Abstract
A detailed and comprehensive understanding of seed reserve accumulation is of great importance for agriculture and crop improvement strategies. This work is part of a research programme aimed at using Brachypodium distachyon as a model plant for cereal grain development and filling. The focus was on the Bd21-3 accession, gathering morphological, cytological, and biochemical data, including protein, lipid, sugars, starch, and cell-wall analyses during grain development. This study highlighted the existence of three main developmental phases in Brachypodium caryopsis and provided an extensive description of Brachypodium grain development. In the first phase, namely morphogenesis, the embryo developed rapidly reaching its final morphology about 18 d after fertilization (DAF). Over the same period the endosperm enlarged, finally to occupy 80% of the grain volume. During the maturation phase, carbohydrates were continuously stored, mainly in the endosperm, switching from sucrose to starch accumulation. Large quantities of β-glucans accumulated in the endosperm with local variations in the deposition pattern. Interestingly, new β-glucans were found in Brachypodium compared with other cereals. Proteins (i.e. globulins and prolamins) were found in large quantities from 15 DAF onwards. These proteins were stored in two different sub-cellular structures which are also found in rice, but are unusual for the Pooideae. During the late stage of development, the grain desiccated while the dry matter remained fairly constant. Brachypodium exhibits some significant differences with domesticated cereals. Beta-glucan accumulates during grain development and this cell wall polysaccharide is the main storage carbohydrate at the expense of starch.
Keywords: Brachypodium, carbohydrates, embryo, endosperm, grain, proteins, seed maturation
Introduction
Seeds are the major resource for human nutrition and animal feed throughout the world as well as raw material for industry and the production of alternative energy. The understanding of seed formation is essential to improve grain properties with respect to yield, nutritional value or industrial usage (e.g. milling or brewing). Among the seed-bearing plants (spermatophytes), cereals and especially wheat, maize, and rice, are highly valued in the human diet and have been selected for agronomical traits over the centuries (Charmet, 2011; Tenaillon and Charcosset, 2011). These plants disseminate their seeds by the way of dry indehiscent fruits (caryopses), which are called grains.
There is strong demographic and economic pressure to increase cereal production by about 50% in the next 40 years, in a sustainable manner (Borlaug, 2007). This increase is expected to come, at least in part, from higher yield and cropping intensities. Therefore, a better understanding of grain development and the molecular and genetic mechanisms involved may help towards this achievement.
Since grains are complex organs composed of several tissues, their formation requires co-ordinated and highly regulated developmental processes, including cellular growth and differentiation, accumulation of storage compounds, acquisition of desiccation tolerance, as well as reversible maintenance of dormancy (Vicente-Carbajosa and Carbonero, 2005; Gutierrez et al., 2007; Santos-Mendoza et al., 2008). Although huge progress has been made during recent years on the understanding of seed development and maturation, the molecular mechanisms involved are far from fully understood (Santos-Mendoza et al., 2008).
Brachypodium distachyon has been proposed as a new model species for grasses (Draper et al., 2001; Bevan et al., 2010; Febrer et al., 2010; Burcu et al., 2011), because it fulfils a number of criteria that make it ideal for laboratory research: small stature, short generation time, easy handling, autogamy, the absence of specific growth requirements (Draper et al., 2001; Opanowicz et al., 2008; Ozdemir et al., 2008; Brkljacic et al., 2011), and an efficient transformation system (Pacurar et al., 2008; Vain et al., 2008; Thole et al., 2009, 2010). A first milestone was the recent sequencing of its 350 Mbp genome (International-Brachypodium-Initiative, 2010). Brachypodium is much more closely related to wheat than to rice or maize (Huo et al., 2009; Abrouk et al., 2010; Qi et al., 2010). Thus, it is of particular interest for wheat research, especially since the hexaploid nature of bread wheat is a major drawback for genetic and molecular approaches. The endosperm contains storage compounds and is the largest compartment of the grain (Opanowicz et al., 2011). To date, information on B. distachyon grain composition is scarce. Due to the overall high protein content and predominance of glutelin-type storage proteins in Brachypodium grain, its composition appears closer to that of oats than wheat (Larré et al., 2010). Brachypodium distachyon is also notable for its high cell wall polysaccharide content relative to starch (Guillon et al., 2011). Several recent articles focused on the development of the endosperm in domesticated grasses (Sabelli and Larkins, 2009) or in the model species Brachypodium (Opanowicz et al., 2011). However, taking into account the potential role of Brachypodium as a model system for grasses, detailed morphological, physiological, and metabolic data for Brachypodium grain during development are still lacking.
This work aims to provide a comprehensive overview of Brachypodium grain development through an exhaustive description from early embryogenesis to late maturation of both the embryo and the endosperm.
Materials and methods
Plant material and growth conditions
B. distachyon variety Bd21-3, a diploid inbred line isolated from the original accession Bd21 (Vogel and Hill, 2008), was used in this study. The grains were first stratified at 4 °C for 4 d on moist paper to promote synchronous germination, then transferred to soil and grown in a growth chamber at 21/18 °C (day/night) at 65% relative humidity under a short day (8/16 h light/dark) photoperiod with a light intensity of 120 μmol m−2 s−1 for a period of 4 weeks. The plants were then transferred into long-day conditions with a 16/8 h light/dark photoperiod with the same light intensity. Plants were irrigated twice a week with a mineral nutrient solution. To harvest grains of defined developmental stages, individual flowers were tagged 7 d before fertilization (i.e. at the beginning of flower development) using coloured tape.
Scanning electron microscopy
Images were produced with a Hirox SH 1500 desktop scanning electron microscope. Dissected embryos were placed directly in the vacuum chamber, frozen to –30 °C with a Pelletier cooler module and imaged without metallization at a tension of 5 kV.
Light microscopy
Pseudo-Schiff-Propidium iodide (PS-PI) staining:
PS-PI staining was conducted as described in Truernit et al. (2008) with minor modifications to the protocol due to the size of the Brachypodium embryo. Seeds were fixed in 4% paraformaldehyde on ice for 60 min followed by an overnight treatment at 4 °C. Samples were then dehydrated in a graded ethanol series (water, 30–50–70% v/v). Samples could then be stored at 4 °C for several months. Tissues were rehydrated in water (three changes, 5 min each) and subjected to a 0.1 N NaOH, 0.5% SDS treatment overnight at room temperature. Grains were then treated with α-amylase (0.2 mg ml−1) at 37 °C overnight following the manufacturer’s instructions. After this final treatment, grains were transferred to 1% periodic acid for 20 min and mounted onto microscope slides as previously described (Truernit et al., 2008).
A Zeiss LSM 710 spectral confocal laser-scanning microscope was used for the acquisition of embryo images. The excitation wavelength for PS-PI-stained samples was 488 nm and emissions were collected at 520 to 720 nm.
Data were processed for two-dimensional orthogonal sections, 3D rendering, and movie exports using the open source software Osirix (Rosset et al., 2004) http://homepage.mac.com/rossetantoine/osirix/) on a quadxeon 2.66-Ghz, 2-GB RAM Apple Mac Pro workstation. Red/Green/Blue stacks of confocal images were imported as DICOM files into Osirix prior to surface rendering, as described in Truernit et al. (2008).
Preparation of resin embedded material for histology and immunocytochemistry:
Samples were prepared as described by Guillon et al. (2011). At the mature stage of grain development, the embryo was excised and the grain then water-soaked between filter paper moistened with distilled water (16 h at 4 °C). Immature grains are naturally hydrated and did not require this treatment. Sections (∼1 mm3) sampled from half grains were fixed in a mixture of paraformaldehyde (3%) and glutaraldehyde (1%) in 0.1 M sodium phosphate buffer (pH 7.4) for 4 h at 4 °C. After washing, the sections were dehydrated in a graded aqueous ethanol series and progressively infiltrated with London Resin White acrylic resin embedded in gelatin capsules. The resin was then polymerized for 4 d at 55 °C. Thin cross-sections (∼1 μm thick) were prepared and mounted on multiwell glass sides pre-treated with VECTABOND reagent (Vector Laboratories, Burlingame, CA, USA). All semi-thin sections (1 μm thick) were observed with a LEICA DMRD microscope (LEICA, www.leica-microsystems.com).
Differential interference contrast (DIC) microscopy:
Cross-sections of B. distachyon were observed using differential interference contrast microscopy (LEICA DMRD) in order to compare maternal tissue organization in B. distachyon with that of domesticated cereals. The microscope was equipped with standard DIC optics using a Plan-APO 40×/0.75 objective. It was coupled to a Nikon DS-1QM camera.
Toluidine blue staining:
Cross-sections were stained with Toluidine blue [1% (w/v) in 2.5% sodium carbonate] for 1 min, washed, dried on a hotplate and examined on a Leica DMRD equipped with a Nikon DS-1QM camera.
Starch and protein staining:
The sections were stained with 0.02 mol l−1 iodine for 2 min, washed with water and stained with Fast Green (1% in ethanol) for 5 min to observe starch and intracellular proteins. They were rinsed extensively with water before examination on the Leica DRD microscope either in bright field or fluorescence mode. Some sections were pretreated with Subtilisin A (Sigma-Aldrich France, St Quentin Fallavier, France), 1% in 1M phosphate buffer, pH 7.2 for 2 h30 at 40°C) in order to degrade proteins prior to Fast Green staining.
