Abstract
The induction of plant defences and their subsequent suppression by insects is thought to be an important factor in the evolutionary arms race between plants and herbivores. Although insect oral secretions (OS) contain elicitors that trigger plant immunity, little is known about the suppressors of plant defences. The Arabidopsis thaliana transcriptome was analysed in response to wounding and OS treatment. The expression of several wound-inducible genes was suppressed after the application of OS from two lepidopteran herbivores, Pieris brassicae and Spodoptera littoralis. This inhibition was correlated with enhanced S. littoralis larval growth, pointing to an effective role of insect OS in suppressing plant defences. Two genes, an ERF/AP2 transcription factor and a proteinase inhibitor, were then studied in more detail. OS-induced suppression lasted for at least 48 h, was independent of the jasmonate or salicylate pathways, and was not due to known elicitors. Interestingly, insect OS attenuated leaf water loss, suggesting that insects have evolved mechanisms to interfere with the induction of water-stress-related defences.
Keywords: Arabidopsis thaliana, defence suppression, gene expression, insect oral secretions, wounding
Introduction
In their continuing battle against herbivorous insects, plants have evolved sophisticated recognition and adaptation mechanisms to withstand insect attack. In response to herbivory, plants produce proteins and metabolites that interfere with insect physiology (Kessler and Baldwin, 2002). For instance, protease inhibitors (PI) inactivate digestive enzymes in the gut and reduce insect performance (Ryan, 1990) whereas chitinases and cysteine proteases target the chitin-rich membrane that lines the gut epithelium (Howe and Jander, 2008). Examples of secondary metabolites that are toxic to insects include nicotine in Solanaceae or glucosinolates in Brassicaceae (Baldwin and Preston, 1999; Halkier and Gershenzon, 2006). In addition, attacked plants emit volatiles that attract predatory mites and parasitic wasps (Paré and Tumlinson, 1999).
Although herbivory leads to tissue damage, plant responses to chewing insects are more complex than a simple wound reaction. Qualitative and quantitative differences between mechanical wounding and insect attack have been documented (Baldwin, 1988; Stout et al., 1994; Alborn et al., 1997; Korth and Dixon, 1997; McCloud and Baldwin, 1997; Lawrence and Novak, 2004; Maffei et al., 2004). At the molecular level, microarray experiments have revealed different gene expression patterns between mechanical wounding and insect herbivory (Reymond et al., 2000; Schittko et al., 2001; Roda et al., 2004; Major and Constabel, 2006; Ralph et al., 2006). During feeding, macerated plant tissues come into contact with insect oral secretions (OS), which contain labial and mandibular saliva mixed with regurgitant. Several studies demonstrated that application of insect OS to artificial wounds can mimic most plant responses to herbivory (Mattiacci et al., 1995; Alborn et al., 1997; Halitschke et al., 2003; Maffei et al., 2004; Reymond et al., 2004; Lawrence et al., 2008; De Vos and Jander, 2009; Erb et al., 2009), suggesting that elicitors in OS constitute the principal source of information by which plants recognize insect attack. Indeed, several elicitors have been isolated from insect OS and trigger plant defences against herbivory. For example, β-glucosidase from Pieris brassicae induces the release of a volatile blend when applied to wounded cabbage leaves, causing the attraction of the parasitoid Cotesia glomerata (Mattiacci et al., 1995). Fatty acid–amino acid conjugates (FACs) are produced by many lepidopteran caterpillars and induce direct and indirect defence responses in plants (Alborn et al., 1997; Pohnert et al., 1999; Halitschke et al., 2001; Mori et al., 2003; Roda et al., 2004). Caeliferins in OS of the American bird grasshopper, Schistocerca americana, are sulphated fatty acids that induce a release of volatiles when applied to damaged leaves of corn seedlings (Alborn et al., 2007). Inceptins from Spodoptera frugiperda OS are proteolytic fragments of a chloroplastic ATP synthase γ-subunit that are generated in the insect midgut and induce defences in cowpea and beans (Schmelz et al., 2006).
Just as plants have evolved specific mechanisms to recognize and fend off herbivores, these have, in turn, developed strategies to cope with plant defences. Apart from their exquisite ability to tolerate or detoxify plant defence compounds (Despres et al., 2007), insects release effectors that suppress defences. However, contrary to the case of bacterial, fungal or oomycete pathogenesis where hundreds of effectors delivered into host cells have been documented (Da Cunha et al., 2007; Ellis et al., 2009), much less is known about defence suppression by insects. Application of OS from Manduca sexta to N. attenuata suppressed both wound-induced expression of a threonine deaminase gene and nicotine accumulation (Kahl et al., 2000; Schittko et al., 2001). This effect was triggered by FACs that inhibited the induction of several transcripts (Halitschke et al., 2001). Another known suppressor is glucose oxidase (GOX), present in the saliva of several caterpillars. GOX application inhibited nicotine production in tobacco (Eichenseer et al., 1999; Musser et al., 2002, 2005) and the induction of a defence gene in alfalfa (Bede et al., 2006). An unknown 10–30 kDa compound in OS of the Colorado potato beetle Leptinotarsa decemlineata reduced wound-induced accumulation of PI transcripts in tomato (Lawrence et al., 2007). However, in most of these examples, an effect of defence suppression on insect performance was not tested. Recently, a study reported that the spider mite Tetranychus evansi suppressed the induction of tomato defences, leading to a better performance of mites that subsequently attacked these plants (Sarmento et al., 2011).
Regarding the suppression of defence responses in Arabidopsis, only one study has suggested that insect OS can down-regulate gene expression. S. exigua caterpillars with impaired salivary secretions triggered a higher expression of defence genes compared with intact caterpillars, but whether this resulted in enhanced larval performance was not tested (Weech et al., 2008). In order to get a deeper insight into the role of insect OS in Arabidopsis, a genome-wide screen was carried out to identify wound-inducible genes whose expression was significantly reduced by OS treatment. It was found that OS from both a specialist insect, Pieris brassicae, and a generalist insect, Spodoptera littoralis suppressed the expression of several genes and that OS-treated plants were more susceptible to insect feeding than wounded plants.
Materials and methods
Plant material and growth conditions
Wild-type Arabidopsis thaliana (L.) Heynh. ecotype Columbia (Col-0) plants were grown for 5–6 weeks in a growth room (22 °C, 65% relative humidity, 100 μmol m−2 s−1, 10/14 h light/dark photoperiod). Arabidopsis coi1-1 (non-glabrous) was obtained from Jane Glazebrook (University of Minnesota, St Paul, MN), sid2-1 from Christiane Nawrath (University of Lausanne, Lausanne, Switzerland), and fad3-2 fad7-2 fad8 from John Browse (Washington State University, Pullman, WA). Homozygous seedlings of male sterile coi1-1 were selected on Murashige and Skoog (MS) medium (2% sucrose, 0.8% agar, and 4.3 g l−1 MS) containing 50 μM jasmonic acid (JA) as described previously (Xie et al., 1998) and were transferred to soil after 10 d of growth.
Insect rearing and sampling of oral secretions
Spodoptera littoralis (Egyptian cotton worm) eggs were obtained from Syngenta (Stein, Switzerland) and were stored at 10 °C until further use. Eggs were placed in a beaker covered with plastic film in a growth chamber (22 °C, 65% relative humidity, 100 μmol m−2 s−1, 10/14 h light/dark photoperiod) to allow hatching. Pieris brassicae (large white butterfly) was reared in a greenhouse on cabbage plants (Brassica oleracea). One day before collecting oral secretions (OS), fourth- to fifth-instar larvae were placed on Arabidopsis plants. OS was collected by gently squeezing larvae manually and placing a pipette tip at their mouth. OS was then stored at –80 °C in Eppendorf tubes.
