Abstract
The phosphoinositol pathway is one of the major eukaryotic signalling pathways. The metabolite of the phosphoinositol pathway, inositol- (1,4,5) trisphosphate (InsP3), is a regulator of plant responses to a wide variety of stresses, including light, drought, cold, and salinity. It was found that the expression of InsP 5-ptase, the enzyme that hydrolyses InsP3, also dramatically affects the levels of inositol phosphate metabolites and the secondary metabolites in transgenic tomato plants. Tomato plants expressing InsP 5-ptase exhibited a reduction in the levels of several important inositol phosphates, including InsP1, InsP2, InsP3, and InsP4. Reduced levels of inositol phosphates accompanied an increase in the accumulation of phenylpropanoids (rutin, chlorogenic acid) and ascorbic acid (vitamin C) in the transgenic fruits of tomato plants. The enhanced accumulation of these metabolites in transgenic tomato plants was in direct correspondence with the observed up-regulation of the genes that express the key enzymes of ascorbic acid metabolism (myo-inositol oxygenase, MIOX; L-galactono-γ-lactone dehydrogenase, GLDH) and phenylpropanoid metabolism (chalcone synthase, CHS1; cinnamoyl-CoA shikimate/quinate transferase, HCT). To understand the molecular links between the activation of different branches of plant metabolism and InsP3 reduction in tomato fruits, the expression of transcription factors known to be involved in light signalling was analysed by real-time RT-PCR. The expression of LeHY5, SIMYB12, and LeELIP was found to be higher in fruits expressing InsP 5-ptase. These results suggest possible interconnections between phosphoinositol metabolism, light signalling, and secondary metabolism in plants. Our study also revealed the biotechnological potential for the genetic improvement of crop plants by the manipulation of the phosphoinositol pathway.
Keywords: Ascorbic acid, LeHY5 transcriptional factor, light signaling, phenylpropanoids, phosphoinositols
Introduction
The discovery of correlations between the stress-signalling pathways and different branches of secondary metabolism is one of the most exciting areas of modern plant biology. The identification of connections between secondary metabolism and stress-signal transduction will not only shed light on the complex biochemical network in plant cells but could also open new perspectives for the genetic improvement of crop plants towards higher nutraceutical value. Light signalling plays an important role in the biosynthesis of various secondary metabolites, including carotenoids, alkaloids, and phenylpropanoids (Mancinelli, 1985; Dixon and Palva, 1995; Vazques-Flota and De Luca, 1998; Hemm et al., 2004; Liu et al., 2004). Many plant secondary metabolites also act as protectors against various environmental stresses including high light. It has been demonstrated that the UV-absorbing characteristics of flavonoids are responsible for their role in UV protection (Winkel-Shirley, 2002). Experiments have shown that many phenylpropanoid genes are light-inducible and that the exposure of plants to light can lead to a higher accumulation of phenylpropanoids in plant tissues (Hemm et al., 2004). As an example, expression of chalcone synthase (CHS), a key enzyme in the phenylpropanoid biosynthetic pathway, can be induced by UV and blue light (Jenkins et al., 2001). Major plant stress signal transduction pathways can be activated and/or regulated by light. It has been documented that the phosphoinositol metabolic pathway is connected with light signalling in plant cells (Salinas-Mondragon et al., 2010). Chen et al. (2008) reported that inositol polyphosphate 5-phosphatase (5ptase13), a key enzyme of the phosphoinositol pathway, is involved in the blue light responses in Arabidopsis thaliana. These studies demonstrated the existence of cross-talk between PHOT1 and 5ptase13 through the regulation of calcium under blue light.
In a previous paper, it was shown that the genetic reduction of inositol triphosphate (InsP3), a major second messenger of the phosphoinositol signalling pathway, through over-expression of the mammalian InsP 5-ptase gene, leads to a significant increase of lycopene in transgenic tomato fruits (Khodakovskaya et al., 2010). It is hypothesized here that the observed increase in lycopene content in InsP 5-ptase-expressing tomatoes is due to the role of InsP3 in mediating light-regulated processes in plants. One of the most successful metabolic engineering approaches to increase the carotenoid and flavonoid contents in tomato fruit was the RNAi suppression of the DET1 transcription factor, a key repressor of several signal-transduction pathways controlled by light (Davuluri et al., 2004; Dixon, 2005). These approaches suggest that genes encoding components of light-signal transduction machinery influence fruit pigmentation and represent genetic tools for the manipulation of fruit quality and nutritional value. Liu et al. (2004) demonstrated that two tomato light signal transduction genes, LeHY5 and LeCOPLIKE, play the role of positive and negative regulators of fruit pigmentation, respectively. LeHY5 transcription factor is able to bind the promoters of light-inducible genes such as CHS (Hardtke et al., 2000). Expression of the HY5 transcription factor is necessary for the regulation of the MYB12 gene expression in response to light and UV (Stracke et al., 2010). There is a tight linkage between the expression level of MYB12 and the flavonoid content in Arabidopsis seedlings (Mehrtens et al., 2005). Genes involved in light signalling, such as HY5, ELIP1, and MYB12, were strongly up-regulated in transgenic Arabidopsis lines expressing the mammalian InsP 5-ptase gene with a decreased level of InsP3 (Salinas-Mondragon et al., 2010).
In this study, it was hypothesized that a reduction of the major second messenger InsP3 can affect different branches of tomato secondary metabolism through regulation of key factors of light signalling. In order to test this hypothesis, the expression of key regulators of light-signalling (LeHY5, SIMYB12, tELF3 and LeELIP) was monitored in tomato fruits expressing the InsP 5-ptase gene. It was found that the expression of these genes was up-regulated in transgenic fruits compared with control tomato fruits. The increase in transcription of light-dependent genes coincided with the accumulation of major flavonoids (chlorogenic acid, rutin) in mature InsP 5-ptase fruits. In addition, it was demonstrated that expression of InsP 5-ptase in transgenic lines resulted in complex perturbations of several metabolic pathways. InsP 5-ptase activity in transgenic tomato lines not only affected the level of its substrate (InsP3) but also resulted in a reduction in the levels of other major phosphoinositol phosphates (InsP1–InsP4). The biosynthetic pathway of ascorbic acid (vitamin C), which is connected with the phosphoinositol pathway through inositol, was also affected in InsP 5-ptase expressing tomato lines. Genes coding for two major enzymes of the ascorbic acid pathway (MIOX and GLDH) were up-regulated in InsP 5-ptase-expressing tomato fruits which resulted in the increased accumulation of ascorbic acid compared with control fruits (wild type, empty vector control). These results indicate how the activity of a key enzyme of one stress-signal transduction pathway can lead to massive changes in other metabolic and stress-signalling pathways. Our study also revealed the potential of genetic manipulations of phosphoinositol pathway for crop improvement.
