Abstract
Sucrose nonfermenting-1 (SNF1)-related protein kinases (SnRKs) form a major family of signalling proteins in plants and have been associated with metabolic regulation and stress responses. They comprise three subfamilies: SnRK1, SnRK2, and SnRK3. SnRK1 plays a major role in the regulation of carbon metabolism and energy status, while SnRKs 2 and 3 have been implicated in stress and abscisic acid (ABA)-mediated signalling pathways. The burgeoning and divergence of this family of protein kinases in plants may have occurred to enable cross-talk between metabolic and stress signalling, and ABA-response-element-binding proteins (AREBPs), a family of transcription factors, have been shown to be substrates for members of all three subfamilies. In this study, levels of SnRK1 protein were shown to decline dramatically in wheat roots in response to ABA treatment, although the amount of phosphorylated (active) SnRK1 remained constant. Multiple SnRK2-type protein kinases were detectable in the root extracts and showed differential responses to ABA treatment. They included a 42 kDa protein that appeared to reduce in response to 3 h of ABA treatment but to recover after longer treatment. There was a clear increase in phosphorylation of this SnRK2 in response to the ABA treatment. Fractions containing this 42 kDa SnRK2 were shown to phosphorylate synthetic peptides with amino acid sequences based on those of conserved phosphorylation sites in AREBPs. The activity increased 8-fold with the addition of calcium chloride, indicating that it is calcium-dependent. The activity assigned to the 42 kDa SnRK2 also phosphorylated a heterologously expressed wheat AREBP.
Keywords: Abscisic acid, calcium, phosphorylation, signalling, stress, transcription factors
Introduction
Sucrose nonfermenting-1 (SNF1)-related protein kinases (SnRKs) form a major family of signalling proteins in plants and have been associated with metabolic regulation and stress responses (Halford and Hey, 2009; Hey et al., 2010). SnRKs have been grouped into three subfamilies: SnRK1, SnRK2, and SnRK3 (Halford, 2006). SnRK1, the homologue of adenosine monophosphate-activated protein kinase (AMPK) from mammals and SNF1 from yeast, has been implicated in the regulation of carbon metabolism and energy status (Halford and Hey, 2009; Smeekens et al., 2010). It controls metabolism at multiple levels; for example, it phosphorylates enzymes such as 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMG-CoA reductase) and sucrose phosphate synthase, leading to their inactivation. It also phosphorylates nitrate reductase, trehalose-phosphate synthase, and 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase, but these enzymes also require the binding of a 14-3-3 protein for inactivation (reviewed by Halford and Hey, 2009). Another key metabolic enzyme in plants, adenosine diphosphate (ADP)-glucose pyrophosphorylase, is regulated by SnRK1 through modulation of its redox state (Tiessen et al., 2003). In addition, SnRK1 causes changes in gene expression in response to nutrient starvation (Baena-González et al., 2007) and, paradoxically, sucrose (Purcell et al., 1998; McKibbin et al., 2006).
The other two SnRK subfamilies, SnRK2 and SnRK3, do not have any counterpart in fungal or animal cells (Halford and Hey, 2009). They both comprise relatively large gene families, with 10 SnRK2 genes described in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) (Boudsocq et al., 2004; Kobayashi et al., 2004) and 25 SnRK3 genes described in Arabidopsis (Halford and Hey, 2009). There is now convincing evidence to associate SnRK2 and SnRK3 with responses to abiotic stresses such as drought, salinity, cold, and osmotic stress (Hey et al., 2010). For example, a SnRK3 family member, SOS2 (salt overly sensitive 2), is involved in phosphorylating and activating SOS1, a Na+/H+ antiporter, in order to maintain ion homeostasis (Guo et al., 2002; Qiu et al., 2002). A number of SnRK2 family members have been shown to be directly up-regulated and activated by osmotic stress and some but not all of these are also activated by abscisic acid (ABA). The fact that not all osmotic stress-regulated SnRK2s are also activated by ABA indicates that activation of SnRK2 in response to osmotic stress and ABA involves different mechanisms (Boudsocq et al., 2004, 2007; Kobayashi et al., 2004). Recent evidence from Arabidopsis shows that these mechanisms involve different patterns of phosphorylation at two serine residues in the activation loop. SnRK2.6, for example, which is an ABA- and osmotic stress-induced kinase, is phosphorylated independently on both of these residues, while for SnRK2.10, which is induced by osmotic stress but not ABA, phosphorylation of one site is required for phosphorylation of the other (Vlad et al., 2010).
Recent studies have shown SnRK2s to be integral components of ABA signalling. Firstly, protein phosphatases type 2C (PP2C) were shown to be negative regulators of ABA signalling and to be involved in SnRK2 inactivation (Gosti et al., 1999; Kuhn et al., 2006). Yeast two-hybrid interactions were described between different PP2Cs and SnRK2 (Cutler et al., 2010) and further work demonstrated that, in the absence of ABA, PP2C inactivated SnRK2 by direct dephosphorylation of one of the serine residues in the activation loop (Umezawa et al., 2009). The elucidation of ABA signalling mechanisms then advanced significantly with the identification of a family of proteins named PYR/PYL/RCAR as ABA receptors (Nishimura et al., 2010). In the presence of ABA, PYR/PYL/RCARs were shown to bind to and inhibit PP2Cs, allowing the accumulation of active SnRK2s and subsequent phosphorylation of ABA-response-element-binding proteins (AREBPs) (Cutler et al., 2010). AREBPs (also known as ABFs) are a family of basic leucine zipper (bZIP) transcription factors that have been known for some time to be substrates for SnRK2 (Kobayashi et al., 2005; Furihata et al., 2006). They recognize the G-box-binding sites known as ABA-response elements present in some ABA-regulated genes (Cutler et al., 2010).
AREBPs also have two potential target sites for phosphorylation by SnRK1 that are conserved throughout the Arabidopsis AREBP family and in all of the AREBPs that have been identified so far in other species (Zhang et al., 2008). Peptides with amino acid sequences based on these sites have been shown to be phosphorylated by purified SnRK1 and by an activity in crude Arabidopsis extracts that is precipitated with SnRK1 antiserum. Furthermore, transgenic Arabidopsis plants over-expressing SnRK1 have been shown to have an ABA-hypersensitive phenotype (Jossier et al., 2009). Arabidopsis extracts also contain a calcium-dependent activity that phosphorylates the AREBP-derived peptides but which does not phosphorylate the AMARA peptide, which is an excellent substrate for SnRK1 and SnRK2. This activity has been tentatively assigned to SnRK3, which is believed to be calcium-dependent, although the involvement of ‘classic’ calcium-dependent protein kinases (CDPKs) cannot be ruled out (Zhang et al., 2008).
These results suggest that AREBPs could be convergence points for multiple signalling pathways involving all three SnRK subfamilies and possibly CDPKs as well, placing them at the interface between metabolic and stress signalling networks (Halford and Hey, 2009; Hey et al., 2010). They also add to the evidence showing calcium to be an important second messenger during ABA signalling. CDPKs are positive regulators of ABA signalling and have been shown to phosphorylate some AREBPs to stimulate gene expression (Choi et al., 2005; Zhu et al., 2007). These protein kinases are activated directly by calcium, but plants contain additional calcium sensors (Boudsocq and Sheen, 2010), including calcineurin B-like proteins (CBL). SnRK3s have been shown to interact with CBLs, and are sometimes referred to as CBL-interacting protein kinases (CIPKs). Two SnRK3s (CIPK15 and CIPK3) have been shown to be negative regulators of ABA signalling (Guo et al., 2002), and SnRK3s have also been shown to interact with ABA-insensitive (ABI) 1 and ABI2, which are also negative regulators of ABA signalling (Zhu et al., 2007).
Their integral role in metabolic and stress signalling pathways makes the transfer of research on SnRKs from Arabidopsis to crop plants particularly important, and this study focuses on wheat (Triticum aestivum). There has been some previous work on SnRKs in this species; for example, transient antisense expression of a SnRK1 gene has been shown to repress the activity of an α-amylase gene (α-Amy2) promoter after cobombardment into cultured wheat embryos (Laurie et al., 2003). In addition, several SnRK2s have been identified in wheat. PKABA1, for example, was isolated from an ABA-treated wheat embryo cDNA library (Anderberg and Walker-Simmons, 1992). It has a predicted molecular weight of 38 kDa and is up-regulated at the transcript level by osmotic, dehydration, and cold treatments in root, shoot and scutellar tissue (Holappa and Walker-Simmons, 1995). PKABA1 appears to be involved in antagonizing gibberellic acid-induced gene expression, down-regulating, for example, α-amylase genes in the barley (Hordeum vulgare) aleurone layer, and in the up-regulation of genes in response to ABA (Gomez-Cadenas et al., 1999; Johnson et al., 2008). TaABF, a wheat seed AREBP, has been shown to interact with PKABA1, and PKABA1 will phosphorylate three peptides with amino acid sequences corresponding to putative phosphorylation sites in TaABF (Johnson et al., 2002).
