Abstract
HvCO9 was characterized to elucidate the barley flowering control mechanisms and to investigate the functional diversification of the barley CONSTANS-like (CO-like) genes in flowering. HvCO9 was located on the same chromosome, 1HL, as Ppd-H2 (HvFT3), which is a positive regulator of short-day (SD) flowering. A phylogenetic analysis showed that HvCO9 was located on the same branch of the CO-like gene tree as rice Ghd7 and the barley and wheat VRN2 genes, which are all negative regulators of flowering. High level HvCO9 expressions were observed under SD conditions, whereas its expression levels were quite low under long-day (LD) conditions. HvCO9 expression correlated with HvFT1 and HvFT2 expression under SD conditions, although no clear effect of HvCO9 on HvFT3 expression, or vice versa, under SD conditions was observed. The over-expression of HvCO9 in rice plants produced a remarkable delay in flowering. In transgenic rice, the expression levels of the flowering-related Ehd1 gene, which is a target gene of Ghd7, and its downstream genes were suppressed, causing a delay in flowering. These results suggest that HvCO9 may act as a negative regulator of flowering under non-inductive SD conditions in barley; this activity is similar to that of rice Ghd7 under non-inductive LD conditions, but the functional targets of these genes may be different. Our results indicate that barley has developed its own pathways to control flowering by using homologous genes with modifications for the timing of expression. Further, it is hypothesized that each pathway may target different genes after gene duplication or species diversification.
Keywords: Barley, CO-like genes, flowering, gene expression, HvCO9, negative regulator, photoperiod
Introduction
Flowering is a crucial developmental phase in the life cycle of seed-propagated plants. A regulatory mechanism that responds to environmental changes (such as day length and temperature) provides a sophisticated control for flowering and ensures that flowering occurs under the most appropriate conditions to maximize seed production and reproductive success.
The molecular mechanisms that control the regulation of flowering have been extensively studied using model plants, such as Arabidopsis thaliana and rice (Oryza sativa L.). Arabidopsis is a long-day (LD) plant: LD plants flower when the days become longer than a critical day length (Thomas and Vince-Prue, 1997). In Arabidopsis, the LD-photoperiodic signals that are mediated by CONSTANS (CO), which encodes a zinc-finger transcriptional activator, induce the transcription of FLOWERING LOCUS T (FT) which encodes a mobile flowering signal, called florigen, to promote flowering (Suárez-López et al., 2001; Imaizumi and Kay, 2006). The CO-FT pathway is conserved in rice, a short-day (SD) plant that flowers when the days become shorter than a crucial length (Thomas and Vince-Prue, 1997). In rice, Hd1, an orthologue of CO, induces the transcription of Hd3a, which is an orthologue of FT (Yano et al., 2000; Hayama et al., 2003; Hayama and Coupland, 2004), to promote flowering under SD conditions. It is interesting to note that photoperiodic pathways containing homologous genes are involved in the regulation of flowering in both Arabidopsis and rice; these pathways are conserved across the differentiation of dicots/monocots, even though these two species show different responses to environmental signals, such as LD and SD flowering induction, respectively.
Although the major components of the regulatory flowering pathways are conserved among distantly related plants, each plant species has evolved its own unique mechanisms to induce flowering under optimal conditions. Recently, the functional differentiation of the barley PEBP genes, including FT- and TFL1 (TERMINAL FLOWER 1)-like genes, was reported in flowering regulation (Kikuchi et al., 2009). Some of these genes perform the same function in flowering as their orthologues in Arabidopsis and rice; for others, however, no evidence was found to support their involvement in flowering. HvFT1 is an integrator of the flowering pathway under both LD and SD conditions, whereas the role of HvFT2 is specifically limited to SD conditions. HvFT3 functions indirectly to promote the expression of HvFT1, which results in flowering under non-inductive (SD) conditions. HvFT3 is also a good candidate gene for Ppd-H2, which is a major barley photoperiod-sensitive gene that promotes flowering under SD conditions. These findings suggest that barley has an adaptive mechanism that adjusts flowering according to photoperiodic changes using a combination of different FT-like genes.
CO is a zinc-finger protein that is necessary for inducing FT expression and is a member of a CO-like gene family in the genomes of higher plants (the family has 17 members in Arabidopsis, 16 members in rice, and nine members in barley; Griffiths et al., 2003). This redundancy of the CO-like genes indicates a possible functional diversification in the regulation of flowering, which is similar to that of the FT-like genes. In fact, although rice Hd1, an orthologue of CO, promotes flowering under SD conditions, another rice CO-like gene, Ghd7 (named OsI), acts as a floral repressor under non-inductive (LD) conditions and suppresses the transcription of Ehd1, a floral activator of multiple flowering signals (Doi et al., 2004; Komiya et al., 2008; Xue et al., 2008). In addition to the nine CO-like genes in barley (Griffiths et al., 2003), VRN-H2 (Yan et al., 2004; Trevaskis et al., 2006) is also included in the CO-like gene family (Greenup et al., 2009). VRN-H2 is a floral repressor in the vernalization response in barley, and this locus consists of three homologous CO-like genes, HvZCCT-Ha, HvZCCT-Hb, and HvZCCT-Hc (Dubcovsky et al., 2005; Karsai et al., 2005).
In this study, the expression pattern of one of the CO-like genes in barley, HvCO9, was analysed under various photoperiodic conditions, and transgenic rice plants that over-expressed this gene were characterized to reveal its functional roles in flowering. The expression pattern of HvCO9 was compared with that of HvCO1, a barley orthologue of Hd1 that promotes flowering (Turner et al., 2005), under different genetic backgrounds with regard to the major photoperiodic response genes, Ppd-H1 and Ppd-H2 (Laurie et al., 1995) to reveal the functional divergence of these two CO-like genes. Furthermore, the functional and evolutionary relationships between HvCO9, rice Ghd7, and barley VRN-H2, which form a distinct clade within the CO-like gene family of cereal plants are discussed (Griffiths et al., 2003; Yan et al., 2004; Dubcovsky et al., 2005). The expression pattern of HvCO9 was compared with those of rice Ghd7 and barley VRN-H2 and the diversification of their possible functional target genes is discussed.
Materials and methods
Plant materials and growth conditions
Two cultivars of barley (Hordeum vulgare), cv. Steptoe and cv. Morex, were used in this study. The doubled haploid (DH) lines that were developed from the F1 cross between Steptoe and Morex have also been used for gene mapping and expression studies (North American Barley Genome Mapping Project; Kleinhofs et al., 1993). The DH lines that were used for the expression analyses of HvCO9 and the FT-like genes are summarized in Table 1. The plants were grown in a growth chamber at 20±2 °C (175 μmol m−2 s−1) under LD (16/8 h light/dark) or SD (12/12 h light/dark) conditions. For the expression analysis, the leaves were harvested in their order of appearance in the middle of the light period. For the diurnal expression analysis, the leaves of plants that were at the two- and three-leaf stages were harvested every 4 h for 2 d under LD and SD conditions.
