast transformations and two transport tests per transformation, respectively.
To test, if Srt1 is a functional sucrose transporter in plant cells, the capacity to import 14C-labelled sucrose of Srt1-RFP or GFP-INT4-expressing protoplasts was compared. From comparative analyses of infection rates obtained with a U. maydis wt strain, a U. maydis Δsrt1 mutant, and a U. maydis Δsrt1 mutant that had been complemented with an srt1-GFP fusion it was known that the fusion of a fluorescent reporter to the Srt1 C-terminus does not affect the functionality of the transporter in the fungus (Wahl et al., 2010). When the capacity to transport 14C-labelled sucrose (initial concentration 0.2 mM) of Srt1-RFP-expressing and GFP-INT4-expressing Arabidopsis protoplasts was compared, significantly larger amounts of sucrose uptake into Srt1-RFP-expressing protoplasts was observed. In summary, these data demonstrate that the Srt1 protein is synthesized from its mRNA, that it is targeted to the plasma membrane, and that it is functionally active in Arabidopsis.
Analyses of recombinant SUC1 and Srt1 proteins in suc2/pSUC2::SUC1 and suc2/pSUC2::srt1 plants
The presence of recombinant proteins in CCs was studied with αSUC1 and αSrt1 antisera. The αSUC1 antiserum has been described before (Feuerstein et al., 2010). For the αSrt1 antiserum, antibodies were raised in rabbits against the 69 C-terminal amino acids of Srt1, which had been fused to the maltose-binding protein. After affinity purification of the raw serum, the αSrt1 fraction was tested on Western blots with membrane proteins from the srt1-expressing yeast (Saccharomyces cerevisiae) strain described by Wahl et al. (2010) and with membrane protein and soluble protein fractions from wt, KW48 and KW41 plants (Fig. 6). KW41 plants represent controls that express srt1 from the 35S promoter in the wt background (p35S; Fig. 2).
Fig. 6.
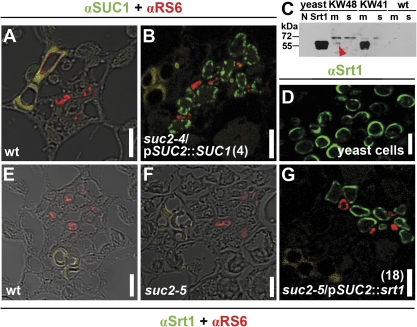
Detection of SUC1 and Srt1 proteins in plants and yeast. (A), (B), and (E–G), Immunohistochemical stainings of 4 μm sections showing leaf veins from wt [(A) and (E)], suc2-4/pSUC2::SUC1 (line 4) (B), suc2-5 (F), and suc2-5/pSUC2::srt1 (line 18) (G) plants. Green fluorescence of anti-rabbit-Cy2 in CCs corresponds to αSUC1 [(A) and (B)] or αSrt1 [(E–G)]. Red fluorescence of anti-mouse-Cy3 corresponds to anti-RS6 labelling of sieve elements. For (A), (E), and (F) the images of the fluorescence signals were merged with the corresponding bright field images. Yellow staining in (A), (B), and (G) shows xylem autofluorescence. (C) Western blot analyses using αSrt1 to detect the 60 kDa Srt1 protein in extracts from yeast strain SEY2102 (Emr et al., 1983) (N, control yeast cells transformed with the empty vector; Srt1, yeast cells expressing srt1) or in extracts from source leaves of wt/pSUC2::srt1 (KW48), wt/p35S::srt1 (KW41), and wt plants (m, membrane fraction; s, soluble fraction). The red arrowhead shows the weak Srt1-derived signal in wt/pSUC2::srt1 plants. (D) Immunostaining of 4 μm sections of srt1-expressing yeast cells. Green fluorescence of anti-rabbit-Cy2 corresponds to αSrt1. Yellowish colour in (A), (B), (E), and (G) shows autofluorescence of cell-wall phenolic compounds in xylem vessels. Bars=5 μm.
The αSrt1 antiserum yielded a strong signal at the expected molecular mass of about 60 kDa in the membrane fraction from srt1-expressing yeast cells but not from control yeast cells (Fig. 6C), and a signal of comparable intensity and of the same size was detected in wt/p35S::srt1 controls (KW41 in Fig. 6C). By contrast, no signals were detected in the soluble-protein fraction of wt/p35S::srt1 controls or in the membrane or soluble fraction from wt plants. In wt/pSUC2::srt1 plants, where srt1 is expected to be expressed in CCs, only a significantly weaker signal could be detected in the membrane extract; no signal was seen in the soluble protein fraction (Fig. 6C).