Another wheat SnRK2-encoding gene, TaSnRK2.4, is up-regulated by ABA, NaCl, polyethylene glycol, and low temperatures in seedling roots (Mao et al., 2010), while another, W55a, is expressed in leaves of seedlings treated with ABA or salt, or subjected to drought (Xu et al., 2009). TaSnRK2.7 is also reported to be involved in abiotic stress responses (Zhang et al., 2011a), while TaSnRK2.8 has been shown to enhance tolerance to drought, salt, and low-temperature stress when over-expressed in Arabidopsis (Zhang et al., 2010). SnRK2, like SnRK1, has also been proposed to be involved in sucrose signalling in wheat (Zhang et al., 2011b). Establishing the degree of functional redundancy between the different SnRK2s will be challenging.
In this study, the interactions between ABA, SnRK1 and SnRK2 were investigated in wheat and contrasting effects of ABA on SnRK1 and SnRK2 protein levels and phosphorylation state were shown in wheat roots. A 42 kDa SnRK2 that was present in wheat roots was shown to phosphorylate a heterologously expressed wheat AREBP considerably more efficiently than SnRK1. Unusually for a SnRK2, evidence was obtained to show this protein kinase to be activated by Ca2+.
Materials and methods
Biological materials and ABA treatments
Wheat (T. aestivum) cultivar Consort was germinated on wet paper towels. After germination, seedlings were transferred to pots containing perlite and allowed to grow for another 5 d under natural photoperiod (10 h light/14 h dark) at 16 °C. ABA treatments were performed by adding the hormone to the irrigation water at a final concentration of 50 μM. Samples were taken and frozen in liquid nitrogen until use.
Extraction of crude protein
Total soluble protein was extracted in ice-cold homogenization buffer [100 mM Tricine/NaOH, 5 mM dithiothreitol (DTT), 0.5 mM ethylene glycol tetra-acetic acid (EGTA), 0.5 mM ethylenediaminetetra-acetic acid (EDTA), and 1 mM benzamidine, pH 8.0]. Immediately before homogenization, phenylmethylsulphonyl fluoride (PMSF; 1 mM final concentration), 1× protease inhibitor cocktail (Calbiochem), phosphatase inhibitors (50 mM sodium fluoride, 25 mM β-glycerolphosphate, 10 mM sodium pyrophosphate and 2 mM sodium orthovanadate) and insoluble polyvinylpyrrolidone (2% w/v) were added. The homogenate was transferred to precooled microfuge tubes and insoluble material was removed by centrifugation (13 000 g) at 4 °C for 20 min. The crude extract was placed into aliquots, frozen in liquid nitrogen, and stored at −80 °C. Determination of protein concentration was as described by Bradford (1976).
Antisera and western blot analysis
Antiserum that had been raised against a peptide from the catalytic domain of SnRK1 (Fragoso et al., 2009) was used in some of the experiments. Antiserum was also raised to a peptide from a highly conserved region in SnRK2 (GYSKSSLLHSQPKST); this antiserum was raised in rabbits and affinity-purified by Eurogentec, Seraing, Belgium. A polyclonal antibody against the phospho-Thr-172 (P-Thr-172) present in the T-loop of AMPK was purchased from Cell Signalling (Danvers, MA, USA). For western blot analysis, 20 μg of protein was separated in 10% Novex NuPAGE BIS-TRIS gels (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Protein transfer onto polyvinylidene fluoride (PVDF) membranes was conducted in an XCell II Blot module (Invitrogen) according to the manufacturer’s instructions. Proteins were detected using an ECL Western Blot Detection Kit (GE Healthcare). Peptide blocking of the SnRK2 antibody reaction was achieved by pretreating the antiserum with 50 μg of the GYSKSSLLHSQPKST peptide.
Isolation of phosphoproteins
Phosphorylated proteins from roots after 10 h of ABA treatment were isolated using a Phosphoprotein Purification Kit (Qiagen), following the manufacturer’s instructions. After elution, fractions were concentrated using a Nanosep ultrafiltration column. Western blot analyses using the phosphoproteins were performed as described above.
Peptide phosphorylation assays
Peptides were synthesized by Biomol International (Exeter, UK): the peptides were AMARA (AMARAASAAALARRR) (Sugden et al., 1999b), AREBP1 (SLGRQSSIYSLTRRR), and AREBP2 (TLPRTLSQKTVDRRR) (Zhang et al., 2008). Assays were conducted in wells of 8×12 microtitre plates, each in a total volume of 25 μl, at 30 °C. The reaction mixture for each assay contained 40 mM HEPES, pH 7.5, 5 mM MgCl2, 200 μM ATP containing 12.5 kBq [γ-33P]ATP (GE Healthcare), 200 μM peptide substrate, 4 mM DTT, 0.5 μM okadaic acid, and 1× protease inhibitor cocktail; the assay was initiated by addition of the extract containing the protein kinase. After 6 min, a 15 μl aliquot of the reaction mixture was transferred to a square (2×2 cm) of phosphocellulose paper (Whatman P81; Whatman, Maidstone, Kent, UK), which was immediately immersed in 1% (v/v) phosphoric acid. The papers were washed with three 800 ml batches of 1% (v/v) phosphoric acid, followed by acetone, and placed onto paper towels to dry. Incorporation of 33P was quantified by liquid scintillation counting (Packard Tri-Carb 2100; Perkin Elmer, Waltham, MA, USA), following immersion of each paper square in scintillation cocktail (Ultima Gold Cocktail, Perkin Elmer). For calcium-dependent activity, 1 mM CaCl2 was included in the reaction mix.
In vitro phosphorylation of wheat AREBP (TaAREBP) and HMG-CoA reductase
The reaction mixture for each assay contained 40 mM HEPES, pH 7.5 (NaOH), 5 mM MgCl2, 200 μM ATP containing 2 μCi [γ-32P]ATP (3000 Ci mmol−1; GE Healthcare), 2 μg of substrate (recombinant TaAREBP protein or HMG-CoA reductase; Sigma), 4 mM DTT, 0.5 μM okadaic acid, 1× protease inhibitor cocktail in a total volume of 20 μl. The reaction was initiated by addition of the extract containing the protein kinase and incubated for 30 min at 30 °C. The reaction was stopped with 5 μl of 4× gel loading buffer. Proteins were heated at 70 °C for 5 min and loaded onto a 4–12% gradient Novex NuPAGE BIS-TRIS gel. Electrophoresis was carried out according to the manufacturer’s instructions. The proteins were transferred onto PVDF membranes using an XCell II Blot module according to manufacturer’s instructions. Radiolabelled proteins were detected by autoradiography at −80 °C using X-ray film (Kodak Biomax MR; Sigma).
Partial separation of SnRK1 and SnRK2
Wheat seedlings were grown and treated with ABA for 10 h as described above. Roots and leaves were separated, frozen in liquid nitrogen and stored at −80 °C. Root tissue (26 g) from control and ABA-treated plants was homogenized with 70 ml of buffer containing 50 mM Tricine, pH 8.0, 50 mM NaF, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 0.1 mM benzamidine, 0.1 mM PMSF, 0.02% Brij 35, 10% glycerol, and 100 mM NaCl. All the steps were performed at 4 °C. The homogenate was centrifuged at 10 000 g for 30 min. Supernatant was precipitated with 50% ammonium sulphate and stirred for 1 h at 4 °C. Proteins were sedimented by centrifugation at 10 000 g for 30 min. The pellet was resuspended in 1.5 ml of extraction buffer, passed through a 0.45 μm filter and frozen in liquid nitrogen until use. An aliquot (1 ml) of the filtered extract was applied to a Sephacryl 200 column (50×1.6 cm), equilibrated and calibrated in the extraction buffer. The column was run at 0.3 ml min−1 and 1 ml fractions were collected and assayed for AMARA, AREBP1, and AREBP2 kinase activities in the presence or absence of 1 mM CaCl2. The column was calibrated using alcohol dehydrogenase (150 kDa), albumin (66 kDa), and carbonic anhydrase (29 kDa) (Sigma).