Plant treatments
Plants were 5–6-weeks-old at the time of treatment. Four holes (1 mm diameter with a cork-borer) were made on five leaves of each of two plants. To each hole, 1 μl of insect OS was immediately applied after perforation and plants were placed in a growth room (22 °C, 65% relative humidity, 100 μmol m−2 s−1, 10/14 h light/dark photoperiod) for different times. At the end of the treatment, treated leaves were collected in liquid nitrogen and stored at –80 °C. Untreated plants were used as controls.
A preliminary experiment showed that undiluted or 2–5-fold diluted OS were as effective in suppressing gene expression (data not shown). For the boiling experiment, OS was boiled for 4 min at 100 °C before use. For the filtering experiment, OS was filtered through 3 kDa cut-off Microcon™ filters (Millipore AG, Zug, Switzerland) for 60 min at 12 000 rpm. The flow-through was applied to leaf holes. For the FACs treatments, N-linolenoyl-L-glutamine (18:3-GLN), N-linolenoyl-L-glutamic acid (18:3-GLU), N-linoleoyl-L-glutamine (18:2-GLN), and N-linoleoyl-L-glutamic acid (18:2-GLN) were obtained from Dr Wilhelm Boland (Max Planck Institute for Chemical Ecology, Jena, Germany). Stock solutions were prepared in EtOH at 100 mM and were diluted to 1 mM in water after sonication. Volicitin was obtained from Peggy Brennan (USDA, Gainesville, FL) and used as a standard. For GOX treatment, glucose oxidase from Aspergillus niger (Sigma-Aldrich, Basel, Switzerland) was dissolved in 10 mM sodium phosphate buffer pH 7.0 and 0.0015 U was applied to each hole.
For experiments with signalling mutants, ERF/AP2 TF and protease inhibitor expression was measured after 6 h and 24 h, respectively, in coi1-1 plants and after 10 h in sid2-1 plants.
Measurement of water loss
For OS effects on dehydration, four holes were made on 7–8 leaves of each of four plants. To each hole, 1 μl of insect OS was immediately applied after perforation. Treated leaves (30 in total per treatment) were cut at the base of the petiole and placed on a filter paper at room temperature in dim light. Leaf fresh weight was recorded at different times after the treatment.
Insect bioassays
Twelve second-instar S. littoralis larvae were weighed and placed on three Arabidopsis plants in one pot. Just before infestation, four holes (1 mm diameter with a cork-borer) were made on five leaves of each of three plants. To each hole, 1 μl of S. littoralis OS was applied and plants were placed in a growth room (25 °C, 50% relative humidity, 110 μmol m−2 s−1, 16/8 h light/dark photoperiod). Untreated plants were used as controls. Each pot was surrounded by a transparent plastic tube, thereby enclosing the larvae, but the top of the tube was left open for aeration. Larvae were fed with treated plants every 24 h for three consecutive days and their final weight was recorded at the end of the experiment. This experiment was repeated eight times.
In the second bioassay, three freshly hatched S. littoralis larvae were placed on each of 12 plants (two plants per pot) that were wounded or treated with P. brassicae OS. Plants were placed in a transparent plastic box and maintained in a growth room for 7 d (22 °C, 65% relative humidity, 100 μmol m−2 s−1, 10/14 h light/dark photoperiod). Larval weight was measured at the end of the treatment. This experiment was repeated three times.
Glucose oxidase activity in OS
For GOX activity, 200 μl of glucose and sucrose solution (33 g l−1 each) was added to glass fibre discs (Whatman). To each disc, 50 μl of insect OS or 50 μl of a GOX solution (0.1 U μl−1 in 10 mM sodium phosphate buffer, pH 7.0) were spotted with 100 μl of horseradish peroxidase (0.016 U μl−1 in 10 mM sodium phosphate buffer, pH 7.0) and 100 μl of 3,3′-diaminobenzidine (DAB). The activity of GOX was indicated by the appearance of a black-brown precipitate.
RNA extraction and cDNA synthesis
Plant tissue was quickly frozen in liquid nitrogen and ground with a cold mortar and pestle. The powder was immediately put into a cold 1.5 ml Eppendorf tube and RNA was extracted with RNeasy Plant Mini Kit (Qiagen, Hombrechtikon, Switzerland) followed by a DNaseI treatment according to the manufacturer’s protocol. For cDNA synthesis, 1 μg of RNA and 1 μg of oligo dT (1 μg μl−1) were completed to a volume of 15.25 μl with nuclease-free water. Samples were heated at 65 °C for 5 min and then placed on ice for 5 min. After that, 5 μl of M-MLV reverse transcriptase buffer (5×), 2.5 μl of DTT (0.1 M), 1.25 μl of dNTPs (10 mM), and 1 μl of M-MLV reverse transcriptase (Invitrogen, Basel, Switzerland) was added to each sample. Reverse transcription was performed in a PCR machine with the following program: 40 °C for 5 min, 50 °C for 50 min, and 70 °C for 15 min. After a brief centrifugation and a dilution in 75 μl water, cDNA was stored at –80 °C until use. cDNA samples were generated in triplicate from each biological replicate.
Real-time quantitative PCR
The expression of At5g61890 and At3g22620 genes was analysed by quantitative real-time PCR (qPCR) using the fluorescent intercalating dye SYBR-Green. Specific qPCR primers were designed with the following criteria: 24-length optimal primer size, 150–300 product size and Tm between 68–74°C with an optimum at 71 °C. The reference gene was the eukaryote translation elongation factor EIF4A1 (At3g13920) whose expression does not changed in response to wounding (Bonaventure et al., 2003). Gene specific primers were the following: EIF4A1, At3g13920-forward (5′-CCAGAAGGCACACAGTTTGAT-3′), At3g13920-reverse (5′-AGACTGAGCCTGTTGAATCAC-3′); PI, At3g22620-forward (5′-TGTCTCACTTCCCCGTGCTTGTAA-3′), At3g22620-reverse (5′-CTCAGA AGGTCGACTGGTGCTTCC-3′); ERF/AP2 TF, At5g61890-forward (5′-GTCTTCTCGGCCCGATCTCAACAC-3′), At5g61890-reverse (5′-CTCGGATTTCAGCTGCCCACT TTC-3′). For each cDNA, qPCR was performed with the FullVelocity SYBR green kit (Agilent Technologies, Basel, Switzerland) in a final volume of 25 μl containing 12.5 μl of 2× SYBR, 3.75 μl of ROX (1/5000 dilution), 4.25 μl of RNAse-free water, 2.5 μl of primer mix (each primer at 1 μM) and 2 μl of cDNA. Reactions were generated in a qPCR machine (MX3000P™, Agilent Technologies, Basel, Switzerland) with the following program: 95 °C for 10 min; then 45 cycles of 10 s at 95 °C, 20 s at 55 °C, and 30 s at 60 °C. Primer efficiencies (E ) were assessed by a five-step dilution regression. The expression level of a target gene (TG) was normalized to the reference gene (RG) and calculated as Normalized Relative Quantity (NRQ) as follows: NRQ=E CtRG/E CtTG.