Table 1.
The DH lines that were used to identify the relationship between HvCO9 and the FT-like genes under SD conditions
| DH line | Genotypesa |
||||
| VRN-H1 | Ppd-H1 | Ppd-H2 | HvCO9 | ||
| (HvFT3) | |||||
| S/M-130 | S | M | M | M | |
| S/M-144 | M | S | M | M | |
| S/M-32 | M | M | S | M | |
| S/M-136 | S | M | M | S | |
| S/M-72 | M | S | M | S | |
| S/M-148 | M | M | S | S | |
| S/M-5 | S | S | S | S | |
| Steptoe | S (VRN1-4b) | S (Ppd-H1) | S (ppd-H2) | S | |
| Morex | M (VRN1-1b) | M (ppd-H1) | M (Ppd-H2) | M | |
S indicates the Steptoe-type genotype and M indicates the Morex-type genotype.
Based on the classification by Hemming et al. (2009); and both genotypes are of the early-heading type without vernalization.
Isolation of HvCO9
Partial sequences of HvCO9 had been reported previously under the accession numbers AY082965 (Griffiths et al., 2003) and FJ767842 and FJ 767852 (Cockram et al., 2010). To isolate the entire coding region of HvCO9, total RNA was extracted from the leaves of cv. Morex using the RNeasy Plant Mini Kit (Qiagen, Germany). To isolate the downstream region of the coding region, the 3′ RACE System for the Rapid Amplification of cDNA Ends (Invitrogen, CA, USA) was used. The primary PCR was performed using a gene-specific primer, HvCO9-F1, and a universal amplification primer (UAP) and was followed by a secondary PCR using the HvCO9-F2 and UAP primer pair.
The first-strand cDNA was synthesized using the TaKaRa RNA PCR Kit (AMV) version 3.0 (Takara Bio, Japan). To isolate the region that was further upstream of part of the coding region, an in silico search was performed of a sequence database that contained full-length barley cDNA libraries that were constructed from the mixed cDNAs of various tissues of the Japanese two-row cultivar, Haruna-Nijo (Matsumoto et al., 2011). One entry, NIASHv1066B04, was identified and, using this information, forward primers were designed to isolate the entire coding region of HvCO9. Lastly, the genomic or cDNA sequences that covered the entire coding region of HvCO9 were amplified using Morex genomic DNA or cDNA as the templates and the HvCO9-F3 and HvCO9-R1 and the HvCO9-F4 and HvCO9-R2 primer pairs for the nested PCR technique. To clone the promoter region of HvCO9, the Morex genome sequence database in the Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) in Germany (http://webblast.ipk-gatersleben.de/barley/indext.php; Mayer et al., 2011) was screened using our HvCO9 sequence as a query. The promoter region was amplified by PCR using a primer pair that consisted of HvCO9P-F and HvCO9-R. The descriptions of the primers that were used in this study are summarized in Table 2.
Table 2.
Sequences of the primers that were used in this study for the isolation, genetic mapping, plasmid construction, and quantitative RT-PCR of HvCO9 and the expression analysis of the transgenic rice plants
| Primer name | Sequence (5′–3′) | Reference |
| For 3′ RACE | ||
| HvCO9-F1 | AAGCTGATGCGGTACAAAGAGA | This study |
| HvCO9-F2 | GTACAAAGAGAAGCGGAAGAGG | This study |
| For isolation of full-length cDNA and genome | ||
| HvCO9-F3 | AAATCGGCCATCACGTGGGGC | This study |
| HvCO9-R1 | GGCAGCCTCCTACGGCAGCAT | This study |
| HvCO9-F4 | TCACGTGGGGCAAGCTGATG | This study |
| HvCO9-R2 | GCACTACGTAGCGTGCGCGTGT | This study |
| HvCO9P-F | ATGGCAATCCCTACTCCTTACAT | Contig1008434a |
| HvCO9P-R | AGGCAGGAGCAGTCCTCAGA | Contig1008434a |
| For mapping of HvCO9 | ||
| HvCO9-F5 | AGCTGATGCGGTACAAAGAGAAGC | This study |
| HvCO9-R3 | ACCCGACCAAGAAATGATCC | This study |
| For plasmid construction | ||
| HvCO9-F4 | TCACGTGGGGCAAGCTGATG | This study |
| HvCO9-R3 | ACCCGACCAAGAAATGATCC | This study |
| For quantitative RT-PCR | ||
| HvCO9-F1 | AAGCTGATGCGGTACAAAGAGA | This study |
| HvCO9-R4 | GAACCACCCGAGGTCGAG | This study |
| HvCO1-F | GGGGCAGAGCAGGCTGCCTC | AF490468 |
| HvCO1-R | TGGCTTCTCTCTCCTTGGAGC | AF490468 |
| HvFT1-F | ATCTCCACTGGTTGGTGACAGA | DQ898520 |
| HvFT1-R | TTGTAGAGCTCGGCAAAGTCC | DQ898520 |
| HvFT2-F | CCTTCTACACCCTGGTGATGGT | DQ297407 |
| HvFT2-R | CCCTCTGGCAGTTGAAGTAGAC | DQ297407 |
| HvFT3-F | GGTTGTGGCTCATGTTATGC | DQ411319 |
| HvFT3-R | CTACTCCCCTTGAGAACTTTC | DQ411319 |
| Actin-F1 | GCCGTGCTTTCCCTCTATG | AY145451 |
| Actin -R1 | GCTTCTCCTTGATGTCCCTTA | AY145451 |
| For expression analysis of transgenic rice plants | ||
| HvCO9-F1 | AAGCTGATGCGGTACAAAGAGA | This study |
| HvCO9-R4 | GAACCACCCGAGGTCGAG | This study |
| Ehd1-F | TCTGAAGTGCAGCTACAAGGTTAC | AB092507 |
| Ehd1-R | TTTCCAACCATGTTATTGTTCTTG | AB092507 |
| Hd1-F | TGAGTACTTTGATCTTGTCGGGTA | AB041838 |
| Hd1-R | TATCACCGTGCTGTCTGGTACTAT | AB041838 |
| Hd3a-F | AGCTAGCAGCTGCAGCTAGTAAGC | AB052944 |
| Hd3a -R | TGCAGCAGATCGATCGGGATCATC | AB052944 |
| RAP1B-F | CCAGTAATCACAAGTTGCAACCT | AB041020 |
| RAP1B -R | TGCCTCTGAATAACAGATGTTTCAA | AB041020 |
| Actin-F2 | GCCGTGCTCTCCCTGTATG | AY145451 |
| Actin -R2 | GCTTCTCCTTGATGTCCCTTA | AY145451 |
Contig name from the Morex genome sequence database in the IPK (Meyer et al., 2011).