(1-3)-1-4)-β-D-glucan and protein immunolabelling:
Cross-sections of B. distachyon grains were pre-incubated for 45 min in phosphate-buffered saline (PBS, pH 7.2) containing 3% (w/v) Bovine Serum Albumin (BSA, Merck, Germany) to block non-specific labelling. The sections were then incubated for 1 h with primary antibodies. Antibody dilutions in PBS containing 0.3% (w/v) BSA (PBS-0.3% BSA) were 1:100 for the monoclonal antibody against (1-3)(1-4)-β-D-glucan (Meikle et al., 1994), Biosupplies, Melbourne, Australia) and 1:2000 for the anti-11S polyclonal antibody (Le Gall et al., 2005). The sections were washed extensively with PBS-0.3% BSA solution before being incubated for 1 h in the dark with goat anti-mouse (for monoclonal antibody) or goat anti-rabbit IgG (for polyclonal sera) antibodies coupled to Alexa 546 (Molecular Probes, Montluçon, France) diluted 1:100 in PBS-0.3% BSA. This was followed by four washes in PBS buffer and two washes with distilled water. Sections were then examined on a LEICA DMRD with a 515–560 nm band-pass filter as excitation light and fluorescence was detected above 570 nm.
In a control experiment, the primary antibody was omitted to test for non-specific secondary antibody binding.
Synchrotron UV fluorescence microspectroscopy:
Transverse sections sampled from half grains at 17 DAF and mature stages of development were transferred into plastic moulds, fixed in formalin for 24 h at 20 °C, and processed for paraffin inclusion as described by Jamme et al. (2008). Microtome sections of 6–8 μm (Microm HM 340E, Thermoscientific, Waltham, USA) were deposited on to glass sides. The paraffin was removed using Histochoice clearing agent (Sigma-Aldrich, http://www.sigmaaldrich.com).
UV fluorescence spectra of cell walls and organelles were recorded by taking advantage of the high stability and brightness of a synchrotron source (SOLEIL, Gif sur Yvette France).
UV excitation of the sample is provided by a bending magnet emission on the DISCO beamline at synchrotron SOLEIL. The white beam from 180 to 600 nm is monochromatized by an iHR320 (Horiba, France) (Giuliani et al., 2009). Synchrotron fluorescence microspectroscopy was performed using an inverted IX71 microscope (Olympus) coupled to a modified Raman spectrometer (Jobin Yvon T6400, Horiba, France) and projected onto a Peltier-cooled iDus CCD (1024×256 pixels, Andor, Belfast, Ireland) (Jamme et al., 2010).The objective used was a glycerin immersion ×40 Ultrafluar (Zeiss, Iena, Germany) with a numerical aperture of 0.6.
Brachypodium sections were covered with UV transparent coverslips. A droplet of glycerin (Zeiss) was added before section positioning onto the objective. The two dimensional rastering was ensured by a nano-positioning XY stage (PI, Karlsruhe, Germany). Spectral images were acquired in the UV energy spectral range using an excitation of 275 nm. Spectra were recorded between 290 nm and 540 nm per step of 0.265 nm. The spatial step size was set to 1×1 μm2.
Carbohydrate analyses
Carbohydrate extraction and quantification were performed with 15–40 grains as described previously (Baud et al., 2002).
(1-3)(1-4)-β-D-glucan analysis:
Grains were hand dissected and the endosperm separated from the outer tissues. The entire grain and endosperm were ground in liquid nitrogen. Small sugars in the samples (e.g. glucose, sucrose) were removed with ethanol. Briefly, 10–20 mg of ground tissue was treated with 1 ml of 80% ethanol in a boiling water bath for 30 min. The supernatant was removed after centrifugation at 10 000 rpm for 5 min and then the residue was treated with 1 ml of 80% ethanol in a boiling water bath for 10 min. After centrifugation, the pellet was successively washed with 1 ml of 80% ethanol then 1 ml of 96% ethanol after which it was dried in an oven at 40 °C overnight. The pellet was then resuspended in water (0.8 ml) supplemented with 0.2 ml of Lichenase (5U ml−1, Megazyme Bray Ireland) and incubated overnight (16 h) at 40 °C. The reaction mixture was then centrifuged and the supernatant heated for 5 min in a boiling water bath. After filtration through a 0.45 μm sieve (Millipore) samples (1/100 dilution in Ultrapure water) supernatants were analysed by HPAEC (High-Performance Anion-Exchange Chromatography). The samples were injected (10 μl aliquots) into a Carbopac PA-1 (4×250 mm) analytical column (Dionex, http://www.dionex.com., temperature: 25C, flow rate: 1 ml min−1). Gluco-oligosaccharides (GOS), 3-O-β-cellobiosyl-D-glucose (BG3, retention time Rt=3.9 min), 3-O-β-cellotriosyl-D-glucose (BG4, Rt=4.7 min), 3-O-β-cellotetraosyl-D-glucose (BG5, Rt=5.9 min) and 3-O-β-cellotpentaosyl-D-glucose (BG6, Rt=6.7 min) were separated by elution using water (70%), 1 M NaOAc (10%), and 0.5 M NaOH (20%). A pulse amperometric detector (PAD, TSP EC2000, http://www.thermo.com) was used for detection. Quantification of peaks was made relative to reference BG3–BG6 standards prepared in the laboratory from barley (1-3)(1-4)-β-D-glucan (Megazyme). Briefly, barley (1-3) (1-4)-β-D-glucan was hydrolysed with Lichenase and GOS were isolated using preparative size-exclusion chromatography on a Bio-Gel P2 column (Bio-Rad). Standards were used to establish a mass response curve with the pulse amperometric detector.
Lipid analyses
Total lipids were extracted from manually dehulled grains following the protocol described by Leonova et al. (2008). Extraction, separation of lipid classes by thin layer chromatography, analyses and preparation of FAMEs (Fatty Acids Methyls Esters) were performed as reported previously (Leonova et al., 2008). Fatty acid (FA) analyses were carried out on an Agilent 7890A gas chromatograph (Agilent Technologies, Santa Clara, USA) coupled to an Agilent 5975C mass spectrometer (Agilent Technologies, Santa Clara, USA) equipped with a BPX70 column (length 30 m with 10 m guard column; inner diameter 0.22 mm; film thickness 0.25 μm; SGE, Austin, USA). Helium at a flow rate of 0.83 ml min−1 was used as the carrier gas. Injections were made in splitless mode at an initial column temperature of 70 °C. The temperature was increased to 160 °C at a rate of 16 °C min−1 then to 240 °C at 4 °C min−1 and finally at 10 °C min−1 to 260 °C for 4 min. Grain lipid contents are given as μg FA per grain determined using methyl-heptadecanoate as an internal standard in GC-MS analysis of the methyl esters (FAME) for all FAs. FA profiles are given as mol% of the total amount of FAs.
Amino acid and protein analyses
Protein extraction and quantification:
Freeze-dried grains were ground to a fine powder in liquid nitrogen using a mortar and pestle. The resulting powder was added to 12 μl mg−1 of 0.062 M TRIS HCl pH 6.8, 2% SDS. Extractable proteins were quantified using the Non-Interfering Protein Assay (Geno Technology, St Louis, MO, USA), according to the supplier’s recommendations.
SDS PAGE electrophoresis:
Samples of extractable proteins were separated on 15% SDS-PAGE gels (13 cm length) according to Laemmli (1970) and stained with Coomassie Brilliant Blue R250 (Sigma, St Louis, MO, USA) according to Devouge et al. (2007).
Total nitrogen and carbon content:
Total carbon and nitrogen contents of the samples were determined in duplicate by the Dumas procedure (Allen et al., 1974) using a Thermo (Carlo Erba) elemental analyser following the manufacturer’s instructions.
Protein identification by mass spectrometry:
Protein bands stained with Coomassie blue were excised from the polyacrylamide gel and prepared for mass spectrometry analysis. Each band underwent the following steps: discolouring, reduction, alkylation by iodoacetamide before in-gel digestion using trypsin hydrolysis, according to Devouge et al. (2007). The resulting peptide mixture was acidified by the addition of 1 μl of aqueous solution of formic acid (1%, vol.), and stored at –20 °C until analysis. Mass spectrometry analysis and protein identification were performed according to Larre et al. (2010).
Results
Grain development
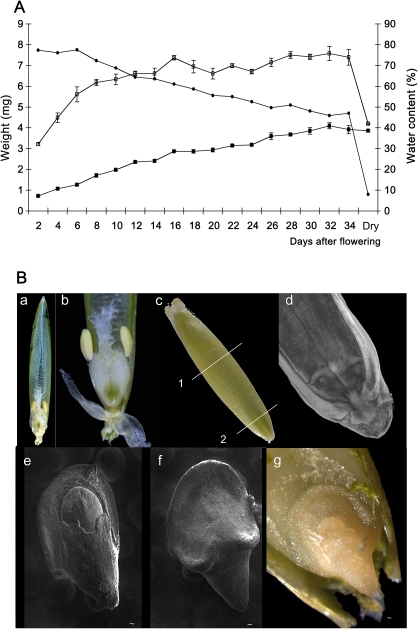
B. distachyon grain development was characterized by weight measurements and microscopic observations. Under the growth conditions used in this study, B. distachyon grain development took between 34 to 36 d and was complete (i.e. dry) in approximately 50 d (Fig. 1A). Between 2 d and 14 d after flowering (DAF), the fresh mass of the grain increased dramatically and then increased slowly to reach its maximum at 36–40 DAF. Between 42 and 54 DAF the fresh weight decreased. The seed dry weight followed a much more linear pattern until the end of seed maturation. The water content decreased almost linearly during seed development with a sharp decrease after 54 DAF. The mature desiccated seed weighed approximately 5.5 mg with a water content of 5%. In mature grains, the embryo occupied about 20% of the grain and was located at the bottom of the grain (Fig. 1B-c, d, g). The mature dissected zygote showed dorso-ventral differentiation with an asymmetrical scutellum surrounding the embryo (Fig. 1A-e, f, g). The coleoptile enveloped the foliage leaves and the meristem (see Supplementary Fig. S1 at JXB online).
Fig. 1.
