Abstract
Double helical RNA has become an attractive target for molecular recognition because many non-coding RNAs play important roles in control of gene expression. Recently, we discovered that short peptide nucleic acids (PNA) bind strongly and sequence selectively to a homopurine tract of double helical RNA via triple helix formation. Herein we tested if the molecular recognition of RNA can be enhanced by α-guanidine modification of PNA. Our study was motivated by the discovery of Ly and co-workers that the guanidine modification greatly enhances the cellular delivery of PNA. Isothermal titration calorimetry showed that the guanidine-modified PNA (GPNA) had reduced affinity and sequence selectivity for triple helical recognition of RNA. The data suggested that in contrast to unmodified PNA, which formed a 1:1 PNA-RNA triple helix, GPNA preferred a 2:1 GPNA-RNA triplex-invasion complex. Nevertheless, promising results were obtained for recognition of biologically relevant double helical RNA. Consistent with enhanced strand invasion ability, GPNA derived from D-arginine recognized the transactivation response element (TAR) of HIV-1 with high affinity and sequence selectivity, presumably via Watson-Crick duplex formation. On the other hand, strong and sequence selective triple helices were formed by unmodified and nucelobase-modified PNAs and the purine rich strand of bacterial A-site. These results suggest that appropriate chemical modifications of PNA may enhance molecular recognition of complex non-coding RNAs.
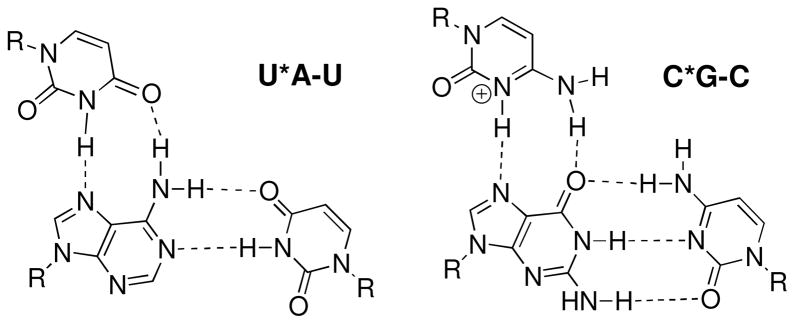
Recent discoveries that non-coding RNAs play important roles in regulation of gene expression stimulate interest in molecular recognition of double helical RNA. However, discovery of small molecules that recognize helical RNA structure and selectively modulate RNA’s function has been a challenging and involved process.1–3 The RNA helix has a relatively uniform and polar surface that presents little opportunity for hydrophobic shape-selective recognition. On the other hand, binding to bulges and internal loops, which are the most common small molecule targets in RNA, is frustrated by the conformational flexibility of non-helical RNA. Hydrogen bond mediated sequence selective triple helix formation could provide a straightforward and effective molecular recognition of double helical RNA.4 Surprisingly, triple helices involving RNA duplex have been little studied. Modestly stable, all RNA triple helices are formed via parallel binding of a pyrimidine rich third strand to a purine rich strand of the double helix.5–7 The molecular recognition of RNA’s sequence occurs via the Hoogsteen hydrogen bonding between uridine and adenosine-uridine base pairs (Figure 1, U*A-U triplet) and between protonated cytidine and guanosine-cytidine base pairs (C*G-C triplet). In contrast to DNA, RNA does not form the pH-independent anti-parallel triplex based on G*G-C, A*A-T and T*A-T triplets.7,8
Figure 1.
Triple helical recognition of RNA via Hoogsteen base triplets.
