Abstract
14-3-3 proteins are found in all eukaryotes where they act as regulators of diverse signalling pathways associated with a wide range of biological processes. In this study the functional characterization of the ZmGF14-6 gene encoding a maize 14-3-3 protein is reported. Gene expression analyses indicated that ZmGF14-6 is up-regulated by fungal infection and salt treatment in maize plants, whereas its expression is down-regulated by drought stress. It is reported that rice plants constitutively expressing ZmGF14-6 displayed enhanced tolerance to drought stress which was accompanied by a stronger induction of drought-associated rice genes. However, rice plants expressing ZmGF14-6 either in a constitutive or under a pathogen-inducible regime showed a higher susceptibility to infection by the fungal pathogens Fusarium verticillioides and Magnaporthe oryzae. Under infection conditions, a lower intensity in the expression of defence-related genes occurred in ZmGF14-6 rice plants. These findings support that ZmGF14-6 positively regulates drought tolerance in transgenic rice while negatively modulating the plant defence response to pathogen infection. Transient expression assays of fluorescently labelled ZmGF14-6 protein in onion epidermal cells revealed a widespread distribution of ZmGF14-6 in the cytoplasm and nucleus. Additionally, colocalization experiments of fluorescently labelled ZmGF14-6 with organelle markers, in combination with cell labelling with the endocytic tracer FM4-64, revealed a subcellular localization of ZmGF14-6 in the early endosomes. Taken together, these results improve our understanding of the role of ZmGF14-6 in stress signalling pathways, while indicating that ZmGF14-6 inversely regulates the plant response to biotic and abiotic stresses.
Keywords: 14-3-3 proteins, abiotic stress, biotic stress, early endosomes, Fusarium verticillioides, drought, Magnaporthe oryzae, Oryza sativa, Zea mays, ZmPR4 promoter
Introduction
Plants are constantly challenged with numerous environmental stresses, both biotic and abiotic. To survive under such conditions, plants have evolved a variety of mechanisms to perceive external stimuli and to transduce the stress signal for activation of the optimal response to each type of stress. A coordinated regulation of plant response requires crosstalk between pathways that are initiated by external cues and orchestrated through a complex network of signalling pathways. There is compelling evidence that stress-responsive genes such as transcription factors or kinases might function in multiple pathways and also facilitate crosstalk between different stress signalling pathways (Ludwig et al., 2004; Fujita et al., 2006; Quilis et al., 2008; Saibo et al., 2009).
The 14-3-3 proteins are a family of highly conserved regulatory proteins originally identified in mammalian brain tissue (Moore et al., 1968). It has become clear that 14-3-3 proteins constitute a protein family present in virtually all eukaryotic organisms (Rosenquist et al., 2000). In plants, 14-3-3 proteins are known to function in protein–protein interactions that mediate signal transduction pathways regulating many biological processes, such as metabolism, hormone and light signalling, transcription, cell-cycle control, protein trafficking, and stress responses (Sehnke et al., 2002b; Roberts, 2003; Morrison, 2009; Gökirmak et al., 2010; Oh, 2010). They function as homo- or heterodimers and each monomer binds to an interacting target protein containing well-defined phosphothreonine or phosphoserine motifs (Ferl, 2004). Plant 14-3-3 proteins are known to interact with a broad range of target proteins, and the consequences of binding are diverse, such as stabilization of the active or inactive proteins, conformational change, and alteration in the intracellular localization of their target proteins (Schultz et al., 1998; Igarashi et al., 2001; Gökirmak et al., 2010).
Multiple 14-3-3 proteins exist in each eukaryotic organism. For instance, the Arabidopsis genome contains 15 14-3-3 genes (Rosenquist et al., 2001). Individual members of the Arabidopsis 14-3-3 gene family are designated by greek letters. However, 14-3-3 isoforms from some plant species are also named GF14, because the first reported plant 14-3-3 isoform, Arabidopsis GF14ω, was identified as a component of the protein/G-box complex and thus designated G-box factor 14-3-3 (Lu et al., 1992). This designation scheme has been maintained in numerous plant species, including Arabidopsis, rice, sorghum, and maize. The presence of complex 14-3-3 gene families in plant species suggests a functional specialization for genes encoding 14-3-3 proteins, which might play diverse roles in developmental processes and stress conditions.
A number of studies have shown that 14-3-3 proteins play a role in plant immunity (Roberts et al., 2002; Chevalier et al., 2009; Gökirmak et al., 2010; Oh, 2010). In Arabidopsis, the 14-3-3 λ protein encoded by the AtGRF6 gene has been shown to be a positive regulator of recognition of powdery mildew 8 (RPW8)-mediated resistance (Yang et al., 2009). A tomato 14-3-3 protein was also found to positively regulate R-protein-mediated resistance and programmed cell death (Oh et al., 2010; Oh and Martin, 2011). Genes encoding 14-3-3s have also been identified as candidate R genes at quantitative trait loci in wheat and rice (Faris et al., 1999; Wu et al., 2004). Levels of 14-3-3 transcripts increase in response to pathogen infection, or treatment with defence-related hormones, in several other plant species, including barley, soybean, poplar, tomato, cotton, wheat, and rice (Brandt et al., 1992; Roberts and Bowles, 1999; Lapointe et al., 2001; Chen et al., 2006; Manickavelu et al., 2010). In other studies, the levels of 14-3-3 transcripts were reported to be regulated by abiotic stresses such as cold, drought, and salt stress (Jarillo et al., 1994; Chen et al., 2006).
Rice is one of the most important cereal crops in the world and a source of food for more than half of the world’s population. Additionally, rice has been adopted as the model plant for functional genomics in cereals. Low and unstable rice productivity is often associated with abiotic stresses such as drought, salinity, and submergence conditions. Rice yield is also severely compromised by diseases caused by pathogens, such as the fungus Magnaporthe oryzae, causing rice blast disease, one of the most important fungal diseases of cultivated rice (Talbot, 2003), or the fungus Fusarium verticillioides (anamorph stage of Gibberella fujikuroi, mating population A), associated with the bakanae disease of rice (Wulff et al., 2010).
The aim of this work was to investigate the function of a particular 14-3-3 protein from maize which is encoded by the ZmGF14-6 gene, in relation to abiotic and biotic stress. We show here that, in maize, ZmGF14-6 gene expression increases in response to fungal infection and salt stress, whereas drought stress results in down-regulation of ZmGF14-6 expression. Transgenic rice plants that express the ZmGF14-6 gene were then produced. Here it is shown that constitutive expression of ZmGF14-6 positively confers tolerance to drought stress, which correlates with the observed higher induction of drought-associated marker genes in roots of ZmGF14-6 plants. Moreover, expression of ZmGF14-6 in rice under the control of either a constitutive or a pathogen-inducible promoter results in enhanced susceptibility to pathogen infection, and is accompanied by a lower induction of genes typically associated to the plant response to pathogen infection. The ZmGF14-6 protein was found to localize at multiple subcellular compartments, cytoplasm, nucleus, as well as in the early endosomes, as determined by coexpression of fluorescently labelled ZmGF14-6 and organelle markers, and experiments with the endocytic tracer FM4-64.
Material and methods
Plant material and treatments
Maize (Zea mays, pure inbred line W64A) seeds were sterilized and germinated on wet filter paper at 27 ± 2 °C with a 16/8 h light/dark cycle at 70% relative humidity. To test the effect of fungal infection, maize embryos (embryonic axis plus scutellum) were manually dissected from dry sterilized seeds and germinated over wet filter paper in the dark. Fusarium verticillioides infection was performed as described previously (Campo et al., 2004). Infected and non-infected control embryos were allowed to continue germination for the indicated times and harvested for total RNA isolation.
For polyethylene glycol (PEG)-induced dehydration treatment, roots of 7-day-old maize seedlings were immersed in PEG-8000 (20% w/v) or water for the indicated times. After treatment, roots were harvested and used for total RNA isolation. For salt stress treatment, roots of maize seedlings were treated with 200 mM NaCl or sterile water for the indicated times.
Rice plants (Oryza sativa L. cv. Senia) were grown at 27 ± 2 °C with a 16/8 h light/dark cycle. For rice transformation, the Agrobacterium tumefaciens EHA105 strain was used to infect embryonic callus derived from mature embryos. Fungi M. oryzae (PR09 isolate; CIRAD Collection, Montpellier, France) and F. verticillioides (isolate collected from rice plants in Spain and supplied by the “Servei de Protecció dels Vegetals”, Generalitat de Catalunya) were grown on rice flour medium (rice flour at 20 g l−1, agar at 15 g l−1, and yeast extract at 2.5 g l−1) and potato dextrose agar medium, respectively. Spores were collected by adding sterile water to the surface of the mycelium. For gene expression analysis of rice abiotic marker genes, roots of 7-day-old rice plants were immersed in PEG-8000 (20%) or water for 24 h. For each sample, roots of 16 individual plants were collected for total RNA isolation. Expression of rice defence genes in Ubi::ZmGF14-6, ZmPR4::ZmGF14-6, and wild-type plants was carried out as previously reported (Quilis et al., 2008). For gene expression analysis of rice biotic marker genes, total RNA was extracted from control and M. oryzae-infected leaves at 24 h after fungal inoculation. Leaves from eight individual plants were collected.
Cloning of the full-length cDNA encoding ZmGF14-6
rapid amplification of cDNA ends (RACE)-PCR was used to clone the full-length sequence of the ZmGF14-6 cDNA from the maize W64A cultivar with the SMART PCR cDNA Synthesis Kit (Clontech, Palo Alto, CA, USA).
Construction of plant expression vectors and rice transformation
For constitutive expression, the ZmGF14-6 cDNA fragment was cloned into the BamHI site of the pAHC17 plasmid DNA (Christensen and Quail, 1996) under the control of the maize ubiquitin 1 (ubi) promoter and nopaline synthase (nos) terminator. Next, the entire cassette for expression of the ZmGF14-6 gene (ubi promoter fused to the ZmGF14-6 gene and the nos terminator) was inserted into the KpnI site of the pCAMBIA1300, resulting in the plasmid pC::ubi::ZmGF14-6::nos (plasmid for expression of the ZmGF14-6 gene) (Supplementary Fig. S1). For the pathogen-inducible expression of ZmGF14-6, a pCAMBIA-derived plasmid containing the Z. mays pathogenesis-related 4 (ZmPR4) promoter was obtained (Moreno et al., 2005). For this, the DNA fragment containing the coding sequence of the ZmGF14-6 gene and the nos terminator was PCR-amplified from the pC::ubi::ZmGF14-6::nos plasmid obtained above. Appropriate restriction sites were introduced into the PCR-amplified DNA suitable for the subsequent cloning steps (SmaI at the 5′ end; EcoRI at the 3′ end). The PCR-amplified ZmGF14-6:nos DNA was then cloned into the SmaI-/EcoRI-digested pC::ZmPR4 to obtain the pC::ZmPR4::ZmGF14-6::nos construct (plasmid for fungal-inducible expression of the ZmGF14-6 gene) (Supplementary Fig. S1). Primers used for cloning are listed in Supplementary Table S1.