Materials and methods
Profiling of inositol phosphates in control and InsP 5-ptase transgenic plants
The profiling of inositol phosphates (InsP1–InsP4) was performed by anion exchange chromatography following [3H] myo-inositol labelling of young tomato seedlings. Wild-type (WT), vector control (EV), transgenic tomato Line 6 (L6), and transgenic Line 7 (L7) seeds were germinated on Murashige and Skoog (MS) basal salt medium (PhytoTechnology Laboratories, Shawnee Mission, KS) for 7 d prior to labelling. Seedlings were then transferred to MS media containing 10 μCi ml−1 [3H] myo-inositol (1 Ci=37 GBq) and incubated for 7 d at 25 °C with a 16 h photoperiod. Each seedling was then weighed and washed 3 times in 3 ml of PBS buffer. Samples were immersed in vials containing 3 ml of stopping buffer and kept on ice for 30 min. Tissues were disrupted in 750 μl of extraction buffer for 2 min using 1.0 mm silica beads as described by Stevenson-Paulik et al. (2005). The extracts were then centrifuged at 13 000 g for 10 min at 4 °C. The soluble layer was immediately processed or stored at –20 °C until further use. The soluble fraction was centrifuged again at 13 000 g for 30 min at 4 °C and the clear supernatant was subjected to anion exchange chromatography on gravity fed columns using Bio-Rad AG-1×8 resin (formate form 200–400 mesh size). Inositol phosphates InsP1, InsP2, InsP3, and InsP4 were then eluted with 12.5 ml of elution buffer (ammonium formate/formic acid) added in 2.5 ml fractions according to the protocol described by Ali et al. (1995). Four types of inositol phosphates were isolated by increasing the concentration of ammonium formate as follows: inositol monophosphates (0.2 M AF/0.1 M FA), inositol bisphosphates (0.4 M AF/0.1 M FA), inositol trisphosphates (0.8 M AF/0.1 M FA), and inositol tetrakisphosphates (1.0 M AF/0.1 M FA). The radioactivity of each eluted fraction was measured by mixing 1 ml of the fraction with 9 ml of Beckman Coulter scintillation cocktail in a LS6500 Beckman Coulter beta liquid scintillation counter.
Phytic acid (InsP6) was measured using a Megazyme kit (Megazyme International, Ireland) for phytic acid (phytate) and total phosphorus in which phytic acid is measured as phosphorus released by phytase and alkaline phosphatase. One gram of leaf tissue was accurately weighed from 4-week-old plants grown in a growth chamber with an approximate light intensity of 200 μmol m−2 s−1. Samples were ground to a fine powder using cold mortars and liquid nitrogen after which they were transferred into 75 ml glass beakers containing 20 ml of 0.66 M hydrochloric acid. Beakers were covered with foil and stirred vigorously overnight at room temperature. 1 ml of extracts were transferred to a 1.5 ml microfuge tubes and centrifuged at 13 000 g for 10 min. 0.5 ml of the resulting extract supernatants were immediately transferred to fresh 1.5 ml microfuge tubes and neutralized by the addition of 0.5 ml of 0.75 M sodium hydroxide solution. Neutralized sample extracts were used in the enzymatic dephosphorylation reaction procedure. Total and free phosphorus contents were measured for each sample. Into a fresh 1.5 ml microfuge tube the following reagents were added: 0.60 ml distilled water, 0.20 ml 200 mM sodium acetate buffer (pH 5.5), 0.05 ml sample extract, 0.02 ml phytase (for total phosphorus), 0.62 ml distilled water, 0.20 ml 200 mM sodium acetate buffer (pH 5.5), and 0.05 ml sample extract (for free phosphorus). Reagents were mixed by vortex and incubated in a water bath set at 40 °C for 10 min. After 10 min, the following reagents were added to each tube respectively; 0.20 ml glycine buffer (pH 10.4) [glycine buffer (400 mM), plus MgCl2 (4 mM), ZnSO4 (0.4 mM)], 0.02 ml alkaline phosphatase (for total phosphorus), 0.02 ml distilled water, 0.20 ml glycine buffer (pH 10.4) [glycine buffer (400 mM), plus MgCl2 (4 mM), ZnSO4 (0.4 mM)] (for free phosphorus). All reagents were mixed by vortex and incubated in a water bath set at 40 °C for 15 min after which reactions were centrifuged at 13 000 g for 10 min. The supernatants were then carefully pipetted for colorimetric determination of phosphorus. In a fresh 1.5 ml microfuge tube, 1 ml of the samples was mixed with 0.5 ml colour reagent solution [1 part 5% ammonium molybdate (Acros Organics, NJ, USA) with 5 parts 10% ascorbic acid (Sigma, St Louis, Mo, USA)]. Reagents were mixed by vortex and incubated in a water bath set at 40 °C for 1 h. After 1 h, reagents were mixed by vortex and 1 ml of each tube was transferred to a semi-micro cuvette and the absorbance was read at 655 nm. ΔAphosphorus was calculated for each sample by subtracting the absorbance of the ‘Free Phosphorus’ samples from the absorbance of the ‘Total Phosphorus’ samples. The phosphorus concentration is expressed in g/100 g sample as follows: Phosphorus (g/100 g)=[ΔAphosphorus×10 000 (conversion from μg g−1 to g/100 g)×1.0 (weight of original sample material)×1.0 (sample volume used in colorimetric step)]/[mean M (mean value of phosphorus standard)×20 (original sample extract volume)×55.6 (dillusion factor)]. Mean value of phosphorus standard was obtained using a standard curve over a dynamic range of 0–7.5 μg phosphorus (R2=1) as follows: MSTD (mean value for each standard)=P (μg phosphorus)/ΔAphosphorus. Mean M=Average of MSTD values. Phytic acid concentration was calculated as follows: Phytic acid (g/100g)=Phosphorus (g/100g)/0.282. The calculation of phytic acid content was based on the assumption that the amount of phosphorus measured is exclusively released from phytic acid and that this comprises 28.2% of phytic acid.
Plant growth and preparation of samples for gene expression and metabolite assays
Transgenic tomato lines expressing InsP 5-ptase were established and characterized early (Khodakovskaya et al., 2010). Tomato (cv. Micro-Tom) seeds of wild-type (WT), empty vector (EV), and of InsP 5-ptase transgenic lines 6 and 7 (L6 and L7) were surface-sterilized and germinated on MS medium in a growth chamber with 100 μmol m−2 s−1 light intensity. After 14 d, the seedlings were transferred into pots containing 25% of sand and 75% of Sun Gro Redi-earth ‘Plug and Seedling’ Mix (Sun Gro Horticulture, Bellevue, WA) under high-light conditions (800 μmol m−2 s−1) with 16 h light (25 °C) and 8 h dark (22 °C). Plants were watered three times a week, and the green/red fruits were harvested after 6/8 weeks of growth under high-light conditions. For real-time quantitative RT-PCR (qRT-PCR) and ascorbic acid assay, fruit samples were snap-frozen in liquid nitrogen and immediately ground to a fine powder. The ground samples were immediately used for total RNA extraction or stored at –80 °C for further analysis. Fruit samples for flavonoid detection were freeze-dried and stored at –80 °C before being used in HPLC analysis.