Practical applications of triple helical recognition of nucleic acids are limited by (1) low stability and slow formation of the triplex caused, at least in part, by electrostatic repulsion between the negatively charged phosphate backbones of the double helix and the incoming third strand oligonucleotide and (2) the requirement for long homopurine tracts, as only U*A-U and C*G-C triplets are used in the common triple helical recognition. Recently, we discovered that short peptide nucleic acids (PNA)9 recognized double helical RNA via highly stable and sequence selective triple helix formation.10–12 PNA, as short as hexamer, formed triple helices with RNA duplex faster and with higher affinity than RNA as the third strand.10 Furthermore, nucleobase modifications allowed recognition of isolated pyrimidine inversions in short polypurine tracts, thus, expanding the potential of recognition to biologically relevant double helical RNA, such as ribosomal RNA and microRNAs.12 These findings inspired a hypothesis that, because of the absence of negatively charged backbone, PNA will be a superior candidate for triple helical recognition of RNA and may overcome the limitations of natural oligonucleotides in triple helical recognition. Interestingly, despite extensive studies on DNA-PNA triplexes,13 binding of PNA to double helical RNA had not been studied before our recent work.10–12 Our results encouraged us to further explore the potential of chemically modified PNA in molecular recognition of double helical RNA.
Despite the excellent chemical and biophysical properties, in vivo applications of unmodified PNA have been limited because of poor uptake by mammalian cells. Recent work on chemically modified PNAs showed that the cellular delivery may be enhanced by attaching cationic cell penetrating peptides.14,15 Ly and co-workers16–18 developed guanidine-modified PNAs (GPNA, the backbone derived from arginine instead of glycine) that maintained strong and sequence selective binding to complementary single stranded DNA and RNA and were efficiently taken up by several cell lines. The enhanced cellular uptake was attributed to the positively charged guanidine groups. We were interested to probe the potential of GPNA in molecular recognition of double helical RNA. We envisioned that combining the high affinity and sequence selectivity of PNA-RNA triplex, as observed in our recent studies,10,12 with the cellular penetration of GPNA would pave the road for in vivo applications of sequence selective recognition of double helical RNA. Herein we used isothermal titration calorimetry and fluorescence spectroscopy to study binding of GPNAs to double helical RNA. The results were further confirmed using circular dichroism (CD) spectroscopy and gel mobility shift assay. We found that guanidine modification reduced the affinity and sequence selectivity of PNA to complementary double helical RNA. The binding stoichiometry increased to 2:1 PNA-RNA complex, suggesting that the most likely mode of binding was a strand invasion triplex. While GPNA did not favor triple helix formation, strong and sequence selective recognition of transactivation response element (TAR) RNA of HIV-1 was achieved using the GPNA derived from D-arginine in a strand invasion mode. Unmodified and nucleobases-modified12 PNA also gave promising results for triple helical recognition of bacterial A-site RNA.
EXPERIMENTAL PROCEDURES
Isothermal Titration Calorimetry
In a typical ITC experiment, RNA hairpin solution (0.95 mL, 5.25 mM) in acetate buffer (100 mM sodium acetate, 1.0 mM EDTA, pH 5.5) was titrated with PNA solution (50 × 5 μL, 96 mM) using a Nano ITC G2 (TA Instruments) calorimeter. For full experimental details and data, see Supporting Information. The titration data (Figures S1–S42) were analyzed using NanoAnalyze software (TA Instruments) using an independent model to obtain the fitting graph and thermodynamic binding data (Table S1).