RNA isolation and Northern blots
Total RNA was extracted from plant tissues using Trizol reagent according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). For Northern blot analysis 10 μg of total RNAs were subjected to 1.2% formaldehyde-containing agarose gel electrophoresis, transferred to nylon membranes (Hybond-N, Amersham, Little Chalfont, Bucks, UK) and fixed by ultraviolet crosslinking. The [α-32P]dCTP-labelled full-length ZmGF14-6 cDNA was used as a probe. Hybridation was performed in phosphate buffer [125 mM Na2HPO4, pH 7.2, 7% (w/v) SDS, 1 mM ethylenediaminetetra-acetic acid (EDTA)] at 65 °C overnight and washed three times for 20 min in 20 mM Na2HPO4 (pH 7.2), 1% (w/v) SDS, and 1 mM EDTA at 65 °C. The hybridized blot was exposed overnight to a phosphor screen, and visualized with a Bio-Rad Personal FX phosphorimager.
Gene expression analysis by quantitative reverse transcriptase-PCR
Reverse transcription reactions were performed using total RNA from maize or rice tissues. The first cDNA was synthesized from DNase-treated total RNA (2 μg) with Transcriptor Reverse Transcriptase (Roche, Indianapolis, IN, USA) and oligo(dT)18 following the manufacturer’s instructions. Quantitative reverse transcriptase-PCR (qRT-PCR) was performed in optical 96-well plates in the Roche Light Cycler 480 instrument using SYBR Green I dye and the primers listed in Supplementary Table S1. Primers were designed using Primer Express software (Applied Biosystems, Foster City, CA, USA). Routinely, three replicate reactions were used for each sample. Data were normalized with OsUbi1 and Z. mays cyclophilin (ZmCyp) as internal controls for rice and maize, respectively. The average cycle threshold (Ct) values from triplicate PCRs were normalized to the average Ct values for the OsUbi1 or ZmCyp gene from the same RNA preparations. Three independent biological replicates were analyzed. Controls of the qRT-PCR reactions without adding the reverse transcriptase enzyme were systematically included in our experiments.
Bacterial expression, purification of the ZmGF14-6 protein, and preparation of the antiserum
The coding sequence of the ZmGF14-6 gene was cloned into the HindIII and BamHI restriction sites of the expression vector PET28a for the expression of a recombinant N-terminal His-tagged protein and expressed in Escherichia coli strain BL21 (DE3). Primers used for cloning are listed in Supplementary Table S1. The cells were grown in Luria–Bertani medium supplemented with 50 μg ml−1 kanamycin to an absorbance of 0.6 at a wavelength of 600 nm. Expression of ZmGF14-6 was induced by adding isopropyl-D-thiogalactopyranoside (IPTG) to Luria–Bertani medium at a final concentration of 0.5 mM, and the incubation was continued for 2 h at 37 °C.
Cells were harvested by centrifugation, re-suspended in PBS buffer (phosphate-buffered saline; 0.05 M sodium phosphate, pH 7.5, and 0.15 M NaCl) and frozen at −20 °C. After thawing, the bacterial suspension was lysed by incubation with lysozyme (1 mg ml−1, 2 h on ice) followed by incubation with 0.1% Triton X-100 for 5 min at room temperature. ZmGF14-6 protein expression was monitored by SDS/PAGE analysis of bacterial extracts. After centrifugation of the bacterial lysate, the recombinant protein was recovered in the supernatant, which was then filtered through a 0.45 μm membrane and loaded on a His-Bind column. Purification of the recombinant protein was carried out under denaturating conditions following the manufacturer’s instructions by metal chelation chromatography with a chelating Sepharose fast-flow resin (Amersham Pharmacia Biotech). The His-tagged ZmGF14-6 protein was recovered after elution with 200 mM imidazole and the purified protein was dialysed against PBS containing 0.1% SDS.
Antibodies were raised in two rabbits by multiple subcutaneous injections of the purified His-tagged ZmGF14-6 protein. Four weekly injections (100 μg each injection) were made and the rabbits were bled 8 days after the third and fourth injections. Blood samples were collected from the marginal ear vein. Preimmune serum was collected from the rabbits 1 week prior to immunization.
Preparation of protein extracts and immunoblotting
Protein extracts were prepared from leaf tissues by using an extraction buffer containing 50 mM Tris/HCl buffer at pH 7.5 containing 0.1% Tween-20, 150 mM NaCl, 10% glycerol, 1 mM dithiothreitol, and a mix of protease inhibitors. All protein concentrations were determined with the Bio-Rad dye reagent and bovine serum albumin as a standard. SDS/PAGE was performed according to the method of (Laemmli, 1970).
Protein extracts were separated by SDS/PAGE (12.5% acryalmide in separation gel) on a miniprotean apparatus for 90 min at 120 V. Proteins separated by SDS/PAGE were stained with Coomassie blue, or were electrophoretically transferred to a nitrocellulose membrane for 1 h at 40 V using a transfer blotter (Invitrogen). Blots were blocked in PBS-T buffer (PBS containing 0.1% Tween) and 10% skimmed dry milk for 1 h at room temperature and incubated overnight at 4 °C in the same buffer containing the anti-ZmGF14-6 antiserum at a dilution of 1:1000. Following that they were washed four times in PBS-T and then incubated with protein A–peroxidase (Sigma) at a dilution of 1:10 000 for 1 h at room temperature. Following four washes in PBS-T, peroxidase activity was made visible by incubating the blot with Enhanced Chemiluminescence Western Blotting Substrate (Pierce, Rockford, IL, USA) for 5 min on an LAS-4000 Image analyzer (Fujifilm).
Disease-resistance assays
Resistance of ZmGF14-6 rice plants to infection by the rice blast fungus M. oryzae (PR9 isolate; CIRAD Collection) was assayed as previously described (Coca et al., 2004). Briefly, the second leaves of 2-week-old soil-grown rice plants were placed into plate dishes with 1% (w/v) water agar containing kinetine at 2 mg l−1. Whatman filter paper discs saturated with a M. oryzae spore suspension (106 spores ml−1) were placed onto the upper face of the leaf for 60 h and then removed. The inoculated leaves were maintained under high-humidity conditions for the required period of time. Development of disease symptoms with time was followed. Three independent T2 homozygous ZmGF14-6 lines and at least 12 plants for each line were assayed. Disease severity was inferred from the lesion size developed at the inoculated spots on the leaves after 4 days of infection. Lesion size was determined on three leaves and three inoculation sites each, and three plants per each transgenic line and wild-type plants.
For assays with F. verticillioides, seeds of ZmGF14-6 and wild-type plants were pre-germinated for 24 h on on wet filter paper and then inoculated with 50 μl of a suspension of F. verticillioides spores (102 spores ml−1), or with sterile water. Seeds were allowed to continue germination (Gómez-Ariza et al., 2007). The length of F. verticillioides-infected seedlings from wild-type and transgenic plants were determined and compared with that of non-infected seedlings using Image J software (seedlings with shoots above 5 cm were considered). Experiments were carried out three times.
Drought treatment of rice plants
Wild-type (36 plants) and transgenic lines (three independent T2 homozygous lines and 36 plants per line) were grown for 15 days. Drought stress was induced by withholding water for up to 18 days. Plants were then irrigated normally for 7 days. Drought tolerance was evaluated by determining the percentage of surviving plants after the period of recovery. Experiments were repeated three times. To measure the water loss under dehydration conditions, leaves of wild-type and homozygous transgenic plants at the three-leaf stage were cut and exposed to air under controlled conditions (27 ± 2 °C). The leaves were weighed 4 h after being cut. Water loss was calculated from the equation:
Confocal microscopy
Onion cells were transformed by particle bombardment using plasmids for the expression of the ZmGF14-6 fused to either the green fluorescent protein (GFP) or the cyan fluorescent protein (CFP) gene. Experiments for coexpression of the fluorescently labelled ZmGF14-6 with organelle markers fused to the yellow fluorescent protein (YFP) were also carried out. The epidermis of onion bulb scales was prepared by peeling the inner epidermis from fresh onion bulbs. Particle bombardment was performed as previously described (Murillo et al., 2003). Confocal laser scanning microscopy was performed with an Olympus FV1000 spectral inverted confocal microscope. For FM4-64 staining, onion epidermis were incubated with FM4-64 (5 μM) for 5 min at room temperature, followed by washing with water three times. The fluorescence was observed immediately after washing. FM4-64 fluorescence was excited with a 543 nm argon ion laser.
Results
Isolation of the full-length cDNA encoding the ZmGF14-6 protein from maize
Differential screening of a cDNA library prepared from germinating maize embryos resulted in the identification of a cDNA clone encoding the C-terminal region and 3′-untranslated region of a 14-3-3 protein (Cordero et al., 1994). In maize, two genes encoding 14-3-3 proteins (also named GF14 proteins) have long been described: the GF14-6 and GF14-12 genes (de Vetten and Ferl, 1994; Rosenquist et al., 2001; Yao et al., 2007). In addition to GF14-6 and GF14-12 genes, other genes encoding 14-3-3 or 14-3-3-like proteins have been described in different maize varieties (Supplementary Fig. S2).
In this work, 5′-RACE was used to obtain the full-length cDNA clone encoding the maize GF14-6 14-3-3 protein. The nucleotide sequence of the full-length cDNA showed 99% identity with the previously described GF14-6 cDNA isolated from maize cell suspension cultures (de Vetten et al., 1992) (results not shown). Changes in the nucleotide sequence between the cDNA clone here characterized (from now on referred to as the ZmGF14-6 gene) and the previously described GF14-6 cDNA most probably reflect variation between the Z. mays W64A (present work) and Z. mays XL80 (de Vetten et al., 1992) lines.