RNA isolation, cDNA synthesis, RT-PCR, and qRT-PCR assays
Total RNA was isolated from tomato leaf and fruit tissues using the RNeasy Plant Mini Kit (Qiagen Sciences, Maryland, USA). cDNA synthesis was performed according to the SuperScript III First Strand Synthesis System Kit protocol (Invitrogen Inc.) using oligo(dT) primers. Following synthesis, cDNA was used for the PCR reaction using gene-specific primers. Amplification of InsP 5-ptase gene and LeCHS1 was carried out by RT-PCR with the following primers: 1.3 kb fragment of the InsP 5-ptase gene was amplified using the primers 5′-GCTCTAGATAACTATGAGAGGATC-3′ (forward primer) and 5′-GCTCTAGAGGCGCTGGCATCTC-3′ (reverse primer); the LeCHS1 gene (X55194) was amplified using 5′-TGGCTGAGAACAACAAGGGTGCTA-3′ (forward primer) and 5′- ATTCACTGGGTCCACGGAACGTAA-3′ (reverse primer). After 25 cycles, PCR products were separated on 1% agarose gels by electrophoresis for 30 min at 5 V cm−1. Quantification of the expression of selected genes by qRT-PCR was carried out using the following primers: for the LeHY5 gene (AJ011914): 5′-ACCATCAGCTGGGACTCAAAGGAA-3′ (forward primer) and 5′-TTCCTCTCCCTTGCTTGTTGTGCT-3′ (reverse primer); for the tELF3 gene (AW093790): 5′-CCTATGTTTCCAAGGCTTCA-3′ (forward primer) and 5′-CTGCTCATAAAGAGCCATT-3′ (reverse primer); for the GLDH gene (AB080690): 5′-TCGAGTTCAGCAGCTTGTGGATGA-3′; (forward primer) and 5′-CACCAACCTGAACAATGCCACCAA-3′ (reverse primer); for the MIOX gene (GQ150167): 5′;-GCTTCATTTGACCGGCCTCATTCA-3′ (forward primer) and 5′-TCTCCAACAACAGCCCATTGAGGA-3′ (reverse primer); for the LeELIP gene (AY547273): 5′-CAAGCCAACACCTGCTAAGCCAAA-3′ (forward primer) and 5′-AGCTCCATACCAATGGCTGCTACA-3′ (reverse primer); for the SIMYB12 gene (EU419748): 5′-TGCCAAATTCTTGGGCAGGACCTA-3' (forward primer) and (5′-TCACCACGTCTGGCATAATCTCCT-3′ (reverse primer); for the tomato HCT (AK324727): 5′-AGGTGAAAAACTCAACGATGGT-3′ (forward primer) and 5′-ACACTAGGCGTGTGGAAATTAG-3′ (reverse primer), for the Actin gene (internal control): 5′-AGGTATTGTGTTGGACTCTGGTGAT-3′ (forward primer) and 5′-ACGGAGAATGGCATGTGGAA-3′ (reverse primer). The qRT-PCR analysis was conducted using SYBR Green PCR master mix (Applied Biosystems, Inc) in an iCycler iQ Multi Color Real Time PCR detection system (Bio-Rad, Hercules, CA, USA). Three independent biological replicates were used in the analysis. The real-time PCR data were generated and analysed by the ‘comparative count’ method to obtain relative mRNA expression in each tissue as described in the iCycler manual (Bio-Rad).
Analysis of flavonoids in mature tomato fruits
Flavonoid analysis in fruits and leaves was performed by high-performance liquid chromatography (HPLC) with ultraviolet (UV) detection (HPLC-UV). 15 mg of freeze-dried tissue powder were transferred into a glass tissue grinder on ice. 1 ml of 80% methanol extraction solvent was added to each sample and ground thoroughly until all of the tissue was extracted. The tissue homogenate was transferred to an Eppendorf tube and mixed by vortexing thoroughly and then kept on ice for 5 min. Samples were sonicated for 30 min in a sonicator bath followed by centrifugation at 13 000 g for 10 min. The supernatant was removed and transferred to a 1 ml syringe capped with a syringe filter. Contents were filtered into a labelled Eppendorf tube on ice. 100 μl of each filtered sample were transferred to an autosampler vial insert. The insert was placed in a labelled autosampler vial. Samples were then run in an HPLC system. HPLC-UV analyses were performed with a Thermo Spectra SYSTEM AS3000 HPLC (Thermo Fisher Scientific Inc.) equipped with a Spectra SYSTEM UV6000 detector. Chromatographic separations were achieved with Alltech columns (Alltech Associates, Inc.) (250 mm length. 4.6 mm ID, 5 lm particle size). The mobile phase consisted of 0.5% H3PO4 (Solvent A) and methanol (MeOH) (Solvent B) at 1 ml min−1 with an initial composition of 95% H3PO4 and 5% MeOH. The gradient elution profile (%B to A) consisted of a linear gradient from 5% B to 25% B (2–4 min), a linear gradient from 25% B to 60% B for the next 16 min, a linear gradient from 60% B to 75% B during the next 14 min, followed by a linear gradient from 75% B to 95% B in 1 min. Spectral data were collected over a range of 240–600 nm and specifically collected at 280, 320, and 360 nm for quantitation of chlorogenic acid and rutin. Quantitation was conducted by comparing peak areas obtained for chlorogenic acid and rutin in experimental samples with a series of reference standards and analysed concurrently with the extracts. Calculations were conducted using UV absorbance at 360 nm; the chromatographic data were processed using Agilent’s EZChrome Elite software.
Ascorbic acid quantification
Ascorbic acid levels were measured by a modified version of the procedure described by Gillespie and Ainsworth (2007) based on an original method developed by Okamura et al. (1980). Frozen tissue (50 mg) was placed in a 2 ml screw-capped tube with beads and 1 ml of ice-cold 6% trichloroacetic acid (TCA) (Sigma) and was then homogenized in a mini-bead beater (Biospec) twice for 1 min each. Samples were next incubated on ice for 10 min and centrifuged for 10 min at 13 000 g at 4 °C. The supernatant was then transferred to a new 1.5 ml tube. A 100 μl aliquot from the supernatant was transferred to a fresh 2 ml tube containing 100 μl 75 mM phosphate buffer (pH 7.4) supplemented with 100 μl of 10 mM dithiothreitol (DTT) (Acros) and incubated at room temperature for 10 min. The following reagents were then added to the assay tube: 100 μl double distilled water, 500 μl 10% TCA, 400 μl 42% H3PO4, 400 μl 2,2'-dipyridyl, and 200 μl 3% FeCl3. The mixtures were vortexed vigorously and incubated at 37 °C for 1 h. 200 μl of sample from the assay tubes were transferred into wells of a clear 96-well microtitre plate, and the absorbance of each well was read at 525 nm. The AsA concentration was expressed in nmol AsA per well according to the standard curve A525–0.2458=nmol AsA per well×0.7837, obtained over a dynamic range of 0–1 μmol AsA (R2=0.9994). The value was then converted to μmol g−1 fresh weight of tissue.
Results
Expression of the InsP 5-ptase gene affects not only the level of InsP3 but also the accumulation of other inositol phosphates
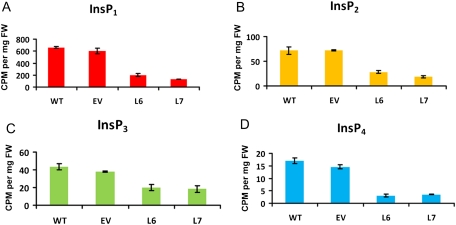
To determine how the expression of InsP 5-ptase affects overall phosphoinositol metabolism, the levels of basic metabolites of the phosphoinositol pathway were measured in wild-type and InsP 5-ptase expressing transgenic tomato seedlings by the introduction of labelled myo-inositol in agar MS growth medium. 3H- labeled inositol phosphates were extracted from leaves of 14-day-old tomato seedlings by anion exchange chromatography as described in the Materials and methods. It was revealed that there is a significant decrease not only in the level of InsP3 but also in levels of other major inositol phosphates, such as InsP1, InsP2, and InsP4 (Fig. 1) in transgenic InsP3 5-ptase lines (L6 and L7) compared with wild-type (WT) seedlings. No changes in the level of InsP6 (phytic acid) between control (WT, EV) and transgenic (L6, L7) tomato lines were found (see Supplementary Fig. S1 at JXB online).
Fig. 1.
Levels of basic inositol phosphates (InsP1, InsP2, InsP3, InsP4) in leaves of transgenic tomato lines expressing the InsP 5-ptase gene (L6, L7) and control lines (WT, EV). Levels of 3H-labelled inositol phosphates (InsP1, InsP2, InsP3, InsP4) expressed as counts per minute (CPM) per mg fresh weight of transgenic tomato seedlings lines expressing the InsP 5-ptase gene (L6, L7) and control lines (WT, EV). (A) Inositol monophosphate (InsP1); (B) inositol bisphosphate (InsP2); (C) inositol trisphosphate (InsP3); and (D) inositol tetrakisphosphate (InsP4). Vertical bars indicate ±SE of three biological replicates.