Microarray experiments and data analysis
For microarray analyses, four 1 mm holes were punctured on five leaves of each of six plants and 1 μl of insect OS was applied to each hole. Treated leaves were harvested 6 h and 24 h after each treatment and immediately stored in liquid nitrogen. Leaves from six untreated plants were collected as controls. Total RNA was extracted and labelled according to a previously published procedure (Bodenhausen and Reymond, 2007). Each experiment was replicated three times independently. A microarray containing 22 473 Arabidopsis gene-specific tags (CATMA; Hilson et al., 2004; Allemeersch et al., 2005) was used for the experiments. Hybridization, scanning, and data analysis were performed as described previously (Reymond et al., 2004). Genes up-regulated by mechanical wounding were selected based on a threshold of a 1.5-fold change in gene expression and a P value <0.05. To select genes suppressed by the application of OS to mechanical wounds, genes were identified whose expression ratio in response to wounding was significantly larger than the expression ratio after OS treatment (Student’s t test, P <0.05).
Results
Identification of genes suppressed by insect OS
Insect feeding by chewing herbivores is a combination of mechanical wounding and contact with OS. It is, however, experimentally difficult to assess the contribution of each factor to gene expression changes. Indeed, in contrast to bacterial pathogens where genes responsible for the secretion of effectors can easily be mutated, generating insects that lack OS, for instance by ablating salivary glands, is technically challenging and does not necessarily remove all OS components (Musser et al., 2006). To identify Arabidopsis genes whose wound-induction might be suppressed by insect OS, a protocol was followed that has been successful in identifying OS-specific responses (Halitschke et al., 2003; Wu et al., 2008). To mimic wounding caused by feeding larvae, small holes were punctured manually in Arabidopsis leaves, whereas, to mimic insect feeding, S. littoralis OS was applied to the holes. We used a microarray containing 22473 Arabidopsis gene-specific tags (Hilson et al., 2004; Allemeersch et al., 2005) and analysed expression changes after wounding or treatment with insect OS. Genes that were significantly induced by wounding were first identified by comparing wounded leaves with untreated leaves (>1.5-fold, Student’s t test, P <0.05). Then, amongst wound-induced genes, a search was made for genes that were suppressed by OS application. Specifically, these genes were selected if their induction by wounding plus OS treatment was significantly lower than the induction by wounding alone (Student’s t test, P <0.05).
Overall, 274 genes were found that were significantly induced by wounding after 6 h and 47 genes after 24 h (see Supplementary Table S1 at JXB online). According to our criteria, eight genes were suppressed after 6 h of OS treatment and five genes were suppressed after 24 h (Table 1). There are two protease inhibitors (At3g22600, At3g22620) that might interfere with proteases in the insect digestive tract and several cell-wall-associated proteins, including two glycine-rich proteins (At3g20470, At4g18280), an hydroxyproline-rich glycoprotein (At5g09530), an arabinogalactan protein (At4g09030), and a protein involved in wax biosynthesis (CER2, At4g24510). Finally, the list contains other proteins of diverse function, including a member of the ERF (ethylene response factor) subfamily B-4 of the ERF/AP2 transcription factor family (At5g61890).
Table 1.
Genes suppressed by Spodoptera littoralis oral secretions (OS) Genes induced by wounding after 6 h or 24 h were selected from microarray data (expression ratio >1.5 and P value <0.05, Student’s t test). Then, genes suppressed by Spodoptera littoralis OS were selected if the expression ratio in response to wounding was significantly larger than the expression ratio in response to OS treatment. Expression ratios (±SE) are the average of six (wounding) and three (OS) independent experiments. Genes in bold were selected for further experiments. P values refer to the comparison between wounding and OS treatment (Student’s t test).
| Expression ratio | ||||
| AGI code | Description | Wounding | S. littoralis OS | P value |
| Treatment 6 h | ||||
| At5g61890 | ERF/AP2 transcription factor | 3.32±0.59 | 1.41±0.28 | <0.001 |
| At3g20470 | Glycine-rich protein (GRP-5) | 2.15±0.15 | 1.22±0.12 | <0.001 |
| At4g18280 | Glycine-rich protein | 2.04±0.15 | 1.22±0.04 | <0.001 |
| At1g73330 | Protease inhibitor (DR4) | 1.96±0.26 | 1.11±0.11 | <0.001 |
| At1g05300 | Metal transporter (ZIP5) | 1.89±0.30 | 0.96±0.10 | <0.01 |
| At4g24510 | Eceriferum protein (CER2) | 1.62±0.09 | 1.07±0.05 | <0.001 |
| At1g75900 | Extracellular lipase 3 (EXL3) | 1.57±0.30 | 0.83±0.12 | <0.05 |
| At1g51090 | Heavy-metal-associated protein | 1.55±0.14 | 1.01±0.15 | <0.01 |
| Treatment 24 h | ||||
| At3g22620 | Protease inhibitor | 5.34±0.62 | 1.60±0.28 | <0.001 |
| At5g09530 | Hydroxyproline-rich glycoprotein | 5.69±0.71 | 1.46±0.32 | <0.001 |
| At5g05340 | Peroxidase | 2.96±0.35 | 1.56±0.18 | <0.001 |
| At1g28400 | Unknown protein | 2.35±0.38 | 1.33±0.06 | <0.001 |
| At4g09030 | Arabinogalactan-protein (AGP10) | 1.90±0.14 | 1.35±0.20 | <0.001 |
To confirm that the observed suppression has biological relevance, our data were compared with similar experiments carried out previously in our laboratory. Using a microarray containing c. 7200 Arabidopsis genes, expression changes in response to feeding by Pieris rapae or S. littoralis larvae, and in response to wounding, had been measured (Reymond et al., 2004). Six of the 13 suppressed genes identified in the present study had probes on this microarray. Although the experimental set-up was different, five genes had a significantly weaker induction in response to insect feeding than in response to wounding, providing an independent confirmation of our results (see Supplementary Table S2 at JXB online).
OS application to wounds enhances insect performance
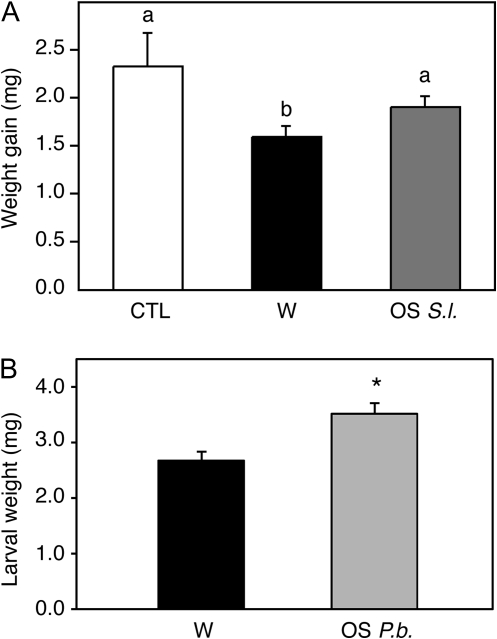
Our observation that OS application to wounds suppressed the induction of several genes potentially involved in defence suggested that chewing insects have evolved this mechanism to counteract the wound response of the plant. It was therefore tested whether OS treatment might enhance the performance of feeding larvae. Second-instar S. littoralis larvae were placed on Arabidopsis plants that were either intact, wounded, or treated with S. littoralis OS. Every day for three consecutive days, larvae were transferred to freshly treated plants. Larvae gained significantly more weight on intact plants than on wounded plants, confirming that plants activate defence genes in response to wounding. However, larvae grew significantly better on plants treated with OS than on wounded plants, suggesting that the suppression of wound-inducible genes was correlated with a higher insect performance (Fig. 1A).