The sequence data for HvCO9 that were obtained in this study were deposited in the DDBJ database under accession numbers AB592331 (cv. Steptoe) and AB592332 (cv. Morex).
Genetic mapping of HvCO9
HvCO9 was mapped in the DH population from the F1 cross between Steptoe and Morex (North American Barley Genome Mapping Project; Kleinhofs et al., 1993) using the cleaved-amplified polymorphic sequence method. The primers are shown in Table 2. The MslI digestion produced fragment sizes of 413 bp for Steptoe, and 304 bp and 109 bp for Morex.
Phylogenetic analysis of the CO-like gene group
A phylogenetic analysis of the amino acid sequence alignment of the CCT (CONSTANS, CO-like, and TOC1) domains of the CO-like genes from barley, wheat, rice (Oryza sativa), and Arabidopsis (Griffiths et al., 2003; this study) was conducted using ClustalX (http://www-igbmc.u-strasbg.fr/BioInfo/ClustalX/Top.html; Thompson et al., 1997) and the Neighbor–Joining method (Saitou and Nei, 1987). A bootstrap analysis for 1000 replicates was performed to provide confidence estimates for the tree topologies using the Neighbor–Joining option in ClustalX. The results were compiled graphically using NJplot (http://pbil.univ-lyon1.fr/software/njplot.html).
Rice transformation and growth conditions for transgenic rice plants
The genomic sequence of HvCO9 was amplified using Morex genomic DNA as a template and gene-specific primers (Table 2), and the amplified fragment was then cloned into the pSTARH302GateA expression vector under the control of the cauliflower mosaic virus (CaMV) 35S promoter (H Ichikawa, H Nakamura, M Hakata, Y Nishizawa, M Kajikawa, unpublished data). The constructed plasmids were used in the Agrobacterium-mediated transformation of the rice plants (Kikuchi et al., 2009).
Fifteen plants (T0 generation) that were transformed with the expression vector containing HvCO9 or a control vector (mock) were transplanted into soil under two types of SD conditions. The first SD condition (SD1) was in a growth chamber at 28 °C during the day and 25 °C at night (9/15 h light/dark; 270 μmol m−2 s−1). The second SD condition (SD2) was in a greenhouse at 28 °C during the day and 24 °C at night under natural light from the middle of September until the end of December (approximately 10.5/13.5 h light/dark). The HvCO9-overexpressing rice plants (line 9) or mock plants of the T1 generations (plants from the seeds of regenerated plants) were sown in a growth chamber under SD conditions at 28 °C during the day and at 25 °C at night (9/15 h light/dark; 270 μmol m−2 s−1) or under LD conditions in a greenhouse at 28 °C during the day and 24 °C at night under natural light from the middle of March until the end of July (approximately 14/10 h light/dark). The number of days that elapsed between the date of transplanting (T0 generations) or sowing (T1 generations) and the appearance of the first panicle was recorded.
Expression analysis
Total RNA was extracted from the leaves using the Get Pure RNA Kit (Dojindo, Japan). The first-strand cDNA was synthesized from 1 μg of each RNA sample in a 20 μl reaction solution using the TaKaRa RNA PCR Kit (AMV) version 3.0 (Takara Bio). For the expression analysis of the flowering-related genes, quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed using gene specific primer pairs for each gene (Table 2). Real-time PCR was carried out using an Mx3000P (Stratagene Products Division, Agilent Technologies, CA, USA) with Brilliant II SYBR Green QPCR Master Mix (Stratagene) according to the manufacturer’s recommendations. A dilution series of the pCR2.1-TOPO vectors (Invitrogen) containing partial fragments of HvCO9 or HvActin and the pTA2 vector (TOYOBO, Japan) that contained a partial fragment of HvCO1 were used to generate the standard curves. The HvCO9 and HvCO1 transcripts were amplified with specific primer pairs (Table 2). The values for HvCO9 and HvCO1 were normalized to HvActin as an internal standard. The real-time PCR results that are presented reflect the results of two independent experiments.
Database analysis for micro-colinearity in the HvCO9 and VRN-H2 regions among members of the grass family
The following sequence resources were used to examine the micro-colinearity among the members of the grass family: (i) the sequence data for the rice genome (RAP-DB, http://rapdb.dna.affrc.go.jp/), the Brachypodium and sorghum genomes (Plant DB provided at the MIPS plant genomics group in the German Research Center for Environmental Health, http://mips.helmholtz-muenchen.de/plant/) and the maize genome (MaizeGDB, http://www.maizegdb.org/) and (ii) mapped DNA marker data (GrainGenes; http://wheat.pw.usda.gov/GG2/index.shtml) (Close et al., 2009; Thiel et al., 2009).
Results
Isolation and characterization of the HvCO9 gene
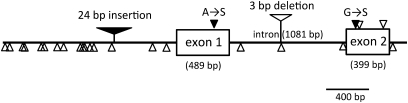
The partial sequences of HvCO9 have previously been reported under the accession numbers, AY082965 (Griffiths et al., 2003) and FJ767842 and FJ 767852 (Cockram et al., 2010). Based on these data, the genomic sequences that covered the entire coding region of HvCO9 from two barley cultivars, Steptoe and Morex, were determined. The former is a late-heading cultivar under field conditions and has the genotype, Ppd-H1/ppd-H2, whereas the latter is an early-heading cultivar that contains the genotype, ppd-H1/Ppd-H2. The genomic sequences of HvCO9 revealed that this gene contains two exons of 489 bp and 399 bp and a single intron (Fig. 1). HvCO9 encodes a protein of 295 amino acids that, similar to other CO-like proteins, contains a CCT domain near its carboxy terminus. The gene structures and the deduced amino acid sequences of HvCO9 were well conserved between the two cultivars, except for two amino acid changes, which are noted in Fig. 1. These results indicate that both Steptoe and Morex contain an intact and functional copy of HvCO9. the Sequences located up to 1.6 kb upstream of the initiation codon of HvCO9 were also obtained from both cultivars; a sequence comparison of the possible promoter regions between Steptoe and Morex revealed the presence of one insertion/deletion of 24 bp in size and several SNPs (Fig. 1).
Fig. 1.
Gene structure of HvCO9 in the barley cultivar Steptoe. The boxes indicate exons. The nucleotide changes that were observed in the Morex cultivar are indicated by the filled and open triangles with and without the amino acid changes, respectively. The filled and open triangles that contain a vertical bar indicate the position of insertions or deletions, respectively, that were found in the Morex HvCO9 gene.