The weak signal in wt/pSUC2::srt1 plants might reflect the comparatively small number of CCs in leaves (probably less than 1% of all cells). To test this hypothesis, immunolocalization studies were performed on sections of methacrylate-embedded, srt1-expressing yeast cells to test the capacity of the αSrt1 antiserum to label Srt1 protein after fixation and embedding (Fig. 6D). To this end, thin sections of embedded cells were treated with αSrt1 and with anti-rabbit-Cy2 2nd antibody. As expected, green Cy2 fluorescence could be observed in the cell periphery of the yeast sections (Fig. 6D) indicating that αSrt1 labels Srt1 in immunolocalizations. No fluorescence was detected in control cells carrying the empty vector (not shown).
Sections from source leaves of suc2/pSUC2::srt1 plants, that had been fixed and embedded essentially as the yeast cells shown in Fig. 6D, were analysed next. These leaf sections were treated with αSrt1, which was expected to label the CCs, and simultaneously with a sieve element (SE)-specific antiserum (αRS6) that was previously shown to label the SEs of Arabidopsis with high specificity (Meyer et al., 2004; Khan et al., 2007; Hoth et al., 2008). The αSrt1 and αRS6 signals were detected with anti-rabbit-Cy2 (green fluorescence) and anti-rabbit-Cy3 (red fluorescence) 2nd antibodies, respectively. In sections from suc2/pSUC2::srt1 source leaves, the Srt1-specific Cy2 fluorescence could be detected in cells that could be classified as CCs as (i) they were in the immediate vicinity of the αRS6-labelled SEs, (ii) they had a significantly larger diameter than these SEs, and (iii) there were more CCs than SEs, which is typical for minor or medium-sized veins (Fig. 6G; Esau, 1969; Schmitt et al., 2008). By contrast, no αSrt1-derived Cy2 fluorescence was detected in wt (Fig. 6E) or suc2-5 plants (Fig. 6F), where the SEs could be labelled by αRS6. These data demonstrate that, as expected by the known specificity of the SUC2 promoter, Srt1 is present in source leaf CCs of suc2/pSUC2::srt1 plants.
A similar result was obtained, when a combination of αSrt1 and αRS6 was used on thin sections of fixed and embedded source-leaf material from suc2/pSUC2::SUC1 and wt plants (Fig. 6A, B). Again, the SUC1 and RS6 antigens were detected by Cy2 and Cy3 fluorescence, and again αRS6 labelled the SEs in sections from all plants analysed. SUC1-specific Cy2 fluorescence, however, was only seen in suc2/pSUC2::SUC1 plants (Fig. 6B), while no SUC1-specific fluorescence could be observed in sections from wt plants (Fig. 6A). Thus, suc2/pSUC2::srt1 and suc2/pSUC2::SUC1 plants have their respective recombinant sucrose transporter specifically localized in the CCs.
Carbohydrate analysis of wt and suc2/pSUC2::SUC1 plants
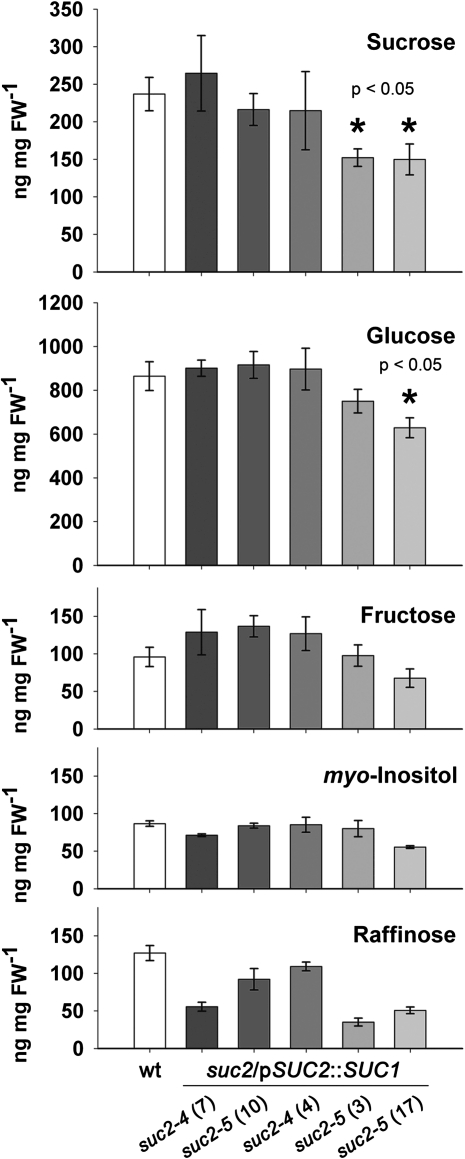
In contrast to Srt1, SUC1 restored wt development in suc2 mutants when expressed in source leaf CCs. However, besides this macroscopically detectable developmental defect, a lack of sucrose export (in the suc2-1 mutant; Gottwald et al., 2000) and, consequently, an accumulation of sucrose (more than 20-fold compared with the wt in the suc2-4 mutant; Srivastava et al., 2008) has been observed in the source leaves of suc2 mutants analysed before. These increased sucrose concentrations lead to the production of protective anthocyanins, a phenotype also clearly visible in the newly characterized suc2-5 mutant (Fig. 1E). It was examined whether SUC1 in the CCs of suc2 null mutants can restore this biochemical phenotype as well. To this end, the carbohydrate content in source leaves of 6-week-old suc2/pSUC2::SUC1 and wt plants was compared by ion exchange chromatography. Figure 7 shows that the levels for glucose, sucrose, myo-inositol, fructose, and raffinose were comparable in most suc2/pSUC2::SUC1 and wt plants. However, in two of the suc2/pSUC2::SUC1 lines [suc2-5 (3) and suc2-5 (17) in Fig. 7] the source-leaf sucrose levels or the sucrose plus glucose levels were significantly (30%) lower than in the wt plants suggesting that phloem loading in these lines might be even more effective than in wt plants.