Grain development. (A) Growth parameters of developing Brachypodium distachyon grain. Each data point is the mean of at least three independent measurements ±SE: (open squares), fresh weight; (closed squares), dry weight; (closed circles) water content. (B) General micrographs of the grain (a) before fertilization; (b) reproductive organs; (c) entire grain at 22 DAP showing the sections made for (c) and for (b); (d) PI image of the region of the grain containing the embryo at 30 DAP; (e, f) SEM micrographs of dissected mature embryo; (e) frontal top, (f) frontal bottom; and (g) bright-field micrograph of the mature embryo in situ.
Embryogenesis and endosperm development
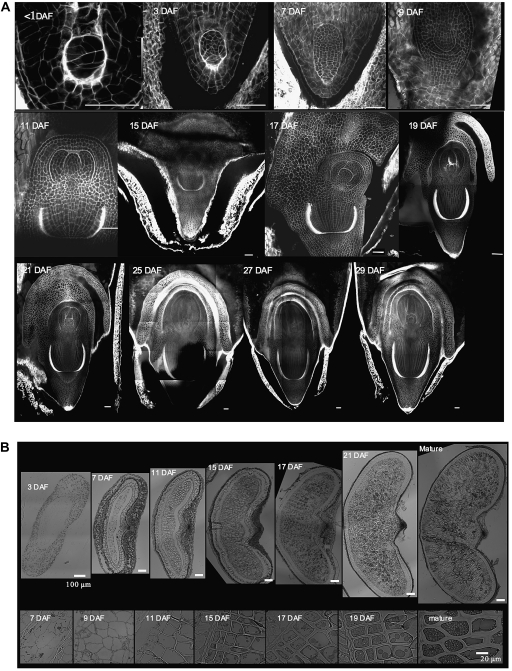
In the first days after fertilization, the zygote underwent a series of divisions with no obvious symmetry (Fig. 2A). The embryo was supported by a circular-shaped cell layer (see Supplementary Fig. S1A-e at JXB online). After 10 DAF, an apical-basal axis emerged with a thickening of the cell walls at the base of the embryo in a cup-shaped pattern (Fig. 2A; see Supplementary Fig. S1A-a, Fig. 2A; see b at JXB online). From 10–14 DAF, an epidermis began to differentiate together with the acquisition of apico-basal symmetry, separating the embryo into two zones. This symmetry led to both apical and root meristems for the upper zone and to the coleorhiza for the lower zone, which became visible at 16 DAF (see Supplementary Fig. S1A-c, d, g, h at JXB online). The upper part was then covered by the coleoptile, which was connected to the coleorhiza, both protecting the future cotyledon, plumule (foliage leaves) and primary root. At 18 DAF the general shape of the embryo was complete and at 26 DAF all the structures of the mature grain were present (see Supplementary Fig. S2 at JXB online). From 27 DAF until the end of maturation, no further morphological changes were observed in the developing embryo.
Fig. 2.
Embryogenesis. (A) Confocal PI images of the embryo during development. Frontal view. (B) B. distachyon grain cross-sections from 3 DAF to mature stages. Bright-field micrographs of toluidine blue-stained sections. Top, morphological development of the grain; Bottom, Aleurone differentiation and development. Bar=50μm (A), 100μm (B-Top), 20μm (B-Bottom).
At 3 DAF the grain consisted of several layers of maternal tissue surrounding the central cell in which the endosperm developed (Fig. 2B). These tissues degenerated as the endosperm enlarged. At 7 DAF the endosperm was fully cellularized. The endosperm cells continued to divide for about another four days. At 11 DAF, the cells of the storage endosperm expanded and their cell walls started to thicken, increasing dramatically up to 36 DAF. Cell divisions still occurred in the outermost cellular layer and the endosperm met the surrounding maternal tissues. At 15 DAF, the aleurone layers, on the outside of the developing endosperm, were recognizable (Fig. 2B bottom; see Supplementary Fig. S3 at JXB online).
Carbohydrate distribution during grain development
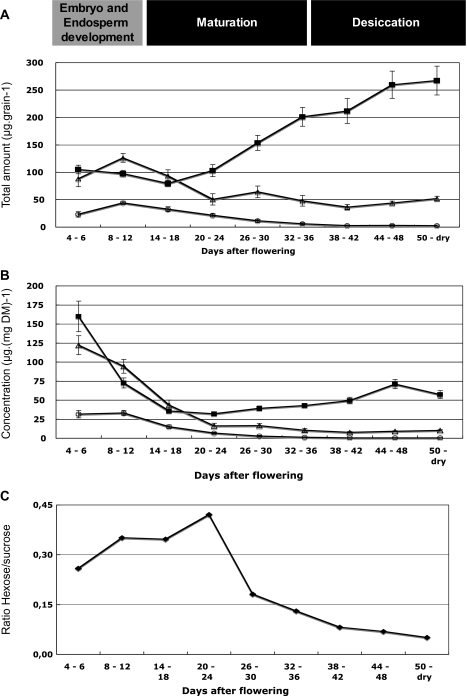
Soluble carbohydrate, starch and β-D glucans were quantified throughout grain development. The young grain, at 4 DAF, contained low levels of hexoses (glucose and fructose) (Fig. 3A). Their concentrations decreased during the first 20 DAF, and in the mature grain, only traces of these hexoses could be found. The total sucrose concentration was at a very low level during the same 20 DAF period, peaking at 8 DAF and then decreasing. The quantity of starch was stable during the first 20 DAF at about 100 μg per grain and then increased to reach 300 μg per mature grain at 56 DAF (Fig. 3A).
Fig. 3.
Distribution of soluble sugar and starch content during Brachypodium grain development. (A) Total amount of carbohydrate and (B) concentration on a dry-weight basis. Data are the mean of three independent measurements ±SE. (C) The hexose-to-sucrose ratio was calculated with the mean concentrations of hexoses and sucrose presented in (A).(open circles) Hexose (Fru+Glc) content; (closed squares) starch content, (open triangles) sucrose content.
When expressed as μg mg−1 of grain dry weight, the hexoses remained very low whereas sucrose decreased sharply from 4 DAF to 20 DAF and remained below 10 μg mg−1 dry weight. From 4–18 DAF, the starch concentration decreased 3-fold from 160 to 45 μg mg−1 dry weight and then increased slowly to peak at 75 μg mg−1 dry weight at 44–48 DAF (Fig. 3B).
The hexose to sucrose ratio was approximately 0.25 at 4 DAF and peaked at about 0.5 between 22–24 DAF. It then decreased sharply over three days to below its initial level and continued to decrease until the end of seed development (Fig. 3C).
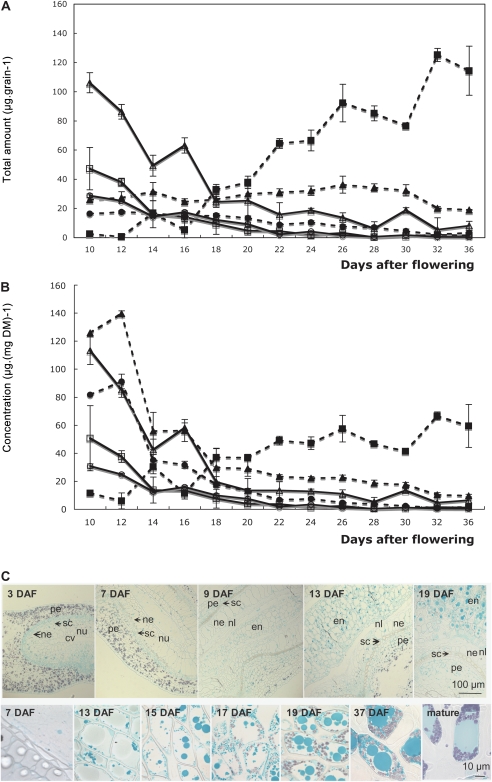
The partition of soluble carbohydrates and starch between the endosperm and the outer layers was investigated using both chemical analyses after grain dissection and light microscopy on grain sections (Fig. 4).
Fig. 4.
Distribution of soluble sugar and starch content during Brachypodium endosperm and embryo development. (A) Total amount of carbohydrates. Endosperm: (closed circles) hexoses (Fru+Glc); (closed squares) starch; (closed triangles) sucrose. Tegument+embryo: (open squares) hexoses (Fru+Glc); (open squares) starch; (open triangles) sucrose. (B) Concentration on a dry-weight basis. Data are the mean of three independent measurements. (C) Bright-field images of grain cross-sections stained with Fast Green and iodine allowing the visualization of both starch (purple) and intracellular proteins (grey). Top, micrographs of peripheral cell layers at the early stages of grain development; bottom, micrographs of storage endosperm cells from the cellularization up to the mature stages; cv, central vacuole; en, endosperm; ne, nucellar epidermis; nl, nucellus lysate; nu, nucellus tissue; pe, pericarp; sc, seed coat.
The total amount of hexoses was relatively low in all tissues tested and a small decrease was observed during grain development. The quantity of sucrose in the outer layers decreased very rapidly from 100 μg per grain at 10 DAF to 20 μg per grain at 20 DAF (Fig. 4A). In the endosperm, the quantity of sucrose remained stable at 30 μg grain−1 until 30 DAF when it decreased slightly to 10 μg. In the outer layers, starch accumulated transiently at the beginning of grain development and then from 16 DAF onwards its amount became negligible. In terms of concentration, between 12 and 14 DAF sucrose and hexoses declined rapidly in the endosperm, dropping from 140 to 50 μg mg−1 DM and from 90 to 35 μg mg−1 DM, respectively. After this initial quick decrease, the concentrations decreased slowly until the end of grain maturation. The same pattern of evolution was observed for the concentration of hexoses and sucrose in the teguments+embryo. By contrast, the starch concentration increased slowly in the endosperm, from 10 μg mg−1 DM to reach 60 μg mg−1 DM at the completion of maturation. In the embryo, the concentration of starch decreased from 50 to 10 μg mg−1 DM between 10 and 14 DAF and then slowly decreased to almost non-detectable levels.