Relationship between HvCO9 and related genes
Using a doubled haploid (DH) population that was derived from the cross between Steptoe and Morex, HvCO9 was mapped to a position that was 9.9 cM proximal to ABC160 and 2.2 cM distal from Glb1 on chromosome 1HL (see Supplementary Fig. S1at JXB online); this is the same chromosome arm on which Ppd-H2 is located, which is nearly consistent with previous mapping studies (Griffiths et al., 2003). Barley chromosome 1H displays partial synteny to segments of rice chromosomes 5 and 10 (Close et al., 2009; Thiel et al., 2009). A phylogenetic analysis of the amino acid sequence alignment of the CCT domains of the CO-like genes from barley, wheat, rice, and Arabidopsis (Griffiths et al., 2003; Karsai et al., 2005; Szücs et al., 2007; this study) revealed that HvCO9 is the most closely related to OsH (Os10g0560400), which is one of the rice CO-like genes, and that these two genes comprised a distinct clade (see Supplementary Fig. S2at JXB online). These results clearly indicate that HvCO9 is the barley orthologue of OsH. This clade includes other cereal CO-like genes, which include OsI (Ghd7) from rice and three HvZCCT genes from the VRN-H2 locus from barley, all of which play repressive roles in flowering (Xue et al., 2008; Greenup et al., 2009). It would be interesting to investigate the functional roles of HvCO9 and OsH in flowering and compare them with OsI and VRN-H2.
The evolutionary relationships between HvCO9 and VRN-H2 was examined to clarify the functional similarities and differences between them. Cockram et al. (2010) revealed that a segment of barley chromosome 1H is duplicated on chromosome 4H and this duplication is conserved in all of the grass family members that have been examined to date. VRN-H2 is located on this collinear duplicated region of the long arm of chromosome 4H (Laurie et al., 1995), and this locus contains three CO-like genes, HvZCCT-Ha, HvZCCT-Hb, and HvZCCT-Hc (Dubcovsky et al., 2005; Karsai et al., 2005). However, no orthologues of the HvZCCTs were identified in these duplicated regions in other grass species, with the exception of wheat (see Supplementary Fig. S3 at JXB online).
Expression of the barley CO-like genes, HvCO9 and HvCO1, during development
To determine whether HvCO9 expression is associated with the photoperiodic flowering response in barley, a quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed using RNA samples extracted from the leaves that were harvested in the middle of the light period from the Steptoe and Morex barley cultivars and the expression of HvCO9 was compared with HvCO1. In barley, the photoperiodic control of flowering depends on two major genes, Ppd-H1 and Ppd-H2, which promote flowering under LD and SD conditions, respectively. Recently, Ppd-H1 and Ppd-H2 have been identified as members of the pseudoresponse regulator family and the FT-like gene family, respectively (Turner et al., 2005; Faure et al., 2007; Kikuchi et al., 2009). Steptoe carries Ppd-H1/ppd-H2 and flowers later than Morex under field conditions (autumn sowing) but earlier than Morex under LD conditions. By contrast, Morex has the ppd-H1/Ppd-H2 genotype and flowers early under field conditions.
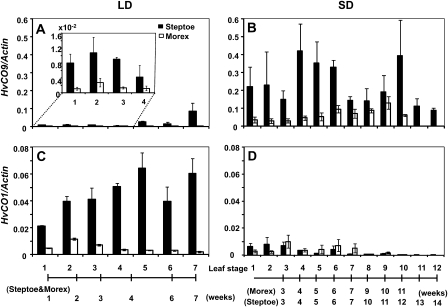
In both Steptoe and Morex, the expression levels of HvCO9 were higher under SD conditions than under LD conditions (Fig. 2A, B), which suggests that the function of HvCO9 is more significant under SD conditions. Varietal differences were also observed for the expression of HvCO9: the expression level of HvCO9 was higher in Steptoe than in Morex (Fig. 2A, B). Similarly, Steptoe displayed a higher level of HvCO1 expression than Morex under LD conditions (Fig. 2C); under SD conditions, however, the level of HvCO1 expression was quite low in both cultivars (Fig. 2D). These results support the hypothesis that the expression of Ppd-H1 under LD conditions in Steptoe increases HvCO1 expression and, thus, promotes flowering. These results are consistent with a previous report (Turner et al., 2005) that showed that the expression of HvCO1 was reduced in ppd-H1 plants.
Fig. 2.
Expression of the barley CO-like genes, HvCO9 (A, B) and HvCO1 (C, D), in Steptoe (Ppd-H1/ppd-H2) and Morex (ppd-H1/Ppd-H2) at each leaf stage under LD conditions (16/8 h light/dark) and SD conditions (12/12 h light/dark), respectively. The inset shows an enlarged graph of the earlier stages. The black bars indicate Steptoe, and the white bars indicate Morex. Each mRNA sample was quantified relative to the HvActin mRNA. The data were standardized with two independent experiments (means ±SE). The numbers at the bottom indicate each developmental stage as a leaf stage, and the time scales show the number of weeks after sowing.
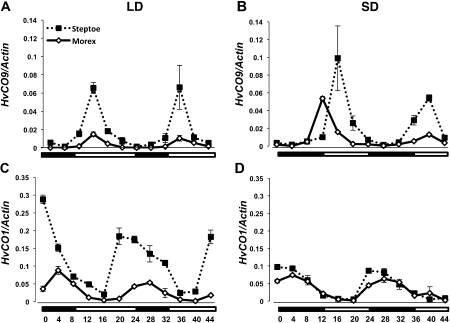
Diurnal oscillation of HvCO9 and HvCO1 expression levels in barley
To investigate the relationship between HvCO9 and HvCO1 in flowering, their diurnal expression patterns were examined by qRT-PCR at the two- and three-leaf stages (during which the shoot apical meristem changes from a vegetative to a reproductive phase in an early flowering variety) under LD and SD conditions. The HvCO9 transcript began to increase in the dark phase and peaked in the early morning under both of the photoperiodic conditions (Fig. 3A, B). This diurnal expression pattern was the same as that of rice Ghd7, although the expression of Ghd7 was much higher under LD conditions than under SD conditions (Xue et al., 2008; Itoh et al., 2010). By contrast, the accumulation of the HvCO1 mRNA was observed at the end of the light phase under LD conditions and at the early dark phase under SD conditions (Fig. 3C, D); nearly identical patterns have been observed for rice Hd1 and wheat WCO1 (Kojima et al., 2002; Shimada et al., 2009). No significant differences in the diurnal expression patterns of HvCO9 and HvCO1 were observed between Steptoe and Morex, although there were differences in the levels of gene expression between these two cultivars (Fig. 3).
Fig. 3.
Diurnal expression of the CO-like genes, HvCO9 (A, B) and HvCO1 (C, D), in Steptoe (Ppd-H1/ppd-H2) and Morex (ppd-H1/Ppd-H2) under LD and SD conditions, respectively. The dotted lines with black squares represent Steptoe and the solid lines with white diamonds represent Morex. The leaves were harvested from the plants at 4 h intervals during the two-leaf and three-leaf stages for 2 d. The vertical axis shows the relative mRNA levels of the CO-like genes that were normalized to HvActin. The mean quantified values ±SE for two independent experiments are shown. The white and black bars at the bottom indicate the light and dark periods, respectively.