Genetic modification of the phosphoinositol pathway can lead to changes in phenylpropanoid biosynthetic pathway in tomato fruits
Based on our working hypothesis with regard to possible changes of the light-dependent pathways of secondary metabolites in InsP 5-ptase expressing tomatoes, it was thought that the phenylpropanoid biosynthetic pathway may be activated in transgenic lines. To investigate this possibility, an RT-PCR analysis was performed for the expression of the genes encoding two key enzymes of the plant phenylpropanoid pathway, chalcone synthase (LeCHS1) and tomato cinnamoyl-CoA shikimate/quinate transferase (HCT), in transgenic and control green tomato fruits (Fig. 2A, C). The analysis revealed that both genes (LeCHS1 and HCT) are expressed at higher levels in green fruits of tested InsP 5-ptase-expressing lines (L6 and L7) compared with control green fruits (wild-type and empty vector control). The LeCHS1 gene expression was especially elevated in fruits of InsP 5-ptase line 7 (L7) which also had the highest level of InsP 5-ptase gene expression (Fig. 2B). To investigate if up-regulation of LeCHS1 gene expression in green transgenic fruits would also result in an increase in major products of LeCHS1 and HCT activities (rutin and chlorogenic acid) in mature (red) transgenic fruits, the levels were quantified by HPLC (Fig. 2C). The accumulation of both of the measured phenylpropanoids in transgenic fruits of line L7 was found to be 2-fold higher than that of control fruits (wild-type and empty vector control). Flavonoid accumulation in transgenic fruits of line L6 was also slightly higher compared with control fruits. These results indicate the existence of a direct link between the activity of InsP 5-ptase, expression of the gene encoding a key enzyme of flavonoid pathway, and production of phenylpropanoid in transgenic fruits.
Fig. 2.
Effect of expression of InsP 5-ptase gene (B) on expression of LeCHS1, tomato chalcone synthase gene (A), expression of HCT, cinnamoyl-CoA shikimate/quinate transferase gene (C) in tomato green fruits and production of chlorogenic acid and rutin (D) in red fruits. Vertical bars (C and D) indicate ±SE of three biological replicates. Analysis of gene expression was performed by RT-PCR (A, B) and real-time PCR (C).
Genetic modification of the phosphoinositol pathway affects the transcription of several gene-regulators of light signalling
In order to understand the link between phosphoinositol metabolism and light signalling, control and transgenic tomato plants were grown under high-light conditions (>800 μmol m−2 s−1) using an in vitro system for 2 months. It was observed that under high-light conditions InsP 5-ptase-expressing tomato plants were able to stay healthier and maintained their leaf chlorophyll content longer compared with wild-type plants (Fig. 3A). Based on this observation, it was hypothesized that light-signalling could be significantly modified by genetic reduction of InsP3 and probably by a decrease in other major phosphoinostol phosphates in transgenic tomato plants. To test this hypothesis, the expression of selected tomato key genes (LeHY5, SlMYB12, tELF3, and LeELIP) involved in photomorphogenesis and light signalling in InsP 5-ptase-expressing and control green fruits was monitored by qRT-PCR (Fig. 3B). Expression analysis revealed significant differences in expression levels of all the genes tested between transgenic and control lines. For example, the expression of the light-regulated positive inducer of fruit pigmentation LeHY5 was significantly enhanced in fruits of transgenic lines expressing the InsP 5-ptase gene compared with fruits of the wild type and the empty vector control. The same tendency was observed for the SIMYB12 gene, a transcription factor which is known as a flavonol-specific activator of flavonoid biosynthesis, as well as for the LeELIP gene, a photoprotective agent known to be expressed mostly in response to light stress. By contrast, expression of the tomato EARLY FLOWERING 3 gene (tELF3) was down-regulated in transgenic plants expressing InsP 5-ptase compared with control lines (Fig. 3B). No statistically significant changes were found in the expression of two negative regulators of light signal transduction and fruit pigmentation: LeCOP1LIKE and LeDET1 (see Supplementary Fig. S2A, B at JXB online). Possible network between InsP3, light signalling factors, phenylpropanoids, and protection against light that may occur in transgenic tomato plants expressing InsP 5-ptase is shown in Fig. 3C.
Fig. 3.
Light signalling is modified in transgenic tomato plants expressing the InsP 5-ptase gene. (A) The phenotype of wild-type and InsP 5-ptase-expressing tomato plants cultivated under high-light conditions (800 μmol m−2 s−1 with 16 h light (25 °C) and 8 h dark (22 °C) for 2 months on incubation in vitro. (B) Expression of genes involved in light signalling in tomato green fruits expressing InsP 5-ptase (L6 and L7) and control lines (WT and EV) analyzed by real-time PCR. Expression levels are given relative to WT for each gene as separate assays. Vertical bars indicate ±SE of three biological replicates. (C) Hypothetical model of suggested network between InsP3, light signalling factors, phenylpropanoids, and protection against light that may occur in transgenic tomato plants expressing InsP 5-ptase.
Reduction of phosphoinositols in InsP 5-ptase tomatoes affects the expression of key enzymes of the ascorbic acid biosynthetic pathway and elevates the ascorbic acid level in transgenic tomato fruits
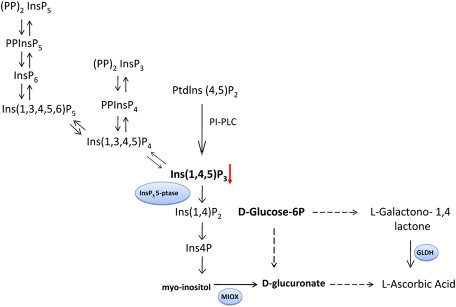
Because the expression of the InsP 5-ptase gene resulted in alterations of the overall phosphoinositol pathway, it was hypothesized that metabolic pathways linked to the phosphoinositol pathway could be modified in transgenic tomatoes, as well. Consecutive dephosphorilation of phosphoinositols leads to the formation of free inositol that plays a role as the first intermediate in the alternative pathway of ascorbic acid biosynthesis in plants (Lorence et al., 2004; Fig. 4). Ascorbate can be synthesized from glucose-6-phosphate via a number of pathways to the last precursor (galactono- 1,4-lactone) in the main biosynthetic pathway (Ioannidi al., 2009). To test if biosynthetic pathways of ascorbic acid were affected by the expression of InsP 5-ptase, expression levels of two key enzymes of pathways (GLDH and MIOX) were monitored by qRT-PCR analysis in transgenic and control green fruits. As shown in Fig. 5A, significantly higher levels of expression of both analysed genes (MIOX and GLDH) were observed in transgenic InsP 5-ptase fruits compared with control lines suggesting a possible increase in the end product, ascorbic acid in transgenic fruits. Therefore, a quantification of ascorbic acid (vitamin C) content was also performed for green and red fruits from two transgenic lines (L6 and L7) and two control lines (WT, EV) using a colorimetric ascorbate assay. As expected, a 40% increase in ascorbate content was observed in both analysed lines of transgenic fruits compared with control fruits (Fig. 5B).
Fig. 4.
Simplified, schematic representation of cross-talk between general phosphoinositol pathway and biosynthetic pathways of ascorbic acid (vitamin C).
Fig. 5.
Effect of genetic reduction of InsP3 in tomato plants on the expression of genes encoding key enzymes of ascorbic acid pathway (A) and the accumulation of ascorbic acid (B) in transgenic and control tomato fruits. Expression analysis (real-time PCR) was performed in green tomato fruits. Expression levels are given relative to WT for each gene as separate assays. Analysis of accumulation of ascorbic acid was performed in green and mature (red) fruits. Vertical bars indicate ±SE of three biological replicates.