This pattern of starch accumulation was confirmed by light microscopy using iodine staining (Fig. 4C). Coloured starch grains were observed in the parenchymatous cells contained in the pericarp at the early stages of grain development. The pericarpic parenchymatous cells were rapidly digested except in the region of the groove where starch was still visible at 13 DAF (Fig. 4C). Starch accumulation in the endosperm started about 14 DAF (Fig. 4A). In grain cross-sections, starch granules were detected from 15 DAF in the endosperm cells (Fig. 4C). At 17 DAF, starch granule formation was well underway (Fig 4C). By this stage, the cells were vacuolated and starch granules concentrated near the cell walls. At 37 DAF, the starch granules were very prominent (Fig. 4C). The granules increased in size and exhibited an ellipsoid form. At 37 DAF, in old and mature grain, the cell volume was mainly occupied by fused vacuoles probably filled with proteins, and even-sized starch granules concentrated in any available free spaces (Fig. 4C).
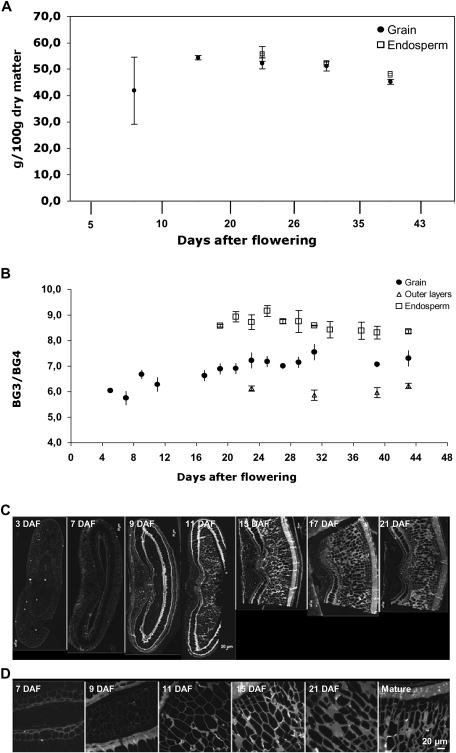
(1-3)(1-4)-β-D glucan was recently reported to be the major carbohydrate in mature grain of B. distachyon (Guillon et al., 2011) and in all likelihood plays a role as a storage compound. Its accumulation was followed by enzymatic determination in whole grains and in the endosperm (Fig. 5). Lichenase is a specific endo (1-3)(1-4)-β-D-glucan 4-glucanohydrolase that cleaves only (1-4) β glucosidic linkages of a three-substituted unit. It solubilizes the (1-3)(1-4)-β-glucan of cereal grains resulting in gluco-oligosaccharides. The gluco-oligosaccharides obtained are mainly 3-O-β-cellobiosyl-D-glucose (BG3), 3-O-β-cellotriosyl-D-glucose (BG4), and oligosaccharides with a higher degree of polymerization (DP5-9). The assay of (1-3)(1-4)-β-D glucan in grain and endosperm tissue is reported in Fig. 5A. Due to the small size of the grain in the early stages of development it was not possible to determine (1-3)(1-4)-β-D glucan in endosperm before 17 DAF. On a dry weight basis, the (1-3)(1-4)-β-D glucan content reached a maximum at 17 DAF and then remained fairly stable, with a small decrease until the end of desiccation. Beta-glucan represented 45% of the grain at maturity corroborating previous results (Guillon et al., 2011). The (1-3)(1-4)-β-D glucan content was slightly higher in endosperm tissue (50% of total weight at maturity) and lower in the outer layers (40% of total weight at maturity). BG3 and BG4 represented 98–99% of the glucan oligosaccharides released by Lichenase. The structure of (1-3)(1-4)-β-D glucan was evaluated using the BG3/BG4 ratio and was fairly stable throughout grain development with a ratio of 7 (Fig. 5B). The ratio observed in the grain was a median value between the ratio measured in endosperm (about 8) and outer layers (6).
Fig. 5.
Spatio-temporal pattern of (1-3)(1-4)-β-D glucan deposition in the developing B. distachyon grain. (A) (1-3)(1-4)-β-D glucan content (expressed as g 100 mg−1 of alcohol insoluble material). whole grain (closed circles); endosperm (open squares). (B) Evolution of weight ratio of tri (BG3)- to tetra-saccharides (BG4) during development; whole grain (closed circles), endosperm (open squares) and outer layers (open triangles). (C, D) Fluorescence micrographs of (1-3)(1-4)-β-D-glucan antibody labelling of developing grain. (C) Grain cross-sections from 3 DAF to 21 DAF including grain outer layers. (D) Endosperm from 7 DAF to the mature stages.
The spatial pattern of (1-3)(1-4)-β-D glucan deposition was observed with light microscopy (Fig. 5C). During the early stages of grain development, (1-3)(1-4)-β-D glucan was present in the outer layers including the nucellar epidermis, integuments, and pericarp (Fig. 5C). While the pericarp parenchyma degenerated at 7 DAF, the nucellar epidermis persisted from 9 DAF to 15 DAF. Intense (1-3)(1-4)-β-D glucan labelling of the outer and inner nucellar cell walls was observed. Glucan labelling in the endosperm appeared simultaneously with the cellularization stage (Fig. 5D). From 7 DAF onwards, (1-3)(1-4)-β-D glucan continued to accumulate and accounted for most of the increase in cell wall thickness.
Protein deposition during grain development
Proteins were measured and localized throughout grain development using biochemical and cytological approaches.
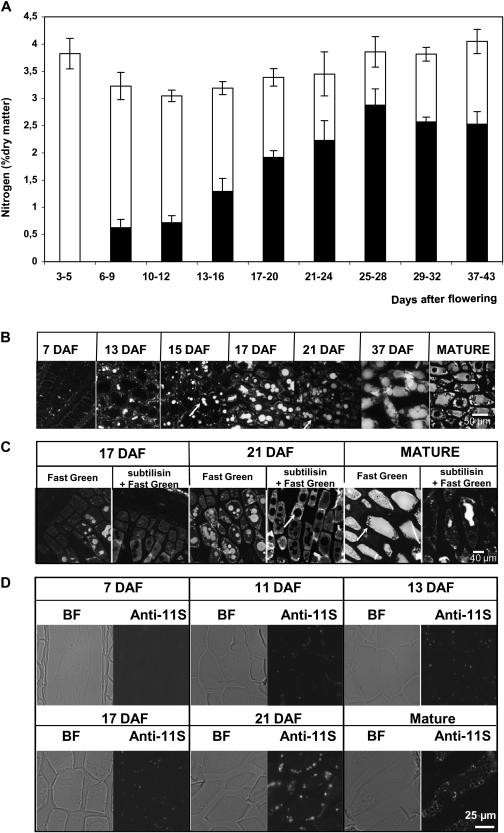
The total nitrogen accumulation was recorded in developing grains. The nitrogen concentration decreased slightly from 3–9 DAF, then gradually increased very slowly to reach a maximum of 3.7% in the mature grain (Fig. 6). Using a conversion factor of 5.7, the total protein content was estimated to be around 21%. Protein content was also estimated from the amount of extractable proteins (Fig. 6A) from 7 DAF to maturity. The amount of protein increased steadily from 9–29 DAF, and then the rate of accumulation decreased slightly reaching 15% of the grain dry weight. Protein accumulation was visible by 13 DAF (Fig. 6B) and reflected the beginning of grain maturation.
Fig. 6.
Protein accumulation during grain development. (A) Changes in the nitrogen content in Brachypodium grains during development. Nitrogen in the form of proteins is shown in black (estimated from the extractable proteins using a conversion factor of 5.7). (B) Fluorescence micrographs of Fast Green-stained cross-sections of B. distachyon grains from 7 DAF to the mature stages without subtilisin pre-treatment. (C) Fluorescence micrographs of Fast Green-stained cross-sections of B. distachyon grains from 7 DAF to the mature stages with subtilisin pre-treatment. (D) Immunofluorescence labelling of protein with anti 11S polyclonal serum in cross-sections of developing grain endosperm.
In a first approach, the pattern of protein deposition in storage endosperm was visualized using Fast Green staining (Fig. 4C). Vesicles of two distinct sizes, both located at the cell periphery, were stained at 13 DAF. From 13 DAF onwards, enlargement of the biggest vesicles was observed and these were distributed within the cytoplasm; these finally coalesced and merged to fill the cell volume in mature grain. The smallest vesicles also increased in size but to a lesser extent. At 37 DAF, they were still visible as individual bodies at the cell periphery. In mature grain these small vesicles were partly fused and surrounded starch granules.
In order to verify that the Fast Green staining was specific to proteins, two alternative experiments were carried out: first, a proteolysis step was introduced prior to Fast Green staining and, secondly, advantage was taken of the synchrotron source to carry out deep ultraviolet (DUV, excitation at 275 nm) fluorescence imaging (see Supplementary Fig. S4 at JXB online). This excitation wavelength allowed aromatic amino acids (tyrosine and tryptophan) present in proteins to be imaged without fluorochromes or probes.
The two types of vesicles were no longer stained with Fast Green after subtilisin proteolysis, confirming that they are filled with proteins (Fig. 6C). Spectral images of grain sections revealed highly fluorescent vesicles in the storage endosperm cells at 17 DAF (see Supplementary Fig. S4A-b at JXB online). At maturity, the fluorescence was distributed throughout the ‘vacuole’ that occupied most of the cell volume (see Supplementary Fig. S4B-b at JXB online). The fluorescence distribution pattern fitted with the structures previously stained with Fast Green. A fluorescence spectrum associated with a pixel in the large ‘vacuole’ (A in Supplementary Fig. S4B-b at JXB online) in the mature grain section was extracted from the data set. The spectrum had a peak centred at 345 nm with two shoulders at 315 nm and 410 nm. The deconvoluted spectrum fitted three main Gaussian functions. The two main Gaussian contributions corresponded to tryptophan (maximum emission at 345 nm) and tyrosine (maximum emission at 315 nm) spectra (Jamme et al., 2010). This confirmed that the fluorescence observed within the endosperm cell can mainly be ascribed to proteins. The third curve with a peak centred around 410 nm corresponded to the fluorescence spectrum of esterified hydroxycinnamic acids, encountered in the endosperm cell walls (Anderson and Akin, 2008).