Fig. 1.
Insect OS suppress Arabidopsis defences. (A) Weight gain of S. littoralis larvae feeding on control plants (white bar), wounded plants (black bar), or on plants treated with S. littoralis OS (dark grey bar) was measured after 3 d. Every day, 12 larvae were transferred to freshly treated plants. Values (±SE) are the mean of eight independent measurements. Bars with different letters differ at P value <0.05 (Student–Newman–Keuls Method). (B) Weight gain of S. littoralis larvae feeding on wounded plants (black bar) or on plants treated with P. brassicae OS (light grey bar) was measured after 7 d. Values (±SE) are the mean of three independent experiments. *P value <0.05 (Student’s t test).
In a similar experiment, it was tested if OS from one insect reduces plant defences that are effective against another insect. Indeed, freshly hatched S. littoralis gained more weight after feeding for 7 d on plants that had been treated once with P. brassicae OS compared with larvae feeding on plants that had been wounded (Fig. 1B). Thus, our data are consistent with the hypothesis that insect OS reduce plant defences by interfering with the induction of wound-responsive genes.
Kinetics of OS-triggered gene suppression
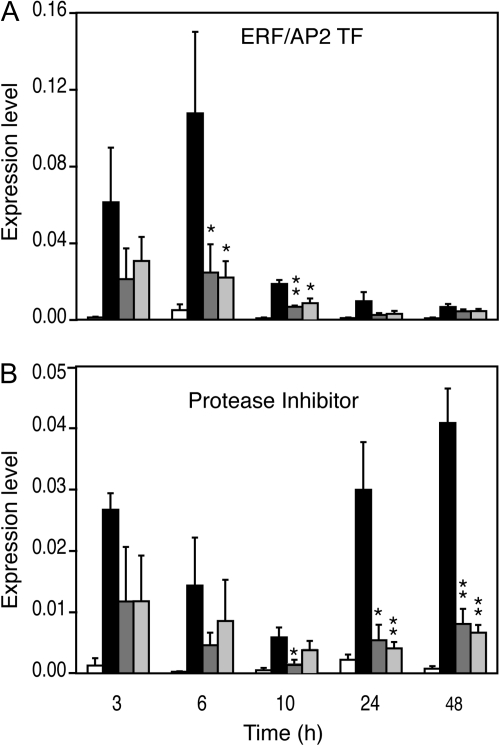
To characterize the suppression of wound-responsive genes by insect OS further, two genes were selected from the list of suppressed genes (Table 1), an ERF/AP2 transcription factor (At5g61890, ERF/AP2 TF) and a protease inhibitor (At3g22620, PI). To test whether the difference in expression between wounding and S. littoralis OS application was really due to a suppression and not to a delay in gene induction between these two treatments, the effect of OS application was measured from 3 h to 48 h by real-time quantitative PCR (qPCR). OS from the specialist lepidopteran herbivore P. brassicae was also used in the same experiment. Suppression of ERF/AP2 TF was observed at all time points and was similar for both insect OS, with a marked effect 6 h and 10 h after treatment (Fig. 2A). For PI, suppression by both OS was relatively weak in the first 6 h but was significant from 10 h to 48 h (Fig. 2B). These data indicate that different insect OS suppress wound-induced gene expression in Arabidopsis.
Fig. 2.
Kinetics of suppression of wound-responsive genes by insect oral secretions (OS). Expression of ERF/AP2 TF (A) and Protease Inhibitor (B) was monitored for several hours after wounding (black bars), and after treatment with S. littoralis OS (dark grey bars), or P. brassicae OS (light grey bars) by qPCR. Untreated plants were used as controls (white bars). Values (±SE) are normalized to the reference gene and each time point is the mean of at least three biological replicates. Statistical differences between wounding and OS treatment are indicated (Student’s t test, *P value <0.05, **P value <0.01).
Suppression of wound-responsive genes is independent of jasmonic acid and salicylic acid pathways
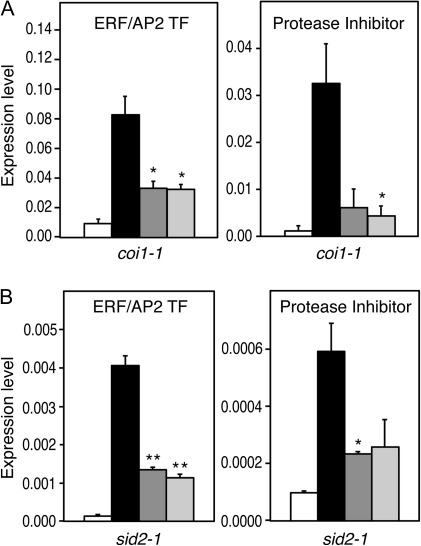
Jasmonic acid (JA) plays a major signalling role in defence against chewing insects in different plant species (McConn et al., 1997; Kessler et al., 2004; Li et al., 2004; Reymond et al., 2004). In N. attenuata, OS from tobacco hornworm Manduca sexta elicits a JA burst that is responsible for the induction of defence genes (Halitschke et al., 2003; Diezel et al., 2009). To test whether ERF/AP2 TF and PI suppression requires a functional JA pathway, an Arabidopsis coi1-1 mutant that is insensitive to JA was used (Xie et al., 1998). OS from S. littoralis and P. brassicae suppressed the wound-induction of ERF/AP2 TF and PI to a similar extent in both wild-type and coi1-1 plants (Fig. 3A), indicating that the suppression of ERF/AP2 TF and PI is JA-independent.
Fig. 3.
Suppression of wound-responsive genes is independent of JA and SA pathways. Expression of ERF/AP2 TF and Protease Inhibitor was monitored by qPCR after wounding (black bars), and after treatment with S. littoralis OS (dark grey bars), or P. brassicae OS (light-grey bars) on the jasmonate-insensitive mutant coi1-1 (A) and the SA-deficient mutant sid2-1 (B). Untreated plants were used as controls (white bars). Values (±SE) are normalized to the reference gene and are the mean of two biological replicates. Statistical differences between wounding and OS treatment are indicated (Student’s t test, *P value <0.05, **P value <0.01).
Salicylic acid (SA) is a potent inducer of pathogenesis-related genes and is involved in resistance against biotrophic pathogens (Glazebrook, 2005). Recently, a study found that OS from beet armyworm (Spodoptera exigua) elicit SA accumulation in N. attenuata and that this might interfere with JA-mediated resistance (Diezel et al., 2009). Similarly, suppression of two defence genes by S. exigua OS was alleviated in Arabidopsis mutants of the SA pathway (Weech et al., 2008). It was therefore tested if suppression of ERF/AP2 TF and PI was affected in the SA-deficient mutant sid2-1 (Nawrath and Metraux, 1999). Again, S. littoralis and P. brassicae OS suppressed the wound-induction of ERF/AP2 TF and PI in both wild-type and sid2-1 plants, suggesting that SA was not required for the suppression of these genes (Fig. 3B).