Discussion
The phosphoinositol pathway is one of the major plant stress-signalling pathways and includes a number of stress-related second messengers, such as inositol (1,4,5) trisphosphate (InsP3), that induce the release of Ca2+ into the cytoplasm, which in turn, causes specific responses to abiotic stresses, such as drought, cold, and high salinity (Taji et al., 2006). The involvement of the phosphoinositol pathway and in particular the InsP3 second messenger in the plant gravity response has been demonstrated and discussed by number of authors. Thus, it was documented that dampening of the InsP3 delays the timing and reduces the magnitude of the gravitropic response in Arabidopsis (Perera et al., 2006). Authors proposed a role of InsP3 as a fundamental component of plant gravisignalling. According to published data, InsP3 signalling is also involved in the control of plant development. Regulation of InsP3 levels is important during germination and early seedling development. Gunesekera et al. (2007) showed that inositol polyphosphate 5-phosphatases 1 and 2 are required for regulating Arabidopsis seedling growth. Evidence of the involvement of InsP3 in ABA signalling have also been reported (Burnette et al., 2003; Taji et al., 2006; Perera et al., 2008). Recently, Salinas-Mondragon et al. (2010) have demonstrated a direct link between the phonosphoinositol pathway and light signalling. By using transgenic Arabidopsis plants expressing the mammalian inositol polyphosphate 5-phosphatase (InsP 5-ptase) gene, they showed that light-induced gene expression is regulated by InsP3-dependent and InsP3-independent signal transduction pathways. Several key regulators of light signalling including HY5, ELIP1, and MYB12 that are involved in the positive regulation of photomorphogenesis and secondary metabolism were up-regulated in Arabidopsis seedlings with reduced InsP3 levels (Salinas-Mondragon et al., 2010). By using previously generated tomato lines expressing the InsP 5-ptase gene (Khodakovskaya et al., 2010), an attempt was made to clarify how genetic reduction of InsP3 in tomato plants can affect the overall phosphoinositol pathway and some pathways of secondary metabolism that are known to be induced by light. In particular, the existence of links between the perturbation of levels of major phosphoinositols, changes in light signalling, and activation of the phenylpropanoid pathway in transgenic tomato fruits expressing the InsP 5-ptase gene were suggested. Flavonoids are a large group of metabolites of the phenylpropanoid pathway. In plants, flavonoids are involved in multiple biological processes including responses to the environment and UV protection (Harborne and Williams, 2000). Up to 70 different flavonoids have been identified in tomato fruits (Moco et al., 2006; Iijima et al., 2008). Chalcone synthase (CHS1) is the first enzyme in the flavonoid pathway that catalyses the formation of naringenin chalcone, the first flavonoid of the pathway in red tomato fruits (Holton and Cornish, 1995; Tanaka et al., 1998; Muir et al., 2001). It has previously been shown that the light-signalling pathway is involved in flavonoid biosynthesis through a bZIP transcriptional regulator: ELONGATED HYPOCOTYL5 (HY5). HY5 protein was able to bind to the promoter of light-inducible gene chalcone synthase (CHS1) in in vitro and in vivo systems (Ang et al., 1998; Lee et al., 2007). Recent studies have demonstrated that Arabidopsis transcription factor HY5 also regulates the transcription of the MYB12 gene in response to light (Stracke et al., 2010). Ballester et al. (2010) have demonstrated that the tomato homologue of MYB12 (SIMYB12) plays an important role in the activation of the flavonoid pathway in tomato fruits through the regulation of naringenin chalcone. In addition, HPLC analysis of myb12 mutants and MYB12 over-expression lines demonstrated a tight linkage between the expression level of MYB12 and the accumulation of flavonol in Arabidopsis seedlings (Mehrtens et al., 2005). Our quantitative expression analysis of tomato homologues of HY5 (LeHY5) and MYB12 (SIMYB12) transcription factors in green fruits of InsP 5-ptase expressing plants grown in high-light conditions showed an increase in the expression of LeHY5 and SIMYB12 compared with control fruits (wild-type, empty vector control) (Fig. 3B). Considering the fact that the expression of LeCHS1 can be regulated by LeHY5, the expression of the LeCHS1 gene was analysed and it was found that LeCHS1 was also up-regulated in both of the tested InsP 5-ptase green fruits compared with the controls (Fig. 2A). All of these data suggest a possible up-regulation of metabolites of the phenylpropanoid pathway in transgenic tomatoes. The levels of chlorogenic acid and rutin were measured in mature (red) tomato fruits using HPLC analysis. Chlorogenic acid is the main phenolic compound besides flavonoids in tomato and has several health-related properties because of its antioxidant and antiviral activity (Farah and Donangelo, 2006). Rutin is the major flavonoid compound in mature tomatoes in most of the cultivars studied (Slimestad and Verheul, 2009). As expected, the accumulation of both phenylpropanoids (chlorogenic acid and rutin) was found to be significantly higher in mature (red) fruits of the two tested transgenic lines (Fig. 2C). The increased level of flavonoids in InsP 5-ptase expressing tomato plants could be the reason that transgenic tomato plants were able to withstand prolonged high-light conditions while control tomato plants developed symptoms of severe leaf bleaching and extensive photooxidative damage (Fig. 3A). The importance of specific types of flavonoids in UV protection has been experimentally demonstrated through a reverse genetic approach in Arabidopsis (Dixon and Palva, 1995). Increased tolerance in InsP 5-ptase tomatoes to high-light can also be attributed to the 3-fold increase in expression of tomato early light-induced protein (LeELIP) in transgenic lines (Fig. 3B). LeELIP has high sequence similarity to the amino acid sequence of Arabidopsis ELIP1 and ELIP2 (Bruno and Wetzel, 2004). Arabidopsis ELIP1 exhibits a strong photoprotective function that could involve either the binding of chlorophylls released during turnover of pigment-binding proteins or the stabilization of the assembly of those proteins under high-light stress (Hutin et al., 2003). The transcriptional factor HY5 promotes the light induction of ELIP1 in Arabidopsis (Harari-Steinberg et al., 2001). Based on previous reports and the data presented here, it is plausible that the up-regulation of the LeHY5 transcription factor has led to an activation of the expression of LeCHS1, SIMYB12, and tomato ELIP1 (LeELIP) genes in green fruits resulting in an activation of the flavonoid pathway in mature fruits thereby increasing the overall tolerance to high-light stress (Fig. 3B, C). It is interesting to note that the tomato homologue of Arabidopsis EARLY FLOWERING 3 gene (tELF3) was down- regulated in InsP 5-ptase fruits compared with control fruits (Fig. 3B). ELF3 is a central component of the signal transduction pathway between the photoreceptors and the oscillator and plays an important role in determining hypocotyls length, flowering time, and circadian rhythms (Tajima et al., 2007; Nefissi et al., 2011). Using mutant analysis, it has been demonstrated that ELF3 works as a floral repressor in Arabidopsis (Zagotta et al., 1996). Thus, by dampening the level of the key second messenger of stress signalling, InsP3, in tomato plants led to a significant perturbation in the light-stress signal transduction machinery and resulted in the activation of the flavonoid biosynthesis pathway to yield tolerance to high-light conditions (Fig. 3A, C). Our results related to the expression of genes involved in light-signal transduction in tomato plants are comparable with the results obtained in Arabidopsis in which the same transgene (InsP 5-ptase) was used for the establishment of transgenic plants (Salinas-Mongdragon et al., 2010). Even so, genes MYB12, HY5, ELIP1, and DET1 showed the same trend in expression between transgenic tomato and Arabidopsis plants expressing the mammalian InsP 5-ptase gene, suggesting the existence of conserved InsP3-dependent and InsP3-independemt light-signalling pathways in these plants. The over-expression of the LeHY5 transcription factor and the observed increase in carotenoids (Khodakovskaya et al., 2010) in InsP 5-ptase transgenic fruits is in agreement with data reported by Liu et al. (2004). These authors described the function of LeHY5 as a positive regulator of tomato fruit pigmentation and carotenoid biosynthesis through the involvement in light signalling.