Fluorescence Spectroscopy
HRP7 (TAR RNA model) solution (2 mL, 0.1 mM) in phosphate buffer (10 mM phosphate, 0.1 mM EDTA, 1 mM MgCl2, pH 6.8) was heated for 6 minutes in 90 °C water bath and then snap-cooled by immediately placing in an ice bath. The sample was placed in a 1 cm path length cuvette and equilibrated at 20 °C using a circulating water bath. The excitation wavelength was set to 305 nm; the emission wavelength was observed at 365 nm. The excitation and emission band width was 10 nm. Titration of the PNA into TAR RNA was done by adding 1–6 μL aliquots of concentrated PNA stock solutions to achieve the required PNA concentration 0.002 to 2 μM. After each addition of PNA, the mixture was stirred for 30 min before fluorescence intensity was measured using a Shimadzu RF-5301pc spectrofluorometer. The data were analyzed by fitting (Figures S44-S50) the change of fluorescence intensity to a single site, two-state binding model as previously described.19
RESULTS
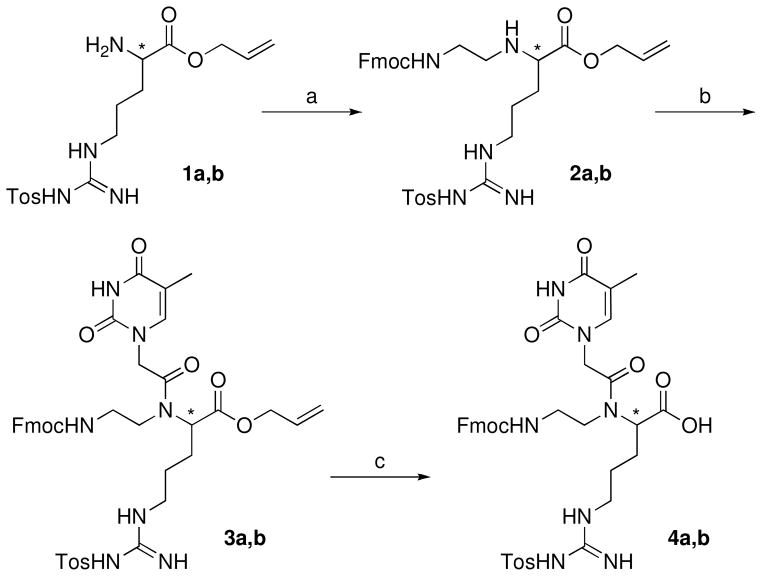
Our study started with modification of the synthetic route designed by Ly and co-workers16,18 to prepare Fmoc protected GPNA monomers that would be compatible with standard PNA synthesis protocols for Expedite 8909 DNA synthesizer. Starting from the known intermediates 1a,b (Scheme 1),16,18 reductive amination with Fmoc-glycinaldehyde20 gave the backbone intermediates 2a,b. The target thymidine GPNA monomers (4a,b, Scheme 1) were prepared by coupling of 2a,b with thymine-1-acetic acid, which was prepared according to established procedures,18 followed by deprotection using N-ethyl aniline and Pd(PPh3)4.16,21
Scheme 1. Synthesis of Fmoc protected GPNA monomers.a.
a The a series has the R stereochemistry (derived from D-arginine) and the b series has the S stereochemistry (derived from L-arginine) at the chiral center (*). Steps (yields for D-series): (a) Fmoc-NHCH2CHO, MeOH, 0 °C, 4h, then acetic acid, NaBH3CN, 30 min, 57%; (b) Thymine-1-acetic acid, 3-hydroxy-1,2,3-benzotriazine-4(3H)-one, N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide, dimethylformamide, 40 °C, 12 h, 60%; (c) Pd(PPh3)4, N-ethyl aniline, tetrahydrofuran, rt, 1h, 81%.