The expression of and their relationship between HvCO9 and Ppd-H2 under SD conditions
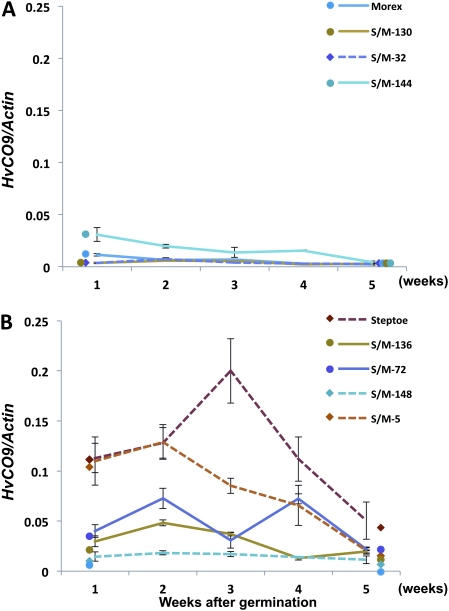
The expression analysis of HvCO9 revealed that HvCO9 expression was under photoperiodic control; therefore, this gene may function preferentially under SD conditions, as mentioned above. Conversely, Ppd-H2 (HvFT3) has been shown to be a positive regulator of flowering under SD conditions (Kikuchi et al., 2009). To identify the relationship between HvCO9 and Ppd-H2 under SD conditions, the expression pattern of HvCO9 was investigated by qRT-PCR using the DH lines, which contained different genotype combinations of HvCO9 and Ppd-H2 and were derived from the F1 cross between Steptoe and Morex (Table 1).
All of the DH lines with the Morex-type HvCO9 (hereafter designated as HvCO9m) that were investigated in this study showed a low level of HvCO9 expression; this level was the same as that of Morex and was irrespective of the Ppd-H2 genotype (Fig. 4A). By contrast, the DH lines that contained the Steptoe-type HvCO9 (hereafter designated as HvCO9s) showed slightly different expression levels than the DH lines that contained HvCO9m (Fig. 4B). Two HvCO9s DH lines that contained Ppd-H2 showed an HvCO9 expression level that was intermediate between that of Morex and Steptoe in the early growth phase; thereafter, the expression level decreased to that of Morex throughout the rest of development. The DH line, S/M-5 (HvCO9s and ppd-H2), exhibited a high level of HvCO9 expression, which was at the same level as Steptoe in the early growth phase. However, the other DH line, S/M-148, which contained the same genotype combination (HvCO9s and ppd-H2) as S/M-5, showed a low HvCO9 expression level that was the same as that in Morex (Fig. 4B).
Fig. 4.
Expression of HvCO9 in the DH lines with different combinations of two genes, HvCO9 and Ppd-H2 (HvFT3), and their parental cultivars, Steptoe and Morex, under SD conditions. (A) The expression patterns of HvCO9 in the DH lines that contained HvCO9m. (B) The expression patterns of HvCO9 in the DH lines that contained HvCO9s. The solid lines indicate the DH line that contained Ppd-H2 and the dotted lines indicate the DH line that contained ppd-H2. Each mRNA was quantified relative to the HvActin mRNA. The data were standardized using two independent experiments (means ± SE).
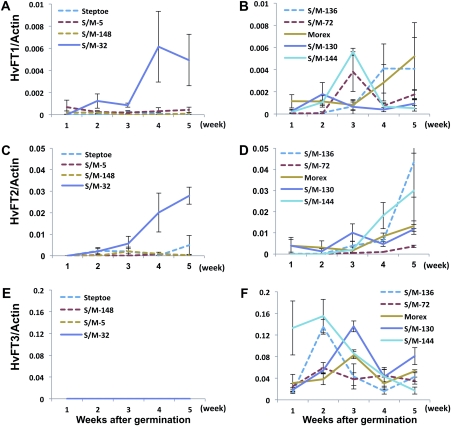
The effect of the HvCO9 genotype on the expression of three FT-like genes, HvFT1, HvFT2, and HvFT3 (Ppd-H2), was also investigated under SD conditions (Fig. 5). No expression of HvFT3 was detected in the DH lines with a Steptoe-type HvFT3 (hereafter designated as HvFT3s) (Fig. 5E). This observation is consistent with our previously published data, in which Steptoe had lost most of the HvFT3 gene, rendering it functionless (Kikuchi et al., 2009). The expression of HvFT3 was observed in the DH lines that contained the Morex-type HvFT3 (hereafter designated as HvFT3m) although there was no obvious difference in the HvFT3 expression patterns between the DH lines that contained HvCO9s (dotted lines) or HvCO9m (solid lines) (Fig. 5F). For the expression levels of HvFT1 and HvFT2, correlations with the HvCO9 genotype were observed in the DH lines that contained HvFT3s; the expression levels of these two FT-like genes in the DH line that contained HvCO9m (S/M-32) were higher than those in the DH line that contained HvCO9s (Fig. 5A, C). In the DH lines that contained HvFT3m, there were no clear differences in the HvFT1 and HvFT2 expression patterns between the DH lines that contained HvCO9s (dotted lines) or HvCO9m (solid lines) (Fig. 5B, D).
Fig. 5.
Expression of HvFT1 (A, B), HvFT2 (C, D), and HvFT3 (Ppd-H2) (E, F) in the DH lines with different combinations of two genes, HvCO9 and Ppd-H2 (HvFT3) and their parental cultivars, Steptoe and Morex. (A, C, E) The expression patterns in the DH lines that contained ppd-H2. (B, D, F) The expression patterns in the DH lines that contained Ppd-H2. The solid lines indicate the DH line that contained HvCO9m and the dotted lines indicate the DH line that contained HvCO9s. Each mRNA was quantified relative to the HvActin mRNA. The data were standardized using two independent experiments (means ±SE).
This observation led to the following hypothesis: Ppd-H2 (HvFT3) and HvCO9 are not directly connected in the flowering pathway under SD conditions, but under limited conditions, they interact indirectly with other associated factor(s) or interact directly with each other. However, other FT-like genes, such as HvFT1 and HvFT2, are probably located downstream of HvCO9, and it is possible that HvCO9 affects their expression levels directly or indirectly.
Over-expression of HvCO9 in rice plants
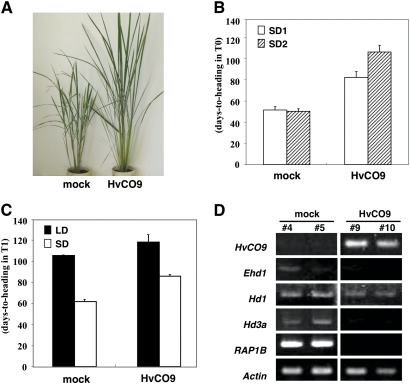
To investigate the function of HvCO9 in flowering, HvCO9, which was under the control of the CaMV 35S promoter, was introduced into rice plants. The T0 generation of the resulting transgenic rice showed a late flowering phenotype under SD conditions compared with the control plants (Fig. 6A, B). The T1 generation of HvCO9-over-expressing line 9 also exhibited significantly late flowering under SD conditions. Late flowering was also observed under LD conditions, but the difference between the transgenic and the control plants was smaller than under the SD conditions (Fig. 6C).