Expression pattern of ZmGF14-6 in response to biotic and abiotic stress
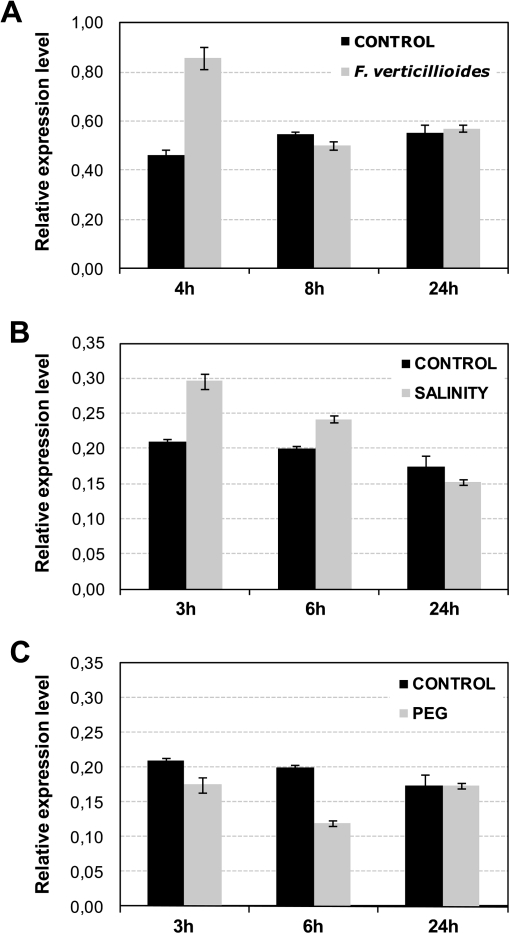
The expression of ZmGF14-6 was measured by qRT-PCR in maize seedlings subjected to biotic and abiotic stress conditions. Responsiveness of ZmGF14-6 to fungal infection was initially investigated. For this study, the fungal pathogen F. verticillioides, which infects seed and root tissues of maize plants, was used (Murillo et al., 1999). Maize embryos were germinated for 24 h and then inoculated with F. verticillioides spores as previously described (Campo et al., 2004). The level of ZmGF14-6 transcripts was monitored in control and fungus-infected germinating maize embryos at different times after inoculation. A transient increase in ZmGF14-6 expression was observed at an early stage of the infection process (4 h after inoculation with fungal spores) (Fig. 1A). ZmGF14-6 gene expression was about two times higher in F. verticillioides-infected embryos relative to that of control non-infected embryos.
Fig. 1.
Expression analysis of ZmGF14-6 in maize. qRT-PCR was used to monitor ZmGF14-6 transcript abundance in maize tissues. The cyclophilin gene (X68678) was used as the internal control for normalization. Data represent mean ± SD (n = 16) from one biological experiment. These experiments were repeated three times with similar results. (A) Expression pattern of ZmGF14-6 in response to fungal infection. Germinating maize embryos were inoculated with spores from the fungus F. verticillioides and harvested at the indicated times after inoculation. (B) ZmGF14-6 gene expression in response to salt treatment. Roots of 7-day-old seedlings were treated with 200 mM NaCl and harvested at the indicated times. (C) Expression pattern of ZmGF14-6 in response to PEG-induced drought stress. Roots of 7-day-old seedlings were treated with 20% PEG 8000, a stress treatment commonly used to mimic drought conditions. ZmGF14-6 transcript levels were measured at the indicated times of treatment.
Next, the expression pattern of ZmGF14-6 in response to salt and drought stress was examined. Salt treatment (200 mM NaCl) increased ZmGF14-6 transcript levels within 3–6 h of treatment and its expression progressively returned to normal levels at 24 h of salt treatment (Fig. 1B). In order to investigate the expression pattern of ZmGF14-6 in relation to drought stress, roots of maize seedlings were stressed in a PEG-8000 solution. The control seedlings were watered as normal and grown under the same conditions. As it is shown in Fig. 1C, ZmGF14-6 expression was down-regulated at 3 and 6 h of treatment with 20% PEG-8000 and returned to normal levels at 24 h of treatment. Overall, results here presented showed that ZmGF14-6 gene expression is rapidly and transiently activated in response to both fungal infection and salt treatment, whereas drought stress results in down-regulation of ZmGF14-6 expression. A rapid and transient increase of ZmGF14-6 expression might indicate a signalling function for ZmGF14-6. These observations also point to a role of ZmGF14-6 in the plant response to different types of stress, both biotic and abiotic stress.
Production and characterization of transgenic rice expressing the ZmGF14-6 gene
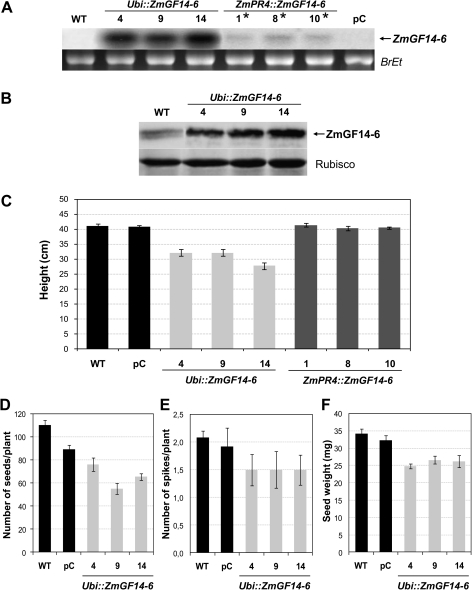
To further investigate the function of ZmGF14-6, transgenic rice lines expressing the ZmGF14-6 gene under the control of either a constitutive promoter or a pathogen-inducible promoter were obtained. Constitutive expression of ZmGF14-6 was carried out by using the ubi promoter (Ubi::ZmGF14-6 lines). For pathogen-induced expression of the ZmGF14-6 gene, the promoter from the maize ZmPR4 gene was selected (ZmPR4::ZmGF14-6 lines). The ZmPR4 promoter was previously reported to be strongly induced in rice tissues in response to fungal infection and wounding (Moreno et al., 2005). The components of the constructs used for the constitutive and pathogen-inducible expression of ZmGF14-6 in rice are shown in Supplementary Fig. S2. The elite japonica rice (O. sativa L.) cultivar was used for rice transformation. Transgenic rice plants were produced by Agrobacterium-mediated transformation. Transgene integration and expression was confirmed in the T0 generation by PCR analysis of genomic DNA and Northern blotting, respectively (results not shown; Northern blot analysis of T2 homozygous lines is shown in Fig. 2A). Three independent lines expressing ZmGF14-6 in a constitutive (Ubi::ZmGF14-6 lines) or inducible manner (ZmPR4::ZmGF14-6 lines) were selected and used as the parental lines to obtain T2 homozygous progeny plants.
Fig. 2.
Molecular and phenotypic characterization of rice plants expressing ZmGF14-6. (A) Northern blot analysis of transgenic rice plants (T2 generation) expressing the ZmGF14-6 gene under the control of the maize constitutive promoter ubiquitin (Ubi::ZmGF14-6) or the pathogen- and wound-inducible promoter of the maize PR4 gene (ZmPR4::ZmGF14-6). In each case, three independently generated transgenic lines are presented (Ubi::ZmGF14-6, lines 4, 9, and 14; ZmPR4::ZmGF14-6, lines 1, 8, and 10). As for ZmPR4::ZmGF14-6 lines, leaves were mechanically wounded prior Northern blot analysis (denoted by asterisks). Samples from wounded leaves were harvested 24 h after wounding. Total RNAs (10 μg) were subjected to formaldehyde-containing agarose gel electrophoresis and hybridized with a 32P-labelled probe. Lane WT, untransformed Senia control. Lane pC, rice plants transformed with the empty pCAMBIA1300 vector. Lower panels show ethidium bromide staining of RNA samples. (B) Immunoblot analysis of total protein extracts from transgenic rice plants constitutively expressing the ZmGF14-6 gene. Leaf protein extracts (20 μg) were separated by 12.5% SDS/PAGE, transferred to nitrocellulose membranes, and probed with the anti-ZmGF14-6 antibody. The RuBisCO protein served as a loading control. (C) Mean height of ZmGF14-6 rice plants (T2 homozygous lines) compared with wild-type (WT) plants. WT and transgenic lines, Ubi::ZmGF14-6, and ZmPR4::ZmGF14-6, as well as vector-transformed (pC) plants, were grown in the greenhouse for 3 weeks. Means were calculated from three independent transgenic lines. The data represent means ± SD (n = 16). (D–F) Productivity of rice plants constitutively expressing ZmGF14-6 (Ubi:ZmGF14-6). Seed production (D), number of spikes (E), and weight of seeds (F) in Ubi:ZmGF14-6, WT, and empty vector (pC) rice plants. For determination of seed weight, seeds from three independent lines were collected from three plants and pooled. Two hundred seeds from each pool were weighted and the mean weight per seed was calculated. The data represent mean ± SD.
Northern blot analysis of selected T2 homozygous Ubi::ZmGF14-6 lines revealed accumulation of ZmGF14-6 transcripts (Fig. 2A, Ubi::ZmGF14-6 lines 4, 9, and 14). Concerning ZmPR4::ZmGF14-6 lines, no accumulation of ZmGF14-6 transcripts occurred in their leaves when these plants are grown under non-inductive conditions (results not shown). These results are in agreement with those previously reported by our group indicating that the ZmPR4 promoter is silent in rice plants under non-inductive conditions (Moreno et al., 2005). Knowing that the ZmPR4 promoter is activated by mechanical wounding in rice leaves (Moreno et al., 2005), the capability of this promoter to drive ZmGF14-6 expression was assessed in mechanically wounded leaves of ZmPR4:ZmGF14-6 plants. For this, leaves of transgenic and control plants were wounded by making perpendicular cuts on the leaf blade. Low levels of ZmGF14-6 transcripts were evident in all independent transgenic lines at 24 h after wounding (Fig. 2A, ZmPR4::ZmGF14-6 lines 1, 8, and 10). The low level of ZmGF14-6 transcript accumulation that is observed in mechanically wounded leaves relative to that of lines constitutively expressing the ZmGF14-6 gene can be explained by the dilution effect of RNA samples extracted from wounded leaves. Namely, the activity of the ZmPR4 promoter was reported to occur only at the cut edge of the wound site (Moreno et al., 2005). As expected, ZmGF14-6 transcripts were not detected in rice lines harbouring the empty vector (Fig. 2A, line pC). Northern blot analysis of ZmGF14-6 rice plants confirmed stable expression and inheritance of the transgene for at least three generations (results not shown).