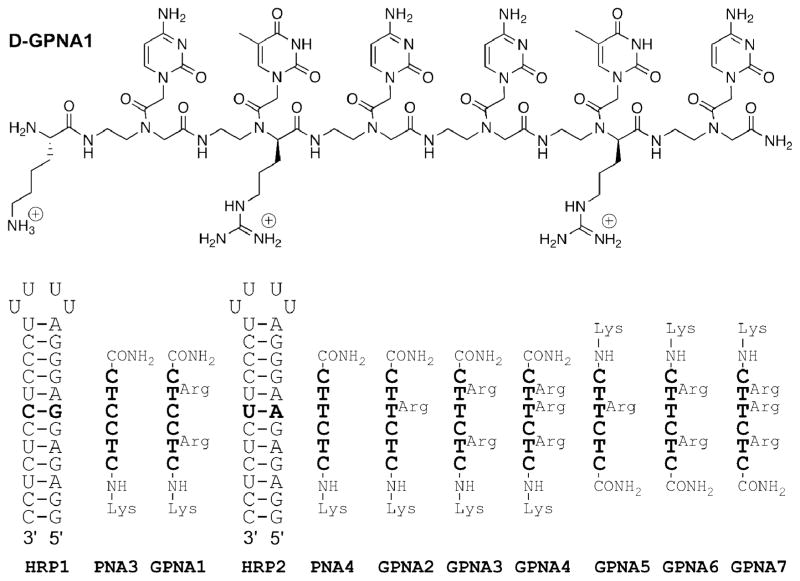
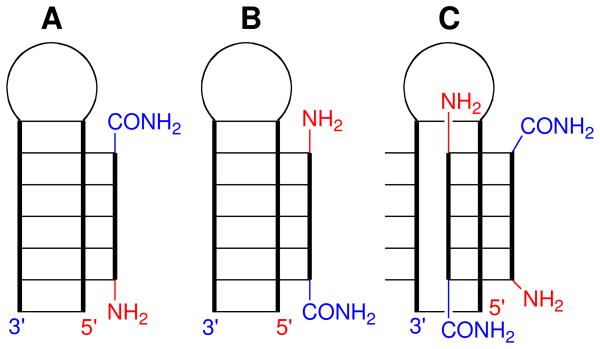
The guanidine-modified PNA oligomers (GPNA, Figure 2) were made using the standard Fmoc synthesis protocols on Expedite 8909 synthesizer and purified by reverse-phase HPLC. Cleavage from the solid support and removal of all protecting groups (including the N-tosyl group) was achieved with a mixture of m-cresol/thioanisole/trifluoromethanesulfonic acid/trifluoroacetic acid (1:1:2:6) as previously reported.16 To allow direct comparison, we prepared the same GPNA sequences (Figure 2), as the PNA used in our previous study on the triple helix formation with double helical RNA.10 The binding of GPNAs to RNA hairpin (HRP1) was studied using isothermal titration calorimetry (ITC, Figure 3).
Figure 2.
Structure of GPNA and sequences of RNA hairpins, PNAs and GPNAs. The numbering of RNA hairpins and PNAs is retained from Ref. 10.
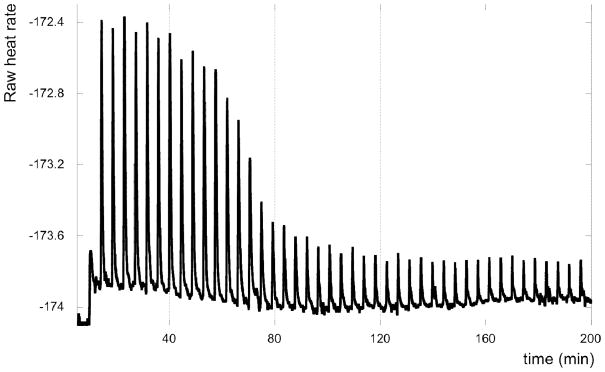
Figure 3.
ITC titration curve of D-GPNA1 (5 μL injections of 96 μM) binding to HRP1 (5.25 μM).