The effect of InsP 5-ptase expression in transgenic tomato plants was found to be more complex than expected and it was not limited only to a reduction in the level of InsP3, the substrate of mammalian InsP 5-ptase. As shown in Fig. 1, the over-expression of transgene resulted in significant perturbation in the entire phosphoinositol pathway. The levels of several major phosphoinositols, such as InsP1, InsP2, InsP3, and InsP4, were dramatically reduced in transgenic InsP 5-ptase expressing tomato seedlings. This decrease in several phosphoinositol metabolites could be due to an overall metabolic shift in substrate-product formation. InsP3 is the first soluble InsP formed as a result of the hydrolysis of the membrane-bound inositol phospholipid, phosphatidylinositol 4,5 bisphosphate, by phosphatidylinositol specific-phosphlipase C (Perera et al., 2002). This InsP3 can be rapidly hydrolysed by InsP 5-ptase to lower InsPs, such as InsP2 and InsP1. InsP3 can also serve as a substrate for the synthesis of higher InsPs, such as InsP4, InsP5, and InsP6. InsP3, when hydrolysed by the over-expressed InsP 5-ptase in question, might cause a sequential conversion of InsP4 into InsP3, InsP5 into InsP4, and InsP6 into InsP5 to compensate for their reduced levels, thus causing an overall metabolite shift. It could also be possible that the over-expression of InsP 5-ptase causes a differential expression of other enzymes of the inositol phosphate metabolic pathway that give rise to the observed phosphoinositol profile. Further experiments, such as assessing the activities and the expression of the other enzymes of phosphoinositol pathways, would be required to offer an adequate explanation.
Myo-inositol is a precursor of the phosphoinositol pathway and is the first metabolite of the alternative biosynthetic pathway of L-ascorbic acid (vitamin C, AsA) (Fig. 4). Several metabolic pathways are involved in the production of AsA in plants (Valpuesta and Botella, 2004), but the Wheeler–Smirnoff pathway is considered to be the primary pathway in the production of ascorbic acid. In the last step of the Wheeler–Smirnoff pathway, L-Galactono-1,4 lactone dehydrogenase (GLDH) catalyses the formation of AsA (Imai et al., 2009). This is a determinant step in the production of AsA by this pathway. Recent results provide supporting evidence for the existence of an animal-like ascorbate biosynthesis pathway based on the conversion of myo-inositol to D-glucuronate (Radzio et al., 2003, Lorence et al., 2004, Zhang et al., 2008) that is considered an alternative route to the Wheeler–Smirnoff pathway. MIOX is a mono-oxygenase that catalyses the cleavage of myo-inositol to D-glucuronate (Radzio et al., 2003; Fig. 4). Although myo-inositol is not a major precursor for AsA biosynthesis, the constitutive expression of the miox4 gene increased AsA accumulation in Arabidopsis leaves 2–3-fold (Lorence et al., 2004). There are many indications that the biosynthetic pathways of ascorbic acid are light-sensitive. For example, GLDH activity can be regulated by light in Brassica campestris (Li et al., 2008). In rice, the AsA content and the expression of L-galactose-1-phosphate phosphatase (GPPase), and GLDH were increased by high light and decreased in the dark (Fukunaga et al., 2010). Taking into account the connection of the phosphoinositol pathway and alternative AsA pathways through myo-inositol and the existence of a link between light signaling and the biosynthesis of AsA, it was logical to suggest that the biosynthesis of ascorbic acid could be modified in transgenic tomato fruits expressing InsP 5-ptase. To test our hypothesis, the expression of key enzymes of both AsA biosynthetic pathways (MIOX and GLDH) were monitored and the level of ascorbic acid was determined in green and mature (red) fruits of control lines and lines expressing InsP 5-ptase (Fig. 5). The expression levels of MIOX and GLDH in green tomato fruits were quantified using qRT-PCR to determine the gene expression pattern controlling AsA biosynthesis in InsP 5-ptase-expressing transgenic plants and to revalidate the role of these enzymes in the pathway. Our data showed higher expression levels of MIOX and GLDH in transgenic lines which suggested an increase in the accumulation of AsA in those fruits. Total AsA measurement in green and red fruit samples showed a significant difference in vitamin C accumulation between wild-type and transgenic lines. Total AsA accumulation in transgenic tomato fruits was increased which is consistent with the up-regulation of MIOX and GLDH genes in green fruits. It is not clear yet whether both genes contributed to the observed increase of vitamin C in transgenic fruits or the effect was associated with the up-regulation of one gene. The possibility to enhance vitamin C production by over-expression of the gene encoding the enzyme of the AsA pathway was demonstrated by Agius et al. (2003). The authors showed that the over-expression of a D-galacturonic acid reductase gene from strawberry in Arabidopsis resulted in an increase in vitamin C content of 2–3-fold. It could be that increased MIOX gene expression and its enzymatic activity cause a metabolic shift in the phosphoinositol pathway to AsA biosynthesis. In transgenic lines expressing InsP 5-ptase, this would lead to an increased accumulation of free myo-inositol as a result of an overall hydrolysis of inositol phosphates into myo-inositol. This excess myo-inositol may enter into the AsA pathway which, upon ring cleavage to D-glucuronate by MIOX, would serve as a precursor for the AsA pathway (Zhang et al., 2008). However, a recent study by Endres and Tenhaken (2009) reports contrasting results. This study reported that excessive myo-inositol feeding to MIOX over-expressing Arabidopsis lines did not alter AsA levels. These conflicting results warrant an assessment of the levels of myo-inositol in InsP 5-ptase expressing lines compared with wild-type tomato plants.
In conclusion, our results suggest that the reduction of second messenger InsP3 in transgenic tomato plants can affect several branches of secondary metabolism through the modification of light signalling. The results support the idea that the second messenger InsP3 of stress signalling may play a role as a negative regulator of photomorphogenetic responses by involvement in light-signal transduction in a way similar to the tomato DE-ETIOLATED 1 (DET1) light regulator. Both carotenoid and phenylpropanoid contents were increased in tomato fruits by the post-transcriptional gene silencing of the DET1 transcription factor that is most likely to control light-regulated gene expression at the level of chromatin remodelling (Davuluri et al., 2004). In our previous paper (Khodakovskaya et al., 2010) and in this study, it has been demonstrated that the reduction of InsP3 in tomato plants correlated with an increase in secondary metabolites that can be regulated by light (carotenoids, flavonoids, and ascorbic acid). Specific pathways related to the involvement of InsP3 in light-signal transduction need to be clarified further. The involvement of other phosphoinositols (InsP1–InsP4) that were reduced in InsP 5-ptase lines in light-signal trasduction also requires further investigation. Our results demonstrated the potential of genetic modifications of the phosphoinositol pathway for the improvement of the nutritional value of crops. As shown, the manipulation of the level of one key messenger of the major stress-signalling pathway can affect the overall pathway and hence multiple branches of secondary metabolism.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Phytic acid (InsP6) concentration in tomato leaves of the wild type, empty vector, and transgenic lines L6 and L7 grown under approximate light intensity of 200 μmol m−1 s−1.