Fig. 7.
Carbohydrate analyses of wt and suc2/pSUC2::SUC1 plants. Source leaves of 6-week-old wt plants and five different suc2/pSUC2::SUC1 lines [suc2-4 (#7), suc2-5 (#10), suc2-4 (#4), suc2-5 (#3), and suc2-5 (#17)] grown under short-day conditions were analysed by ion exchange chromatography to determine amounts of the indicated carbohydrates (n=3 ±SE). Asterisks show significantly decreased sucrose or glucose concentrations based on Student’s t tests.
Discussion
Arabidopsis knockout mutants harbouring a T-DNA insertion in their SUC2 gene or potato plants expressing antisense constructs for their SUT1 gene, fail to export photoassimilates from their source leaves, show feedback inhibition of their photosynthetic activity, and form stunted, often tiny plants (Kühn et al., 1996; Gottwald et al., 2000; Srivastava et al., 2008; Srivastava et al., 2009; this paper). In the present study, Arabidopsis lines were generated and analysed that had SUC2, their companion cell-specific phloem loader, replaced either with SUC1, another Arabidopsis sucrose transporter that is usually expressed in pollen, anther connective tissue, developing ovules, or roots of seedlings (Stadler et al., 1999; Sivitz et al., 2008; Feuerstein et al., 2010), or with Srt1 from the corn smut fungus U. maydis (Wahl et al., 2010). Transcription and translation of the transgenes were followed by RT-PCR and with specific antisera, respectively. CC-specific expression was confirmed in immunohistochemical analyses. Although both SUC1 and Srt1 were synthesized specifically and exclusively in CCs, only SUC1 complemented the strong developmental defects of the suc2 mutant lines.
SUC1 complements all defects described for suc2 mutants
SUC1-complemented suc2 mutants developed and flowered like wt plants (Fig. 4D, E) and showed no accumulation of carbohydrates in their source leaves (Fig. 7). For several reasons, this successful replacement of SUC2 by a non-phloem sucrose transporter was not predictable. Firstly, although SUC1 and SUC2 have comparable affinities for their substrate sucrose, they respond differently to changes in the extracellular pH. This had already been demonstrated during the initial characterization of these proteins (Sauer and Stolz, 1994) and confirmed in more detail during the present study (see Supplementary Fig. S1 at JXB online). Secondly, in different publications, phloem loading by SUC2-type transporters was reported to be regulated by physical interaction with other transporters, for example, with SUC3 (synonym SUT2) and SUC4 (synonym SUT4) in Arabidopsis (Reinders et al., 2002; Schulze et al., 2003; Kühn and Grof, 2010). While an interaction with SUC4 can, meanwhile, be excluded, as SUC4-transporters were characterized as tonoplast proteins (Endler et al., 2006; Schulz et al., 2011), the regulatory interaction of SUC2 and SUC3 (SUT2) is still under discussion.
Our results demonstrate that, quite obviously under the growth conditions analysed, the different pH-sensitivities of SUC1 and SUC2 are of no or only of minor importance. It may well be, however, that this altered pH-sensitivity of SUC1 becomes important during the adaptation of phloem loading to environmental changes or to certain stress conditions. In fact, it has been discussed only recently that changes in the extracellular pH might represent a tool to regulate the competition for sucrose at the host/pathogen interface (Wippel et al., 2010).