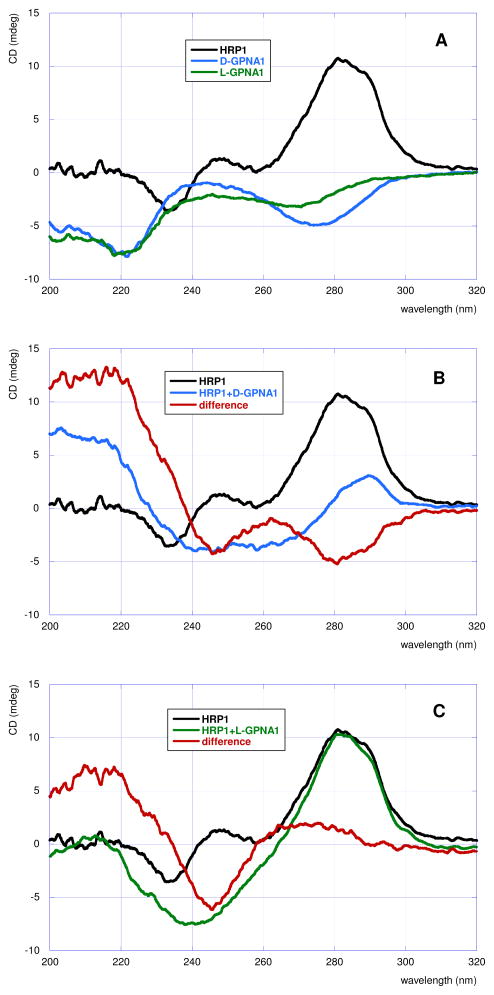
The ITC results, summarized in Table 1, showed that modification of both T monomers in the hexamer CTCCTC (PNA3) with guanidine residue derived from either D- or L-arginine lowered the affinity for double stranded RNA (cf., entry 1 with 2 and 3). Interestingly, the binding order (PNA:RNA stoichiometry) increased from one (observed in our previous study) to two, suggesting that a more complex binding mode, presumably a duplex invasion forming a GPNA-RNA-GPNA triple helix, was taking place (Figure 4). The complex formation was further confirmed using circular dichroism (CD) spectroscopy. Wittung et al. have shown that strand invasion of DNA duplex by PNA resulted in a decrease of CD signal at 240–250 nm and an increase of CD signal at around 280 nm.22,23 In contrast to unmodified PNA, GPNAs exhibited weak but notable CD signal (Figure 5A, blue and green lines) at the concentration used in ITC experiments. Addition of 4.5 equivalents (conditions mimicking the end point of ITC titration) of D-GPNA1 and L-GPNA1 to HRP1 induced a decrease in CD signal at 240–250 nm, which was clearly visible in the difference spectra (red lines in Figure 5B and 5C, respectively). However, we did not observe significant increase around 280 nm. In contrast, the binding of D-GPNA1 showed a decrease of signal at that wavelength. Overall, the CD spectra confirmed the formation of GPNA-RNA complexes.
Table 1.
Thermodynamic data for binding of PNA and GPNA to RNA and DNA hairpins.a
| Ent ry | Sequence | Ka × 106 M−1 | −ΔH kcal/mol | −ΔS eu | −ΔG kcal/mol | Binding order | |
|---|---|---|---|---|---|---|---|
| 1b | PNA3 | NH2-CTCCTC | 84 ± 80 | 29.4 ± 5.3 | 63 ± 17 | 10.6 ± 0.5 | 1.1 ± 0.2 |
| Symmetric sequence, optimal for both parallel triple helix and anti-parallel duplex (strand invasion) | |||||||
| 2 | D-GPNA1 | NH2-CTD-ArgCCTD-ArgC | 4.6 ± 1.6 | 27.3 ± 2.5 | 61 ± 9 | 9.1 ± 0.2 | 1.8 ± 0.1 |
| 3 | L-GPNA1 | NH2-CTL-ArgCCTL-ArgC | 2.2 ± 1.6 | 24.2 ± 9.6 | 53 ± 34 | 8.6 ± 0.5 | 2.0 ± 0.1 |
| 4c | D-GPNA1 | NH2-CTD-ArgCCTD-ArgC | 0.4 | 29.1 | 72 | 7.7 | 2.1 |
| 5c | L-GPNA1 | NH2-CTL-ArgCCTL-ArgC | 2.5 | 21.0 | 41 | 8.7 | 2.2 |
|
| |||||||
| 6b | PNA4 | NH2-CTCTTC | 47 ± 22 | 26.4 ± 3.2 | 54 ± 12 | 10.4 ± 0.3 | 1.3 ± 0.1 |
| Sequence optimal for parallel triple helix | |||||||