Role of known OS elicitors in suppressing wound-responsive genes
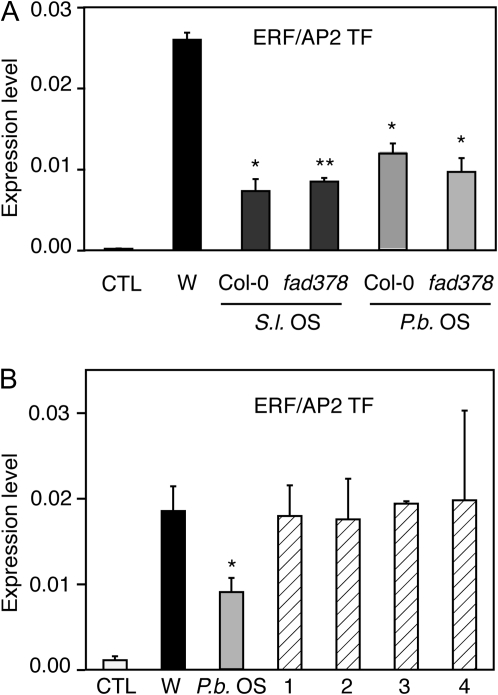
Fatty acid–amino acid conjugates (FACs) are present in the OS of several lepidopteran caterpillars (Pohnert et al., 1999) and modulate the expression of defence genes (Halitschke et al., 2003). The fatty acid moiety originates from plant membranes and is conjugated to either GLN or GLU in the insect midgut (Paré et al., 1998). The FACs composition of S. littoralis and P. brassicae OS was analysed by HPLC. Major FACs found in S. littoralis OS were 18:3-GLN and 17-hydroxy-18:3-GLN (volicitin), while 18:2-GLN and 18:2-GLU were present at much lower levels (see Supplementary Fig. S1 at JXB online). For P. brassicae OS, only a small peak corresponding to 18:3-GLN could be detected. To assess the contribution of 18:3-derived FACs in the suppression of ERF/AP2 TF, caterpillars were fed with Arabidopsis fatty-acid desaturase triple mutant fad3fad7fad8 that lacks 18:3 fatty acids (McConn and Browse, 1996). Consequently, OS from insects feeding on fad3fad7fad8 contained no 18:3-derived FACs, but had increased levels of 18:2-derived FACs (not shown). Suppression of ERF/AP2 TF by OS from insects fed with Col-0 or fad3fad7fad8 plants was similar, indicating that the absence of 18:3-derived FACs did not diminish OS suppressive activity (Fig. 4A). In addition, when purified FACs were applied to wounded leaves, no suppression of ERF/AP2 TF was observed, suggesting that these elicitors are not involved in this phenomenon (Fig. 4B). Similarly, fad3fad7fad8-derived OS also suppressed PI wound-induction whereas purified FACs did not (not shown).
Fig. 4.
Role of FACs in the suppression of wound-responsive genes. Expression of ERF/AP2 TF was monitored by qPCR after wounding (black bars), and after treatment with S. littoralis OS (dark grey bars), P. brassicae OS (light grey bars), or FACs (hatched bars) for 10 hr. Untreated plants were used as controls (white bars). (A) Larvae were fed with Col-0 or fad378 mutant plants lacking 18:3. (B) 1, 18:3-GLN; 2, 18:3-GLU; 3, 18:2-GLN; 4, 18:2-GLU. Values (±SE) are normalized to the reference gene and are the mean of two biological replicates. Statistical differences between wounding and OS treatment are indicated (Student’s t test, *P value <0.05, **P value <0.01).
Glucose oxidase (GOX) is found in the saliva of lepidopteran larvae and is involved in the suppression of herbivore-induced resistance in Solanaceous plants and in Medicago truncatula (Musser et al., 2002, 2005; Bede et al., 2006). GOX activity was detected in S. littoralis OS but not in P. brassicae OS (see Supplementary Fig. S2 at JXB online), although both secretions equally suppressed wound-induced gene expression. When purified GOX was applied to wounded Arabidopsis leaves, no suppression of ERF/AP2 TF was observed (see Supplementary Fig. S2 at JXB online). In addition, P. brassicae OS that were boiled or filtered through a 3 kDa cut-off filter kept their suppressive activity (see Supplementary Fig. S2 at JXB online), suggesting that it is caused by small non-enzymatic compounds.
Most lepidopteran larvae have an alkaline pH in the midgut. P. brassicae and S. littoralis OS used in our experiments have a pH of 8.5–9.0 and this could affect the expression of defence genes. However, intact P. brassicae OS and P. brassicae OS that were passed through an ion-exchange column and had a pH of 3.5 inhibited ERF/AP2 TF and PI expression to a similar extent, thus excluding that a pH effect was the cause of this suppression (not shown).
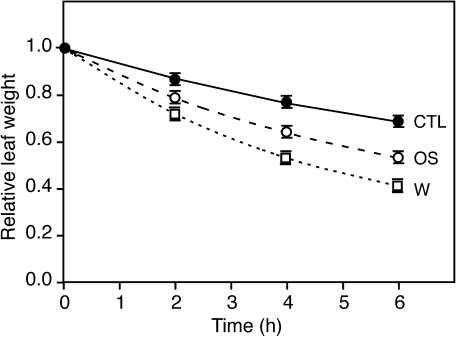
OS treatment attenuates wound-induced leaf water loss
An analysis of expression profiles of suppressed genes (Table 1) in publicly available microarray databases, carried out with the Genevestigator tool (http://www.genevestigator.com) (Zimmermann et al., 2004), indicated that some of these genes are induced in response to drought. It was therefore reasoned that the suppressive effect of OS might be related to a difference in water loss between wounded and OS-treated leaves. To test this hypothesis, the water loss of detached Arabidopsis leaves, that were either intact, wounded, or treated with OS, was measured. Whereas intact leaves lost only 31±2% of their initial fresh weight over a period of 6 h, wounded leaves lost 59±2% of their weight (Fig. 5). Interestingly, leaves treated with OS lost significantly less water than wounded leaves (47±2%), but still more than intact leaves, indicating that OS application on the wound site was attenuating leaf water loss. This effect was found with both P. brassicae and S. littoralis OS (see Supplementary Fig. S3 at JXB online).
Fig. 5.
Insect OS reduce wound-induced water loss. Leaves were wounded (open squares) or treated with P. brassicae OS (open circles). Untreated plants were used as controls (filled circles). Fresh weight of detached leaves was measured for 6 h. Values (±SE) are the mean of 30 leaves and are expressed relative to the initial fresh weight.
Discussion
In this study, several Arabidopsis genes were identified that had a significantly lower induction in response to insect OS treatment than in response to wounding. In addition, larvae gained more weight when feeding on OS-treated plants than on wounded plants. The most straightforward interpretation of these results is that compounds in the OS and/or some properties of these secretions are able to suppress the wound-induced expression of defence genes, leading to a better larval growth. This implies that the resistance provided by mechanical wounding is equivalent to the resistance caused by insect-induced mechanical damage, but that the latter is counteracted by the suppressive activity of OS. Although it is experimentally difficult to separate the wound response inflicted by insect feeding from the effects of OS, this hypothesis is corroborated by studies showing that the macroscopical damage caused by mechanical wounding is very similar to that caused by insect feeding (Maffei et al., 2006), that the majority of genes induced by herbivory are also induced by mechanical wounding (Reymond et al., 2004), and that feeding by insects with ablated spinnerets triggers a higher resistance to larvae than feeding by insects with intact spinnerets (Musser et al., 2002). Once the chemical nature of insect suppressors is known, further experiments will be necessary to demonstrate that plant treatment with purified suppressors is indeed able to attenuate wound-induced resistance caused by insect feeding.