To verify that the ZmGF14-6 protein was produced correctly in transgenic rice, Western blot analyses of leaf extracts were carried out. To this end, the ZmGF14-6 protein was produced in E. coli and used for the obtention of polyclonal antibodies in rabbits as described in the Material and Methods section. Next, the anti-ZmGF14-6 antibody was used to examine the accumulation of the 14-3-3 protein in protein extracts obtained from leaves of transgenic rice lines. In agreement with results obtained by Northern blot analysis, all the transgenic lines constitutively expressing ZmGF14-6 accumulated higher levels of 14-3-3 protein than wild-type plants (Fig. 2B). The anti-ZmGF14-6 antisera recognized the endogenous 14-3-3 protein(s) (Fig. 2B, lane WT). This was expected because 14-3-3 proteins are highly conserved in eukaryotes (Ferl, 1996).
Constitutive expression of ZmGF14-6 in rice results in delayed growth and reduced seed production
The developmental performance of rice plants constitutively expressing ZmGF14-6 differed from that of control plants. Thus, transgenic Ubi::ZmGF14-6 rice plants grown in the greenhouse showed growth retardation and reduced height compared to either wild-type plants or plants transformed with the empty vector (Fig. 2C). Contrary to this, rice plants expressing ZmGF14-6 in a pathogen-inducible manner exhibited no obvious phenotypic difference from the wild-type and empty vector-expressing plants (Fig. 2C).
Constitutive expression of ZmGF14-6 in rice also had a penalty on seed productivity. Ubi::ZmGF14-6 plants produced fewer seeds than control plants (Fig. 2D). The observed reduction in seed productivity was the consequence of both a lower number of spikes produced per plant and a decrease in seed weight (Fig. 2E and F). Together, these studies revealed that plant growth and productivity are adversely affected by constitutive expression of ZmGF14-6 in rice.
Constitutive expression of ZmGF14-6 in rice enhances drought tolerance
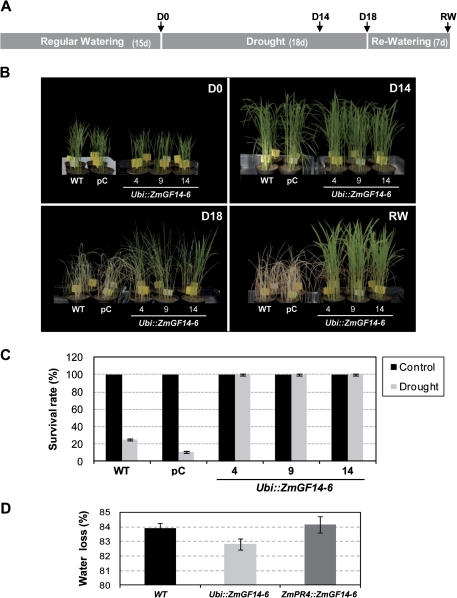
Transgenic ZmGF14-6-rice plants were tested for tolerance to drought stress. For this, transgenic plants (three independent T2 homozygous lines), wild-type, and transgenic lines expressing the empty vector were assayed (at least 36 plants per line). To investigate whether transgenic expression of ZmGF14-6 has an effect on drought tolerance, 15-day-old plants were deprived of irrigation for 18 days and then water was supplied for 7 days (Fig. 3A). Visual differences between transgenic and control plants were already evident at 15 days of growth, the control plants exhibiting higher shoot biomass than transgenic plants (Fig 3B, panel D0). However, when water was withheld, control plants exhibited a delayed growth compared to transgenic lines which continued growing normally. Empty vector transgenic plants were very similar to wild-type plants in performance. By 14 days without water, control plants showed visual symptoms of drought-induced damage, such as leaf rolling and wilting. By this time transgenic plants remained green and healthier than control plants (Fig. 3B, panel D14). After 18 days of drought treatment, wild-type and empty vector plants were severely affected by drought stress while the ZmGF14-6-expressing plants exhibited substantially less wilting and showed less leaf damage than control plants did (Fig. 3B, panel D18). Upon rewatering, all independent transgenic plants recovered (Fig. 3B, panel RW and Fig. 3C). Only 25 and 10.42% of the wild-type and empty vector plants, respectively, survived under drought treatment (Fig. 3C). During the course of this work, control and transgenic plants at different developmental stages were exposed to drought stress. In all experiments, transgenic plants exhibited enhanced tolerance to drought stress (results not shown).
Fig. 3.
Phenotype of ZmGF14-6 rice plants under drought stress conditions. (A) Diagram showing conditions used for drought tolerance assay. Plants were grown under well-watered conditions for 15 days. For drought stress treatment, water was withheld for 18 days (starting at D0) and then plants were supplied with water again for 7 days. In each experiment, wild-type (WT), empty-vector (pC), and transgenic lines (lines 4, 9 and 14), and at least 36 plants per line, were assayed. Assays to determine the tolerance to drought stress were carried out three times and at different developmental stages with similar results. (B) Phenotype of non-transformed (WT), lines transformed with the empty vector (pC), and lines constitutively expressing the ZmGF4-6 gene (lines 4, 9 and 14) grown under normal irrigation conditions for 15 days (panel D0). The pictures were taken at 14 and 18 days of drought stress (D14 and D18, respectively), and after 7 days after rewatering (RW). (C) Survival of ZmGF14-6 rice plants after 7 days of recovery from drought stress. Transgenic plants show 100% survival. Each column represents the mean ± SD of triplicate experiments (n = 36 in each experiment) (D) Water loss in wild-type and transgenic rice plants. Leaves were harvested from wild-type and ubi::ZmGF14-6 rice plants (n = 15) at the three-leaf stage. Fresh weight of detached leaves was measured (time 0). Water loss was calculated from the decrease in fresh weight compared to time 0. Each column represents data from three independent transgenic lines, ubi::ZmGF14-6 or ZmPR4::ZmGF14-6 lines (15 plants per line). (This figure is available in colour at JXB online.)
Water loss was measured in Ubi::ZmGF14-6 and ZmPR4::ZmGF14-6 transgenic lines and wild-type plants. After 4 h of treatment, leaves from the Ubi::ZmGF14-6 rice lines had slight lower water loss than wild-type plants (Fig. 3D). Water loss of detached leaves from ZmPR4::ZmGF14-6 plants was similar to that of leaves from wild-type plants. These results further showed that constitutive expression of ZmG14-6 in rice reduces water losses in leaves under drought conditions (Fig. 3D).
ZmGF14-6 positively regulates the expression of drought-associated marker genes in rice roots
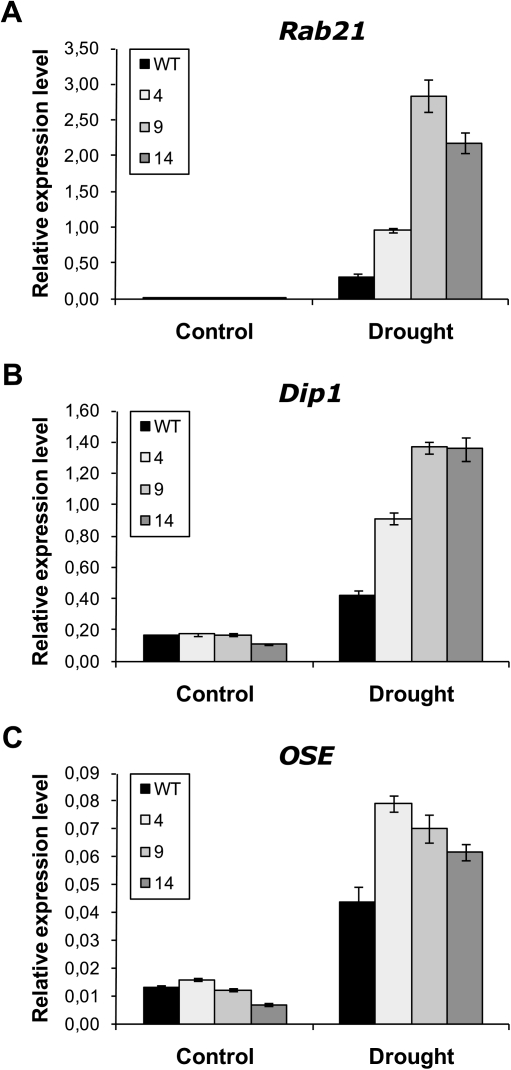
The expression of the endogenous drought-regulated rice genes was examined in roots of transgenic Ubi:ZmGF14-6 and wild-type rice plants by qRT-PCR. Rice genes whose expression has been reported to be up-regulated in response to drought stress were selected for this study. They were rab21 (responsive to abscisic acid 21, a rice dehydrin), dip1 (dehydration-stress inducible protein 1), and OSE2 (a bZIP transcription factor) (Rabbani et al., 2003; Yi et al., 2010). The expression of these marker genes was analysed in roots of rice plants grown either under regular watering conditions or under drought-mimic stress application (20% PEG-8000). Under normal growth conditions there were no relevant differences in the expression of the drought marker genes between wild-type and Ubi:ZmGF14-6 roots (Fig. 4). Under PEG-induced dehydration, however, the expression of all three genes (rab21, dip1, and OSE2) was induced at higher levels in transgenic roots compared to wild-type roots (Fig. 4). Overall, these results suggest that constitutive expression of ZmGF14-6 in rice leads to an enhanced capacity to activate the expression of genes that are normally activated by drought stress in roots of the host plant, which is consistent with the observed phenotype of drought tolerance in the transgenic plants.
Fig. 4.
Expression of drought-associated genes in roots of wild-type (WT) and transgenic lines. Three independent T2 homozygous Ubi::ZmGF14-6 rice plants (lines 4, 9, and 14; 16 plants per line) were grown hydroponically in water for 7 days and then exposed to 20% PEG-8000 (drought) or water (control) for 24 h. qRT-PCR was performed using 2 μg of total RNA with specific primers for (A) rab21 (Os11g26750), (B) dip1 (Os02g44870), and (C) OSE (Os01g64730). Relative expression levels were calculated and normalized with respect to the rice ubiquitin mRNA (OsUbi1, Os06g46770). Each column represents an average of three replicates and bars indicate SDs.
Expression of ZmGF14-6 increases susceptibility to pathogen infection in rice plants
In this work we examined whether expression of ZmGF14-6 in rice has an influence on pathogen resistance in the host plant. Transgenic ZmGF14-6 plants, both Ubi:ZmGF14-6 and ZmPR4::ZmGF14-6 plants, were tested for resistance to infection with the fungal pathogens F. verticillioides and M. oryzae, the causal agents of the bakanae and blast diseases of rice, respectively.