The results obtained with Fast Green staining and fluorescence imaging were compared with those observed using immunocytochemical labelling. Figure 6D shows the spatial and temporal patterns of deposition of storage proteins labelled with anti-11S globulin serum. By 13 DAF, a few small vesicles at the cytoplasm periphery were labelled. From 17 DAF onwards their number and size increased. In mature grain, labelling was detected, surrounding starch granules, but surprisingly no labelling was observed within the large ‘vacuoles’ that were previously shown to be filled with proteins.
Further characterization of the grain proteome
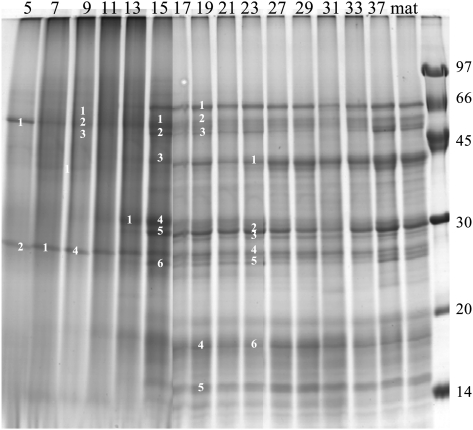
In order to characterize the accumulation of storage proteins during seed development, SDS-PAGE analyses were performed at various stages from 5 DAF to maturity (Fig. 7). Some of the main bands were analysed by mass spectrometry (Table 1; see Supplementary Table S1 at JXB online). Peptide identification was performed using the available putative Brachypodium protein database. Since these sequences are not yet annotated, tentative functional annotations were assessed by running a BLAST homology search against the NCBI nr database (Altschul et al., 1997). The results are summarized in Table 1 and the complete peptide sequence for each protein is reported in the supplementary data (see Supplementary Table S1 at JXB online). Comparison of the protein profiles obtained for each developmental stage reveals large changes between 13 and 17 DAF with a higher resolution and an increase in the number of detectable bands following electrophoresis (Fig. 7),as proteins began to accumulate (see Fig. 6A).
Fig. 7.
A representative SDS PAGE electrophoresis of extracted proteins at various stages of Brachypodium grain development (days after flowering, as indicated in each well and the bands analysed by mass spectrometry are numbered in each row).
Table 1.
List of proteins identified from SDS-PAGE displayed in Figure 3
| No | Instrument | Bradi_1.0 | MW | Nb Uni pep | % Cov | Score | e-value | Best homologue protein name | |
| 1 | Q-TOF | Bradi1g25440.1 | 55160 | 14 | 45 | 1604 | P93594 β-amylase | QT95 | |
| Bradi1g25440.2 | 50007 | 13 | 45 | 1541 | P93594 β-amylase | 31AG | |||
| 2 | Q-TOF | Bradi1g09690.1 | 31817 | 4 | 16 | 337 | Q76BW5 Xyloglucan endotransglycosylase/ hydrolase protein 8 | QT95 32AG | |
| 4 | Q-TOF | Bradi2g38060.1 | 55113 | 15 | 34 | 1056 | 11S globulin [Avena sativa] | QT95 | |
| Bradi1g25440.1 | 55160 | 15 | 41 | 1248 | P93594 β-amylase | 35AG | |||
| Bradi1g05910.1 | 56331 | 7 | 21 | 574 | Globulin 2 [Zea mays] | ||||
| 5 | Q-TOF | Bradi2g38060.1 | 55113 | 18 | 49 | 1721 | 11S globulin [Avena sativa] | QT95 | |
| Bradi2g40840.1 | 55679 | 10 | 23 | 804 | 11S globulin [Avena sativa] | 36AG | |||
| Bradi4g28220.1 | 54630 | 8 | 20 | 529 | Seed storage globulin [Avena sativa] | ||||
| 6 | Q-TOF | Bradi2g38060.1 | 55113 | 9 | 29 | 861 | 11S globulin [Avena sativa] | QT95 | |
| Bradi1g13040 | 64973 | 9 | 22 | 681 | Globulin 3 [Triticum aestivum] | 37AG | |||
| Bradi2g02140 | 39841 | 7 | 31 | 499 | Xylanase inhibitor 7250S [Triticum aestivum] | ||||
| Bradi3g05220.1 | 42206 | 7 | 29 | 524 | Phosphoglycerate kinase cytosolic STEP2/5 glycolysis EC=2.7.2.3 | ||||
| Bradi3g14040.1 | 36623 | 5 | 29 | Glyceraldehyde-3-phosphate dehydrogenase, | |||||
| cytosolic, EC=1.2.1.12 [Hordeum | |||||||||
| vulgare] STEP1/5 glycolysis | |||||||||
| 7 | Q-TOF | Bradi1g09690.1 | 31817 | 11 | 45 | 1147 | Xyloglucan endotransglycosylase/hydrolase | QT95 | |
| protein 8; EC=2.4.1.207, Cell wall | 38A | ||||||||
| biogenesis/degradation | |||||||||
| Bradi2g38060.1 | 55113 | 7 | 20 | 686 | 11S globulin [Avena sativa] | ||||
| Polypeptide alpha (1 to 291) |
All analyses were performed with a Q-TOF instrument. The columns correspond to: the assigned developmental stage-protein band number (DAF-N), the protein identity as referred to in the Bradi_1.0 (release May 2009: http://www.brachypodium.org) (Bradi_1.0), its theoretical molecular weight (MW), the number of unique peptides matched (NUP), the per cent sequence coverage (% Cov), the MASCOT protein score (Score), the NCBI database best homologue protein name and the E-value resulting of the BLAST, Gene Ontology Biological process (in the case of storage proteins the Molecular function is indicated).
In the first developmental stages (5–13 DAF), eight bands were analysed which resulted in the identification of seven expressed proteins all with putative metabolic functions, therefore reflecting the high metabolic activity at these stages. The most represented were the products encoded by Bradi1g25440.1 and Bradi1g09690.1 (Table 1). These peptides are homologous to β-amylase and xyloglucan endotransglycosidase, which are involved in carbohydrate metabolism. ATPase accumulation probably reflected important mitochondrial activities during seed maturation.
The developmental stage at 15 DAF can be considered as a change in direction in protein synthesis and function from high metabolic activity to storage. Two types of storage globulins were identified, four homologues of the glutelin-type (e.g. Bradi2g38060.1) which constitute the bulk of storage proteins (Larre et al., 2010) and two 7S globulin homologues (Bradi1g05910.1 and Bradi1g13040.1) considered as ‘embryoglobulins’ (Kriz, 1989; Laudencia-Chingcuanco and Vensel, 2008; Larre et al., 2010). The protein encoded by Bradi2g02140.1, which is homologous to xylanase inhibitors, was also identified at this stage. The expression of these proteins, usually accumulated in the mature seed, started at 15 DAF, increasing upon development and peaking at 19 DAF and 23 DAF, as shown in Supplementary Table S1 at JXB online. One S-rich type prolamin protein was also identified between 15 and 19 DAF. Proteins involved in glycolysis such as glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase and enolase were detected in the early developmental stages suggesting a strong requirement for glycolysis as a source of metabolites and energy for supplying many metabolic pathways. Many other proteins were also identified with putative functions in various biological processes and these are listed in Supplementary Table S1at JXB online.
Fatty acid and lipid composition and content
Total fatty acid content and composition were measured in dry mature grains. The total amount of fatty acids was 80.89 μg grain−1, representing 1.64% of the total grain weight (see Supplementary Table S2 top at JXB online). The grain contained mostly C16:0 (palmitic acid, 15.92 mol%), C18:1 (oleic acid, 35.71 mol%), and C18:2 (linoleic acid, 36.97 mol%) plus small amounts of C12:0 (lauric acid, 2.13 mol%), C14:0 (myristic acid, 2.78 mol%), and C18:3 (α-linolenic acid, 3.86 mol%). Minor quantities of C14:1 (myristoleic acid), C16:1 (palmitoleic acid), C18:0 (stearic acid; 0.86 mol%), and very long chain fatty acids (>20 carbons) were also detected.
The proportion of total fatty acids acylated into PL and TAG molecules was also analysed and represented 13.6% and 57.4%, respectively, of total fatty acids (data not shown).
Discussion
With the emergence of Brachypodium distachyon as a model plant for temperate cereals and forage grasses, our aim was to draw a more comprehensive overview of Brachypodium seed development. In doing so, some features of embryo development and reserve deposition were highlighted. Interestingly, under the growth conditions used in this study, B. distachyon grain developed in approximately 36 d and dry seed was obtained in about 50 DAF. This is a bit longer than reported in previous studies (Opanowicz et al., 2011), probably due to the differences in growth conditions. The day/night cycle used in our study was more closely related to ‘natural conditions’ than the long day photoperiod previously used to shorten Brachypodium grain production. The average duration for embryogenesis, maturation, and dehydration in our study is similar to those of wheat grown under standard photoperiods (Rogers and Quatrano, 1983).
Cytological analysis of embryo development
Using a modified pseudo-Schiff propidium iodide (PS-PI) staining technique (Truernit et al., 2008) a very precise visualization of the internal embryo structures during development was obtained compared with previous studies. The cellular organization of the embryo and surrounding tissues could be observed through image stacks taken along the z-axis of the samples, generating a novel 3D reconstruction of the entire embryo. One limitation was the large size of the embryo, which at more than 1200×550 μM (including the coleorhiza and the scutellum), was too large to image in one go with this technique. The images presented are thus stacks of mosaics (of up to nine combined images). (See additional data for accessing files at JXB online). These images allowed the cereal embryo to be rotated and virtually re-sectioned revealing highly detailed structures from soon after fertilization to late embryo development (numerical files are provided as supplementary data at JXB online).