Supplementary Fig. S2. Expression of LeDET1(A) and COP1LIKE (B) genes in tomato green fruits expressing InsP 5-ptase (L6 and L7) and control lines (WT and EV) analysed by real-time PCR; vertical bars indicate ±SE of three biological replicates.
Acknowledgments
Authors thank the Graduate Institute of Technology, University of Arkansas at Little Rock, for providing graduate assistantships to Mohammad Alimohammadi and Kanishka de Silva. The authors are grateful to Dr Stephen Grace for help with the analysis of flavonoids in tomato fruits by HPLC. This work was supported by EPSCoR-NSF-P3 Center (Grant P3-202 for MVK). The editorial assistance of Dr Marinelle Ringer is also acknowledged.
Glossary
Abbrevations
- CHS1
chalcone synthase
- HCT
cinnamoyl-CoA shikimate/quinate transferase
- GLDH
L-galactono-γ-lactone dehydrogenase
- LeHY5
Lycopersicon esculentum hypocotyl elongated transcriptional factor
- MIOX
myo-inositol oxygenase
- ELF3
early flowering 3 transcriptional factor
- ELIP
early light-induced protein
- SlMYB12
Solanum lycopersicum MYB12 transcriptional factor
- AsA
ascorbic acid
- GLDH
L-galactono-1,4 lactone dehydrogenase
References
- Agius F, González-Limothe R, Caballero JL, Muñoz-Blanco J, Botella MA, Valpuesta V. Engineering increased vitamin C levels in plants by over-expression of a D-galacturonic acid reductase. Nature Biotechnology. 2003;21:177–181. doi: 10.1038/nbt777. [DOI] [PubMed] [Google Scholar]
- Ali N, Craxton A, Summer M, Shears SB. Effects of aluminum upon hepatic multiple inositol polyphosphate phosphatase. Biochemical Journal. 1995;305:557–561. doi: 10.1042/bj3050557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Molecular Cell. 1998;1:213–222. doi: 10.1016/s1097-2765(00)80022-2. [DOI] [PubMed] [Google Scholar]
- Ballester AR, Molthoff J, de Vos R, et al. Biochemical and molecular analysis of pink tomatoes: deregulated expression of the gene encoding transcription factor SlMYB12 leads to pink tomato fruit color. Plant Physiology. 2010;152:71–84. doi: 10.1104/pp.109.147322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno AK, Wetzel CM. The early light inducible protein (ELIP) gene is expressed during the chloroplast-to-chromoplast transition in ripening tomato fruit. Journal of Experimental Botany. 2004;55:2541–2548. doi: 10.1093/jxb/erh273. [DOI] [PubMed] [Google Scholar]
- Burnette RN, Gunesekera BM, Gillaspy GE. An Arabidopsis inositol 5-phosphatase gain-of-function alters abscisic acid signaling. Plant Physiology. 2003;132:1011–1019. doi: 10.1104/pp.019000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Lin WH, Wang Y, Luan S, Xue HW. An inositol polyphosphate 5-phosphatase functions in PHOTOTROPIN1 signaling in Arabidopsis by altering cytosolic Ca2+ The Plant Cell. 2008;20:353–366. doi: 10.1105/tpc.107.052670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri GR, van Tuinen A, Mustilli AC, et al. Manipulation of DET1 expression in tomato results in photomorphogenic phenotypes caused by post-transcriptional gene silencing. The Plant Journal. 2004;40:344–354. doi: 10.1111/j.1365-313X.2004.02218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA. A two-for-one in tomato nutritional enhancement. Nature Biotechnology. 2005;23:825–826. doi: 10.1038/nbt0705-825. [DOI] [PubMed] [Google Scholar]
- Dixon RA, Palva N. Stress-induced phenylpropanoid metabolism. The Plant Cell. 1995;7:1085–1097. doi: 10.1105/tpc.7.7.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres S, Tenhaken R. Myo-inositol oxygenase controls the level of myo-inositol in Arabidopsis, but does not increase ascorbic acid. Plant Physiology. 2009;149:1042–1049. doi: 10.1104/pp.108.130948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah A, Donangelo CM. Phenolic compounds in coffee. Brazilian Journal of Plant Physiology. 2006;18:23–36. [Google Scholar]
- Fukunaga K, Fujikawa Y, Esaka M. Light regulation of ascorbic acid biosynthesis in rice via light responsive cis-elements in genes encoding ascorbic acid biosynthetic enzymes. Bioscience, Biotechnology, and Biochemistry. 2010;74:888–891. doi: 10.1271/bbb.90929. [DOI] [PubMed] [Google Scholar]
- Gillespie KM, Ainsworth EA. Measurement of reduced, oxidized, and total ascorbate content in plants. Nature Protocols. 2007;2:871–874. doi: 10.1038/nprot.2007.101. [DOI] [PubMed] [Google Scholar]
- Gunesekera B, Torabinejad J, Robinson J, Gillaspy GE. Inositol polyphosphate 5-phosphatases 1 and 2 are required for regulating seedling growth. Plant Physiology. 2007;143:1408–1417. doi: 10.1104/pp.106.089474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborne JB, Williams CA. Advances in flavonoid research since 1992. Phytochemistry. 2000;55:481–504. doi: 10.1016/s0031-9422(00)00235-1. [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Gohda K, Osterlund MT, Oyama T, Okada K, Deng XW. HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. The EMBO Journal. 2000;19:4997–5006. doi: 10.1093/emboj/19.18.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harari-Steinberg O, Ohad I, Chamovitz DA. Dissection of the light signal transduction pathways regulating the two early light-induced protein genes in Arabidopsis. Plant Physiology. 2001;127:986–997. [PMC free article] [PubMed] [Google Scholar]
- Hemm MR, Rider SD, Ogas J, Murry DJ, Chapple C. Light induces phenylpropanoid metabolism in Arabidopsis roots. The Plant Journal. 2004;38:765–778. doi: 10.1111/j.1365-313X.2004.02089.x. [DOI] [PubMed] [Google Scholar]
- Holton TA, Cornish EC. Genetics and biochemistry of anthocyanin biosynthesis. The Plant Cell. 1995;7:1071–1083. doi: 10.1105/tpc.7.7.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutin C, Nussaume L, Moise N, Moya I, Kloppstech K, Havaux M. Early light-induced proteins protect Arabidopsis from photooxidativestress. Proceedings of the National Academy of Sciences, USA. 2003;100:4921–4926. doi: 10.1073/pnas.0736939100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima Y, Nakamura Y, Ogata Y, Tanaka K, Sakurai N, Suzuki T. Metabolite annotations based on the integration of mass spectral information. The Plant Journal. 2008;54:949–962. doi: 10.1111/j.1365-313X.2008.03434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, Niwa M, Ban Y, Hirai M, Ôba K, Moriguchi T. Importance of the L-galactonolactone pool for enhancing the ascorbate content revealed by L-galactonolactone dehydrogenase-over-expressing tobacco plants. Plant Cell, Tissue and Organ Culture. 2009;96:105–112. [Google Scholar]
- Ioannidi E, Kalamaki MS, Engineer C, Pateraki I, Alexandrou D. Expression profiling of ascorbic acid-related genes during tomato fruit development and ripening and in response to stress conditions. Journal of Experimental Botany. 2009;60:663–678. doi: 10.1093/jxb/ern322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GI, Long JC, Wade HK, Shenton MR, Bibikova TN. UV and blue light signaling: pathways regulating chalcone synthase gene expression in Arabidopsis. New Phytologist. 2001;151:121–131. doi: 10.1046/j.1469-8137.2001.00151.x. [DOI] [PubMed] [Google Scholar]