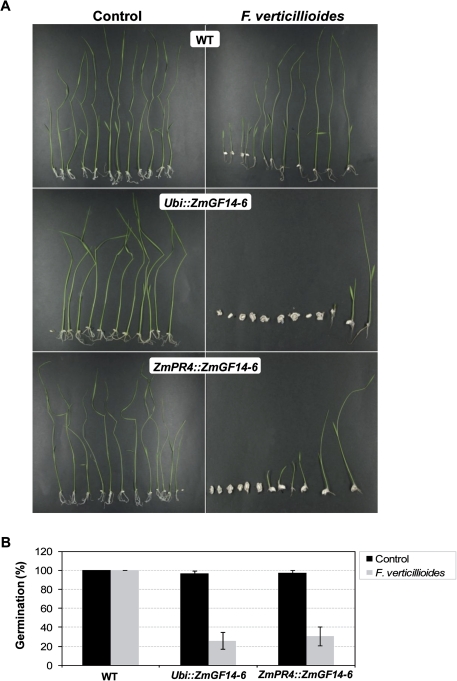
F. verticillioides is a seedborne and soil-transmitted pathogen. Under the experimental conditions here assayed, the F. verticillioides-infected seeds from wild-type lines germinated and developed (Fig. 5A, upper panels). Contrary to this, the fungus-infected seeds from plants constitutively expressing ZmGF14-6 showed extensive colonization by the fungus and most of them did not germinate (Fig. 5A, middle panels). Equally, most of the F. verticillioides-infected seeds from rice lines expressing ZmGF14-6 under an inducible regime (ZmPR4::ZmGF14-6 lines) were not able to overcome infection by F. verticillioides (Fig. 5A, lower panels). The survival for Ubi::ZmGF14-6 and ZmPR4::ZmGF14-6 transgenic lines was 25.71 and 30.56%, respectively (Fig. 5B).
Fig. 5.
Susceptibility or rice plants expressing the ZmGF14-6 gene to infection by F. verticillioides. Three independently T2 homozygous lines expressing ZmGF14-6 under the control of the constitutive promoter (Ubi::ZmGF14-6 lines 4, 9, and 14) or the pathogen-inducible ZmPR4 promoter (ZmPR4::ZmGF14-6 lines 1, 8, and 10) were assayed in disease-resistance assays. Representative results obtained for one line for each transformation event are shown (similar results were observed with the three independent ZmGF14-6 lines for each transformation event). (A) Seeds were germinated for 24 h, inoculated with 50 μl of sterile water (Control, left) or 50 μl of fungal spores (102 spores ml−1) (F. verticillioides, right), and then allowed to continue germination. Picture was taken 10 days after inoculation. Pictures shown are from one of three experiments that gave similar results. (B) Percentage of seeds that overcome infection by the fungus F. verticillioides relative to non-infected seeds. Seedling survival was scored for each line at 10 days after inoculation with fungal spores (the histograms show the mean ± SD from three independent replicates). (This figure is available in colour at JXB online.)
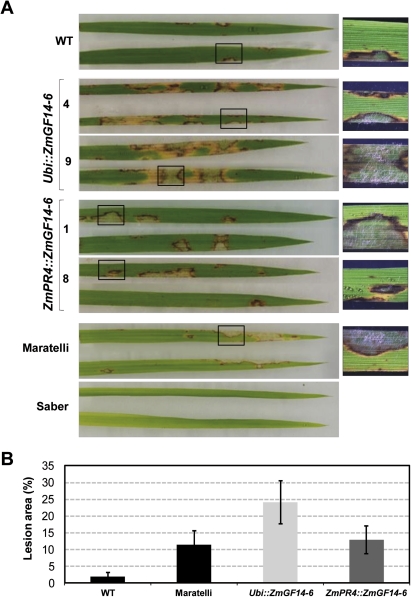
For studies on resistance to the rice blast fungus M. oryzae the detached-leaf assay was used (Coca et al., 2004). Leaves from ZmGF14-6 and wild-type plants were inoculated with a M. oryzae spore suspension. Two rice cultivars, Maratelli and Saber (susceptible and resistant to M. oryzae), were used as reference cultivars in these assays (Koutroubas et al., 2009). Differences in the degree and development of disease symptoms caused by M. oryzae between ZmGF14-6 and wild-type plants were clearly observed. Using an inoculum of 106 spores ml−1, all the transgenic Ubi::ZmGF14-6 and ZmPR4::ZmGF14-6 lines, as well as the susceptible cultivar Maratelli, exhibited clear symptoms of infection at 4 days post-inoculation (Fig. 6A). Sporulating lesions were also visible in the inoculated leaves from transgenic plants. By this time, disease symptoms were less evident in leaves of wild-type (Senia) plants. As expected, the resistant cultivar Saber did not develop disease symptoms.
Fig. 6.
Susceptibility of rice plants expressing the ZmGF14-6 gene to infection by M. oryzae. Disease resistance was tested in transgenic lines expressing the ZmGF14-6 gene under the control of either the constitutive ubiquitin (ubi) promoter or the pathogen-inducible ZmPR4 promoter using the detached-leaf assay (Coca et al., 2004). In each case, three independent transgenic lines were assayed with similar results (results for only two of these lines are shown). Reference cultivars used in this study were Maratelli (susceptible) and Saber (resistant). (A) Leaves were locally inoculated with a M. oryzae spore suspension (106 spores ml−1). Disease symptoms 4 days after inoculation of leaves are shown. Results shown are from one of four experiments that produced similar results. (B) Size of lesions (in cm2) produced on the leaves of the control and transgenic ZmGF14-6 rice lines 4 days after inoculation with M. oryzae spores (106 spores ml−1). The average lesion was determined from spot infections by using the APS Assess 2.0 programe (three independent transgenic lines, 12 plants per line, and five inoculations per leaf). Error bars represent SDs. (This figure is available in colour at JXB online.)
The observed susceptibility of GF14-6 plants to M. oryzae infection was further assessed by measuring the average lesion size on leaf surface. As shown in Fig. 6B, the inoculated leaves from Ubi:ZmGF14-6 and ZmPR4::ZmGF14-6 plants exhibited broader areas of lesion on their surface compared to control non-transformed rice plants. Overall this study demonstrated that expression of ZmGF14-6 in transgenic rice led to an enhanced disease susceptibility to fungal infection.
Transgenic expression of ZmGF14-6 negatively regulates the expression of pathogen-regulated rice genes
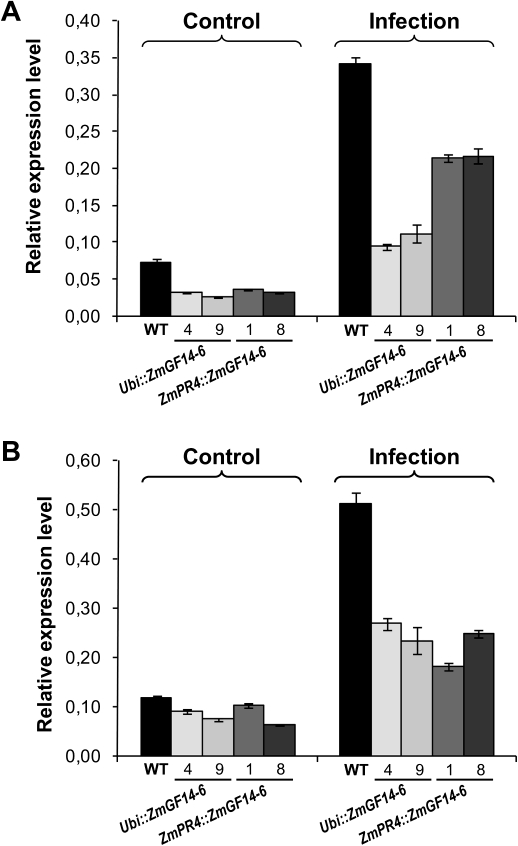
Knowing that ZmGF14-6 rice plants exhibited increased susceptibility to infection by fungal pathogens, it was of interest to determine whether transgenic expression of ZmGF14-6 in rice has an effect on the expression of endogenous defence-related genes. PR genes whose expression is known to be activated by M. oryzae infection in rice leaves were selected and their expression examined in wild-type and transgenic lines (Gómez-Ariza et al., 2007; Quilis et al., 2008). They were PBZ1 (probenazole inducible 1) and PR5 (pathogenesis-related 5; thaumatin-like protein) genes. In particular, PBZ1 is considered a marker of the activation of the rice defence response to pathogen infection (Midoh and Iwata, 1996). PR gene expression was examined by qRT-PCR in wild-type, Ubi::ZmGF14-6, and ZmPR4::ZmGF14-6 plants. The fungus-induced expression of control and transgenic plants was also examined. As expected, PBZ1 and PR5 expression was induced in M. oryzae-infected leaves of wild-type plants (Fig. 7A and B, black bars). Induction of PBZ1 and PR5 expression in response to fungal infection was also observed in transgenic lines, albeit to a much lower extent compared to the fungus-induced expression that occurs in wild-type plants (Fig. 7A and B, grey bars). Overall, this study demonstrated that expression of ZmGF14-6 in rice reduces the intensity of the fungus-induced expression of the PBZ1 and PR5 genes. A reduced capacity to activate the endogenous defence response would also explain the observed phenotype of susceptibility to infection by the rice blast fungus M. oryzae in transgenic ZmGF14-6 plants.
Fig. 7.
Expression of the rice defence genes PBZ1 (A) and PR5 (B) in ZmGF14-6 and wild-type (WT) rice plants. Expression analyses were carried out by qRT-PCR in 24 h M. oryzae-infected and non-infected leaves using specific primers for the PBZ1 (Os12g36880) and PR5 (Os3g46070) rice genes. Each RNA was prepared from a pool of leaves from eight plants at the three-leaf stage. Relative expression levels were calculated and normalized with respect to the rice ubiquitin mRNA (OsUbi1, Os06g46770). Results for Ubi::ZmGF14-6 (lines 4 and 9) and ZmPR4::ZmGF14-6 (lines 1 and 8) are shown.