Fig. 6.
The phenotype (A), flowering time (B, C), and expression pattern of flowering-related genes (D) in transgenic rice plants that over-expressed HvCO9. (A) The photographs show a transgenic plant (HvCO9) and a control plant (mock) at the heading stage under SD conditions. (B) The average number of days to heading ±SE of 15 transgenic T0 plants under two SD conditions; the white bars and bars with diagonal lines indicate the SD1 and SD2 conditions, respectively. Both of the conditions are described in the Materials and methods. (C) The average number of days to heading ±SE of the T1 generations of transgenic line 9 under LD (black bars) and SD conditions (white bars). (D) The expression of flowering-related genes in the transgenic rice plants and mock controls. The leaves were harvested 5 weeks after sowing under LD conditions. Mock indicates the transgenic plant that contained only the vector construct (negative control).
The expression profiles of several rice genes that are related to flowering, including Ehd1, Hd1, Hd3a, and RAP1B, were examined by RT-PCR using two HvCO9-overexpressing lines (lines 9 and 10) and two lines of control plants (mock, lines 4 and 5) under LD conditions. The days to heading for the control plants, lines 4 and 5, and the transgenic plants, lines 9 and 10, were 30, 46, 85, and 62 d, respectively. Ehd1, Hd3a, and RAP1B were not expressed at detectable levels in the transgenic lines (Fig. 6D), which indicated that the heading delay of the HvCO9-overexpressing plants may be due to the suppression of Ehd1 and its downstream genes. The expression profile that was observed in the HvCO9-overexpressing plants was the same as that of Ghd7, which acts as an LD-specific floral repressor in rice (Xue et al., 2008).
Discussion
The CONSTANS-FLOWERING LOCUS T (CO-FT) interaction is a key pathway in the photoperiodic regulation of flowering and is conserved among distantly related plants (Greenup et al., 2009; Yanovsky and Kay, 2003).
In this study, the focus was on HvCO9, a barley CO-like gene, rather than HvCO1, in order to clarify the functional role of the CO-like gene family in barley. The previous phylogenetic analyses revealed that HvCO9 was located in a subgroup with two related rice genes, OsH and OsI (Ghd7), and Triticeae VRN2, which is a floral repressor of the vernalization response (Griffiths et al., 2003; Yan et al., 2004; Hemming et al., 2008; Greenup et al., 2009). However, this subfamily does not have an Arabidopsis counterpart; in other words, it is a grass species-specific CO-like subfamily. Furthermore, two genes in this subfamily, rice Ghd7 and Triticeae VRN2, have been identified as floral repressors (Xue et al., 2008; Greenup et al., 2009). Our analysis using transgenic HvCO9-over-expressing rice plants suggests that HvCO9 has the same function as Ghd7, namely, the negative regulation of flowering (Fig. 6). It is interesting that grass species have developed systems for flowering repression that are different from those of Arabidopsis, which contains its own CO-like gene subfamily.
Our transgenic studies revealed the possible regulatory targets of HvCO9 in the photoperiodic gene pathway. The over-expression of HvCO9 in rice under LD conditions apparently suppresses the expression of Ehd1 and such downstream genes as Hd3a, a rice FT orthologue, but it did not affect Hd1 expression (Fig. 6D). This observation is similar to the case of Ghd7, which has been shown to suppress Ehd1 specifically under LD conditions (Xue et al., 2008). From these results, there is speculation that a possible function of HvCO9 in barley is the suppression of the function of an Ehd1-like gene and that HvCO9 acts as a floral suppressor. However, a fundamental issue with this hypothesis is that Ehd1 is unique to rice, and to date, no obvious orthologous genes have been identified in Arabidopsis or temperate cereals, including barley (Doi et al., 2004; Greenup et al., 2009; Higgins et al., 2010). It will be of great interest to identify the target gene of HvCO9 in barley and to determine how the pathway containing HvCO9 functions for the regulation of flowering in barley.
To characterize the function of HvCO9 further, an extensive expression analyses was conducted of HvCO9. The co-ordinated expression analyses of HvCO9 and HvCO1 revealed that these two CO-like genes displayed contrasting expression patterns. HvCO9 was highly expressed under SD conditions and showed very low expression under LD conditions (Fig. 2A, B). Under SD conditions, Morex, a cultivar that contains Ppd-H2, showed a lower expression level than Steptoe, a cultivar that contains ppd-H2 (Fig. 2B). By contrast, high HvCO1 expression levels were observed under LD conditions, and the expression levels were extremely low under SD conditions (Fig. 2C, D). Under LD conditions, Steptoe (with Ppd-H1) showed a higher expression of HvCO1 than Morex (with ppd-H1) (Fig. 2C). The diurnal expression patterns of HvCO9 and HvCO1 were conserved under both LD and SD conditions, and these diurnal expression patterns were similar in the two cultivars, although the expression levels were different (Fig. 3). The analysis of HvCO9 and HvCO1 expression using two barley cultivars that contain different combinations of photoperiod-sensitive genes suggests that these two CO-like genes act in different photoperiodic response signalling pathways: HvCO9 acts in the SD signalling pathway, and HvCO1 acts in the LD pathway. In addition, these two genes have opposite effects on flowering: HvCO9 acts as a repressor, and HvCO1 acts as an inducer.
If HvCO9 is situated in the SD signalling pathway, then it would be interesting to investigate the relationship between HvCO9 and Ppd-H2, which is the SD signal mediator that promotes flowering. Our previous study (Kikuchi et al., 2009) revealed that the Morex cultivar contains an active form of Ppd-H2 (HvFT3m) that promotes flowering under SD conditions, and it also revealed that ppd-H2 in Steptoe is a truncated form of HvFT3 (HvFT3s) and does not produce a functional protein. Using the DH lines with different genotype combinations of HvCO9 and Ppd-H2, the influence of the Ppd-H2 genotype on the expression of HvCO9 was investigated (Fig. 4). The impact of Ppd-H2 on the expression of HvCO9 differed with respect to the genotype of HvCO9. All of the DH lines that contained HvCO9m showed a low level of expression, regardless of the Ppd-H2 genotype (Fig. 4A). However, the HvCO9 expression in the DH lines that contained HvCO9s was affected by the Ppd-H2 genotype, in which a higher expression under ppd-H2 and a lower expression under Ppd-H2 were observed (Fig. 4B). These results indicate that the regulation of HvCO9 expression differs by cultivar and that a clear hierarchical relationship does not exist between HvCO9 and Ppd-H2. One large insertion/deletion and several SNPs were identified in the possible promoter region (up to 1.6 kb upstream of the HvCO9 initiation codon) between Steptoe and Morex (Fig. 1). Database searches (PlantPAN, http://plantpan.mbc.nctu.edu.tw/index.php; Chang et al., 2008) using the upstream sequence of Morex revealed that the 24 bp insertion/deletion region contained a GAMYB or Opaque-2 motif, both of which are described as cis-regulatory elements for seed development. These sequence variations may cause differences in expression, although the functional roles of these sequence variations in the regulation of HvCO9 expression could not be specified.