Although there is some discussion about the exact amount of OS that are in contact with wounded tissue during insect feeding (Peiffer and Felton, 2009), our results unequivocally show that insect OS can suppress wound-induced gene expression in Arabidopsis. In addition, the finding that OS from two unrelated insects, a specialist and a generalist, have similar suppressive activity suggests that this corresponds to a general property of insect OS. There is substantial information on OS elicitors that amplify the wound response in plants, including FACs, hydrolytic enzymes, proteolytic degradations products, and sulphated fatty acids (Zhu-Salzman et al., 2005; Howe and Jander, 2008). However, much less is known about the nature of OS that can suppress defence gene activation. It is shown here that FACs and GOX are not responsible for suppressing wound-inducible genes in Arabidopsis. Moreover, the finding that boiled OS as well as OS components smaller than 3 kDa are still active in suppressing gene expression excludes other known salivary elicitors of proteinaceous nature, including a β-glucosidase present in P. brassicae OS (Mattiacci et al., 1995), or a heat-sensitive protein from Colorado potato beetle OS (Lawrence et al., 2007).
Using coi1-1 and sid2-1 mutants, it has been shown that the suppressive activity of insect OS in Arabidopsis was independent of the JA and SA signalling pathways. That the suppression is independent of the JA pathway is rather surprising, given the importance of this pathway in defence against herbivory. Indeed, JA controls the expression of a majority of insect-inducible genes (Reymond et al., 2004) and mutant plants deficient in JA signalling, including coi1-1, are more susceptible to insect pests than wild-type plants (Kessler et al., 2004, Li et al., 2004; Reymond et al., 2004). Similarly to bacterial effectors that inhibit signalling components of the innate immune response (Da Cunha et al., 2007), it would have been reasonable to postulate that insect OS inhibit the JA pathway. Although it is possible that insect OS target signalling components that act downstream of COI1, for example, JAZ repressors or MYC transcription factors, it is likely that the suppression operates through another mechanism. Thus, further studies will be needed to identify the exact nature of suppressors in P. brassicae and S. littoralis OS and to unravel signalling events leading to the inhibition of defence gene expression in Arabidopsis. In addition, it would be interesting to test whether a wound-induced resistance is observed in coi1-1 plants to assess the contribution of JA-independent gene expression in defence against herbivores.
Interestingly, leaves treated with OS lost significantly less water than wounded leaves. Insect OS is a complex mixture composed of saliva mixed with ingested leaf material. A study of the physico-chemical properties of several lepidopteran OS has shown that they have ampiphilic properties, independent of insect diet, and spread on hydrophobic glass surfaces (Rostas and Blassmann, 2009). OS intrinsic detergent properties might modify plant membranes at the wound site and either ‘seal’ the surface opened to the air as a result of biting or interfere with water stress signalling pathway. However, a different water loss was not observed between wounded leaves and leaves treated with a detergent (0.01% Triton X-100) or with a FAC (1 mM 18:3-GLN) (data not shown), excluding this hypothesis. Insect OS generate channel-like pores of varying conductivity in plant membranes (Luhring et al., 2007). These pores might be responsible for the early events that follow insect attack, including membrane depolarization and ion fluxes (Maffei et al., 2004, 2006; Maischak et al., 2007). Whether OS reduce water stress through physico-chemical effects or through signalling events will be the subject of future research.
Although there is convincing evidence for a suppression of defence genes by insect OS, a direct effect on insect performance has rarely been demonstrated (Musser et al., 2002; Sarmento et al., 2011). It was found that S. littoralis larvae grew better on plants treated with OS than on wounded plants, suggesting that OS efficiently inhibited plant defences. The list of suppressed genes contains two protease inhibitors (At3g22600, At3g22620) that might inhibit larval digestion or development. Other genes encode proteins located in the extracellular matrix and might either participate in cell wall reinforcement or modify the digestibility of its components. The role of the ERF/AP2 transcription factor is unknown but it will be interesting to test in the future whether it regulates the expression of defence genes, for instance, by testing transgenic lines that over-express this gene. However, our findings that the suppression of wound-responsive genes by insect OS is correlated with an increased larval performance strongly suggests that these genes play a role in defence against herbivory.
Chewing herbivores are not the only types of insects that suppress defences. Aphids ingesting phloem sap from sieve tubes trigger a calcium-dependent plugging mechanism by the plant to block sap loss. Saliva from Megoura viciae contains calcium-binding proteins that prevent sieve tube plugging and thus provides aphids with a continuous flow of nutrients (Will et al., 2007). The spider mite Tetranychus urticae displays intraspecific variation and some lines were shown to suppress the defences of tomato plants and to perform much better on their host (Kant et al., 2008; Sarmento et al., 2011). Since mites inject saliva into the cells of their host plants, it is plausible that saliva from these lines contains suppressors. Thus, there is increasing evidence that insects, like microbial and fungal pathogens, are equipped with diverse strategies and effectors to inhibit plant defence responses.
In conclusion, it was found that OS from two lepidopteran species suppress the induction of wound-responsive genes, by potentially inhibiting plant water-stress responses, and that this is correlated to an enhanced performance of feeding larvae. Our results are consistent with the notion that expression of plant defence may result from the antagonistic actions of inductive and suppressive signals, illustrating the long-lasting evolutionary arms race between plants and insects.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Table S1. List of genes up-regulated 6 h or 24 h after wounding.
Supplementary Table S2. Suppression of wound-responsive genes by insect OS or insect herbivory.
Supplementary Fig. S1. HPLC chromatogram of insect OS.
Supplementary Fig. S2. OS suppression of wound-responsive genes does not require an enzymatic activity.
Supplementary Fig. S3. P. brassicae and S. littoralis OS reduce wound-induced water loss.
Acknowledgments
We thank Wilhelm Boland, Mathias Funke, and Peggy Brennanfor for the gift of FACs. Roland Reist (Syngenta, Stein, Switzerland) kindly provided Spodoptera littoralis eggs. Luc Beauverd, Olivier Emery, and Wouter Kegge helped with the experiments. This work was supported by the Swiss National Science Foundation (grant number 31003A_132915 to PR).