Despite their different pH responses, SUC1 and SUC2 share about 80% identical amino acids. It might, therefore, well be that CC-specific regulatory mechanisms that possibly modulate the activity of SUC2 in WT plants will also act on SUC1. Such regulatory mechanisms could be post-translational modifications or protein/protein interactions.
In summary, the successful replacement of SUC2 by SUC1 indicates that the phloem loader SUC2 does not contain a specific domain or a special functional property that puts it in a unique position compared with all other non-phloem plant sucrose transporters. The important role of the SUC2 gene for plant growth and development rather depends on its promoter that directs and limits the function of the SUC2 protein to the CCs of WT plants or of SUC1 in the transgenic lines analysed in the present study.
Srt1 cannot complement the defects of suc2 mutant lines
In a second approach we replaced SUC2 by the U. maydis Srt1 protein. It was shown that the srt1 gene is transcribed (Fig. 3B), checked that the Srt1 protein is synthesized and made specifically in CCs (Fig. 6G), confirmed that it is targeted to the plasma membrane (Fig. 5D), and demonstrated that recombinant Srt1-RFP acts as functional sucrose transporter in Arabidopsis protoplasts (Fig. 5E). Nevertheless, and in contrast to SUC1, Srt1 cannot replace SUC2 (Fig. 4B, C).
The extracellular sucrose concentrations at the mesophyll/CC interface should be more than sufficient to drive sucrose uptake by Srt1. In apoplastic loaders like Arabidopsis, bulk apoplastic sucrose concentrations were found to be 2–6 mM (López-Millán et al., 2000; Voitsekhovskaja et al., 2000; Lohaus et al., 2001), concentrations that are nearly saturating for SUC2 (Km 0.8–1.4 mM) and completely saturating for Srt1 (Km 26 μM). Nonetheless, the lack of complementation might result from this much lower Km value of Srt1, as low Km values (or high affinities) typically come along with low transport capacities or vice versa. Therefore, low-affinity/high-capacity (LAHC) transporters and high-affinity/low-capacity (HALC) transporters have been described in numerous systems (Delrot and Bonnemain, 1981; Maynard and Lucas, 1982; Russell, 1990; Weise et al., 2000; Geiger, 2011). Although SUC2-type transporters are usually described as HALC transporters [although LAHC activities were measured in planta (Delrot and Bonnemain, 1981; Maynard and Lucas, 1982), sucrose transporters with LAHC activities have not been identified so far], the 50-fold lower Km of Srt1 clearly characterizes Srt1 as a transporter with very low capacity, which may be too low to replace the missing activity of SUC2.
Alternatively, the lack of complementation might result from a specific difference in the transport properties reflecting different physiological roles of Srt1 in U. maydis and of SUC2 in Arabidopsis CCs. Although fungal sucrose transporters belong to the major facilitator superfamily (MFS) of transporters described almost 20 years ago by Marger and Saier (1993), Srt1 and SUC2 differ significantly with respect to their physiological tasks. Whereas sucrose imported by Srt1 into U. maydis cells is used for cellular metabolism, sucrose loaded by SUC2 into CCs is accumulated to generate the osmotic driving force for long-distance mass flow (Münch, 1930). Therefore, although Srt1 is an energy-dependent H+-symporter and although it can accumulate sucrose to intracellular concentrations that exceed the concentrations in the extracellular lumen, it catalyses the permanent exchange of accumulated sucrose already at relatively low concentrations (Wahl et al., 2010). This exchange flux is a well-known property of transporters that do not accumulate their substrates under physiological conditions (Komor et al., 1972; Eddy, 1982).
Together, the low transport capacity of Srt1 (predicted from its high affinity) and the catalysis of an exchange flux already at low intracellular concentration might be the reason for the unsuccessful complementation of suc2 mutants. Both factors will reduce the capacity to accumulate sucrose inside CCs to concentrations that are high enough to initiate long-distance transport and to remove photoassimilates from the source leaves.
Srt1 and SUC1 are differentially distributed in CCs
A direct comparison of the immunohistochemical images of leaf sections obtained after the treatment with αSrt1 (Fig. 6G) or αSUC1 (Fig. 6B) antisera revealed a difference in the distribution of Srt1 and SUC1. Whereas αSrt1-decoration of Srt1 results in a uniform labelling of the CCs (Fig. 6G), quite likely showing the plasma membrane (Fig. 5D), αSUC1-decoration of SUC1 results in a patchy distribution of the fluorescence at the surface of the CCs (Fig. 6B). This resembles the similarly patchy distribution observed for other plant sucrose transporters in CCs (Sauer, 2007; Schmitt et al., 2008), and may point towards a concentration of these proteins within large subdomains of the CC plasma membrane. As SUC1–GFP fusions show uniform labelling of the plasma membrane in Arabidopsis mesophyll protoplasts (Feuerstein et al., 2010), just like the Srt1–RFP fusion (Fig. 5D, E), this may be a CC-specific phenomenon. Whether or not this contributes to the successful complementation of suc2 plants remains to be analysed.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Differences in the pH-dependences of SUC1 and SUC2.