Subcellular localization of ZmGF14-6
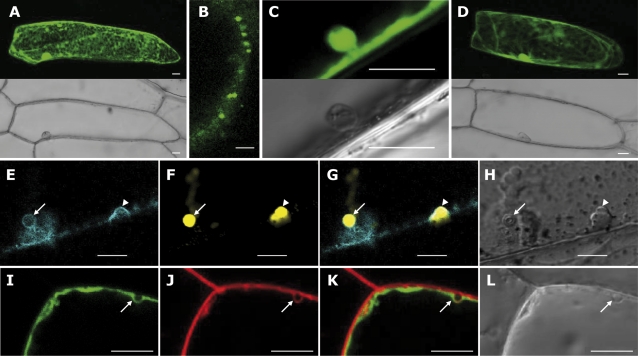
There are numerous studies that indicate that 14-3-3 proteins have multiple subcellular localizations (Paul et al., 2005). To address the subcellular localization of ZmGF14-6, a construct containing the ZmGF14-6 gene fused in-frame with the GFP gene (ZmGF14-6::GFP) under the control of the 35S CaMV promoter was prepared, and the fusion gene was transiently expressed in onion epidermal cells. In agreement with results previously reported for 14-3-3 proteins from barley and Arabidopsis (Paul et al., 2005; Li and Dhaubhadel, 2011), confocal laser scanning microscopy revealed ZmGF14-6-specific labelling distributed in the cytoplasm and nucleus of transformed onion cells (Fig. 8A). Noticeably, ZmGF14-6 fluorescence was also observed in vesicle structures near the plasma membrane (Fig. 8B and C). Control experiments in which onion cells were transformed with the GFP alone, showed a diffused distribution of green fluorescence throughout the cell (Fig. 8D).
Fig. 8.
Subcellular localization of the ZmGF14-6 protein in onion cells. Onion cells were transformed with ZmGF14-6::GFP (A–C, I–L), GFP (D), or ZmGF14-6::CFP (E–H) via particle bombardment. Confocal images were taken 24 h post-bombardment. Projection images (A, D) and individual sections (B, C, E–L) are shown. Scale bars = 20 μm. (A–C) Cell transformed with ZmGF14-6::GFP. A magnification image of a ZmGF14-6-expressing onion cell is shown in (B) and (C). Fluorescence and transmission images are shown. (D) Onion epithelial cell expressing GFP. (E–H) Co-expression of ZmGF14-6::CFP and VAMP727::YFP, a marker of early endosomes. CFP fluorescence image (E) and YFP fluorescence image (F) are merged in (G). Transmission image is shown in (H). (I–L) Onion epithelial cell expressing ZmGF14-6::GFP were labelled with FM4-64. GFP fluorescence image (I) and FM4-64 fluorescence image (J) are merged in (K). (L) Transmission image of the onion cell.
To obtain a better understanding of the subcellular localization of ZmGF14-6, coexpression experiments using fluorescent-tagged soluble NSF-attachment protein receptor (SNARE) proteins as organelle markers were carried out. The Golgi bodies were visualized with YFP-SYP31 (cis-Golgi marker; where SYP is syntaxin of plants) and YFP-SYP41 (trans-Golgi marker), the prevacuolar compartment with YFP-SYP21, and early endosomes with YFP-VAMP727 (Uemura et al., 2004; where VAMP is vesicle-associated membrane protein). For colocalization experiments, a fusion gene of ZmGF14-6 and the cyan fluorescent protein gene (ZmGF14-6-CFP) was transiently expressed. No colocalization was observed between ZmGF14-6-CFP and any of the two YFP-SYP21/YFP-SYP31 markers, and only occasionally some of the YFP-SYP41-labelled organelles colocalized with ZmGF14-6-CFP in some cells (results not shown). Of interest, ZmGF14-6-GFP correctly localized on the same vesicles as the VAMP727 yellow marker (Fig. 8E–H). This study also revealed invaginations of the plasma membrane in which ZmGF14-6 and VAMP727 colocalized (Fig. 8E–H, arrowheads). VAMP27 is an R-SNARE protein widely used as a marker for early endosomes (Uemura et al., 2004).
The lipophilic styril dye FM4-64 is known to follow the endocytic pathway, from the plasma membrane via endosomes to the vacuole in various eukaryotes including plants (Ueda et al., 2001). In living cells, FM4-64 does not permeate cell membranes but, instead, intercalates into the plasma membrane and is then taken into the cells by endocytosis (Vida and Emr, 1995). Epithelial onion cells expressing ZmGF14-6-GFP were stained with the endocytic marker FM4-64 (red). This study revealed a clear colocalization of ZmGF14-6-GFP and FM4-64 fluorescence in plasma membrane-derived vesicles. ZmGF14-6 and FM4-64 fluorescence, however, did not colocalize at the plasma membrane suggesting that ZmGF14-6 is recruited during formation of endocytic vesicles (Fig. 8I–L, arrows). From these results it is concluded that, in addition to the nucleus and cytoplasm, ZmGF14-6 localizes at the early endosomes. These observations support a role for this maize 14-3-3 protein in the plant endocytic pathway.
Discussion
Plant 14-3-3 proteins have recently attracted much interest due to growing evidence that they play important roles in the mediation of cellular responses to environmental stresses. Compelling evidence exists that distinct 14-3-3 proteins mediate resistance to pathogen infection in Arabidopsis and tomato plants (Yang et al., 2009; Oh et al., 2010; Oh and Martin, 2011). In the literature there are also examples of plant genes encoding 14-3-3 proteins whose expression is regulated by abiotic stresses such as salinity, drought, and wounding (Lapointe et al., 2001; Chen et al., 2006). However, no functional evidence on the involvement of most of these genes in conferring tolerance to abiotic stress has been demonstrated. Only the Arabidopsis gene GF14λ, when introduced into cotton plants, has been shown to improve tolerance under moderate drought conditions (Yan et al., 2004).
Here we report the functional characterization of the maize ZmGF14-6 gene encoding a 14-3-3 protein in transgenic rice. In maize, ZmGF14-6 gene expression is transcriptionally regulated in response to biotic and abiotic stress. These transcriptional responses are rapid and transient. Moreover, ZmGF14-6 expression is regulated in opposite directions depending on the type of stress. Whereas ZmGF14-6 is activated in response to pathogen infection and salinity, its expression is down-regulated by drought stress.
Definitive proof that ZmGF14-6 is a mediator of the plant response to both biotic and abiotic stresses came from studies in transgenic rice. Thus, the results presented here show that expression of the maize ZmGF14-6 in rice plants confers tolerance to drought stress. Under drought stress conditions, a strong activation of drought-associated rice genes, namely rab21, dip1, and OSE, occurs in roots of ZmGF14-6 rice plants. Notably, the expression of drought marker genes remains at normal levels in well-watered transgenic roots, and only when plants are subjected to drought stress conditions does a higher expression of these marker genes occur in transgenic roots compared to wild-type roots. Presumably, the concerted action of ZmGF14-6 with other drought-responsive components is needed for maximal rab21, dip1, and OSE activation. In this respect, 14-3-3 proteins have been proposed to play a scaffolding role in the assembly of transcriptional complexes controlling the expression of drought-associated genes. For instance, 14-3-3 proteins have been shown to interact with viviparous-1 (VP1) and Em-binding protein 1 (EmBP1), two transcription factors controlling abscisic acid-responsive gene expression, and hence drought-responsive gene expression (Schultz et al., 1998).
Contrary to previously reported results indicating that 14-3-3 proteins positively regulate disease resistance in several plant/pathogen interactions, results here presented demonstrate that expression of ZmGF14-6 in transgenic rice increases susceptibility to infection by the fungal pathogens F. verticillioides and M. oryzae in rice. This observation was already indicative of a negative role for the ZmGF14-6 gene in the plant defence response to pathogen infection. By examining the expression of the endogenous rice defence genes, it is shown that ZmGF14-6 negatively regulates expression of the defence marker genes PBZ1 and PR5. The same phenotype of susceptibility was observed in rice plants expressing the ZmGF14-6 gene under a constitutive or an inducible regime, thus ruling out the possibility of artifacts caused by the constitutive expression and over accumulation of ZmGF14-6 in rice tissues.
Clearly, under natural conditions, the signal transduction pathways that are activated in response to different environmental stresses must interconnect to allow plants to coordinate and prioritize their responses. The observed phenotypes of ZmGF14-6-rice plants under different types of stress suggest that this particular 14-3-3 protein might be a modulator of different stress-induced signalling pathways. It can be reasoned that ZmGF14-6 exerts its regulatory role through the concerted action on positive and/or negative regulators associated to each type of stress. Positive effects of ZmGF14-6 on drought tolerance might result from activation of positive regulators (or repression of negative regulators) of drought-associated responses whereas negative effects of ZmGF14-6 on immunity might result from activating negative regulators (or repressing positive regulators) of defence responses to pathogen infection. Specific interactions of the ZmGF14-6 protein with different target proteins might occur which might well explain its involvement in different signalling pathways. All independent transgenic lines used in the present study exhibited the same phenotype and behaviour under biotic and abiotic stress conditions, and transgene-mediated alterations in host gene expression, excluding the possibility of an insertional effect caused by transgene integration into the rice genome.
On the other hand, it is well established that plant responses and signalling pathways activated by drought and salt stress are largely overlapping (Rabbani et al., 2003). Results presented here indicate that ZmGF14-6 inversely responds to each one of these abiotic stresses in maize plants, its expression being up-regulated by salt stress and down-regulated by drought stress. There is then the possibility that ZmGF14-6 functions in a stress-specific manner. Alternatively, ZmGF14-6 might be a common component of the two signalling pathways that positively or negatively regulates the plant response to one or another stress. If so, ZmGF14-6 might represent a key component in crosstalk between the signalling pathways induced by drought and salt stress. However, the consequences of ZmGF14-6 expression in rice plants under salt conditions have not been examined in this work.
In addition to changes in 14-3-3 gene expression, other mechanisms by which 14-3-3 proteins might exert their regulatory functions are interactions with different client proteins. Moreover, multiple 14-3-3 protein isoforms exist in each organism, each of them controlling the activity of target proteins through different modes of action, such as inhibition of target protein degradation, conformational changes of a target protein, and scaffolding of two target proteins (Oh, 2010). Knowing that 14-3-3 proteins can regulate the activities of many different client proteins and with different modes of action, it is not surprising to find that 14-3-3 proteins play roles at multiple levels, not only in the plant response to stress, but also in the regulation of a wide range of physiological and developmental processes. In line with this, constitutive expression of ZmGF14-6 had important consequences in terms of the ‘physiological costs’ for the rice plant. Thus, transgenic plants constitutively expressing ZmGF14-6 had reduced growth and lower grain yields even when grown in well-watered conditions. Contrary to this, transgenic expression of ZmGF14-6 under the control of the pathogen-inducible ZmPR4 promoter has no penalty on plant growth (seed productivity in ZmPR4::ZmGF4-6 lines was not examined in this work).