| 7 | D-GPNA2 | NH2-CTCTD-ArgTC | 0.5 | 70.0 | 209 | 7.8 | 0.5 |
| 8 | D-GPNA3 | NH2-CTD-ArgCTTD-ArgC | 0.5 | 41.8 | 114 | 7.8 | 0.8 |
| 9 | D-GPNA4 | NH2-CTD-ArgCTD-ArgTD-ArgC | 0.6 | 22.9 | 50 | 7.9 | 1.2 |
| 10 | L-GPNA2 | NH2-CTCTL-ArgTC | 1.8 | 39.6 | 104 | 8.5 | 0.7 |
| 11 | L-GPNA3 | NH2-CTL-ArgCTTL-ArgC | 0.8 | 58.8 | 170 | 8.0 | 0.5 |
| 12 | L-GPNA4 | NH2-CTL-ArgCTL-ArgTL-ArgC | 0.5 | 61.6 | 181 | 7.8 | 0.4 |
|
| |||||||
| Sequence optimal for anti-parallel duplex (strand invasion) | |||||||
| 13d | D-GPNA5 | NH2-CTTD-ArgCTC | 10.5 | 19.8 | 34 | 9.6 | 1.4 |
| 14d | D-GPNA6 | NH2-CTD-ArgTCTD-ArgC | 5.0 | 13.4 | 14 | 9.1 | 2.0 |
| 15d | D-GPNA7 | NH2-CTD-ArgTD-ArgCTD-ArgC | 0.4 ±0.01 | 25.8 ± 3.7 | 61 ± 13 | 7.6 ± 0.0 | 2.3 ± 0.0 |
| 16d | L-GPNA7 | NH2-CTL-ArgTL-ArgCTL-ArgC | 3.8 ± 0.1 | 13.0 ± 7.3 | 14 ± 25 | 8.9 ± 0.3 | 2.1 ± 0.1 |
Average association constants Ka (± standard deviation) in 100 mM sodium acetate, 1.0 mM EDTA, pH 5.5. Entries 1–3 binding to HRP1, entries 4 and 5 binding to DNA version of HRP1 and entries 6–16 binding to HRP2.
From our previous study, reference 10.
Binding to DNA version of HRP1.
PNA is anti-parallel to purine tract of HRP2 (Figure 4B).
Figure 4.
Schematic representation of the binding modes: A parallel triple helix (amino end of PNA aligned with 5′ end of RNA); B antiparallel triple helix; C strand invasion triplex that combines antipareallel PNA binding via Watson-Crick hydrogen bonds and parallel PNA binding via Hoogsteen hydrogen bonds.
Figure 5.
CD spectra of D-GPNA1, L-GPNA1 and HRP1 (A), binding of D-GPNA1 to HRP1 (B) and binding of L-GPNA1 to HRP1 (C). The red lines in B and C are the arithmetic difference of the complex spectra (GPNA+HRP1, blue and green in B and C, respectively) minus sum of GPNA (blue and green in A) and HRP1 (black) spectra. RNA concentration is 5.25 μM, GPNAs 24 μM.
In accord with our previous study,10 binding of D-GPNA1 to HRP1 hairpin made of deoxynucleotides (DNA version of HRP1) was weaker by about an order of magnitude (cf., entries 2 and 4). In contrast, the affinity of L-GPNA1 for either RNA or DNA hairpin was practically the same (cf., entries 3 and 5). At physiologically relevant conditions (37 °C, in 2 mM MgCl2, 90 mM KCl, 10 mM NaCl, 50 mM potassium phosphate at pH 7.4) we observed no binding of D-GPNA1 to HRP1.