References
- Alborn HT, Hansen TV, Jones TH, Bennett DC, Tumlinson JH, Schmelz EA, Teal PEA. Disulfoxy fatty acids from the American bird grasshopper Schistocerca americana, elicitors of plant volatiles. Proceedings of the National Academy of Sciences, USA. 2007;104:12976–12981. doi: 10.1073/pnas.0705947104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alborn HT, Turlings TCJ, Jones TH, Stenhagen G, Loughrin JH, Tumlinson JH. An elicitor of plant volatiles from beet armyworm oral secretion. Science. 1997;276:945–949. [Google Scholar]
- Allemeersch J, Durinck S, Vanderhaeghen R, et al. Benchmarking the CATMA microarray. A novel tool for Arabidopsis transcriptome analysis. Plant Physiology. 2005;137:588–601. doi: 10.1104/pp.104.051300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin IT. The alkaloidal responses of wild tobacco to real and simulated herbivory. Oecologia. 1988;77:378–381. doi: 10.1007/BF00378046. [DOI] [PubMed] [Google Scholar]
- Baldwin IT, Preston CA. The eco-physiological complexity of plant responses to insect herbivores. Planta. 1999;208:137–145. [Google Scholar]
- Bede JC, Musser RO, Felton GW, Korth KL. Caterpillar herbivory and salivary enzymes decrease transcript levels of Medicago truncatula genes encoding early enzymes in terpenoid biosynthesis. Plant Molecular Biology. 2006;60:519–531. doi: 10.1007/s11103-005-4923-y. [DOI] [PubMed] [Google Scholar]
- Bodenhausen N, Reymond P. Signaling pathways controlling induced resistance to insect herbivores in. Arabidopsis. Molecular Plant–Microbe Interactions. 2007;20:1406–1420. doi: 10.1094/MPMI-20-11-1406. [DOI] [PubMed] [Google Scholar]
- Bonaventure G, Salas JJ, Pollard MR, Ohlrogge JB. Disruption of the FATB gene in Arabidopsis demonstrates an essential role of saturated fatty acids in plant growth. The Plant Cell. 2003;15:1020–1033. doi: 10.1105/tpc.008946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Cunha L, Sreerekha MV, Mackey D. Defense suppression by virulence effectors of bacterial phytopathogens. Current Opinion in Plant Biology. 2007;10:349–357. doi: 10.1016/j.pbi.2007.04.018. [DOI] [PubMed] [Google Scholar]
- De Vos M, Jander G. Myzus persicae (green peach aphid) salivary components induce defence responses in Arabidopsis thaliana. Plant, Cell and Environment. 2009;32:1548–1560. doi: 10.1111/j.1365-3040.2009.02019.x. [DOI] [PubMed] [Google Scholar]
- Despres L, David JP, Gallet C. The evolutionary ecology of insect resistance to plant chemicals. Trends in Ecology and Evolution. 2007;22:298–307. doi: 10.1016/j.tree.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Diezel C, von Dahl CC, Gaquerel E, Baldwin IT. Different lepidopteran elicitors account for cross-talk in herbivory-induced phytohormone signaling. Plant Physiology. 2009;150:1576–1586. doi: 10.1104/pp.109.139550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenseer H, Mathews MC, Bi JL, Murphy JB, Felton GW. Salivary glucose oxidase. multifunctional roles for Helicoverpa zea? Archives of Insect Biochemistry and Physiology. 1999;42:99–109. doi: 10.1002/(SICI)1520-6327(199909)42:1<99::AID-ARCH10>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Ellis JG, Rafiqi M, Gan P, Chakrabarti A, Dodds PN. Recent progress in discovery and functional analysis of effector proteins of fungal and oomycete plant pathogens. Current Opinion in Plant Biology. 2009;12:399–405. doi: 10.1016/j.pbi.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Erb M, Flors V, Karlen D, de Lange E, Planchamp C, D’Alessandro M, Turlings TCJ, Ton J. Signal signature of above-ground-induced resistance upon belowground herbivory in maize. The Plant Journal. 2009;59:292–302. doi: 10.1111/j.1365-313X.2009.03868.x. [DOI] [PubMed] [Google Scholar]
- Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annual Review of Phytopathology. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- Halitschke R, Gase K, Hui D, Schmidt DD, Baldwin IT. Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. VI. Microarray analysis reveals that most herbivore-specific transcriptional changes are mediated by fatty acid–amino acid conjugates. Plant Physiology. 2003;131:1894–1902. doi: 10.1104/pp.102.018184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halitschke R, Schittko U, Pohnert G, Boland W, Baldwin IT. Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. III. Fatty acid–amino acid conjugates in herbivore oral secretions are necessary and sufficient for herbivore- specific plant responses. Plant Physiology. 2001;125:711–717. doi: 10.1104/pp.125.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkier BA, Gershenzon J. Biology and biochemistry of glucosinolates. Annual Review of Plant Biology. 2006;57:303–333. doi: 10.1146/annurev.arplant.57.032905.105228. [DOI] [PubMed] [Google Scholar]
- Hilson P, Allemeersch J, Altmann T, et al. Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Research. 2004;14:2176–2189. doi: 10.1101/gr.2544504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GA, Jander G. Plant immunity to insect herbivores. Annual Review of Plant Biology. 2008;59:41–66. doi: 10.1146/annurev.arplant.59.032607.092825. [DOI] [PubMed] [Google Scholar]
- Kahl J, Siemens DH, Aerts RJ, Gabler R, Kuhnemann F, Preston CA, Baldwin IT. Herbivore-induced ethylene suppresses a direct defense but not a putative indirect defense against an adapted herbivore. Planta. 2000;210:336–342. doi: 10.1007/PL00008142. [DOI] [PubMed] [Google Scholar]
- Kant MR, Sabelis MW, Haring MA, Schuurink RC. Intraspecific variation in a generalist herbivore accounts for differential induction and impact of host plant defences. Proceedings of the Royal Society B, Biological Sciences. 2008;275:443–452. doi: 10.1098/rspb.2007.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler A, Baldwin IT. Plant responses to insect herbivory: the emerging molecular analysis. Annual Review of Plant Biology. 2002;53:299–328. doi: 10.1146/annurev.arplant.53.100301.135207. [DOI] [PubMed] [Google Scholar]
- Kessler A, Halitschke R, Baldwin IT. Silencing the jasmonate cascade: Induced plant defenses and insect populations. Science. 2004;305:665–668. doi: 10.1126/science.1096931. [DOI] [PubMed] [Google Scholar]
- Korth KL, Dixon RA. Evidence for chewing insect-specific molecular events distinct from a general wound response in leaves. Plant Physiology. 1997;115:1299–1305. doi: 10.1104/pp.115.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence SD, Novak NG. Maize genes induced by herbivory and volicitin. Journal of Chemical Ecology. 2004;30:2543–2557. doi: 10.1007/s10886-004-7949-8. [DOI] [PubMed] [Google Scholar]
- Lawrence SD, Novak NG, Blackburn MB. Inhibition of proteinase inhibitor transcripts by Leptinotarsa decemlineata regurgitant in Solanum lycopersicum. Journal of Chemical Ecology. 2007;33:1041–1048. doi: 10.1007/s10886-007-9285-2. [DOI] [PubMed] [Google Scholar]
- Lawrence SD, Novak NG, Ju CJ-T, Cooke JEK. Potato, Solanum tuberosum, defense against Colorado potato beetle, Leptinotarsa decemlineata (Say): microarray gene expression profiling of potato by Colorado potato beetle regurgitant treatment of wounded leaves. Journal of Chemical Ecology. 2008;34:1013–1025. doi: 10.1007/s10886-008-9507-2. [DOI] [PubMed] [Google Scholar]