Additionally, 14-3-3 proteins have multiple subcellular locations such as the nucleus, cytoplasm, endoplasmic reticulum, chloroplast, and mitochondria, which is consistent with their multifunctional roles in cellular processes (Bihn et al., 1997; Sehnke et al., 2002a; Paul et al., 2005). The subcellular localization of ZmGF14-6, and by extension its interaction with client proteins, might be also important in determining the role of this protein in the plant response to biotic and abiotic stress. Transient expression of the ZmGF14-6-GFP fusion gene in onion cells revealed a broad distribution of the ZmGF14-6 protein at the cytoplasm and nucleus. Concerning its nuclear localization, the ZmGF14-6 protein was originally identified as being associated with the G-box-binding complex (de Vetten and Ferl, 1994). It is then not surprising to find a fraction of the ZmGF14-6 protein accumulating at the nucleus. Of particular interest was the finding that ZmGF14-6 localizes at early endosomes, as determined by its clear colocalization with VAMP727, a marker of early endosomes. Studies using the red styryl marker dye FM4-64 unambiguously demonstrated that ZmGF14-6 localizes at plasma-derived early endosomes (Vida and Emr, 1995).
In other studies, the interaction of 14-3-3 proteins with plasma membrane proteins, such as H+-ATPases, was reported. The regulation of reactive oxygen species production by the activity of a plasma membrane-bound nicotinamide adenosine dinucleotide phosphate oxidase, which is itself regulated by its interaction with a 14-3-3 protein, has been also described in tobacco cells treated with criptogein (Elmayan et al., 2007). Although the ZmGF14-6 protein itself does not show plasma membrane localization, an interaction of this 14-3-3 protein with proteins undergoing endocytosis during the plant response to biotic and/or abiotic stress can be postulated.
In plant cells, the early endosome is the first endosomal compartment that receives endocytosed cargo from the plasma membrane (Ueda et al., 2004). Concerning the contribution of the endocytic machinery in signalling processes in plants, it is now widely accepted that endocytosis of plasma membrane receptors plays a crucial role in plant–microbe interactions (Leborgne-Castel et al., 2010). Thus, surface receptors, such as the plant defence-related flagellin-sensing 2 (FLS2) receptor kinase and the brassinosteroid receptor BRI1 appear to signal from endosomes upon ligand binding and internalization. Although a comprehensive understanding of how plant endocytosis impacts stress-induced signalling pathways is still lacking, the localization of ZmGF14-6 to the early endosomes strengthens the prospect that this particular 14-3-3 protein interacts with target proteins in the endocytic pathway. Whether ZmGF14-6 interacts with and regulates the activity of endocytosed proteins remains to be determined. It will be also of interest to investigate whether the ZmGF14-6 protein is involved in shuttling of its client proteins in the plant cells either from the early endosomes to the cytosol, during endosome to plasma membrane recycling, or in nucleoplasmic shuttling.
Collectively, the results presented here demonstrate the complexity of the roles played by the ZmGF14-6 protein in plant adaptation to stress conditions while illustrating the antagonistic relationships that occur between signalling pathways controlling the plant response to biotic and abiotic stresses. Clearly, tolerance to drought stress in rice with no undesirable agronomic characteristics is desirable. Taking into account that transgenic expression of ZmGF14-6 in rice is associated with a penalty during normal growth as well as under infection conditions, the practical question is whether the benefits observed by transgenic expression of ZmGF14-6 in rice in terms of drought tolerance could be used to engineer drought tolerance in rice plants under field conditions. Using drought-inducible promoters with low basal activity might allow the improvement of drought tolerance while minimizing disturbance to plant development in the absence of stress or during pathogen infection. These findings also suggest possible research opportunities to explore the function of plant 14-3-3 proteins while also having a bearing on future biotechnological attempts to improve tolerance to drought stress in other crops.
Supplementary material
Supplementary material is available at JXB online.
Supplementary Fig. S1. pCAMBIA1300-derived plasmids used for rice transformation and expression of the ZmGF14-6 gene in rice.
Supplementary Fig. S2. Comparison of the deduced amino acid sequences of 14-3-3 proteins and 14-3-3-like proteins so far described in maize.
Supplementary Table S1. List of primers used for expression analysis of maize and rice genes by qRT-PCR and cloning purposes.
Acknowledgments
We thank Dr M. Sato (Kyoto University, Japan) for fluorescent organelle markers. This work was supported by grants BIO2006-05583 and BIO2009-08719 from the Spanish Ministry of Science and Innovation, as well as by the Consolider-Ingenio 2010 Programme CSD2007-00036 “Centre for Research in Agrigenomics”. We also thank the “Departament d’Innovació, Universitats i Empresa” from the Generalitat de Catalunya (Xarxa de Referencia en Biotecnología and SGR 09626) for substantial support. SC carried out most of the experimental work, and BSS coordinated the design and execution of this study. CPP and LM performed infection experiments. SC and BSS wrote the paper. GP and JM collaborated in rice transformation.
Glossary
Abbreviations
- CFP
cyan fluorescent protein
- Ct
cycle threshold
- Dip1
dehydration-stress inducible protein 1
- EDTA
ethylenediaminetetra-acetic acid
- GFP
green fluorescent protein
- PBS
phosphate-buffered saline
- PBS-T
phosphate-buffered saline containing Tween 20
- PBZ1
probenazole inducible 1
- PEG
polyethylene glycol
- PR5
pathogenesis-related 5
- qRT-PCR
quantitative reverse transcriptase-PCR
- Rab21
responsive to abscisic acid 21
- RACE
rapid amplification of cDNA ends
- SNARE
soluble NSF-attachment protein receptor
- ubi
ubiquitin
- VAMP
vesicle-associated membrane protein
- YFP
yellow fluorescent protein
- ZmPR4
Zea mays pathogenesis-related 4
References
- Bihn EA, Paul A-L, Wang SW, Erdos GW, Ferl RJ. Localization of 14-3-3 proteins in the nuclei of Arabidopsis and maize. The Plant Journal. 1997;12:1439–1445. doi: 10.1046/j.1365-313x.1997.12061439.x. [DOI] [PubMed] [Google Scholar]
- Brandt J, Thordal-Christensen H, Vad K, Gregersen PL, Collinge DB. A pathogen-induced gene of barley encodes a protein showing high similarity to a protein kinase regulator. The Plant Journal. 1992;2:815–820. [PubMed] [Google Scholar]
- Campo S, Carrascal M, Coca M, Abian J, San Segundo B. The defense response of germinating maize embryos against fungal infection: a proteomics approach. Proteomics. 2004;4:383–396. doi: 10.1002/pmic.200300657. [DOI] [PubMed] [Google Scholar]
- Chen F, Li Q, Sun L, He Z. The rice 14-3-3 gene family and its involvement in responses to biotic and abiotic stress. DNA Research. 2006;13:53–63. doi: 10.1093/dnares/dsl001. [DOI] [PubMed] [Google Scholar]
- Chevalier D, Morris ER, Walker JC. 14-3-3 and FHA domains mediate phosphoprotein interactions. Annual Review of Plant Biology. 2009;60:67–91. doi: 10.1146/annurev.arplant.59.032607.092844. [DOI] [PubMed] [Google Scholar]
- Christensen AH, Quail PH. Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Research. 1996;5:213–218. doi: 10.1007/BF01969712. [DOI] [PubMed] [Google Scholar]
- Coca M, Bortolotti C, Rufat M, Peñas G, Eritja R, Tharreau D, del Pozo A, Messeguer J, San Segundo B. Transgenic rice plants expressing the antifungal AFP protein from Aspergillus giganteus show enhanced resistance to the rice blast fungus Magnaporthe grisea. Plant Molecular Biology. 2004;54:245–259. doi: 10.1023/B:PLAN.0000028791.34706.80. [DOI] [PubMed] [Google Scholar]
- Cordero MJ, Raventós D, San Segundo B. Expression of a maize proteinase-inhibitor gene is induced in response to wounding and fungal infection - systemic wound-response of a monocot gene. The Plant Journal. 1994;6:141–150. doi: 10.1046/j.1365-313x.1994.6020141.x. [DOI] [PubMed] [Google Scholar]
- de Vetten NC, Ferl RJ. Two genes encoding GF14 (14-3-3) proteins in Zea mays (structure, expression, and potential regulation by the G-box-binding complex) Plant Physiology. 1994;106:1593–1604. doi: 10.1104/pp.106.4.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vetten NC, Lu G, Ferl RJ. A maize protein associated with the G-box binding complex has homology to brain regulatory proteins. The Plant Cell Online. 1992;4:1295–1307. doi: 10.1105/tpc.4.10.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmayan T, Fromentin J, Riondet C, Alcaraz G, Blein J-P, Simon-Plas F. Regulation of reactive oxygen species production by a 14-3-3 protein in elicited tobacco cells. Plant, Cell & Environment. 2007;30:722–732. doi: 10.1111/j.1365-3040.2007.01660.x. [DOI] [PubMed] [Google Scholar]
- Faris JD, Li WL, Liu DJ, Chen PD, Gill BS. Candidate gene analysis of quantitative disease resistance in wheat. Theoretical and Applied Genetics. 1999;98:219–225. [Google Scholar]
- Ferl RJ. 14-3-3 proteins and signal transduction. Annual Review of Plant Physiology and Plant Molecular Biology. 1996;47:49–73. doi: 10.1146/annurev.arplant.47.1.49. [DOI] [PubMed] [Google Scholar]
- Ferl RJ. 14-3-3 proteins: regulation of signal-induced events. Physiologia Plantarum. 2004;120:173–178. doi: 10.1111/j.0031-9317.2004.0239.x. [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Current Opinion in Plant Biology. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Gökirmak T, Paul A-L, Ferl RJ. Plant phosphopeptide-binding proteins as signaling mediators. Current Opinion in Plant Biology. 2010;13:527–532. doi: 10.1016/j.pbi.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Gómez-Ariza J, Campo S, Rufat M, Estopa M, Messeguer J, San Segundo B, Coca M. Sucrose-mediated priming of plant defense responses and broad-spectrum disease resistance by overexpression of the maize pathogenesis-related PRms protein in rice plants. Molecular Plant-Microbe Interactions. 2007;20:832–842. doi: 10.1094/MPMI-20-7-0832. [DOI] [PubMed] [Google Scholar]