Acknowledgments
We thank Matthew Thompson (University of Maryland) for providing the pBSUC2 and pSOUP vectors, Brian Ayre (University of Texas) for suc2-4 seeds and Susanne Wolfenstetter and Sabine Schneider for Arabidopsis protoplast transformation. This work was supported by a grant of the Deutsche Forschungsgemeinschaft to NS (Research Unit 666; Sa 382/21).
References
- Abel S, Theologis A. Transient transformation of Arabidopsis leaf protoplasts: a versatile experimental system to study gene expression. The Plant Journal. 1994;5:421–427. doi: 10.1111/j.1365-313x.1994.00421.x. [DOI] [PubMed] [Google Scholar]
- Ayre B. Membrane-transport systems for sucrose in relation to whole-plant carbon partitioning. Molecular Plant. 2011;4:377–394. doi: 10.1093/mp/ssr014. [DOI] [PubMed] [Google Scholar]
- Burnette WN. ‘Western blotting’: electrophoretic transfer of proteins from sodium dodecyl sulfate–polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radio-iodinated protein A. Analytical Biochemistry. 1981;112:195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Chandran D, Reinders A, Ward JM. Substrate specificity of the Arabidopsis thaliana sucrose transporter AtSUC2. Journal of Biological Chemistry. 2003;278:44320–44325. doi: 10.1074/jbc.M308490200. [DOI] [PubMed] [Google Scholar]
- Chincinska IA, Liesche J, Krügel U, Michalska J, Geigenberger P, Grimm B, Kühn C. Sucrose transporter StSUT4 from potato affects flowering, tuberization, and shade avoidance response. Plant Physiology. 2008;146:515–528. doi: 10.1104/pp.107.112334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deblaere R, Bytebier B, De Greve H, Deboeck F, Schell J, Van Montagu M, Leemans J. Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic Acids Research. 1985;13:4777–4788. doi: 10.1093/nar/13.13.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delrot S, Bonnemain J-L. Involvement of protons as a substrate for the sucrose carrier during phloem loading in Vicia faba leaves. Plant Physiology. 1981;67:560–564. doi: 10.1104/pp.67.3.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsel G, Bergler J, Wippel K, Sauer N, Vogelmann K, Hoth S. C-terminal armadillo repeats are essential and sufficient for association of the plant U-box armadillo E3 ubiquitin ligase SAUL1 with the plasma membrane. Journal of Experimental Botany. 2010;62:775–785. doi: 10.1093/jxb/erq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. Gateway-compatible vectors for plant functional genomics and proteomics. The Plant Journal. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- Eddy AA. Mechanisms of solute transport in selected eukaryotic micro-organisms. Advances in Microbial Physiology. 1982;23:71–78. doi: 10.1016/s0065-2911(08)60335-5. [DOI] [PubMed] [Google Scholar]
- Emr SD, Schekman R, Flessel MC, Thorner J. An MF alpha 1-SUC2 (alpha-factor-invertase) gene fusion for study of protein localization and gene expression in yeast. Proceedings of the National Academy of Sciences, USA. 1983;80:7080–7084. doi: 10.1073/pnas.80.23.7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler A, Meyer S, Schelbert S, Schneider T, Weschke W, Peters SW, Keller F, Baginsky S, Martinoia E, Schmidt UG. Identification of a vacuolar sucrose transporter in barley and Arabidopsis mesophyll cells by a tonoplast proteomic approach. Plant Physiology. 2006;141:196–207. doi: 10.1104/pp.106.079533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau K. The phloem. In: Zimmermann W, Ozenda P, Wulff HD, editors. Encyclopedia of plant anatomy. Vol. V. Berlin, Stuttgart: Gebr Bornträger; 1969. pp. 154–163. [Google Scholar]
- Feuerstein A, Niedermeier M, Bauer K, Engelmann S, Hoth S, Stadler R, Sauer N. Expression of the AtSUC1 gene in the female gametophyte, and ecotype-specific expression differences in male reproductive organs. Plant Biology 12, Supplement 1. 2010:S105–S114. doi: 10.1111/j.1438-8677.2010.00389.x. [DOI] [PubMed] [Google Scholar]
- Geiger D. Plant sucrose transporters from a biophysical point of view. Molecular Plant. 2011;4:395–406. doi: 10.1093/mp/ssr029. [DOI] [PubMed] [Google Scholar]