The sequence of PNA3 was symmetric and thus provided optimal binding for both parallel triple helix and anti-parallel duplex formation, as required for triplex-invasion (Figure 4). To gain more insight into different binding modes, we prepared guanidine-modified variants of hexamer CTCTTC (PNA4) and studied their binding to HRP2. All the sequences designed to bind in a parallel mode to the polypurine tract of HRP2 (GPNA2-GPNA4, Figure 2, the amino terminus aligned with the 5′-end of RNA, see also Figure 4A) had similar affinity for the RNA target that was about two orders of magnitude lower than the affinity of the unmodified PNA4 (Table 1, cf., entry 6 and entries 7–12). While there was very little dependence on the number of modifications in the D-series (entries 7–9), in the L-series (entries 10–12) the affinity appeared to decrease somewhat with increasing number of guanidine modifications. Interestingly, increasing the number of modifications in the D-series was followed by decrease of both binding enthalpy and entropy (ΔH and ΔS in Table 1) while the reverse was true in the L-series. This result suggested that the stereoisomeric guanidine modifications had distinct interactions with the RNA target. The PNA:RNA stoichiometry (binding order in Table 1) suggested that the parallel GPNAs maintained the original PNA-RNA-RNA triple helical mode of recognition.
In contrast, the sequences designed to bind in an anti-parallel mode (Figure 4B) to the polypurine tract of HRP2 (GPNA5-GPNA7, Figure 2, the amino terminus aligned with the 3′-end of RNA) had significant differences in binding affinity depending on the number and stereochemistry of guanidine modifications. In the D-series (entries 13–15) the binding affinity decreased more than 20 times going from one (entry 13) to three guanidine modifications (entry 15). The binding order increased to two with two and three guanidine modifications, suggesting that the anti-parallel sequences may favor triplex-invasion (GPNA-RNA-GPNA, Figure 4C), as observed for GPNA1 (entries 2 and 3). GPNA7 derived from L-arginine (entry 16) had significantly higher affinity than the D-isomer (entry 15). Binding of GPNA7 to HRP2 produced similar changes in CD spectra as observed for GPNA1/HRP1 (Figure S43). While D-GPNA5 had the highest binding affinity (Ka ~107, entry 13) among all guanidine-modified PNAs tested at pH 5.5, we observed no binding when the experiment in entry 13 was repeated in acetate buffer at pH 7.3.
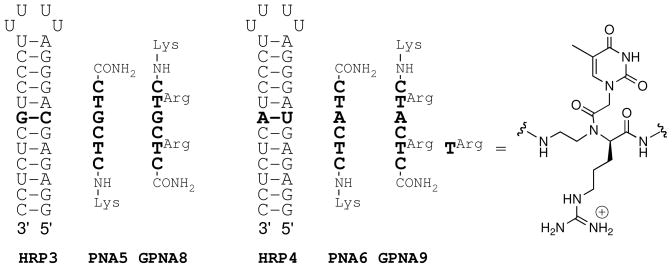
Next we checked the sequence selectivity of GPNA1 binding to all four RNA hairpins having a variable central base pair (HRP1-HRP4, Figures 2 and 6). The results in Table 2 showed that the affinity of unmodified PNA3 and either D- or L-isomer of GPNA1 for the “mismatched” hairpins (HRP2-HRP4) was approximately the same (D-GPNA1 vs. HRP4 was the only notable exception). Thus, the sequence specificity of GPNA was reduced compared to unmodified PNA because of lower affinity for the “matched” target (highlighted bold in Table 2). We also studied the sequence selectivity of anti-parallel (Figure 4B) GPNA8 and GPNA9 in comparison with PNA5 and PNA6, respectively (Figure 6). These sequences all have a “mismatched” central Hoogsteen base triplet, however, because two molecules of D-GPNA8 and D-GPNA9 bind to an RNA hairpin (binding order = 2, see Supporting Information), the combinations D-GPNA8/HRP3 and D-GPNA9/HRP4 would have a matched Watson-Crick base pair if the binding were following the triplex-invasion mode (Figure 4C). Indeed, D-GPNA8 had higher binding affinity than PNA5 to HRP3, consistent with formation of G-C base pair upon invasion, and to HRP4, possibly due to a stabilizing G-U wobble base pair (highlighted bold in Table 2). Similar result, consistent with formation of A-U base pair, was obtained for the D-GPNA9/HRP4 combination, supporting our hypothesis that the binding order of two indicated triplex-invasion and formation of GPNA-RNA-GPNA complex.