- Igarashi D, Ishida S, Fukazawa J, Takahashi Y. 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. The Plant Cell. 2001;13:2483–2497. doi: 10.1105/tpc.010188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo JA, Capel J, Leyva A, Martínez-Zapater JM, Salinas J. Two related low-temperature-inducible genes of Arabidopsis encode proteins showing high homology to 14-3-3 proteins, a family of putative kinase regulators. Plant Molecular Biology. 1994;25:693–704. doi: 10.1007/BF00029607. [DOI] [PubMed] [Google Scholar]
- Koutroubas SD, Katsantonis D, Ntanos DA, Lupotto E. Blast fungus inoculation reduces accumulation and remobilization of pre-anthesis assimilates to rice grains. Phytopathologia Mediterranea. 2009;48:240–252. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:380–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lapointe G, Luckevich MD, Cloutier M, Séguin A. 14-3-3 gene family in hybrid poplar and its involvement in tree defence against pathogens. Journal of Experimental Botany. 2001;52:1331–1338. [PubMed] [Google Scholar]
- Leborgne-Castel N, Adam T, Bouhidel K. Endocytosis in plant–microbe interactions. Protoplasma. 2010;247:177–193. doi: 10.1007/s00709-010-0195-8. [DOI] [PubMed] [Google Scholar]
- Li X, Dhaubhadel S. Soybean 14-3-3 gene family: identification and molecular characterization. Planta. 2011;233:569–582. doi: 10.1007/s00425-010-1315-6. [DOI] [PubMed] [Google Scholar]
- Lu G, DeLisle AJ, de Vetten NC, Ferl RJ. Brain proteins in plants: an Arabidopsis homolog to neurotransmitter pathway activators is part of a DNA binding complex. Proceedings of the National Academy of Sciences, USA. 1992;89:11490–11494. doi: 10.1073/pnas.89.23.11490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig AA, Romeis T, Jones JDG. CDPK-mediated signalling pathways: specificity and cross-talk. Journal of Experimental Botany. 2004;55:181–188. doi: 10.1093/jxb/erh008. [DOI] [PubMed] [Google Scholar]
- Manickavelu A, Kawaura K, Oishi K, Shin IT, Kohara Y, Yahiaoui N, Keller B, Suzuki A, Yano K, Ogihara Y. Comparative gene expression analysis of susceptible and resistant near-isogenic lines in common wheat infected by Puccinia triticina. DNA Research. 2010;17:211–222. doi: 10.1093/dnares/dsq009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midoh N, Iwata M. Cloning and characterization of a probenazole-inducible gene for an intracellular pathogenesis-related protein in rice. Plant and Cell Physiology. 1996;37:9–18. doi: 10.1093/oxfordjournals.pcp.a028918. [DOI] [PubMed] [Google Scholar]
- Moore BW, Perez VJ, Gehring M. Assay and regional distribution of a soluble protein characteristic of the nervous system. Journal of Neurochemistry. 1968;15:265–272. doi: 10.1111/j.1471-4159.1968.tb11610.x. [DOI] [PubMed] [Google Scholar]
- Moreno AB, Peñas G, Rufat M, Bravo JM, Estopa M, Messeguer J, San Segundo B. Pathogen-induced production of the antifungal AFP protein from Aspergillus giganteus confers resistance to the blast fungus Magnaporthe grisea in transgenic rice. Molecular Plant-Microbe Interactions. 2005;18:960–972. doi: 10.1094/MPMI-18-0960. [DOI] [PubMed] [Google Scholar]
- Morrison DK. The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends in Cell Biology. 2009;19:16–23. doi: 10.1016/j.tcb.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murillo I, Cavallarín L, San Segundo B. Cytology of infection of maize seedlings by Fusarium moniliforme and immunolocalization of the pathogenesis-related PRms protein. Phytopathology. 1999;89:737–747. doi: 10.1094/PHYTO.1999.89.9.737. [DOI] [PubMed] [Google Scholar]
- Murillo I, Roca R, Bortolotti C, Segundo BS. Engineering photoassimilate partitioning in tobacco plants improves growth and productivity and provides pathogen resistance. The Plant Journal. 2003;36:330–341. doi: 10.1046/j.1365-313x.2003.01880.x. [DOI] [PubMed] [Google Scholar]
- Oh CS. Characteristics of 14-3-3 proteins and their role in plant immunity. Plant Pathology Journal. 2010;26:1–7. [Google Scholar]
- Oh C-S, Martin GB. Tomato 14-3-3 protein TFT7 interacts with a MAP kinase kinase to regulate immunity-associated programmed cell death mediated by diverse disease resistance proteins. Journal of Biological Chemistry. 2011;286:14129–14136. doi: 10.1074/jbc.M111.225086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh C-S, Pedley KF, Martin GB. Tomato 14-3-3 protein 7 positively regulates immunity-associated programmed cell death by enhancing protein abundance and signaling ability of MAPKKK α. The Plant Cell. 2010;22:260–272. doi: 10.1105/tpc.109.070664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A-L, Sehnke PC, Ferl RJ. Isoform-specific Subcellular Localization among 14-3-3 Proteins in Arabidopsis Seems to be Driven by Client Interactions. Molecular Biology of the Cell. 2005;16:1735–1743. doi: 10.1091/mbc.E04-09-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilis J, Peñas G, Messeguer J, Brugidou C, Segundo BS. The Arabidopsis AtNPR1 inversely modulates defense responses against fungal, bacterial, or viral pathogens while conferring hypersensitivity to abiotic stresses in transgenic rice. Molecular Plant-Microbe Interactions. 2008;21:1215–1231. doi: 10.1094/MPMI-21-9-1215. [DOI] [PubMed] [Google Scholar]
- Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiology. 2003;133:1755–1767. doi: 10.1104/pp.103.025742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MR. 14-3-3 Proteins find new partners in plant cell signalling. Trends in Plant Science. 2003;8:218–223. doi: 10.1016/S1360-1385(03)00056-6. [DOI] [PubMed] [Google Scholar]
- Roberts MR, Bowles DJ. Fusicoccin, 14-3-3 proteins, and defense responses in tomato plants. Plant Physiology. 1999;119:1243–1250. doi: 10.1104/pp.119.4.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MR, Salinas J, Collinge DB. 14-3-3 proteins and the response to abiotic and biotic stress. Plant Molecular Biology. 2002;50:1031–1039. doi: 10.1023/a:1021261614491. [DOI] [PubMed] [Google Scholar]
- Rosenquist M, Sehnke P, Ferl RJ, Sommarin M, Larsson C. Evolution of the 14-3-3 protein family: does the large number of isoforms in multicellular organisms reflect functional specificity? Journal of Molecular Evolution. 2000;51:446–458. doi: 10.1007/s002390010107. [DOI] [PubMed] [Google Scholar]
- Rosenquist M, Alsterfjord M, Larsson C, Sommarin M. Data mining the arabidopsis genome reveals fifteen 14-3-3 genes. Expression is demonstrated for two out of five novel genes. Plant Physiology. 2001;127:142–149. doi: 10.1104/pp.127.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saibo NJM, Lourenço T, Oliveira MM. Transcription factors and regulation of photosynthetic and related metabolism under environmental stresses. Annals of Botany. 2009;103:609–623. doi: 10.1093/aob/mcn227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz TF, Medina J, Hill A, Quatrano RS. 14-3-3 proteins are part of an abscisic acid VIVIPAROUS1 (VP1) response complex in the Em promoter and interact with VP1 and EmBP1. The Plant Cell. 1998;10:837–848. doi: 10.1105/tpc.10.5.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnke PC, DeLille JM, Ferl RJ. Consummating signal transduction: the role of 14-3-3 proteins in the completion of signal-induced transitions in protein activity. The Plant Cell. 2002a;14:S339–S354. doi: 10.1105/tpc.010430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnke PC, Rosenquist M, Alsterfjord M, DeLille J, Sommarin M, Larsson C, Ferl RJ. Evolution and isoform specificity of plant 14-3-3 proteins. Plant Molecular Biology. 2002b;50:1011–1018. doi: 10.1023/a:1021289127519. [DOI] [PubMed] [Google Scholar]
- Talbot NJ. On the trail of a cereal killer. Annual Review of Microbiology. 2003;57:177. doi: 10.1146/annurev.micro.57.030502.090957. [DOI] [PubMed] [Google Scholar]
- Ueda T, Yamaguchi M, Uchimiya H, Nakano A. Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO Journal. 2001;20:4730–4741. doi: 10.1093/emboj/20.17.4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Uemura T, Sato MH, Nakano A. Functional differentiation of endosomes in Arabidopsis cells. The Plant Journal. 2004;40:783–789. doi: 10.1111/j.1365-313X.2004.02249.x. [DOI] [PubMed] [Google Scholar]
- Uemura T, Ueda T, Ohniwa RL, Nakano A, Takeyasu K, Sato MH. Systematic analysis of SNARE molecules in Arabidopsis: Dissection of the post-Golgi network in plant cells. Cell Structure and Function. 2004;29:49–65. doi: 10.1247/csf.29.49. [DOI] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. Journal of Cell Biology. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JL, Sinha PK, Variar M, Zheng KL, Leach JE, Courtois B, Leung H. Association between molecular markers and blast resistance in an advanced backcross population of rice. Theoretical and Applied Genetics. 2004;108:1024–1032. doi: 10.1007/s00122-003-1528-1. [DOI] [PubMed] [Google Scholar]
- Wulff EG, Sørensen JL, Lübeck M, Nielsen KF, Thrane U, Torp J. Fusarium spp. associated with rice Bakanae: ecology, genetic diversity, pathogenicity and toxigenicity. Environmental Microbiology. 2010;12:649–657. doi: 10.1111/j.1462-2920.2009.02105.x. [DOI] [PubMed] [Google Scholar]
- Yan J, He C, Wang J, Mao Z, Holaday SA, Allen RD, Zhang H. Overexpression of the Arabidopsis 14-3-3 protein GF14λ in cotton leads to a “stay-green” phenotype and improves stress tolerance under moderate drought conditions. Plant & Cell Physiology. 2004;45:1007–1014. doi: 10.1093/pcp/pch115. [DOI] [PubMed] [Google Scholar]
- Yang X, Wang W, Coleman M, Orgil U, Feng J, Ma X, Ferl R, Turner JG, Xiao S. Arabidopsis 14-3-3 lambda is a positive regulator of RPW8-mediated disease resistance. The Plant Journal. 2009;60:539–550. doi: 10.1111/j.1365-313X.2009.03978.x. [DOI] [PubMed] [Google Scholar]
- Yao Y, Du Y, Jiang L, Liu JY. Molecular analysis and expression patterns of the 14-3-3 gene family from Oryza sativa. Journal of Biochemistry and Molecular Biology. 2007;40:349–357. doi: 10.5483/bmbrep.2007.40.3.349. [DOI] [PubMed] [Google Scholar]
- Yi N, Kim Y, Jeong M-H, Oh S-J, Jeong J, Park S-H, Jung H, Choi Y, Kim J- K. Functional analysis of six drought-inducible promoters in transgenic rice plants throughout all stages of plant growth. Planta. 2010;232:743–754. doi: 10.1007/s00425-010-1212-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.