- Gil L, Yarin I, Shalitin D, Sauer N, Turgeon R, Wolf S. Sucrose transporter plays a role in phloem loading of CMC-infected melon plants that are defined as symplastic loaders. The Plant Journal. 2011;66:366–374. doi: 10.1111/j.1365-313X.2011.04498.x. [DOI] [PubMed] [Google Scholar]
- Gottwald JR, Krysan PJ, Young JC, Evert RF, Sussman MR. Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proceedings of the National Academy of Sciences, USA. 2000;97:13979–13984. doi: 10.1073/pnas.250473797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackel A, Schauer N, Carrari F, Fernie AR, Grimm B, Kühn C. Sucrose transporter LeSUT1 and LeSUT2 inhibition affects tomato fruit development in different ways. The Plant Journal. 2006;45:180–192. doi: 10.1111/j.1365-313X.2005.02572.x. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. Journal of Molecular Biology. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Molecular Biology. 2000;42:819–832. doi: 10.1023/a:1006496308160. [DOI] [PubMed] [Google Scholar]
- Holsters M, Silva B, Van Vliet F, et al. The functional organization of the nopaline A. tumefaciens plasmid pTiC58. Plasmid. 1980;3:212–230. doi: 10.1016/0147-619x(80)90110-9. [DOI] [PubMed] [Google Scholar]
- Hoth S, Niedermeier M, Feuerstein A, Hornig J, Sauer N. An ABA-responsive element in the AtSUC1 promoter is involved in the regulation of AtSUC1 expression. Planta. 2010;232:911–923. doi: 10.1007/s00425-010-1228-4. [DOI] [PubMed] [Google Scholar]
- Hoth S, Stadler R, Sauer N, Hammes UZ. Differential vascularization of nematode-induced feeding sites. Proceedings of the National Academy of Sciences, USA. 2008;105:12617–12622. doi: 10.1073/pnas.0803835105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Khan JA, Wang Q, Sjolund RD, Schulz A, Thompson GA. An early nodulin-like protein accumulates in the sieve element plasma membrane of Arabidopsis. Plant Physiology. 2007;143:1576–1589. doi: 10.1104/pp.106.092296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor E, Haaß D, Tanner W. Unusual features of the active hexose uptake system of. Chlorella vulgaris. Biochimica et Biophysica Acta. 1972;266:649–660. doi: 10.1016/0006-3002(72)90008-x. [DOI] [PubMed] [Google Scholar]
- Kühn C, Grof CPL. Sucrose transporters of higher plants. Current Opinion in Plant Biology. 2010;13:288–298. doi: 10.1016/j.pbi.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Kühn C, Quick WP, Schulz A, Riesmeier JW, Sonnewald U, Frommer WB. Companion cell-specific inhibition of the potato sucrose transporter SUT1. Plant, Cell and Environment. 1996;19:1115–1123. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lei M, Liu Y, Zhang B, Zhao Y, Wang X, Zhou Y, Raghothama KG, Liu D. Genetic and genomic evidence that sucrose is a global regulator of plant responses to phosphate starvation in Arabidopsis. Plant Physiology. 2011 doi: 10.1104/pp.110.171736. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohaus G, Pennewiss K, Sattelmacher B, Hussmann M, Muehling KH. Is the infiltration–centrifugation technique appropriate for the isolation of apoplastic fluid? A critical evaluation with different plant species. Physiologia Plantarum. 2001;111:457–465. doi: 10.1034/j.1399-3054.2001.1110405.x. [DOI] [PubMed] [Google Scholar]
- López-Millán AF, Morales F, Abadía A, Abadía J. Effects of iron deficiency on the composition of the leaf apoplastic fluid and xylem sap in sugar beet. Implications for iron and carbon transport. Plant Physiology. 2000;124:873–884. doi: 10.1104/pp.124.2.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marger MD, Saier MH., Jr. A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends in Biochemical Science. 1993;18:13–20. doi: 10.1016/0968-0004(93)90081-w. [DOI] [PubMed] [Google Scholar]
- Maynard JW, Lucas WJ. A reanalysis of the two-component phloem loading system in. Beta vulgaris. Plant Physiology. 1982;69:734–739. doi: 10.1104/pp.69.3.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Melzer M, Truernit E, Hümmer C, Besenbeck R, Stadler R, Sauer N. AtSUC3, a gene encoding a new Arabidopsis sucrose transporter, is expressed in cells adjacent to the vascular tissue and in a carpel cell layer. The Plant Journal. 2000;24:869–882. doi: 10.1046/j.1365-313x.2000.00934.x. [DOI] [PubMed] [Google Scholar]