Further separation of the putative calcium-dependent SnRK2 from other activities was achieved by pooling the activity from a Sephacryl column and loading it onto a Mono Q column (HR 5/5; Pharmacia). The column was equilibrated with buffer A (50 mM Tricine, pH 8.0, 50 mM NaF, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 1 mM benzamidine, 1 mM PMSF, 10% glycerol, and 50 mM NaCl) and non-binding protein was washed using the same buffer. Binding protein was then eluted using a linear gradient from 50 to 500 mM NaCl in the same buffer. Fractions with activity were identified using AREBP1 as substrate.
Molecular cloning and heterologous expression of a wheat AREBP (TaAREBP)
Frozen plants (100 mg) were ground into a fine powder under liquid nitrogen. RNA was extracted using the RNeasy Plant RNA Mini Kit (Qiagen) following the manufacturer’s instructions. cDNA synthesis was performed using a RETROscript Kit (Ambion) according to manufacturer’s instructions. Primers for amplifying AREBP cDNA from wheat were designed based on the published nucleotide sequences of three wheat cDNAs (Kobayashi et al., 2008):
Forward: 5′-ATGGACTTCAGGAGCAGCAACGGC-3′
Reverse: 5′-TCACCAGGGGCCGGTCAACGTTCT-3′
PCR reactions were performed in a 20 μl total volume containing 1 μl wheat cDNA template, 0.1 μl 100 μM forward and reverse primers, 0.2 units Phusion DNA polymerase (Finnzymes), 0.4 μl 10 mM dNTP mix, and 4 μl 5× Phusion HF buffer. The reaction tubes were placed in an Eppendorf Mastercycler using a 60–80 °C temperature gradient to establish optimal conditions. PCR products were initially cloned in the pGEM-T Easy vector (Promega UK, Southampton, UK) but for heterologous expression and purification of the wheat AREBP the PCR product was subcloned into the pET28 vector so that the protein would have a 6× His tag at the N-terminal end. Protein expression was performed in the protease-deficient Escherichia coli host strain BL21 and purification of the wheat AREBP in its native conformation was performed using the Ni-NTA Spin Kit (Qiagen) following the manufacturer’s instructions.
Results
SnRK1 and SnRK2 are differentially regulated by ABA in wheat roots
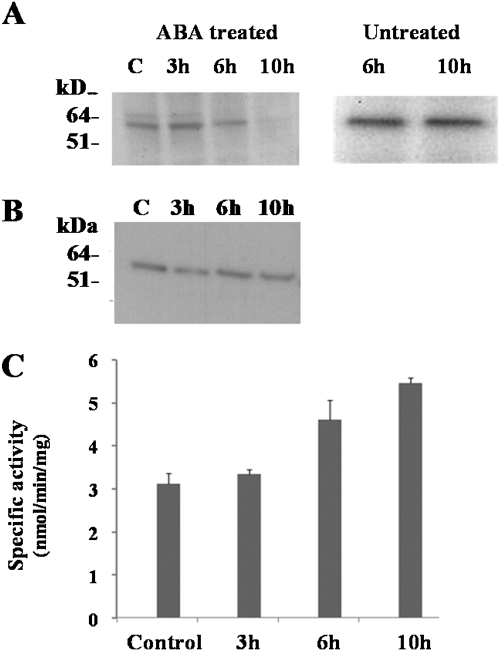
Western analysis was used to determine the amount of SnRK1 protein in extracts from roots of wheat plants treated with 50 μM ABA for 3, 6, and 10 h, using an antibody against a peptide from the protein kinase catalytic domain of SnRK1 (Fragoso et al., 2009). The results showed that the amount of SnRK1 protein decreased dramatically after 10 h of ABA treatment (Fig. 1A, left-hand panel). No such reduction was seen in untreated roots (Fig. 1A, right-hand panel). SnRK1 is activated by phosphorylation at a threonine residue in the so-called T-loop (Sugden et al., 1999a), equivalent to Thr-172 of its mammalian homologue, AMPK, and when the relative amount of phosphorylated (activated) SnRK1 protein in the treated roots was determined using an antibody that specifically recognizes the phosphorylated form of the protein kinase (the P-Thr-172 antibody) there was little difference between the extracts from control plants and those from plants treated with ABA (Fig. 1B). Furthermore, when SnRK activity in the extracts was measured using the AMARA peptide as a substrate it was found to have increased as a result of 6 h of ABA treatment and to have increased further after 10 h of treatment (Fig. 1C). In other words, despite the dramatic reduction in total SnRK1 protein, the amount of active SnRK1 remained the same and AMARA peptide kinase activity actually increased. Note that recognition of the phosphorylated form of SnRK1 by the P-Thr-172 antibody was first demonstrated in spinach (Sugden et al., 1999a). The amino acid sequences of the T-loop regions of spinach and wheat SnRK1 are aligned in Fig. 2 along with those of Arabidopsis SnRK1, human AMPK, and yeast (Saccharomyces cerevisiae) SNF1: the sequences are all extremely similar and those of wheat, spinach, and Arabidopsis are identical.
Fig. 1.
Regulation of SnRK1 during ABA treatment. (A) Left-hand panel: proteins isolated from a time-course of ABA-treated and control (untreated) roots were fractionated on a 10% Novex NuPAGE BIS-TRIS gel and transferred to PVDF membranes. SnRK1 was detected by western blot using a specific antibody. Right-hand panel: Western blot analysis of proteins from untreated roots after 6 h and 10 h. C, control. (B) Western blot analysis of phosphorylated SnRK1 detected using an antibody raised against the P-Thr-172 from mammalian AMPK. (C) AMARA peptide kinase activity in protein extracts from a time-course of ABA-treated and control (untreated) roots.
Fig. 2.
Alignment of the amino acid residues in the regulatory T-loop region of human AMPK (GenBank accession no. AAB32732), yeast (S. cerevisiae) protein kinase SNF1 ( M13971), and SnRK1 from wheat (AJ431365), Arabidopsis (M93023), and spinach (Crawford et al., 2001). The P-Thr-172 antibody recognizes the phosphorylated forms of AMPK, SNF1, and SnRK1 (Sugden et al., 1999b). The phosphorylated threonine residue is indicated with an arrow.