Carbohydrate metabolism during grain development
The hexose-to-sucrose ratio peaked between 22–24 DAF and then decreased sharply. In several plant species a developmentally regulated shift in the ratio of hexose to sucrose appears to occur. A high hexose-to-sucrose ratio correlates with cell division and morphogenesis, while a low ratio signals the deposition of reserves and a maturation phase (Weber et al., 1997; Fox and Guerinot, 1998; Baud et al., 2002; Borisjuk et al., 2002; Hill et al., 2003). The data presented in this paper suggest that such a correlation also exists in Brachypodium. Thus, the drop in the hexose-to-sucrose ratio observed from about 24 DAF may trigger an active phase of reserve accumulation, consisting mainly in storage carbohydrate deposition. At 24 DAF embryo morphogenesis is complete and the endosperm has reached its final size. It is likely that a change in metabolism occurs at this step of grain development.
In the outer layers, starch accumulated transiently at the beginning of grain development where it contributed to carbon storage during the earlier phases. In the endosperm, starch starts to accumulate at the beginning of cell differentiation. A few small starch granules were seen to concentrate at the periphery of the endosperm cells. Although the granules increased in size during development, a bimodal granule population differing in size, as reported for wheat, was not observed. In mature grain, the starch content of the endosperm was low (less than 10% of the whole grain) compared with most domesticated cereals (50–70%).
In Brachypodium, as in other cereals (Wilson et al., 2006, Philippe et al., 2006; Robert et al., 2011) (1-3)(1-4)-β-D glucan appears in the endosperm walls at the cellularization stage of endosperm development. At first the (1-3)(1-4)-β-D glucan in growing grain may act as a structural compound but then it rapidly accumulates during development to become the predominant polysaccharide in the mature grain endosperm (Guillon et al., 2011). The amount of (1-3)(1-4)-β-D-glucan was unusually high (more than 70% of total stored glucose) compared with most other domesticated cereals (18% of the total glucose; Morall and Briggs, 1978; Buckeridge et al., 2004). The overall high (1-3)(1-4)-β-D glucan and low starch content strongly suggests that (1-3)(1-4)-β-D glucan plays a storage function.
In Brachypodium the amount of (1-3)(1-4)-β-D-glucan is also high in the grain outer layers. Beta-glucan was detected in the nucellar epidermis in the early stages of grain development. The nucellar epidermis was still persistent at 22 DAF. As previously reported by Opanowicz et al., (2011), very thick nucellar epidermis cell walls were observed with cells compressed to the side of lobes. In wheat, rye, oat, and barley, the nucellar epidermis appears as a single crushed layer of empty cells, the walls of which are depleted in (1-3)(1-4)-β-D glucan (see Supplementary Fig. S5 at JXB online). Immunolabelling of wheat grain clearly indicated a decline of (1-3)(1-4)-β-D glucan when the aleurone cells differentiate and the nucellar epidermis cells compress (Robert et al., 2011). In mature wheat grain, glucose accounts for less than 5% of nucellar epidermis weight while arabinose and xylose reach up to 67% together (Barron et al., 2007). In the absence of modified aleurone cells and reduced nucellar projection, the nucellar epidermis of B. distachyon is suspected of being involved in nutrient transport into the endosperm (Opanowicz et al., 2011).
Differences in the chemical structure of the (1-3)(1-4)-β-D glucan were observed between B. distachyon and other cereals. The BG3/BG4 ratio in endosperm is higher in B. distachyon (about 8) than in wheat (3–4.5), barley (2.3–3.4), and oat (1.5–2.3) (Izydorczy and Dexter, 2008). This ratio was also higher in the endosperm than in the outer layers. Depending on the nature of the tissues, differences in BG3/BG4 ratios were also observed in barley (Izydorczy et al., 2003; Cui et al., 2000), with the (1-3)(1-4)-β-D glucan of the outer layers exhibiting higher BG3/BG4 values than their counterparts in the starchy endosperm. The structural variations in the (1-3)(1-4)-β-D glucan were associated with differences in their solubility and extractability (Izydorczy and Dexter, 2008). The impact of these structural variations in planta is still unknown, as well as the detailed biosynthesis process of (1-3)(1-4)-β-D glucan.
In a current model for cell wall formation there is a turnover of cell wall components with synthesis and degradation/remodelling events occurring simultaneously. Synthesis is likely to dominate during growth while degradation and remodelling mechanisms are enhanced during maturation (Brummell, 2006; Obel et al., 2006). In seeds accumulating cell wall polysaccharides as storage carbohydrates, several changes occur: biosynthesis and hydrolysis are temporally displaced and the proportion of one polysaccharide increases with, potentially, some changes to its chemical structure (Buckeridge, 2000; Hoch, 2007).
The shift from structural to storage functions with the specific deposition of higher than normal levels of (1-3)(1-4)-β-D glucan demands some key transformations in cell wall metabolism that require further investigation. Additional studies of this organ could therefore help identify new members of the metabolic pathway. A detailed investigation of the regulation of the genes encoding biosynthetic and hydrolytic enzymes may provide some clues for understanding the mobilization of storage cell wall polysaccharides in grain.
Fatty acid and lipid composition and content
Large variations in fatty acid content are found among Poacea. The total amount of fatty acids varies from 17–38.8% of the seed weight in Z. mays, 16% in O. sativa, 10% in T. sativum, and 1.4% in A. fatua (Ucciani, 1995; Dubois et al., 2008) (see Supplementary Table S2 bottom at JXB online). The quantity found in B. distachyon (1.6% of seed weight) is thus closest to that of A. fatua. Although Brachypodium and cereals are close relatives, their seeds have different fatty acid compositions. The lipid composition of B. distachyon grain, with mainly C16:0 (15.9 mol%), C18:1 (35.7 mol%), C18:2 (37 mol%), and some C18:3 (3.9 mol%) resembles that of Avena sativa (Banas et al., 2007). The respective proportions of PL and TAG in the whole grain of Brachypodium are also close to those of Avena sativa (Banas et al., 2007).
Protein deposition
Globulins and prolamins were the two main families of storage proteins identified in mature B. distachyon seeds. Although the number of genes of the two families was similar, the detected proteins were mostly glutelins (i.e. salt-insoluble globulins) (Larré et al., 2010). In this study, the expression of storage proteins was observed as early as 15 DAF. At this stage, members of both families were identified. Their accumulation persisted until the grain reached maturity. The globulins identified during development have previously been characterized in mature seeds (Larré et al., 2010). However, in this study, new prolamins belonging to a list of nine in silico predicted prolamins (Larré et al., 2010) were found for the first time to be expressed in mature seed. On the other hand two prolamins previously identified in the mature grain were not found in this experiment possibly due to their weak accumulation. Overall, among the nine in silico predicted prolamin genes, four appear to be expressed in the grain. The storage proteins in B. distachyon resemble those in rice, which also has a large amount of glutelins. By contrast, the temporal pattern of deposition is different. Globulins and prolamins appeared simultaneously in B. distachyon while, in rice, glutenins are detected first (Tanaka et al., 1980).
The identification of storage proteins from 15 DAF was consistent with an initial increase in the amount of total protein and the first observation of protein bodies at 13 DAF. This study establishes that two populations of storage structures exist in maturing B. distachyon endosperm: small protein bodies mostly located at the periphery of the endosperm cells and close to starch granules, and large vacuoles filling endosperm cell lumen. The globulin-type proteins were detected in the small protein bodies. Altogether these results suggest that the large vacuoles are mainly filled with glutelins. In rice, two types of protein bodies were also described (Tanaka et al., 1980; Krishnan and White, 1995) but both were small-sized and similar to the size of the protein bodies in B. distachyon. The larger vacuoles, which may sometimes fill a large part of the endosperm cells, resembled vacuoles usually found in dicotyledonous cotyledons (Hoh et al., 1995). The presence of two types of storage structures, in addition to the nature of its seed storage proteins, suggests that B. distachyon is closer to rice than to wheat. Nevertheless, at maturity, a continuous proteinaceous matrix was observed as in wheat grains (Bechtel et al., 1991).
Before 15 DAF several enzymes involved in carbohydrate metabolism were identified, among which was a β-amylase with homology to Hordeum β-amylase (BAM2). This enzyme is probably involved in the degradation of starch to maltose (daSilva et al., 2005; Fulton et al., 2008). Another protein homologous to xyloglucan endotransglucosylase/hydrolases (XTH1) was found in abundance before the accumulation of storage proteins. These enzymes are involved in the cleavage and/or rearrangement of xyloglucan backbones in muro. XTHs are thought to play an important role in turgor-driven cell expansion (Eklof and Brumer, 2010; Hernandez-Nistal et al., 2010). The accumulation of this enzyme is surprising because no xyloglucan was detected in developing B. distachyon grain (data not shown). Pettolino et al. (2010) reported the expression of a large number of XET genes in barley. The physiological and biochemical functions of this enzyme in cereal grain require further investigation.
Conclusion
This paper provides a morphological and biochemical description of Brachypodium grain development in the Bd21.3 reference strain. This analysis of grain development indicates that Brachypodium exhibits some significant differences with domesticated cereals. (1-3)(1-4)-β-glucan accumulates during grain development and this cell wall polysaccharide is the main storage carbohydrate at the expense of starch in the mature grain. This feature is unique amongst the small grain cereals examined. The accumulation of storage proteins differs as well and the presence of two types of storage structures, in addition to the nature and content of the grain storage proteins, suggests that B. distachyon is closer to rice than wheat, nevertheless with a different temporal pattern of deposition. Lastly, both lipid content and composition is closer to that of Avena fatua grain. Altogether, these data indicate that type and content of reserve compounds in Brachypodium grain varies and is not specifically close to any domestic cereal.