- Meyer S, Lauterbach C, Niedermeier M, Barth I, Sjolund RD, Sauer N. Wounding enhances expression of AtSUC3, a sucrose transporter from Arabidopsis sieve elements and sink tissues. Plant Physiology. 2004;134:684–693. doi: 10.1104/pp.103.033399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch E. Die Stoffbewegungen in der Pflanze. Gustav Fischer Verlag: Jena; 1930. [Google Scholar]
- Payyavula RS, Tay KH, Tsai CJ, Harding SA. The sucrose transporter family in Populus: the importance of a tonoplast PtaSUT4 to biomass and carbon partitioning. The Plant Journal. 2011;65:757–770. doi: 10.1111/j.1365-313X.2010.04463.x. [DOI] [PubMed] [Google Scholar]
- Reinders A, Schulze W, Kühn C, Barker L, Schulz A, Ward JM, Frommer WB. Protein–protein interactions between sucrose transporters of different affinities colocalized in the same enucleate sieve element. The Plant Cell. 2002;14:1567–1577. doi: 10.1105/tpc.002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JB. Low-affinity, high-capacity system of glucose transport in the ruminal bacterium Streptococcus bovis: evidence for a mechanism of facilitated diffusion. Applied and Environmental Microbiology. 1990;56:3304–3307. doi: 10.1128/aem.56.11.3304-3307.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer N. Molecular physiology of higher plant sucrose transporters. FEBS Letters. 2007;581:2318–2324. doi: 10.1016/j.febslet.2007.03.048. [DOI] [PubMed] [Google Scholar]
- Sauer N, Ludwig A, Knoblauch A, Rothe P, Gahrtz M, Klebl F. AtSUC8 and AtSUC9 encode functional sucrose transporters, but the closely related AtSUC6 and AtSUC7 genes encode aberrant proteins in different Arabidopsis ecotypes. The Plant Journal. 2006;40:120–130. doi: 10.1111/j.1365-313X.2004.02196.x. [DOI] [PubMed] [Google Scholar]
- Sauer N, Stadler R. A sink-specific H+/monosaccharide co-transporter from Nicotiana tabacum: cloning and heterologous expression in baker's yeast. The Plant Journal. 1993;4:601–610. doi: 10.1046/j.1365-313x.1993.04040601.x. [DOI] [PubMed] [Google Scholar]
- Sauer N, Stolz J. SUC1 and SUC2: two sucrose transporters from Arabidopsis thaliana; expression and characterization in baker's yeast and identification of the histidine-tagged protein. The Plant Journal. 1994;6:67–77. doi: 10.1046/j.1365-313x.1994.6010067.x. [DOI] [PubMed] [Google Scholar]
- Sauer N, Stolz J. Expression of foreign transport proteins in yeast. In: Baldwin SA, editor. Membrane transport. Oxford University Press; 2000. pp. 79–105. [Google Scholar]
- Schmitt B, Stadler R, Sauer N. Immunolocalization of solanaceous SUT1 proteins in companion cells and xylem parenchyma: new perspectives for phloem loading and transport. Plant Physiology. 2008;148:187–199. doi: 10.1104/pp.108.120410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S, Beyhl D, Hedrich R, Sauer N. Functional and physiological characterization of Arabidopsis INOSITOL TRANSPORTER1, a novel tonoplast-localized transporter for myo-inositol. The Plant Cell. 2008;20:1073–1087. doi: 10.1105/tpc.107.055632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S, Hulpke S, Schulz A, et al. Vacuoles release sucrose via tonoplast-localized SUC4-type transporters. Plant Biology. 2011 doi: 10.1111/j.1438-8677.2011.00506.x. in press. [DOI] [PubMed] [Google Scholar]
- Schneider S, Schneidereit A, Konrad KR, Hajirezaei M-R, Gramann M, Hedrich R, Sauer N. Arabidopsis INOSITOL TRANSPORTER4 mediates high-affinity H+ symport of myoinositol across the plasma membrane. Plant Physiology. 2006;141:565–577. doi: 10.1104/pp.106.077123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz A, Beyhl D, Marten I, Wormit A, Neuhaus HE, Poschet G, Büttner M, Schneider S, Sauer N, Hedrich R. Proton-driven sucrose symport and antiport is provided by the vacuolar transporters SUC4 and TMT1/2. The Plant Journal. 2011 doi: 10.1111/j.1365-313X.2011.04672.x. in press. [DOI] [PubMed] [Google Scholar]