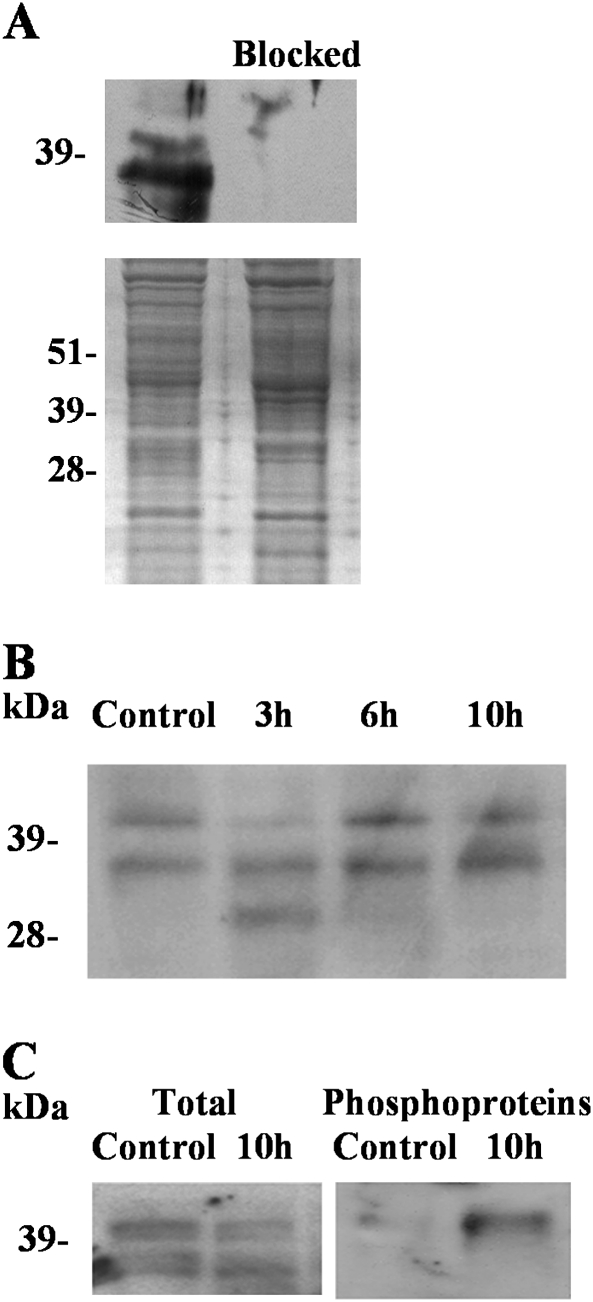
The AMARA peptide is regarded as a specific substrate for SnRK1 and SnRK2 (Sugden et al., 1999b; Zhang et al., 2008). The approximate doubling of AMARA peptide kinase activity in the absence of a clear increase in the levels of activated SnRK1 and the findings of studies that have revealed a direct connection between ABA signal transduction and SnRK2 family members therefore prompted an investigation of SnRK2. Antiserum was raised against a peptide with amino acid sequence GYSKSSLLHSQPKST, corresponding to a conserved region in the catalytic domain of SnRK2. A BLAST search (conducted on 27 January 2011) of the non-redundant protein sequence database using the National Centre for Biotechnology Information (NCBI) portal, with default settings, matched this sequence with approximately 200 plant SnRK2-type protein kinase sequences in the database. The list of 1000 best matches contained no other sequences with fewer than four mismatches and no other protein kinases. In western analyses, this antiserum recognized two proteins, of 38 and 42 kDa, in the control roots (Fig. 3A); this was consistent with the predicted size for SnRK2-type protein kinases, which in Arabidopsis ranges from 38 to 42 kDa (Hrabak et al., 2003). The reaction of the antiserum with this protein was completely blocked by pretreatment with the GYSKSSLLHSQPKST peptide. The amount of the 42 kDa protein appeared to decline in response to 3 h of ABA treatment but to recover to the levels in the controls after 6 and 10 h of treatment (Fig. 3B), whereas the 38 kDa protein was unchanged. A third protein, of approximately 34 kDa, appeared after 3 h of treatment but disappeared after 6 and 10 h (Fig. 3B).
Fig. 3.
Regulation of SnRK2 during ABA treatment. (A) Western blot analysis of proteins extracted from wheat roots. SnRK2 proteins were detected using an antibody raised to peptide GYSKSSLLHSQPKST (top panel, left). An identical blot was incubated with the antiserum after the antiserum had been pre-treated with an excess of the peptide (top panel, Blocked). A stained gel of the protein extracts is shown in the lower panel. (B) Western blot analysis showing SnRK2 in proteins extracted from wheat roots treated with no ABA (Control) or with ABA for 3, 6 or 10 h. (C) Western blot analysis showing SnRK2 in proteins extracted from control roots and roots treated for 10 h with ABA. Left-hand panel, crude extracts; right-hand panel, phosphoprotein fraction.
Like SnRK1, SnRK2 is activated by phosphorylation. Phosphoproteins were therefore purified from the extracts of control roots and roots that had been treated with ABA for 10 h and western blot analysis was performed on the total proteins and phosphoprotein fractions using the anti-SnRK2 antibody (Fig. 3C). This analysis confirmed that the levels of the 42 and 38 kDa proteins detected by the antibody in the total protein fractions were not affected by the treatment. However, the 42 kDa protein was clearly present in the phosphoprotein-enriched fraction of the ABA-treated roots, while there was only a trace in the phosphoprotein-enriched fraction of the untreated roots. The 38 kDa protein was not detected at all in the phosphoprotein-enriched fractions.
Evidence that the 42 kDa SnRK2 is calcium-dependent and that it phosphorylates AREBP1 and AREBP2 peptides
It has been demonstrated that SnRK2 plays an integral role in ABA signalling by phosphorylating some of the AREBPs that are responsible for ABA-induced transcription (Kobayashi et al., 2005). To test whether the 42 kDa SnRK2 that was activated in wheat roots in response to ABA could be involved in AREBP regulation, two peptides, AREBP1 and 2, were tested as substrates for the protein kinase. These peptides contain target sites for SnRK1 and 2 that are conserved in AREBPs from plant species including Arabidopsis, wheat, barley, and rice and had been shown previously to be substrates for both SnRK1 and SnRK2 in Arabidopsis extracts, and for highly purified SnRK1 from spinach (Spinacea oleracea) (Zhang et al., 2008).
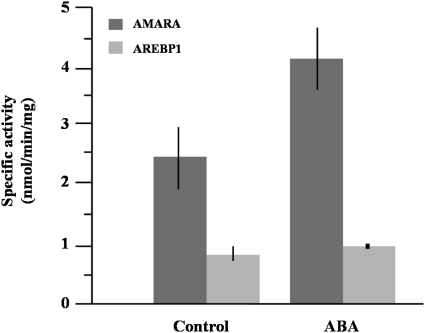
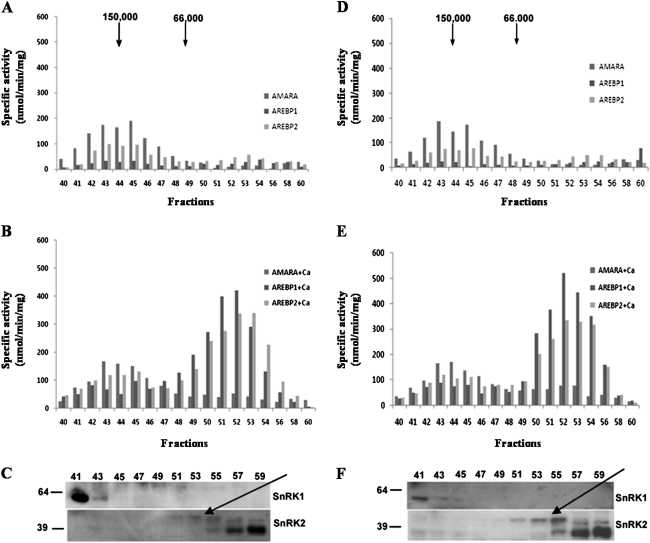
Using the AREBP1 peptide as a substrate with crude protein obtained from control and ABA-treated roots, less activity was found than with the AMARA peptide as substrate (Fig. 4), and there was no indication of an increase in phosphorylation of the AREBP1 peptide in response to ABA treatment, while phosphorylation of AMARA increased as before. To investigate further, a partial fractionation through a Sephacryl S-200 column was performed to separate SnRK1 from SnRK2. Loading the same amount of protein obtained from the control and 10 h ABA-treated roots, 1 ml fractions were collected and assayed for AMARA, AREBP1, and AREBP2 phosphorylation activity. The profile of activity obtained for untreated roots is shown in Fig. 5A. The main peak of activity for all three peptides was around 150 kDa, which is consistent with the approximate size of heterotrimeric SnRK1. This was confirmed by western blot analysis of the fractions (Fig. 5C), which showed that SnRK1 antiserum reacted with an approximately 58 kDa protein in these fractions; the predicted size of the catalytic subunit of SnRK1 is 58 kDa. There appeared to be more protein in slightly higher-molecular-mass fractions than the fractions with most activity. This may indicate that inactive protein was complexed with different interacting factors than active protein. Consistent with this was the observation that ABA reduced the amount of protein in the higher but not the lower-molecular-mass fractions containing this activity (Fig. 5C and F, fractions 41–45). The ‘preferred’ substrate for this activity was AMARA, followed by AREBP2 and AREBP1. There was a smaller peak of activity in lower-molecular-mass fractions. The preferred substrate for this activity was AREBP2, followed by AREBP1 and AMARA.
Fig. 4.
Activity of SnRK1 measured using AMARA and AREBP1 peptides. Proteins obtained from control and 10 h ABA-treated roots were used to measure the kinase activity with the AMARA and AREBP1 peptides as substrates. Bars represent means ±SD (n=3).
Fig. 5.