With a fully sequenced genome, a short life-cycle, and the genetic tools available for mutagenesis/transformation, Brachypodium appears to be a good model for dissecting numerous aspects of grass biology and development. In particular, it could be used to get a better understanding of the metabolic switches responsible for carbon partitioning during grain filling.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Table S1. Full list of proteins identified from SDS-PAGE displayed in Fig. 3. All analyses were performed with a Q-TOF instrument.
Supplementary Table S2. Fatty acid composition and content of B. distachyon grain.
Supplementary Fig.S1. Multiplanar reconstructions of the embryo.
Supplementary Fig. S2. The Brachypodium embryo at 28 DAF; longitudinal section.
Supplementary Fig. S3. Bright field micrograph of cross-sections from a B. distachyon grain at 22 DAF.
Supplementary Fig. S4. Synchrotron UV microspectroscopy (wavelength excitation 275 nm).
Supplementary Fig. S5. DIC (left panel) and immunofluorescence (right panel) micrographs of grain cross-sections from diverse cereals.
Acknowledgments
This work was performed thanks to the protein database produced by the US Department of Energy Joint Genome Institute http://www.jgi.doe.gov/. We would like to acknowledge G Deshayes (UR1268 Biopolymères Interactions Assemblages, F-44300 Nantes, France) for technical assistance for electrophoresis analysis. We are indebted to S Daniel (UR1268 Biopolymères Interactions Assemblages, F-44300 Nantes, France) for his technical support in (1-3)(1-4)-β-D glucan analysis. We are thankful to S Durant for technical assistance with microscopy. We thank Paul Robert for useful discussions and DISCO Beamline (SOLEIL synchrotron, France).
References
- Abrouk M, Murat F, Pont C, et al. Palaeogenomics of plants: synteny-based modelling of extinct ancestors. Trends in Plant Science. 2010;15:479–487. doi: 10.1016/j.tplants.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Allen WM, Patterson DS, Slater TF. A biochemical study of experimental Johne's disease. III. Protein metabolism in sheep and mice. Journal of Comparative Pathology. 1974;84:391–398. doi: 10.1016/0021-9975(74)90014-0. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson WF, Akin DE. Structural and chemical properties of grass lignocelluloses related to conversion for biofuels. Journal of Industrial Microbiology and Biotechnology. 2008;35:355–366. doi: 10.1007/s10295-007-0291-8. [DOI] [PubMed] [Google Scholar]
- Banas A, Debski H, Banas W, et al. Lipids in grain tissues of oat (Avena sativa): differences in content, time of deposition, and fatty acid composition. Journal of Experimental Botany. 2007;58:2463–2470. doi: 10.1093/jxb/erm125. [DOI] [PubMed] [Google Scholar]
- Barron C, Surget A, Rouau X. Relative amounts of tissues in mature wheat (Triticum aestivum L.) grain and their carbohydrate and phenolic acid composition. Journal of Cereal Science. 2007;45:88–96. [Google Scholar]
- Baud S, Boutin JP, Miquel M, Lepiniec L, Rochat C. An integrated overview of seed development in Arabidopsis thaliana ecotype Ws. Plant Physiology and Biochemistry. 2002;40:151–160. [Google Scholar]
- Bechtel DB, Wilson JD, Shewry PR. Immunocytochemical localization of the wheat storage protein triticin in developing endosperm tissue. Cereal Chemistry. 1991;68:573–577. [Google Scholar]
- Bevan MW, Garvin DF, Vogel JP. Brachypodium distachyon genomics for sustainable food and fuel production. Current Opinion in Biotechnology. 2010;21:211–217. doi: 10.1016/j.copbio.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Borisjuk L, Walenta S, Rolletschek H, Mueller-Klieser W, Wobus U, Weber H. Spatial analysis of plant metabolism: sucrose imaging within Vicia faba cotyledons reveals specific developmental patterns. The Plant Journal. 2002;29:521–530. doi: 10.1046/j.1365-313x.2002.01222.x. [DOI] [PubMed] [Google Scholar]
- Borlaug N. Feeding a hungry world. Science. 2007;318:359. doi: 10.1126/science.1151062. [DOI] [PubMed] [Google Scholar]
- Brkljacic J, Grotewold E, Scholl R, et al. Brachypodium as a model for the grasses: today and the future. Plant Physiology. 2011;157:3–13. doi: 10.1104/pp.111.179531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummell DA. Primary cell wall during fruit ripening. New Zealand Journal of Forestry Science. 2006;36:99–111. [Google Scholar]
- Buckeridge MS, Rayon C, Urbanowicz B, Tiné MAS, Carpita NC. Mixed (1→3)(1→4)-β-D-glucans of grasses. Cereal Chemistry. 2004;81:115–127. [Google Scholar]
- Buckeridge MS, Santos HP, Tiné MAS. Mobilisation of storage cell wall polysaccharides in seeds. Plant Physiology and Biochemistry. 2000;38:141–156. [Google Scholar]
- Burcu K, Yordem BK, Conte SS, Ma JF, Yokosho K, Vasques KA, Gopalsamy SN, Walker EL. Brachypodium distachyon as a new model system for understanding iron homeostasis in grasses: phylogenetic and expression analysis of Yellow Stripe-Like (YSL) transporters. Annals of Botany. 2011 doi: 10.1093/aob/mcr200. doi:10.1093/aob/mcr200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charmet G. Wheat domestication: lessons for the future. Comptes Rendus Biologie. 2011;334:212–220. doi: 10.1016/j.crvi.2010.12.013. [DOI] [PubMed] [Google Scholar]
- Cui W, Wood PJ, Blackwell B, Nikiforuk J. Physicochemical properties and structural characterization by two-dimensional NMR spectroscopy of wheat β-D-glucans: comparison with other cereal β-D-glucans. Carbohydrate Research. 2000;35:225–243. [Google Scholar]
- daSilva LL, Taylor JP, Hadlington JL, Hanton SL, Snowden CJ, Fox SJ, Foresti O, Brandizzi F, Denecke J. Receptor salvage from the prevacuolar compartment is essential for efficient vacuolar protein targeting. The Plant Cell. 2005;17:132–148. doi: 10.1105/tpc.104.026351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devouge V, Rogniaux H, Nesi N, Tessier D, Gueguen J, Larre C. Differential proteomic analysis of four near-isogenic Brassica napus varieties bred for their erucic acid and glucosinolate contents. Journal of Proteome Research. 2007;6:1342–1353. doi: 10.1021/pr060450b. [DOI] [PubMed] [Google Scholar]
- Draper J, Mur LAJ, Jenkins G, Ghosh-Biswas GC, Bablak P, Hasterok R, Routledge APM. Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiology. 2001a;127:1539–1555. [PMC free article] [PubMed] [Google Scholar]
- Dubois V, Breton S, Linder M, Fanni J, Parmentier M. Proposition de classement des sources végétales d'acides gras en fonction de leur profil nutritionnel. Oleagineux Corps Gras Lipides. 2008;15:56–75. [Google Scholar]
- Eklof JM, Brumer H. The XTH gene family: an update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiology. 2010;153:456–466. doi: 10.1104/pp.110.156844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Febrer M, Goicoechea JL, Wright J, et al. An integrated physical, genetic and cytogenetic map of Brachypodium distachyon, a model system for grass research. PLoS One. 2010 doi: 10.1371/journal.pone.0013461. 5, e13461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox TC, Guerinot ML. Molecular biology of cation transport in plants. Annual Review of Plant Physiology and Plant Molecular Biology. 1998;49:669–696. doi: 10.1146/annurev.arplant.49.1.669. [DOI] [PubMed] [Google Scholar]
- Fulton DC, Stettler M, Mettler T, et al. Beta-AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active beta-amylases in Arabidopsis chloroplasts. The Plant Cell. 2008;20:1040–1058. doi: 10.1105/tpc.107.056507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliani A, Jamme F, Rouam V, et al. DISCO: a low-energy multipurpose beamline at synchrotron SOLEIL. Journal of Synchrotron Radiation. 2009;16:835–841. doi: 10.1107/S0909049509034049. [DOI] [PubMed] [Google Scholar]
- Guillon F, Bouchet B, Jamme F, Robert P, Quemener B, Barron C, Larre C, Dumas P, Saulnier L. Brachypodium distachyon grain: characterization of endosperm cell walls. Journal of Experimental Botany. 2011;62:1001–1015. doi: 10.1093/jxb/erq332. [DOI] [PubMed] [Google Scholar]
- Gutierrez L, Van Wuytswinkel O, Castelain M, Bellini C. Combined networks regulating seed maturation. Trends in Plant Science. 2007;12:294–300. doi: 10.1016/j.tplants.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Hernandez-Nistal J, Martin I, Labrador E, Dopico B. The immunolocation of XTH1 in embryonic axes during chickpea germination and seedling growth confirms its function in cell elongation and vascular differentiation. Journal of Experimental Botany. 2010;61:4231–4238. doi: 10.1093/jxb/erq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill LM, Morley-Smith ER, Rawsthorne S. Metabolism of sugars in the endosperm of developing seeds of oilseed rape. Plant Physiology. 2003;131:228–236. doi: 10.1104/pp.010868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch G. Cell-wall hemicellulose as mobile carbon stores in non-reproductive plant tissues. Functional Ecology. 2007;21:823–834. [Google Scholar]
- Hoh B, Hinz G, Jeong BK, Robinson DG. Protein storage vacuoles form de novo during pea cotyledon development. Journal of Cell Science. 1995;108:299–310. doi: 10.1242/jcs.108.1.299. [DOI] [PubMed] [Google Scholar]