- Schulze WX, Reinders A, Ward J, Lalonde S, Frommer WB. Interactions between co-expressed Arabidopsis sucrose transporters in the split-ubiquitin system. BMC Biochemistry. 2003;4:3. doi: 10.1186/1471-2091-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze W, Weise A, Frommer WB, Ward JM. Function of the cytosolic N-terminus of sucrose transporter AtSUT2 in substrate affinity. FEBS Letters. 2000;485:189–194. doi: 10.1016/s0014-5793(00)02180-3. [DOI] [PubMed] [Google Scholar]
- Sivitz AB, Reinders A, Johnson ME, Krentz AD, Grof CPL, Perroux JM, Ward JM. Arabidopsis sucrose transporter AtSUC9. High-affinity transport activity, intragenic control of expression, and early flowering mutant phenotype. Plant Physiology. 2007;143:188–198. doi: 10.1104/pp.106.089003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivitz AB, Reinders A, Ward JM. Arabidopsis sucrose transporter AtSUC1 is important for pollen germination and sucrose-induced anthocyanin accumulation. Plant Physiology. 2008;147:92–100. doi: 10.1104/pp.108.118992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava AC, Dasgupta K, Ajieren E, Costilla G, McGarry RC, Ayre BG. Arabidopsis plants harbouring a mutation in AtSUC2, encoding the predominant sucrose/proton symporter necessary for efficient phloem transport, are able to complete their life cycle and produce viable seed. Annals of Botany. 2009;104:1121–1128. doi: 10.1093/aob/mcp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava AC, Ganesan S, Ismail IO, Ayre BG. Functional characterization of the Arabidopsis thaliana AtSUC2 Suc/H+ symporter by tissue-specific complementation reveals an essential role in phloem loading but not in long-distance transport. Plant Physiology. 2008;148:200–211. doi: 10.1104/pp.108.124776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R, Brandner J, Schulz A, Gahrtz M, Sauer N. Phloem loading by the PmSUC2 sucrose carrier from Plantago major occurs into companion cells. The Plant Cell. 1995;7:1545–1554. doi: 10.1105/tpc.7.10.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R, Sauer N. The Arabidopsis thaliana AtSUC2 gene is specifically expressed in companion cells. Botanica Acta. 1996;109:299–306. [Google Scholar]
- Stadler R, Truernit E, Gahrtz M, Sauer N. The AtSUC1 sucrose carrier may represent the osmotic driving force for anther dehiscence and pollen tube growth in. Arabidopsis. The Plant Journal. 1999;19:269–278. doi: 10.1046/j.1365-313x.1999.00527.x. [DOI] [PubMed] [Google Scholar]
- Thompson MV, Wolniak SM. A plasma membrane-anchored fluorescent protein fusion illuminates sieve element plasma membranes in Arabidopsis and tobacco. Plant Physiology. 2008;146:1599–1610. doi: 10.1104/pp.107.113274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon R, Beebe DU. The evidence of symplasmic phloem loading. Plant Physiology. 1991;96:349–354. doi: 10.1104/pp.96.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voitsekhovskaja OV, Pakhomova MV, Syutkina AV, Gamalei YV, Heber U. Compartmentation of assimilate fluxes in leaves. II. Apoplastic sugar levels in leaves of plants with different companion cell types. Plant Biology. 2000;2:107–112. [Google Scholar]
- Wahl R, Wippel K, Goos S, Kämper J, Sauer N. A novel high-affinity sucrose transporter is required for virulence of the plant pathogen. Ustilago maydis. PLoS Biology. 2010 doi: 10.1371/journal.pbio.1000303. 8, e1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise A, Barker L, Kühn C, Lalonde S, Henrik Buschmann H, Frommer WB, Ward JM. A new subfamily of sucrose transporters, SUT4, with low affinity/high capacity localized in enucleate sieve elements of plants. The Plant Cell. 2000;12:1345–1356. doi: 10.1105/tpc.12.8.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wippel K, Wittek A, Hedrich R, Sauer N. Inverse pH regulation of plant and fungal sucrose transporters: a mechanism to regulate competition for sucrose at the host/pathogen interface? PLoS One. 2010;26 doi: 10.1371/journal.pone.0012429. 5(8): e12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J-J, Theodoulou F, Sauer N, Sanders D, Miller AJ. A kinetic model with ordered cytoplasmic dissociation for SUC1, an Arabidopsis H+/sucrose cotransporter expressed in Xenopus oocytes. Journal of Membrane Biology. 1997;159:113–125. doi: 10.1007/s002329900275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.