Profile of activity of proteins isolated from ABA-treated (10 h) and untreated roots after fractionation through a Sephacryl S-200 column and western blot analysis evaluating the presence of SnRK1 and SnRK2 in fractions from the column. Activity was evaluated using AMARA, AREBP1, and AREBP2 as substrates in the absence or presence of 1 mM CaCl2. SnRK1 antiserum was raised against a peptide from the catalytic domain of SnRK1 (Fragoso et al., 2009) while SnRK2 antiserum was raised to a peptide from a highly conserved region in SnRK2. (A) Activity profile of proteins from ABA-untreated roots, no calcium added. (B) As for (A) with calcium added. (C) Western blot analysis evaluating the presence of SnRK1 and SnRK2 in ABA-untreated roots. (D) Activity profile of proteins from ABA-treated roots, no calcium added. (E) As (D) with calcium added. (F) Western blot analysis evaluating the presence of SnRK1 and SnRK2 in ABA-treated roots. The 42 kDa SnRK2 is indicated with an arrow. Fraction numbers are indicated and the positions of 39 and 64 kDa molecular-mass markers are shown in (C) and (F).
Surprisingly, when protein extracted from ABA-treated roots was used, the profile of activity did not change (Fig. 5D). However, when CaCl2 was added to the reactions to a concentration of 1 mM a different picture emerged. In both control (untreated) and ABA-treated roots (Fig. 5B and E, respectively) the high-molecular-mass fractions had higher activity towards both AREBP1 and AREBP2, while activity towards the AMARA peptide did not change. This indicated that SnRK1 activity did not change but that these fractions also contained a calcium-dependent protein kinase that preferred the AREBP peptides as substrates. In a previous study a calcium-dependent activity in Arabidopsis seedlings that phosphorylated AREBP1 and 2 but not AMARA was tentatively assigned to a SnRK3-type protein kinase (Zhang et al., 2008) and it is possible that a SnRK3 was responsible for the calcium-dependant activity in these fractions from wheat roots. This activity did not appear to be affected by ABA treatment. However, the activity peak in the lower-molecular-mass fractions showed a dramatic (eight-fold) increase when Ca2+ was present and the AREBP peptides were used as substrates. Furthermore, the activity was higher in the extracts from ABA-treated roots: the total activity from fractions 49–56 was 1760 nmol min−1 mg−1 for the control and 2230 nmol min−1 mg−1 for the ABA samples, a difference of almost 30% (Fig. 5B and E). Western blot analysis with the SnRK2 antiserum showed the presence of the 42 kDa SnRK2 in the fractions containing this calcium-dependent activity (Fig. 5C and F). In this peak, there appeared to be more protein in slightly lower-molecular-mass fractions than the fractions with most activity, again possibly indicating that active and inactive protein was complexed with different interacting factors. The 38 kDa SnRK2 was also detected by the antiserum but not until fraction 55.
This represented strong immunological evidence that the 42 kDa SnRK2 was responsible for the major calcium-dependent activity in the extracts. The possibility that another protein kinase could have been copurified with the 42 kDa SnRK2 and be responsible for the calcium-dependent activity in the fractions was considered. However, the other possible candidates—SnRK3, CDPKs and CDPK-related protein kinases (CRKs)—are all larger than 42 kDa. In Arabidopsis, CDPKs range from 54 to 68 kDa and CRKs from 64 to 68 kDa (Hrabak et al., 2003). SnRK3s may be smaller but few have been characterized in detail. Hrabak et al. (2003), for example, reported the size of SnRK3.17, accession number At2g26980, as 43 kDa, but the most recent GenBank database entry gives a predicted size of 50 kDa. The smallest SnRK3 in the GenBank database now is SnRK3.4 at 46 kDa but this too is derived from genome nucleotide sequence data with no further experimental confirmation of the protein’s size. The other SnRK3s range from 48 to 58 kDa. As an additional check, the activity was pooled and loaded onto a Mono Q column. The activity that was recovered in fractions from this column was measured using the AREBP2 peptide as substrate and is shown in Fig. 6A. Some of the activity and protein was lost in this step, but Western blot analysis of the sample before loading onto the column and of the protein eluted in the fraction containing the highest activity confirmed the presence of the 42 kDa SnRK2 (Fig. 6B). The 38 kDa SnRK2 was not detectable in this fraction at all. The activity was completely eradicated by the addition of EGTA to chelate the Ca2+.
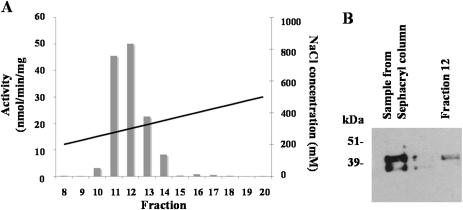
Fig. 6.
(A) Calcium-dependent activity peak recovered from a Mono Q column loaded with pooled fractions from a Sephacryl column containing the activity assigned to 42 kDa SnRK2 (Fig. 5). Activity (left-hand y-axis; bars) was measured using AREBP2 as a substrate. Salt concentration (right-hand y-axis) is also plotted (solid diagonal line). (B) Western blot analysis showing the presence of the 42 kDa SnRK2 in the sample loaded onto the column and in the recovered fraction containing most of the activity (fraction 12).
Phosphorylation of a heterologously expressed wheat AREBP by the 42 kDa calcium-dependent SnRK2
Evidence of the involvement of the calcium-dependent activity assigned to the 42 kDa SnRK2 in ABA signalling pathways was sought by testing its ability to phosphorylate a heterologously expressed AREBP from wheat. Primers from a conserved region of three wheat AREBPs described previously (Kobayashi et al., 2008) were designed and used to amplify by gradient reverse transcriptase PCR a 1200 bp product using wheat seedling RNA as template. The nucleotide and derived amino acid sequence of the complete PCR product showed 100% identity with Wabi5-3 (Kobayashi et al., 2008). Note that ABI5 is probably the best-characterized of the Arabidopsis AREBPs; it gets its name from the ability of null mutants lacking the ABI5 protein to germinate in the presence of concentrations of ABA that prevent germination of wild-type Arabidopsis (Finkelstein, 1994). However, ABI5 is only one of at least 14 AREBPs in Arabidopsis (Zhang et al., 2008) that show differential regulation of expression and even, in some cases, antagonistic functions. A number of AREBPs that have been indentified in other species have been given the name ABI5 or similar without clear evidence that they are more closely related to Arabidopsis ABI5 than any of the other Arabidopsis AREBPs. We will therefore refer to the wheat AREBP simply as TaAREBP. The cDNA was subcloned into pET28 to enable the protein to be expressed with a His tag and the nucleotide sequence of the recombinant plasmid was determined and checked to ensure that the TaAREBP coding sequence was inserted in frame correctly. The size of the expressed protein was confirmed by SDS-PAGE and it was purified on a nickel column (not shown).
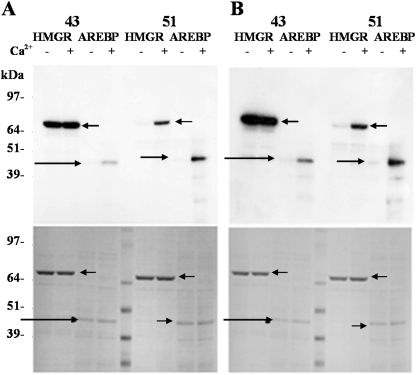
SnRK1- and SnRK2-enriched fractions (Fig. 5, fractions 43 and 51, respectively) were used to phosphorylate HMG-CoA reductase, a well-established substrate for SnRK1 and SnRK2 (Ball et al., 1995; Barker et al., 1996), and the recombinant TaAREBP transcription factor. The results showed that SnRK1 from both control (Fig. 7A) and ABA-treated roots (Fig. 7B) was able to phosphorylate HMG-CoA reductase very efficiently, but phosphorylated the TaAREBP protein poorly in comparison. In the presence of Ca2+, phosphorylation of the TaAREBP protein increased, confirming that a calcium-dependent protein kinase in this fraction could phosphorylate TaAREBP. There was no apparent increase in HMG-CoA reductase phosphorylation by this fraction with the addition of Ca2+. With the fraction containing the 42 kDa SnRK2, the phosphorylation of both substrates was greatly enhanced by the presence of Ca2+, indeed it was barely detectable in the absence of Ca2+, and TaAREBP phosphorylation was comparable with that of HMG-CoA reductase. In this experiment, no clear effect of the ABA treatment was evident.
Fig. 7.
Phosphorylation of HMG-CoA reductase and TaAREBP by wheat SnRKs. Activities assigned to SnRK1 and SnRK2 (Fig. 5, fractions 43 and 51, respectively) from control roots (A) and 10 h ABA-treated roots (B) were used to phosphorylate HMG-CoA reductase (HMGR) and TaAREBP (AREBP) proteins in vitro in the presence (+) or absence (−) of 1 mM CaCl2. After incubation, the proteins were separated using 4–12% gradient Novex NuPAGE BIS-TRIS gels and transferred to PVDF membranes. Phosphoproteins were detected by autoradiography (top), while total protein was detected by Coomassie staining of similar gels (bottom). HMG-CoA reductase and TaAREBP are indicated with arrows.