VRN-H2 (HvZCCTs) is the gene that is most closely related to HvCO9. Both HvCO9 and VRN-H2 have a common function as a repressor of flowering, that is, they act as floral repressors. VRN-H2 expression is clearly regulated in a daylength-dependent manner in the vernalization-responsive accessions: it is highly expressed in the leaves under LD conditions, but the expression is very low or absent under SD conditions (Trevaskis et al., 2006). This finding is completely opposite to the photoperiodic pattern of HvCO9 expression (Fig. 2). The over-expression of VRN-H2 in transgenic barley plants delays flowering by 4 weeks, compared with the controls (Hemming et al., 2008). HvFT1 expression was significantly lower in the transgenic plants than in the control plants, but the expression levels of VRN-H1, a key gene for floral induction in the vernalization pathway, did not differ between the plants that were over-expressing VRN-H2 and the control lines (Hemming et al., 2008). The effects on flowering by the over-expression of both HvCO9 and VRN-H2 were nearly same, although the plant materials that were used in the transgenic studies were different: rice was used for HvCO9, and barley was used for VRN-H2. These results suggest that these two genes encode proteins that share a common function in repressing flowering and that their expression is regulated by day length, although the response to the daylength condition is opposite for the two genes.
VRN2 is located on barley chromosome 4H and the wheat homoeologous chromosome group 5 (Laurie et al., 1995; Dubcovsky et al., 1998). The surrounding regions of VRN-H2 originated from the duplication of the HvCO9 region on barley 1H (see Supplementary Figs S1 and S2 at JXB online), and this duplication has been observed in many species of grass, suggesting that the duplication occurred in the common ancestor to all grass species (Cockram et al., 2010; see Supplementary Fig. S4 at JXB online). However, the VRN2 gene is specific to the Triticeae, and no VRN2 homologues were identified in other species. Cockram et al. (2010) suggested that several deletions of the VRN2 locus have occurred during the evolution of the grass family, but it is more likely that the VRN2 locus was created from the targeted duplication of HvCO9 to the homologous region after the divergence of Triticeae. If so, the diversification of the regulation of gene expression in response to different daylengths is estimated to occur in a short time after the gene duplication.
Based on the results for the relationship between HvCO9 and VRN-H2, it can be hypothesized that the functional targets of these genes may be the same. As shown in Fig. 6, the over-expression of HvCO9 in rice clearly suppresses rice Ehd1, which is a key activator for multiple flowering signal pathways; however, no orthologueous gene has been found in the barley genome thus far (Greenup et al., 2009; Higgins et al., 2010). Conversely, the target gene of VRN-H2 has been postulated to be HvFT1 (by a transgenic study) (Hemming et al., 2008) or HvFT3 (by an extensive expression analysis) (Casao et al., 2011). These data led us to postulate that the target gene of HvCO9 may be an FT-like gene, as has been found for VRN-H2. Our previous study has revealed that two FT-like genes, HvFT1 and HvFT2, function together in floral induction under SD conditions (Kikuchi et al., 2009). Therefore, the influence of HvCO9 on the expression of these FT-like genes in barley was investigated (Fig. 5). Because Ppd-H2 had a strong effect under SD conditions on the regulation of flowering-related genes which included FT-like genes, the DH lines that contained ppd-H2 (HvFT3s) were used for the expression analyses of the FT-like genes to avoid the influence of Ppd-H2. As shown in Fig. 5A and C, the expression levels of HvFT1 and HvFT2 were correlated with the HvCO9 genotypes, which suggests that the possible targets of HvCO9 under SD conditions are FT-like genes rather than an Ehd1-homologue, a case that is similar to VRN-H2 under LD conditions. As a limited number of lines were used for our expression analysis in the present study, it will be interesting to investigate the relationship between the genotypes and the gene expression levels in future studies using additional lines or different populations.
Gene duplication and any ensuing functional diversification are thought to be the major driving forces for each plant species to construct its own unique mechanisms for flowering. In addition to the case of the CO-like genes in barley that are reported in the present study, an example in rice has previously been published. Rice has two related FT-like genes, Hd3a and RFT1, which were derived from a local gene duplication after the diversification of rice from the other cereal species (Komiya et al., 2008, 2009). Both of the FT-like genes function as major floral activators, but Hd3a is specific to SD conditions, whereas RFT1 is LD-specific. These results suggest that, although each plant species is dependent on commonly conserved functions, they have evolved their own unique mechanisms by integrating specific factors with a duplicated origin in their individual system.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1.Chromosomal location of HvCO9 and the comparative mapping of the HvCO9 region of barley chromosome 1H with rice chromosomes 5 and 10.
Supplementary Fig. S2. Phylogenetic tree of the CO-like proteins from barley, wheat, rice, and Arabidopsis.
Supplementary Fig. S3. (a–d) Micro-collinearity among the Poaceae at the VRN2 locus and the OsH region.
Supplementary Fig. S4. Putative evolutionary history of the duplicated region, including the orthologueues of HvCO9 in the grass family.
Acknowledgments
We thank Dr Takuji Tonooka for his valuable suggestions and for providing the plant materials. We also thank Drs Takashi Matsumoto and Hiroaki Ichikawa for providing the information regarding the full-length barley cDNA library and the pSTARH302GateA expression vector, respectively. This work was partly supported by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Integrated research project for plant, insect, and animal using genome technology GD-3005).