- Li L, Zhao YF, McCaig BC, Wingerd BA, Wang JH, Whalon ME, Pichersky E, Howe GA. The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. The Plant Cell. 2004;16:126–143. doi: 10.1105/tpc.017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhring H, Nguyen VD, Schmidt L, Rose USR. Caterpillar regurgitant induces pore formation in plant membranes. FEBS Letters. 2007;581:5361–5370. doi: 10.1016/j.febslet.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Maffei M, Bossi S, Spiteller D, Mithofer A, Boland W. Effects of feeding Spodoptera littoralis on lima bean leaves. I. Membrane potentials, intracellular calcium variations, oral secretions, and regurgitate components. Plant Physiology. 2004;134:1752–1762. doi: 10.1104/pp.103.034165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffei ME, Mithofer A, Arimura GI, et al. Effects of feeding Spodoptera littoralis on lima bean leaves. III. Membrane depolarization and involvement of hydrogen peroxide. Plant Physiology. 2006;140:1022–1035. doi: 10.1104/pp.105.071993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maischak H, Grigoriev PA, Vogel H, Boland W, Mithofer A. Oral secretions from herbivorous lepidopteran larvae exhibit ion channel-forming activities. FEBS Letters. 2007;581:898–904. doi: 10.1016/j.febslet.2007.01.067. [DOI] [PubMed] [Google Scholar]
- Major IT, Constabel CP. Molecular analysis of poplar defense against herbivory: comparison of wound- and insect elicitor-induced gene expression. New Phytologist. 2006;172:617–635. doi: 10.1111/j.1469-8137.2006.01877.x. [DOI] [PubMed] [Google Scholar]
- Mattiacci L, Dicke M, Posthumus MA. Beta-glucosidase: an elicitor of herbivore-induced plant odor that attracts host-searching parasitic wasps. Proceedings of the National Academy of Sciences, USA. 1995;92:2036–2040. doi: 10.1073/pnas.92.6.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloud ES, Baldwin IT. Herbivory and caterpillar regurgitants amplify the wound- induced increases in jasmonic acid but not nicotine in. Nicotiana sylvestris. Planta. 1997;203:430–435. [Google Scholar]
- McConn M, Browse J. The critical requirement for linolenic acid is pollen development, not photosynthesis in an Arabidopsis mutant. The Plant Cell. 1996;8:403–416. doi: 10.1105/tpc.8.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn M, Creelman RA, Bell E, Mullet JE, Browse J. Jasmonate is essential for insect defense in. Arabidopsis. Proceedings of the National Academy of Sciences, USA. 1997;94:5473–5477. doi: 10.1073/pnas.94.10.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori N, Yoshinaga N, Sawada Y, Fukui M, Shimoda M, Fujisaki K, Nishida R, Kuwahara Y. Identification of volicitin-related compounds from the regurgitant of lepidopteran caterpillars. Bioscience, Biotechnology and Biochemistry. 2003;67:1168–1171. doi: 10.1271/bbb.67.1168. [DOI] [PubMed] [Google Scholar]
- Musser RO, Cipollini DF, Hum-Musser SM, Williams SA, Brown JK, Felton GW. Evidence that the caterpillar salivary enzyme glucose oxidase provides herbivore offense in solanaceous plants. Archives of Insect Biochemistry and Physiology. 2005;58:128–137. doi: 10.1002/arch.20039. [DOI] [PubMed] [Google Scholar]
- Musser RO, Farmer E, Peiffer M, Williams SA, Felton GW. Ablation of caterpillar labial salivary glands: technique for determining the role of saliva in insect–plant interactions. Journal of Chemical Ecology. 2006;32:981–992. doi: 10.1007/s10886-006-9049-4. [DOI] [PubMed] [Google Scholar]
- Musser RO, Hum-Musser SM, Eichenseer H, Peiffer M, Ervin G, Murphy JB, Felton GW. Herbivory: caterpillar saliva beats plant defences. Nature. 2002;416:599–600. doi: 10.1038/416599a. [DOI] [PubMed] [Google Scholar]
- Nawrath C, Metraux JP. Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. The Plant Cell. 1999;11:1393–1404. doi: 10.1105/tpc.11.8.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paré PW, Alborn HT, Tumlinson JH. Concerted biosynthesis of an insect elicitor of plant volatiles. Proceedings of the National Academy of Sciences, USA. 1998;95:13971–13975. doi: 10.1073/pnas.95.23.13971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paré PW, Tumlinson JH. Plant volatiles as a defense against insect herbivores. Plant Physiology. 1999;121:325–331. [PMC free article] [PubMed] [Google Scholar]
- Peiffer M, Felton GW. Do caterpillars secrete ‘oral secretions’? Journal of Chemical Ecology. 2009;35:326–335. doi: 10.1007/s10886-009-9604-x. [DOI] [PubMed] [Google Scholar]
- Pohnert G, Jung V, Haukioja E, Lempa K, Boland W. New fatty acid amides from regurgitant of lepidopteran (Noctuidae, Geometridae) caterpillars. Tetrahedron. 1999;55:11275–11280. [Google Scholar]
- Ralph SG, Yueh H, Friedmann M, et al. Conifer defence against insects: microarray gene expression profiling of Sitka spruce (Picea sitchensis) induced by mechanical wounding or feeding by spruce budworms (Choristoneura occidentalis) or white pine weevils (Pissodes strobi) reveals large-scale changes of the host transcriptome. Plant, Cell and Environment. 2006;29:1545–1570. doi: 10.1111/j.1365-3040.2006.01532.x. [DOI] [PubMed] [Google Scholar]
- Reymond P, Bodenhausen N, Van Poecke RM, Krishnamurthy V, Dicke M, Farmer EE. A conserved transcript pattern in response to a specialist and a generalist herbivore. The Plant Cell. 2004;16:3132–3147. doi: 10.1105/tpc.104.026120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE. Differential gene expression in response to mechanical wounding and insect feeding in. Arabidopsis. The Plant Cell. 2000;12:707–720. doi: 10.1105/tpc.12.5.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roda A, Halitschke R, Steppuhn A, Baldwin IT. Individual variability in herbivore-specific elicitors from the plant's perspective. Molecular Ecology. 2004;13:2421–2433. doi: 10.1111/j.1365-294X.2004.02260.x. [DOI] [PubMed] [Google Scholar]
- Rostas M, Blassmann K. Insects had it first: surfactants as a defence against predators. Proceedings of the Royal Society B, Biological Sciences. 2009;276:633–638. doi: 10.1098/rspb.2008.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CA. Protease inhibitors in plants: genes for improving defenses against insects and pathogens. Annual Review of Phytopathology. 1990;28:245–249. [Google Scholar]
- Sarmento RA, Lemos F, Bleeker PM, Schuurink RC, Pallini A, Oliveira MGA, Lima ER, Kant M, Sabelis MW, Janssen A. A herbivore that manipulates plant defence. Ecology Letters. 2011;14:229–236. doi: 10.1111/j.1461-0248.2010.01575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittko U, Hermsmeier D, Baldwin IT. Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. II. Accumulation of plant mRNAs in response to insect-derived cues. Plant Physiology. 2001;125:701–710. doi: 10.1104/pp.125.2.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz EA, Carroll MJ, LeClere S, Phipps SM, Meredith J, Chourey PS, Alborn HT, Teal PEA. Fragments of ATP synthase mediate plant perception of insect attack. Proceedings of the National Academy of Sciences, USA. 2006;103:8894–8899. doi: 10.1073/pnas.0602328103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout MJ, Workman J, Duffey SS. Differential induction of tomato foliar proteins by arthropod herbivores. Journal of Chemical Ecology. 1994;20:2575–2594. doi: 10.1007/BF02036193. [DOI] [PubMed] [Google Scholar]
- Weech M-H, Chapleau M, Pan L, Ide C, Bede JC. Caterpillar saliva interferes with induced Arabidopsis thaliana defence responses via the systemic acquired resistance pathway. Journal of Experimental Botany. 2008;59:2437–2448. doi: 10.1093/jxb/ern108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will T, Tjallingii WF, Thonnessen A, van Bel AJE. Molecular sabotage of plant defense by aphid saliva. Proceedings of the National Academy of Sciences, USA. 2007;104:10536–10541. doi: 10.1073/pnas.0703535104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JQ, Hettenhausen C, Schuman MC, Baldwin IT. A comparison of two Nicotiana attenuata accessions reveals large differences in signaling induced by oral secretions of the specialist herbivore. Manduca sexta. Plant Physiology. 2008;146:927–939. doi: 10.1104/pp.107.114785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- Zhu-Salzman K, Bi J-L, Liu T- X. Molecular strategies of plant defense and insect counter-defense. Insect Science. 2005;12:3–15. [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.