Discussion
ABA regulates numerous adaptive responses in plants and SnRK2s have been shown to be key components of the ABA signalling pathway. SnRK1 and SnRK3 have also been implicated in ABA signalling, although SnRK1 is better known as a regulator of carbon metabolism. The initial aim of this study was to investigate further the interaction between ABA and SnRK1 in wheat to increase our understanding of how stress and ABA signalling pathways cross-talk with metabolic signalling pathways (Halford and Hey, 2009; Hey et al., 2010). This has implications for improving grain yield under suboptimal conditions and for mitigating the effects of stress on grain composition, processing properties, and the formation of contaminants such as acrylamide in wheat products (Muttucumaru et al., 2006; Curtis et al., 2009). As the study progressed we also investigated a SnRK2 and obtained evidence that it was calcium-dependent.
It has been demonstrated that application of ABA to wheat roots brought about a dramatic decrease in SnRK1 protein. Phosphate starvation has been shown to cause a similar reduction in one of the SnRK1 isoenzymes of Arabidopsis, AKIN11 (Fragoso et al., 2009), while the other SnRK1 isoenzyme of Arabidopsis, AKIN10, interacts with the CUL4-DDB1-E3 ligase receptor, PRL1, leading to its degradation through the proteosome (Lee et al., 2008). The data therefore add to the evidence that protein turnover is an important mechanism in the regulation of SnRK1 activity.
Although SnRK1 protein was dramatically reduced, the amount of phosphorylated (and therefore presumably active) SnRK1 remained constant. So why are SnRK1 levels reduced while the amount of SnRK1 in the active state remains the same; in other words why is the amount of inactive SnRK1 reduced in response to ABA? It is hypothesized that unstressed wheat roots have the capacity to increase SnRK1 activity many-fold through the phosphorylation of a large pool of inactive SnRK1 enzyme. The stimulus that would bring this about has not been identified but, given what is known about SnRK1 function, it could involve carbon/energy status. Activation would probably involve phosphorylation by an upstream kinase, possibly SnAK1/2 (Hey et al., 2007), and reduced activity of protein phosphatase PP2C, which dephosphorylates and inactivates SnRK1 (Sugden et al., 1999a). PP2C is now known to be inhibited in the presence of ABA by binding of the PYR/PYL/RCAR ABA receptor, resulting in the activation of SnRK2 (Cutler et al., 2010; Nishimura et al., 2010). In our model, an undesirable increase in SnRK1 activity as a result of PP2C inhibition would be avoided by the breakdown of the inactive SnRK1 pool. The fact that transgenic Arabidopsis plants over-expressing SnRK1 have been shown to be ABA-hypersensitive (Jossier et al., 2009) is consistent with this model in that it shows that a combination of ABA and ‘too much’ SnRK1 activity could have a deleterious effect on the plant.
The increase in AMARA peptide kinase activity in response to ABA treatment and the evidence from other studies that SnRK2 is an integral part of ABA signalling prompted an investigation of SnRK2 in the ABA-treated wheat roots. There are 10 SnRK2s in Arabidopsis and rice and they are activated in response to environmental stresses such as drought, salinity, and cold. Some, but not all, are also regulated by ABA, indicating the presence of ABA-dependent and -independent responses (Boudsocq et al., 2004; Kobayashi et al., 2004). It is not yet clear how redundant the different SnRK2s are, but, given the differential regulation by ABA, it may be expected that there are some differences in function. The SnRK2 family of wheat has not been characterized in detail. Using antibodies raised against a conserved site in SnRK2s, at least three different SnRK2 isoforms could be identified after separation by SDS-PAGE, and it is possible that each ‘band’ was made up of more than one individual SnRK2. One of these had an estimated molecular mass of 38 kDa, whereas another had an estimated molecular mass of 42 kDa; the 42 kDa SnRK2 became almost undetectable after 3 h of ABA treatment but levels recovered after 6 and 10 h of treatment. It is possible that one 42 kDa SnRK2 was reduced by ABA treatment while another of the same size was increased.
Activation of SnRK2 involves phosphorylation of two conserved serines in the activation loop (Vlad et al., 2010). It was possible to detect the 42 kDa SnRK2 in the phosphoprotein-enriched fraction of extracts from roots that had been treated with ABA for 10 h, suggesting that ABA brings about activation of this 42 kDa SnRK2. By contrast, the 38 kDa protein was not detected in the phosphoprotein fraction at all.
SnRK2s phosphorylate and activate transcription factors of the AREBP family and regulation of an AREBP, TaABF, by the SnRK2 PKABA1 has been suggested to be important in regulating seed dormancy and germination in wheat (Johnson et al., 2002). Arabidopsis SnRK1 has also been shown to phosphorylate peptides (AREBP1 and 2) with amino acid sequences based on putative phosphorylation sites in AREBPs (Zhang et al., 2008). In this study, it was shown first that peptide AREBP1 was phosphorylated by a protein kinase or kinases in crude extracts from wheat roots, but that this activity was not increased by ABA treatment. Separation of SnRK1 and SnRK2 using a gel filtration column showed a peak of activity using the AMARA peptide as a substrate (the AMARA peptide is an excellent substrate for SnRK1 and 2; Sugden et al., 1999b; Crawford et al., 2001; Zhang et al., 2008) in high-molecular-mass fractions (approximately 150 kDa) which could correspond to SnRK1 complexes. This activity did not change with ABA treatment. This SnRK1 activity was able to phosphorylate both AREBP1 and AREBP2 but ‘preferred’ the AMARA peptide. By contrast, while lower-molecular-mass fractions showed relatively little activity either with or without ABA treatment, this activity showed a preference for the AREBP peptides over AMARA, a clear distinction in substrate specificity. The relatively low activity in these fractions was puzzling until the effect of adding Ca2+ to the assay was investigated. There was an increase in activity in the high-molecular-mass fractions with the AREBP peptides as substrates; we tentatively assign this activity to SnRK3. In contrast, the activity in the lower-molecular-mass fractions increased dramatically and western blot analysis showed that detection of the 42 kDa SnRK2 coincided with this activity peak. Note that the increase in activity induced by Ca2+ in this experiment was considerably greater than that induced by ABA on its own.
The ability of these activities to phosphorylate recombinant wheat AREBP (TaAREBP) was also tested using HMG-CoA reductase, an established substrate for SnRK1 and SnRK2, as a control. The activity assigned to SnRK1 was able to phosphorylate HMG-CoA reductase in the presence or absence of Ca2+ in control or ABA-treated roots, but it phosphorylated TaAREBP relatively poorly. As with the AREBP peptides, a calcium-dependent activity that phosphorylated recombinant TaAREBP was present in the same high-molecular-mass fractions. The activity assigned to the 42 kDa SnRK2 phosphorylated both HMG-CoA reductase and TaAREBP in a calcium-dependent manner and the phosphorylation increased in samples treated with ABA.
This TaAREBP, which has previously been assigned the name Wabi5-3, is induced in response to cold, drought, and exogenous ABA application and its expression has been shown to peak after 10 h of ABA treatment application, coinciding with the time of higher SnRK2 activation. Its coexpression with promoter-GUS constructs from cold-responsive genes caused an increase in GUS activity for some of the genes, suggesting that the TaAREBP was responsible for their activation (Kobayashi et al., 2008). The data from our study and these previous studies suggest that expression of these genes is regulated by an ABA-induced, calcium-dependent SnRK2 that phosphorylates TaAREBP.