- Khodakovskaya M, Sword C, Perera I, Boss W, Brown C, Sederoff H. Expression of inositol-1,4,5-triphosphate metabolism affects drought tolerance, carbohydrate metabolism, and phosphate-sensitive biomass increases in tomato. Plant Biotechnology Journal. 2010;8:170–183. doi: 10.1111/j.1467-7652.2009.00472.x. [DOI] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. The Plant Cell. 2007;19:731–749. doi: 10.1105/tpc.106.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Ren X, Song Y, Hou X. Enhancement of GLDHase activity by light in maintaining AsA content in leaves of Brassica campestris ssp. chinensis. Proceedings of the international symposium on endogenous and exogenous plant bioregulators, IHC 2006, Seoul, Korea, 13–19 August 2006. 2008 International Society for Horticultural Science (ISHS) [Google Scholar]
- Liu Y, Roof S, Ye Z, Barry C, van Tuinen A, Vrebalov J, Bowler C, Giovannoni J. Manipulation of light signal transduction as a means of modifying fruit nutritional quality in tomato. Proceedings of the National Academy of Sciences, USA. 2004;101:9897–9902. doi: 10.1073/pnas.0400935101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorence A, Chevone BI, Mendes P, Nessler CL. Myo-inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiology. 2004;134:1200–1205. doi: 10.1104/pp.103.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancinelli AL. Light-dependent anthocyanin synthesis: a model system for the study of plant photomorphogenesis. Botanical Reviews. 1985;51:107–157. [Google Scholar]
- Mehrtens F, Kranz H, Bednarek P, Weisshaar B. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiology. 2005;138:1083–1096. doi: 10.1104/pp.104.058032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moco S, Bino RJ, Vorst O, Verhoeven HA, de Groot J, van Beek TA, Vervoot J, de Vos CHR. A liquid chromatography-mass spectrometry-based metabolome database for tomato. Plant Physiology. 2006;141:1205–1218. doi: 10.1104/pp.106.078428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir SR, Collins GJ, Robinson S, Hughes S, Bovy S, De Vos CH, van Tunen AJ, Verhoeyen ME. Overexpression of petunia chalcone isomerase in tomato results in fruit containing increased levels of flavonols. Nature Biotechnology. 2001;19:470–474. doi: 10.1038/88150. [DOI] [PubMed] [Google Scholar]
- Nefissi R, Natsui Y, Miyata K, Oda A, Hase Y, Nakagawa M, Ghorbel A, Mizoguchi T. Double loss-of-function mutation in EARLY FLOWERING 3 and CRYPTOCHROME 2 genes delays flowering under continuous light but accelerates it under long days and short days: an important role for Arabidopsis CRY2 to accelerate flowering time in continuous light. Journal of Experimental Botany. 2011;62:2731–2744. doi: 10.1093/jxb/erq450. [DOI] [PubMed] [Google Scholar]
- Okamura M. An improved method for determination of L-ascorbic acid and L-dehydroascorbic acid in blood plasma. Clinica Chimica Acta. 1980;103:259–268. doi: 10.1016/0009-8981(80)90144-8. [DOI] [PubMed] [Google Scholar]
- Perera I, Heilmann I, Stevenson-Paulik JM, Boss W. The phosphoinositide (PI) pathway and signaling in plants. In: Sopory SK, Oelmüller R, Maheswari SC, editors. Signal transduction in plants: current advances. The Netherlands: Kluwer Academic Publishers; 2002. pp. 83–92. [Google Scholar]
- Perera I, Hung C-Y, Brady S, Muday GK, Boss W. A universal role for inositol 1,4,5-triphosphate-mediated signaling in plant gravitropism. Plant Physiology. 2006;140:746–760. doi: 10.1104/pp.105.075119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera I, Hung C-Y, Moore CD, Stevenson-Paulik J, Boss WF. Transgenic Arabidopsis plants expressing the type 1 inositol 5-phosphatase exhibit increased drought tolerance and altered abscisic acid signaling. The Plant Cell. 2008;20:2876–2893. doi: 10.1105/tpc.108.061374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radzio JA, Lorence A, Chevone BI, Nessler CL. L-Gulono-1,4-lactone oxidase expression rescues vitamin C-deficient Arabidopsis (vtc) mutants. Plant Molecular Biology. 2003;53:837–844. doi: 10.1023/B:PLAN.0000023671.99451.1d. [DOI] [PubMed] [Google Scholar]
- Salinas-Mondragon RE, Kajla JD, Perera IY, Brown CS, Sederoff HW. Role of inositol 1,4,5-triphosphate signalling in gravitropic and phototropic gene expression. Plant, Cell and Environment. 2010;33:2041–2055. doi: 10.1111/j.1365-3040.2010.02204.x. [DOI] [PubMed] [Google Scholar]
- Slimestad R, Verheul M. Review of flavonoids and other phenolics from fruits of different tomato (Lycopersicon esculentum Mill.) cultivars. Journal of the Science of Food and Agriculture. 2009;89:1255–1270. [Google Scholar]
- Stevenson-Paulik J, Bastidas RJ, Chiou ST, Frye RA, York JD. Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proceedings of the National Academy of Sciences, USA. 2005;102:12612–12617. doi: 10.1073/pnas.0504172102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Favory JJ, Gruber H, Bartelniewoehner L, Bartels S, Binkert M, Funk M, Weisshaar B, Ulm R. The Arabidopsis bZIP transcriptional regulator HY5 activates expression of the PFG1/MYB12 gene in response to light and UV-B radiation. Plant, Cell and Environment. 2010;33:88–103. doi: 10.1111/j.1365-3040.2009.02061.x. [DOI] [PubMed] [Google Scholar]
- Taji T, Takahashi S, Shinozaki K. Inositols and their metabolites in abiotic and biotic stress responses. In: Lahiri Majumder A, Biswas BB, editors. Biology of inositols and ohosphoinositols. Springer; 2006. pp. 239–264. [DOI] [PubMed] [Google Scholar]
- Tajima T, Oda A, Nakagawa M, Kamada H, Mizoguchi T. Natural variation of polyglutamine repeats of a circadian clock gene ELF3 in Arabidopsis. Plant Biotechnology. 2007;24:237–240. [Google Scholar]
- Tanaka Y, Tsuda S, Kusumi T. Metabolic engineering to modify flower color. Plant and Cell Physiology. 1998;39:1119–1126. [Google Scholar]
- Valpuesta V, Botella MA. Biosynthesis of L-ascorbic acid in plants: new pathways for an old antioxidant. Trends in Plant Science. 2004;9:573–577. doi: 10.1016/j.tplants.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Vazquez-Flota FA, De Luca V. Jasmonate modulates development and light-regulated alkaloid biosynthesis in Catharanthus roseus. Phytochemistry. 1998;49:395–402. doi: 10.1016/s0031-9422(98)00176-9. [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. Biosynthesis of flavonoids and effects of stress. Current Opinion in Plant Biology. 2002;5:218–223. doi: 10.1016/s1369-5266(02)00256-x. [DOI] [PubMed] [Google Scholar]
- Zagotta MT, Hicks KA, Jacobs CI, Young JC, Hangarter RP, Meeks-Wagner DR. The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. The Plant Journal. 1996;10:691–702. doi: 10.1046/j.1365-313x.1996.10040691.x. [DOI] [PubMed] [Google Scholar]
- Zhang W, Gruszewski HA, Chevone BI, Nessler CL. An Arabidopsis purple acid phosphatase with phytase activity increases foliar ascorbate. Plant Physiology. 2008;146:431–440. doi: 10.1104/pp.107.109934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.