References
- Casao MC, Igartua E, Karsai I, Lasa JM, Gracia MP, Casas AM. Expression analysis of vernalization and day-length response genes in barley (Hordeum vulgare L.) indicates that VRNH2 is a repressor of PPDH2 (HvFT3) under long days. Journal of Experimental Botany. 2011;62:1939–1949. doi: 10.1093/jxb/erq382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang WC, Lee TY, Huang HD, Huang HY, Pan RL. PlantPAN: plant promoter analysis navigator, for identifying combinatorial cis-regulatory elements with distance constraint in plant gene group. BMC Genomics. 2008;9:561. doi: 10.1186/1471-2164-9-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close TJ, Bhat PR, Lonardi S, et al. Development and implementation of high-throughput SNP genotyping in barley. BMC Genomics. 2009;10:582. doi: 10.1186/1471-2164-10-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockram J, Howells RM, O’Sullivan DM. Segmental chromosomal duplications harboring group IV CONSTANS-like genes in cereals. Genome. 2010;53:231–240. doi: 10.1139/g09-101. [DOI] [PubMed] [Google Scholar]
- Doi K, Izawa T, Fuse T, Yamonouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A. Ehd1a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes and Development. 2004;18:926–936. doi: 10.1101/gad.1189604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J, Chen C, Yan L. Molecular characterization of the allelic variation at the VRN-H2 vernalization locus in barley. Molecular Breeding. 2005;15:359–407. [Google Scholar]
- Dubcovsky J, Lijavetzky D, Appendino L, Tranquilli G. Comparative RFLP mapping of Triticum monococcum genes controlling vernalization requirement. Theoretical and Applied Genetics. 1998;97:968–975. [Google Scholar]
- Faure S, Higgins J, Turner A, Laurie DA. The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare) Genetics. 2007;176:599–609. doi: 10.1534/genetics.106.069500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenup A, Peacock WJ, Dennis ES, Trevaskis B. The molecular biology of seasonal flowering-response in Arabidopsis and the cereals. Annals of Botany. 2009;103:1165–1172. doi: 10.1093/aob/mcp063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths S, Dunford RP, Coupland G, Laurie DA. The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiology. 2003;131:1855–1867. doi: 10.1104/pp.102.016188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R, Coupland G. The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and rice. Plant Physiology. 2004;135:677–684. doi: 10.1104/pp.104.042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K. Adaptation of photoperiodic control pathway produces short-day flowering in rice. Nature. 2003;422:719–722. doi: 10.1038/nature01549. [DOI] [PubMed] [Google Scholar]
- Hemming MN, Fieg S, Peacock WJ, Dennis ES, Trevaskis B. Regions associated with repression of the barley (Hordeum vulgare) VERNALIZATION1 gene are not required for cold induction. Molecular Genetics and Genomics. 2009;282:107–117. doi: 10.1007/s00438-009-0449-3. [DOI] [PubMed] [Google Scholar]
- Hemming MN, Peacock WJ, Dennis ES, Trevaskis B. Low-temperature and daylength cues are integrated to regulate FLOWERING LOCUS T in barley. Plant Physiology. 2008;147:355–366. doi: 10.1104/pp.108.116418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JA, Bailey PC, Laurie DA. Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. PLoS one. 2010 doi: 10.1371/journal.pone.0010065. 5, e10065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Kay SA. Photoperiodic control of flowering: not only by coincidence. Trends in Plant Science. 2006;11:550–558. doi: 10.1016/j.tplants.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Itoh H, Nonoue Y, Yano M, Izawa T. A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nature Genetics. 2010;42:635–638. doi: 10.1038/ng.606. [DOI] [PubMed] [Google Scholar]
- Karsai I, Szücs P, Mészáros K, Filichkina T, Hayes PM, Skinner JS, Láng L, Bedő Z. The VRN-H2 locus is a facultative x winter growth habit barley (Hordeum vulgare L.) mapping population. Theoretical and Applied Genetics. 2005;110:1458–1466. doi: 10.1007/s00122-005-1979-7. [DOI] [PubMed] [Google Scholar]
- Kikuchi R, Kawahigashi H, Ando T, Tonooka T, Handa H. Molecular and functional characterization of PEBP genes in barley reveal the diversification of their roles in flowering. Plant Physiology. 2009;149:1341–1353. doi: 10.1104/pp.108.132134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinhofs A, Kilian A, Saghai Maroof MA, et al. A molecular, isozyme and morphological map of the barley (Hordeum vulgare) genome. Theoretical and Applied Genetics. 1993;86:705–712. doi: 10.1007/BF00222660. [DOI] [PubMed] [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant and Cell Physiology. 2002;43:1096–1105. doi: 10.1093/pcp/pcf156. [DOI] [PubMed] [Google Scholar]
- Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K. Hd3a and RFT1 are essential for flowering in rice. Development. 2008;135:767–774. doi: 10.1242/dev.008631. [DOI] [PubMed] [Google Scholar]
- Komiya R, Yokoi S, Shimamoto K. A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development. 2009;136:3443–3450. doi: 10.1242/dev.040170. [DOI] [PubMed] [Google Scholar]
- Laurie DA, Pratchett N, Bezant JH, Snape JW. RFLP mapped of five major genes and eight quantitative trait loci controlling flowering time in a winter×spring barley (Hordeum vulgare L.) cross. Genome. 1995;38:575–585. doi: 10.1139/g95-074. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Tanaka T, Sakai H, et al. Comprehensive sequence analysis of 24 783 barley full-length cDNAs derived from twelve clone libraries. Plant Physiology. 2011;156:20–28. doi: 10.1104/pp.110.171579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KFX, Martis M, Hedley PE, et al. Unlocking the barley genome by chromosomal and comparative genomics. The Plant Cell. 2011;23:1249–1263. doi: 10.1105/tpc.110.082537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei N. A Neighbor–Joining method: a new method for constructing phylogenetic tree. Molecular Biology and Evolution. 1987;44:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shimada S, Ogawa T, Kitagawa S, et al. A genetic network of flowering-time genes in wheat leaves, in which an APETALA1/FRUITFULL-like gene, VRN1, is upstream of FLOWERING LOCUS T. The. Plant Journal. 2009;58:668–681. doi: 10.1111/j.1365-313X.2009.03806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410:1116–1120. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- Szücs P, Skinner JS, Karsai I, Cuesta-Marcos A, Haggard KG, Corey AE, Chen THH, Hayes PM. Validation of the VRN-H2/VRN-H1 epistatic model in barley reveals that intron length in VRN-H1 may account or a continuum of vernalization sensitivity. Molecular Genetics and Genomics. 2007;277:249–261. doi: 10.1007/s00438-006-0195-8. [DOI] [PubMed] [Google Scholar]
- Thiel T, Graner A, Waugh R, Grosse I, Close TJ, Stein N. Evidence and evolutionary analysis of ancient whole-genome duplication in barley predating the divergence from rice. BMC Evolutionary Biology. 2009;9:209. doi: 10.1186/1471-2148-9-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B, Vince-Prue D. Photoperiodism in plants. 2nd edn. San Diego: Academic Press Inc; 1997. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL-X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–7882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B, Hemming MN, Peacock WJ, Dennis ES. HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiology. 2006;140:1397–1405. doi: 10.1104/pp.105.073486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, DunfordRP Laurie DA. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science. 2005;310:1031–1034. doi: 10.1126/science.1117619. [DOI] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weng X, et al. Natural variation in Ghd7 is as important regulator of heading date and yield potential in rice. Nature Genetics. 2008;40:761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dobcovsky J. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science. 2004;303:1640–1644. doi: 10.1126/science.1094305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, et al. Hd1a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. The Plant Cell. 2000;12:2473–2483. doi: 10.1105/tpc.12.12.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA. Living by the calendar: how plants know when to flower. Nature Reviews Molecular Cell Biology. 2003;4:265–276. doi: 10.1038/nrm1077. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.