The SnRK2 family of proteins have generally been regarded as calcium-independent and activities assigned to SnRK2 in cauliflower, spinach, barley, and Arabidopsis have not increased with the addition of Ca2+ (Ball et al., 1994; Barker et al., 1996; Sugden et al., 1999b; Crawford et al., 2001; Zhang et al., 2008). As a further check that the 42 kDa SnRK2 in the wheat root extracts was responsible for the calcium-dependent activity, another separation step was undertaken, this time on a Mono Q column. Once again, the 42 kDa SnRK2 was detectable in the calcium-dependent activity peak. We therefore consider the evidence that wheat roots contain a 42 kDa calcium-dependent SnRK2 to be compelling, but concede that, since this is based on coelution of an activity and a protein, the possibility cannot entirely be precluded that another protein kinase coeluted with the 42 kDa SnRK2. However, other well-known calcium-dependent protein kinases, such as SnRK3, CDPKs, and CRKs, are all significantly larger than 42 kDa (Hrabak et al., 2003). Furthermore, a protein kinase that was amplified from rice endosperm cDNA and called rice endosperm kinase (REK) has been shown to autophosphorylate in response to Ca2+ (Hotta et al., 1998). The authors of that study described REK as a calcium-dependent protein kinase, but did recognize a close similarity to SnRKs, particularly the wheat SnRK2 PKABA1 (Anderberg and Walker-Simmons, 1992; Holoppa and Walker-Simmons, 1995; Gomez-Cadenas et al., 1999). REK is, in fact, quite clearly a SnRK2, but the significance of this result has been overlooked. The authors suggested that the acidic ‘patch’ (a short stretch of acidic amino acids) in the C-terminal region of REK might bind Ca2+ and Harmon (2003) has also suggested that this region of SnRK2s might have such a role and provide a mechanism for regulation.
In this study, the effect of ABA on the phosphorylation state of the 42 kDa SnRK2 was clear, but the effect on its activity was not so marked; indeed, Ca2+ had a much greater effect. The interaction of ABA, stress and in this case Ca2+ in regulating SnRK2 activity is clearly complex, and the situation may be complicated further by subtle differences in substrate preference for different SnRK2s. It is notable that previous studies that have not detected calcium-dependent SnRK2 activity have not used ABA-treated tissues as a source. Although calcium dependence has been demonstrated previously for REK, a protein kinase that can now be identified as a SnRK2, this report is the first to state unequivocally that a SnRK2 may be calcium-dependent.
Acknowledgments
PC and EM-B were supported at Rothamsted Research through sabbatical fellowships from Dirección General de Asuntos del Personal Académico, Universidad Nacional Autónoma de México (DGAPA-UNAM). Rothamsted Research receives grant-aided support from the Biotechnology and Biological Sciences Research Council (BBSRC) of the UK.
Glossary
Abbreviations
- SNF1
sucrose non-fermenting-1
- SnRK
SNF1-related protein kinase
- AMPK
adenosine monophosphate-activated protein kinase
- ABA
abscisic acid
- AREBP
ABA-response-element-binding protein
- HMG-CoA reductase
3-hydroxy-3-methylglutaryl-coenzyme A reductase
- PP2C
protein phosphatase type 2C
- CDPK
calcium-dependent protein kinase
- CRK
CDPK-related protein kinase
- CBL
calcineurin B-like protein
- ABI
ABA-insensitive
- DTT
dithiothreitol
- EDTA
ethylenediaminetetra-acetic acid
- EGTA
ethylene glycol tetra-acetic acid
- PMSF
phenylmethylsulphonyl fluoride
- PVDF
polyvinylidene fluoride
- REK
rice endosperm kinase
References
- Anderberg RJ, Walker-Simmons MK. Isolation of a wheat cDNA clone for an abscisic acid-inducible transcript with homology to protein kinases. Proceedings of the National Academy of Sciences, USA. 1992;89:10183–10187. doi: 10.1073/pnas.89.21.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J. A central integrator of transcription networks in plant stress and energy signalling. Nature. 2007;448:938–942. doi: 10.1038/nature06069. [DOI] [PubMed] [Google Scholar]
- Ball KL, Dale S, Weekes J, Hardie DG. Biochemical characterization of two forms of 3-hydroxy-3-methyl-gluteryl CoA reductase kinase from cauliflower (Brassica oleracea) European Journal of Biochemistry. 1994;219:743–750. doi: 10.1111/j.1432-1033.1994.tb18553.x. [DOI] [PubMed] [Google Scholar]
- Ball KL, Barker JHA, Halford NG, Hardie DG. Immunological evidence that HMG-CoA reductase kinase-A is the cauliflower homologue of the RKIN1 subfamily of plant protein kinases. FEBS Letters. 1995;377:189–192. doi: 10.1016/0014-5793(95)01343-1. [DOI] [PubMed] [Google Scholar]
- Barker JAH, Slocombe SP, Ball KL, Hardie DG, Shewry PR, Halford NG. Evidence that barley 3-hydroxy-3-methylglutaryl coenzyme A reductase kinase is a member of the sucrose non-fermenting-1 related protein kinase family. Plant Physiology. 1996;112:1141–1149. doi: 10.1104/pp.112.3.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M, Sheen J. Calcium sensing and signaling. In: Pareek A, Sopory Bohnert HJ, Govindjee, editors. Abiotic stress adaptation in plants: physiological, molecular and genomic foundation. Dordrecht: Springer; 2010. pp. 75–90. [Google Scholar]
- Boudsocq M, Barbier-Brygoo H, Lauriere C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. Journal of Biological Chemistry. 2004;279:41758–41766. doi: 10.1074/jbc.M405259200. [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Droillard MJ, Barbier-Brygoo H, Lauriere C. Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting-1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Molecular Biology. 2007;63:491–503. doi: 10.1007/s11103-006-9103-1. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Choi H, Park HJ, Park JH, Kim S, Im MY, Seo HH, Kim YW, Kim SY. Arabidopsis calcium-dependent protein kinase AtCPK32 interacts with ABF4, a transcriptional regulator of abscisic acid-responsive gene expression, and modulates its activity. Plant Physiology. 2005;139:1750–1761. doi: 10.1104/pp.105.069757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford RM, Halford NG, Hardie DG. Cloning of DNA encoding a catalytic subunit of SNF1-related protein kinase-1 (SnRK1-α1) and immunological analysis of multiple forms of the kinase in spinach leaf. Plant Molecular Biology. 2001;45:731–741. doi: 10.1023/a:1010600603276. [DOI] [PubMed] [Google Scholar]
- Curtis TY, Muttucumaru N, Shewry PR, Parry MA, Powers SJ, Elmore JS, Mottram DS, Hook S, Halford NG. Evidence for genetic and environmental effects on free amino acid levels in wheat grain: implications for acrylamide formation during processing. Journal of Agricultural and Food Chemistry. 2009;57:1013–1021. doi: 10.1021/jf8031292. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finklestein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annual Review of Plant Biology. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR. Mutations at two new Arabidopsis ABA response loci are similar to the ABI3 mutations. The Plant Journal. 1994;5:765–771. [Google Scholar]
- Fragoso S, Espíndola L, Páez-Valencia J, Gamboa A, Camacho Y, Martinez-Barajas E, Coello P. SnRK1 isoforms AKIN10 and AKIN11 are differentially regulated in Arabidopsis plants under phosphate starvation. Plant Physiology. 2009;149:1906–1916. doi: 10.1104/pp.108.133298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proceedings of the National Academy of Sciences, USA. 2006;103:1988–1993. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Cadenas A, Verhey SD, Holappa LD, Shen Q, Ho T-HD, Walker-Simmons MK. An abscisic acid-induced protein kinase, PKABA1, mediates abscisic acid-suppressed gene expression in barley aleurone layers. Proceedings of the National Academy of Sciences, USA. 1999;96:1767–1772. doi: 10.1073/pnas.96.4.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AA, Vartanian N, Giraudat J. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. The Plant Cell. 1999;11:1897–1910. doi: 10.1105/tpc.11.10.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xiong L, Song CP, Gong D, Halfter U, Zhu JK. A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Developmental Cell. 2002;3:233–244. doi: 10.1016/s1534-5807(02)00229-0. [DOI] [PubMed] [Google Scholar]
- Halford NG. Regulation of carbon and amino acid metabolism: roles of sucrose nonfermenting-1-related protein kinase-1 and general control nonderepressible-2-related protein kinase. Advances in Botanical Research Incorporating Advances in Plant Pathology. 2006;43:93–142. [Google Scholar]
- Halford NG, Hey SJ. SNF1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochemical Journal. 2009;419:247–259. doi: 10.1042/BJ20082408. [DOI] [PubMed] [Google Scholar]