- Huo N, Vogel JP, Lazo GR, You FM, Ma Y, McMahon S, Dvorak J, Anderson OD, Luo MC, Gu YQ. Structural characterization of Brachypodium genome and its syntenic relationship with rice and wheat. Plant Molecular Biology. 2009;70:47–61. doi: 10.1007/s11103-009-9456-3. [DOI] [PubMed] [Google Scholar]
- International-Brachypodium-Initiative. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463:763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- Izydorczy K, Dexter JE. Barley glucans and arabinoxylans: molecular structure, physicochemical properties, and uses in food products—a review. Food Research International. 2008;41:850–868. [Google Scholar]
- Izydorczy K, Dexter JE, Desjardin RG, Rossnagel BG, Lagassé SL, Hatcher DX. Roller milling of Canadia hull-less b: optimization of roller milling conditions and composition of mill streams. Cereal Chemistry. 2003;80:637–644. [Google Scholar]
- Jamme F, Robert P, Bouchet B, Saulnier L, Dumas P, Guillon F. Aleurone cell walls of wheat grain: high spatial resolution investigation using synchrotron infrared microspectroscopy. Applied Spectroscopy. 2008;62:895–900. doi: 10.1366/000370208785284448. [DOI] [PubMed] [Google Scholar]
- Jamme F, Villette S, Giuliani A, Rouam V, Wien F, Lagarde B, Refregiers M. Synchrotron UV fluorescence microscopy uncovers new probes in cells and tissues. Microscopy and Microanalysis. 2010;16:507–514. doi: 10.1017/S1431927610093852. [DOI] [PubMed] [Google Scholar]
- Krishnan HB, White JA. Morphometric analysis of rice seed protein bodies (implication for a significant contribution of prolamine to the total protein content of rice endosperm) Plant Physiology. 1995;109:1491–1495. doi: 10.1104/pp.109.4.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriz AL. Characterization of embryo globulins encoded by the maize Glb genes. Biochemistry and Genetics. 1989;27:239–251. doi: 10.1007/BF02401804. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larre C, Penninck S, Bouchet B, Lollier V, Tranquet O, Denery-Papini S, Guillon F, Rogniaux H. Brachypodium distachyon grain: identification and subcellular localization of storage proteins. Journal of Experimental Botany. 2010;61:1771–1783. doi: 10.1093/jxb/erq050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudencia-Chingcuanco DL, Vensel WH. Globulins are the main seed storage proteins in. Brachypodium distachyon. Theoretical and Applied Genetics. 2008;117:555–563. doi: 10.1007/s00122-008-0799-y. [DOI] [PubMed] [Google Scholar]
- Le Gall M, Quillien L, Gueguen J, Rogniaux H, Seve B. Identification of dietary and endogenous ileal protein losses in pigs by immunoblotting and mass spectrometry. Journal of Nutrition. 2005;135:1215–1222. doi: 10.1093/jn/135.5.1215. [DOI] [PubMed] [Google Scholar]
- Leonova S, Shelenga T, Hamberg M, Konarev AV, Loskutov I, Carlsson AS. Analysis of oil composition in cultivars and wild species of oat (Avena sp.) Journal of Agricultural and Food Chemistry. 2008;56:7983–7991. doi: 10.1021/jf800761c. [DOI] [PubMed] [Google Scholar]
- Meikle PJ, Hoogenraad NJ, Bonig I, Clarke AE, Stone BA. A (1→3,1→4)-beta-glucan-specific monoclonal antibody and its use in the quantitation and immunocytochemical location of (1→3,1→4)-beta-glucans. The Plant Journal. 1994;5:1–9. doi: 10.1046/j.1365-313x.1994.5010001.x. [DOI] [PubMed] [Google Scholar]
- Morrall P, Briggs DE. Changes in cell wall polysaccharides of germinating barley grains. Phytochemistry. 1978;17:1495–1502. [Google Scholar]
- Obel N, Neumetzler L, Pauly M. Hemicelluloses and cell expansion. In: Verbelen J-P, Vissenberg K, editors. The expanding cell. Plant Cell Monograph. Berlin, Heidelberg: Springer Verlag; 2006. pp. 56–88. [Google Scholar]
- Opanowicz M, Vain P, Draper J, Parker D, Doonan JH. Brachypodium distachyon: making hay with a wild grass. Trends in Plant Science. 2008;13:172–177. doi: 10.1016/j.tplants.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Opanowicz M, Hands P, Betts D, Parker ML, Toole GA, Mills EN, Doonan JH, Drea S. Endosperm development in Brachypodium distachyon. Journal of Experimental Botany. 2011;62:735–748. doi: 10.1093/jxb/erq309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdemir BS, Hernandez P, Filiz E, Budak H. Brachypodium genomics. International Journal of Plant Genomics. 2008;2008:536104. doi: 10.1155/2008/536104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacurar DI, Thordal-Christensen H, Nielsen KK, Lenk I. A high-throughput Agrobacterium-mediated transformation system for the grass model species Brachypodium distachyon L. Transgenic Research. 2008;17:965–975. doi: 10.1007/s11248-007-9159-y. [DOI] [PubMed] [Google Scholar]
- Pettolino FA, Wilson S, Hrmova M, Farkas M, Harver A, Fincher GB, Bacic A. What are gyloglucan-endotransglycosylas enzymes doing in barley? XII cell Wall meeting, Porto. 2010 [Google Scholar]
- Philippe S, Saulnier L, Guillon F. Arabinoxylan and (1→3),(1→4)-beta-glucan deposition in cell walls during wheat endosperm development. Planta. 2006;224:449–461. doi: 10.1007/s00425-005-0209-5. [DOI] [PubMed] [Google Scholar]
- Qi L, Friebe B, Wu J, Gu Y, Qian C, Gill BS. The compact Brachypodium genome conserves centromeric regions of a common ancestor with wheat and rice. Functional and Integrative Genomics. 2010;10:477–492. doi: 10.1007/s10142-010-0190-3. [DOI] [PubMed] [Google Scholar]
- Robert P, Jamme J, Barron C, Bouchet B, Saulnier L, Dumas P, Guillon F. Change in wall composition of transfer and aleurone cells during wheat grain development. Planta. 2011;233:393–406. doi: 10.1007/s00425-010-1306-7. [DOI] [PubMed] [Google Scholar]
- Rogers S, Quatrano R. Morphological staging of wheat caryopsis development. American Journal of Botany. 1983;70:308–311. [Google Scholar]
- Rosset A, Spadola L, Ratib O. OsiriX: an open-source software for navigating in multidimensional DICOM images. Journal of Digital Imaging. 2004;17:205–216. doi: 10.1007/s10278-004-1014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli PA, Larkins BA. The development of endosperm in grasses. Plant Physiology. 2009;149:14–26. doi: 10.1104/pp.108.129437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L. Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. The Plant Journal. 2008;54:608–620. doi: 10.1111/j.1365-313X.2008.03461.x. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Sugimoto T, Ogawa M, Kassai Z. Isolation and characterization of two types of protein bodies in rice endosperm. Agricultural and Biological Chemistry. 1980;44:1633–1639. [Google Scholar]
- Tenaillon MI, Charcosset A. A European perspective on maize history. Comptes Rendus Biologie. 2011;334:221–228. doi: 10.1016/j.crvi.2010.12.015. [DOI] [PubMed] [Google Scholar]
- Thole V, Alves AC, Worland B, Bevan MW, Vain P. A protocol for efficiently retrieving and characterizing flanking sequence tags (FSTs) in Brachypodium distachyon T-DNA insertional mutants. Nature+ Protocols. 2009;4:650–661. doi: 10.1038/nprot.2009.32. [DOI] [PubMed] [Google Scholar]
- Thole V, Worland B, Wright J, Bevan MW, Vain P. Distribution and characterization of more than 1000 T-DNA tags in the genome of Brachypodium distachyon community standard line Bd21. Plant Biotechnology Journal. 2010;8:734–747. doi: 10.1111/j.1467-7652.2010.00518.x. [DOI] [PubMed] [Google Scholar]
- Truernit E, Bauby H, Dubreucq B, Grandjean O, Runions J, Barthelemy J, Palauqui JC. High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. The Plant Cell. 2008;20:1494–1503. doi: 10.1105/tpc.107.056069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ucciani E. Nouveau dictionnaire des huiles végétales. Paris: Lavoisier; 1995. [Google Scholar]
- Wilson SM, Burton RA, Doblin MS, Stone BA, Newbigin EJ, Fincher GB, Bacic A. Temporal and spatial appearance of wall polysaccharides during cellularization of barley (Hordeum vulgare) endosperm. Planta. 2006;224:655–667. doi: 10.1007/s00425-006-0244-x. [DOI] [PubMed] [Google Scholar]
- Vain P, Worland B, Thole V, McKenzie N, Alves SC, Opanowicz M, Fish LJ, Bevan MW, Snape JW. Agrobacterium-mediated transformation of the temperate grass Brachypodium distachyon (genotype Bd21) for T-DNA insertional mutagenesis. Plant Biotechnology Journal. 2008;6:236–245. doi: 10.1111/j.1467-7652.2007.00308.x. [DOI] [PubMed] [Google Scholar]
- Vicente-Carbajosa J, Carbonero P. Seed maturation: developing an intrusive phase to accomplish a quiescent state. International Journal of Developmental Biology. 2005;49:645–651. doi: 10.1387/ijdb.052046jc. [DOI] [PubMed] [Google Scholar]
- Vogel J, Hill T. High-efficiency Agrobacterium-mediated transformation of Brachypodium distachyon inbred line Bd21-3. Plant Cell Reports. 2008;27:471–478. doi: 10.1007/s00299-007-0472-y. [DOI] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Heim U, Sauer N, Wobus U. A role for sugar transporters during seed development: molecular characterization of a hexose and a sucrose carrier in fava bean seeds. The Plant Cell. 1997;9:895–908. doi: 10.1105/tpc.9.6.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.