- Harmon AC. Calcium-regulated protein kinases of plants. Gravitational and Space Biology Bulletin. 2003;16:1–8. [PubMed] [Google Scholar]
- Hey S, Mayerhofer H, Halford NG, Dickinson JR. DNA sequences from Arabidopsis which encode protein kinases and function as upstream regulators of Snf1 in yeast. Journal of Biological Chemistry. 2007;282:10472–10479. doi: 10.1074/jbc.M611244200. [DOI] [PubMed] [Google Scholar]
- Hey SJ, Byrne E, Halford NG. The interface between metabolic and stress signalling. Annals of Botany. 2010;105:197–203. doi: 10.1093/aob/mcp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holappa LD, Walker-Simmons MK. The wheat abscisic acid-responsive protein kinase messenger RNA, PKABA1, is up-regulated by dehydration, cold temperature and osmotic stress. Plant Physiology. 1995;108:1203–1210. doi: 10.1104/pp.108.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta H, Aoki N, Matsuda T, Adachi T. Molecular analysis of a novel protein kinase in maturing rice seed. Gene. 1998;213:47–54. doi: 10.1016/s0378-1119(98)00207-8. [DOI] [PubMed] [Google Scholar]
- Hrabak EM, Chan CWM, Gribskov M, et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiology. 2003;132:666–680. doi: 10.1104/pp.102.011999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RR, Wagner RL, Verhey SD, Walker-Simmons MK. The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiology. 2002;130:837–846. doi: 10.1104/pp.001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RR, Shin M, Shen JQ. The wheat PKABA1-interacting factor TaABF1 mediates both abscisic acid-suppressed and abscisic acid-induced gene expression in bombarded aleurone cells. Plant Molecular Biology. 2008;68:93–103. doi: 10.1007/s11103-008-9354-0. [DOI] [PubMed] [Google Scholar]
- Jossier M, Bouly J-P, Meimoun P, Arjmand A, Lessard P, Hawley S, Hardie DG, Thomas M. SnRK1 (SNF1-related kinase 1) has a central role in sugar and ABA signalling in Arabidopsis thaliana. The Plant Journal. 2009;59:316–328. doi: 10.1111/j.1365-313X.2009.03871.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi F, Maeta E, Terashima A, Takumi S. Positive role of a wheat HVABI5 ortholog in abiotic stress response of seedlings. Physiologia Plantarum. 2008;134:74–86. doi: 10.1111/j.1399-3054.2008.01107.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Yamamoto S, Minami H, Kagaya Y, Hattori T. Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. The Plant Cell. 2004;16:1163–1177. doi: 10.1105/tpc.019943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Murata M, Minami H, Yamamoto S, Kayaga Y, Hobo T, Yamamoto A, Hattori T. Abscisic acid-activated SnRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. The Plant Journal. 2005;44:939–949. doi: 10.1111/j.1365-313X.2005.02583.x. [DOI] [PubMed] [Google Scholar]
- Kuhn JM, Boisson-Dernier A, Dizon MB, Maktabi MH, Schroeder JI. The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiology. 2006;140:127–139. doi: 10.1104/pp.105.070318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie S, McKibbin RS, Halford NG. Antisense SNF1-related (SnRK1) protein kinase gene represses transient activity of an α-amylase (α-Amy2) gene promoter in cultured wheat embryos. Journal of Experimental Botany. 2003;54:739–747. doi: 10.1093/jxb/erg085. [DOI] [PubMed] [Google Scholar]
- Lee J-H, Terzaghi W, Gusmaroli G, Charron J-BF, Yoon H-J, Chen H, He YJ, Xiong Y, Deng XW. Characterisation of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. The Plant Cell. 2008;20:152–167. doi: 10.1105/tpc.107.055418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Zhang H, Tian S, Chang X, Jing R. TaSnRK2.4, an SNF1-type serine/threonine protein kinase of wheat (Triticum aestivum L.), confers enhanced multistress tolerance in Arabidopsis. Journal of Experimental Botany. 2010;61:683–696. doi: 10.1093/jxb/erp331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKibbin RS, Muttucumaru N, Paul MJ, Powers SJ, Burrell MM, Coates S, Purcell PC, Tiessen A, Geigenberger P, Halford NG. Production of high starch, low glucose potatoes through over-expression of the metabolic regulator, SnRK1. Plant Biotechnology Journal. 2006;4:409–418. doi: 10.1111/j.1467-7652.2006.00190.x. [DOI] [PubMed] [Google Scholar]
- Muttucumaru N, Halford NG, Elmore JS, Dodson AT, Parry M, Shewry PR, Mottram DS. The formation of high levels of acrylamide during the processing of flour derived from sulfate-deprived wheat. Journal of Agricultural and Food Chemistry. 2006;54:8951–8955. doi: 10.1021/jf0623081. [DOI] [PubMed] [Google Scholar]
- Nishimura N, Sarkeshik A, Nito K, et al. PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. The Plant Journal. 2010;61:290–299. doi: 10.1111/j.1365-313X.2009.04054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell PC, Smith AM, Halford NG. Antisense expression of a sucrose nonfermenting-1-related protein kinase sequence in potato results in decreased expression of sucrose synthase in tubers and loss of sucrose-inducibility of sucrose synthase transcripts in leaves. The Plant Journal. 1998;14:195–202. [Google Scholar]
- Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proceedings of the National Academy of Science, USA. 2002;99:8436–8441. doi: 10.1073/pnas.122224699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S, Ma J, Hanson J, Rolland F. Sugar signals and molecular networks controlling plant growth. Current Opinion in Plant Biology. 2010;13:274–279. doi: 10.1016/j.pbi.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Sugden C, Crawford RM, Halford NG, Hardie DG. Regulation of spinach SNF1-related (SnRK1) kinases by protein kinases and phosphatases is associated with phosphorylation of the T loop and is regulated by 5'-AMP. The Plant Journal. 1999a;19:433–439. doi: 10.1046/j.1365-313x.1999.00532.x. [DOI] [PubMed] [Google Scholar]
- Sugden C, Donaghy P, Halford NG, Hardie DG. Two SNF1-related protein kinases from spinach leaf phosphorylate and inactivate 3-hydroxy-3-methylglutaryl-coenzyme A reductase, nitrate reductase and sucrose phosphate synthase in vitro. Plant Physiology. 1999b;120:257–274. doi: 10.1104/pp.120.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiessen A, Prescha K, Branscheid A, Palacios N, McKibbin R, Halford NG, Geigenberger P. Evidence that SNF1-related kinase and hexokinase are involved in separate sugar-signalling pathways modulating post-translational redox activation of ADP-glucose pyrophosphorylase in potato tubers. The Plant Journal. 2003;35:490–500. doi: 10.1046/j.1365-313x.2003.01823.x. [DOI] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2009;106:17588–17593. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F, Droillard M-J, Valot B, Khafif M, Rodrigues A, Brault M, Zivy M, Rodriguez PL, Merlot S, Laurière C. Phospho-site mapping, genetic and in planta activation studies reveal key aspects of the different phosphorylation mechanisms involved in activation of SnRK2s. The Plant Journal. 2010;63:778–790. doi: 10.1111/j.1365-313X.2010.04281.x. [DOI] [PubMed] [Google Scholar]
- Xu Z, Liu L, Ni ZY, Liu P, Chen M, Li LC, Chen YF, Ma YZ. W55a encodes a novel protein kinase that is involved in multiple stress responses. Journal of Integrative Plant Biology. 2009;51:58–66. doi: 10.1111/j.1744-7909.2008.00776.x. [DOI] [PubMed] [Google Scholar]
- Zhang H, Mao X, Wang C, Jing R. Overexpression of a common wheat gene TaSnRK2.8 enhances tolerance to drought, salt and low temperature in Arabidopsis. PLoS One. 2010;5:1–12. doi: 10.1371/journal.pone.0016041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Mao X, Jing R, Chang X, Xie H. Characterization of a common wheat (Triticum aestivum L.) TaSnRK2.7 gene involved in abiotic stress responses. Journal of Experimental Botany. 2011a;62:975–988. doi: 10.1093/jxb/erq328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Mao X, Jing R. SnRK2 acts within an intricate network that links sucrose metabolic and stress signaling in wheat. Plant Signaling and Behavior. 2011b;6:652–654. doi: 10.4161/psb.6.5.14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Andralojc PJ, Hey SJ, Primavesi LF, Specht M, Koehler J, Parry MAJ, Halford NG. Arabidopsis SNF1-related protein kinase-1 and calcium-dependent protein kinase phosphorylate conserved target sites in ABA response element binding proteins. Annals of Applied Biology. 2008;153:401–409. [Google Scholar]
- Zhu WSY, Yu XC, Wang XJ, et al. Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. The Plant Cell. 2007;19:3019–3036. doi: 10.1105/tpc.107.050666. [DOI] [PMC free article] [PubMed] [Google Scholar]