Figure 6.
Structure of RNA hairpins, PNAs and GPNAs used in sequence selectivity study.
Table 2.
Sequence selectivity of GPNA binding to RNA hairpins.a
| PNA (variable base) | HRP1a(G-C) | HRP2a (A-U) | HRP3a (C-G) | HRP4a (U-A) |
|---|---|---|---|---|
| PNA3 (C)b | 84 | 0.4 | 0.5 | 0.2 |
| D-GPNA1 | 4.6 | 0.3 | 0.7 | 1.3 |
| L-GPNA1 | 2.2 | 0.8 | 0.6 | 0.4 |
|
| ||||
| PNA5 (G)b | 1.5 | 0.4 | 0.2 | 0.1 |
| D-GPNA8c | 0.8 | 0.6 | 1.2 | 0.7 |
|
| ||||
| PNA6 (A)b | 6.0 | 1.6 | 0.7 | 0.05 |
| D-GPNA9c | NDd | 0.6 | NDd | 0.97 |
Average association constants Ka × 106 M−1 in sodium acetate buffer, pH 5.5.
From our previous study, reference 10.
PNA is anti-parallel to purine tract of RNA.
Not determined.
Next, we tested if PNA and GPNA could recognize more complex biologically relevant RNA, such as the ribosomal A-site. Model hairpins (Figure 7) were designed containing the secondary structure of ribosomal A-sites (sequences from http://www.rna.ccbb.utexas.edu/) of H. Sapiens (HRP5) and M. Tuberculosis (HRP6) and closing at one end with a stable RNA tetraloop and on the other end with a couple of C-G base pairs. The structures of the A rich bulge, which is the target of aminoglycoside antibiotics, of bacterial and human A-sites are remarkably similar. However, significant differences occur in the helical region just above the A rich bulge (bold in Figure 7). Interestingly, the A-site helices feature short polypurine tracts interrupted by single pyrimidine, U in HRP5 and C in HRP6, which prompted us to explore if the A-site RNA may be recognized via triplex or triplex-invasion mode (Figure 7, Table 3).
Figure 7.
Structure of ribosomal A-site model RNA hairpins and the complementary PNAs and GPNAs.
Table 3.
| Entry | PNA | Sequence | M. Tuberculosis (HRP6) | H. Sapiens (HRP5) |
|---|---|---|---|---|
| 1 | PNA10 | NH2-CCCTGCTT | 1.2 (1.7) | 0.4 (2.0) |
| 2 | GPNA10 | NH2-CCCTL-ArgTL-ArgGCT | 0.5 (2.9) | 2.8 (5.0) |
| 3 | PNA11 | NH2-CCTGCTT | 0.2 (1.1) | 0.04 (0.9) |
| 4 | PNA12 | NH2-CTGCTT | 0.2 (1.6) | 0.06 (0.9) |
| 5 | GPNA12 | NH2-CTL-ArgGCTT | 0.4 (1.8) | 0.8 (2.1) |
| 6 | GPNA13 | NH2-CTL-ArgGCTL-ArgT | 2.9 (8.0) | 8.1 (8.8) |
| 7 | PNA13 | NH2-CCCTPCTT | 1.5 (1.1) | NBb |
| 8 | PNA14 | NH2-CCCTPexCTT | 2.0 (1.1) | NBb |
Average association constants Ka × 106 M−1 in sodium acetate buffer, pH 5.5; binding order is given in parenthesis.
No binding, Ka < 103 M−1.
Next, we tested if PNA and GPNA could recognize more complex biologically relevant RNA, such as the ribosomal A-site. Model hairpins (Figure 7) were designed containing the secondary structure of ribosomal A-sites (sequences from http://www.rna.ccbb.utexas.edu/) of H. Sapiens (HRP5) and M. Tuberculosis (HRP6) and closing at one end with a stable RNA tetraloop and on the other end with a couple of C-G base pairs. The structures of the A rich bulge, which is the target of aminoglycoside antibiotics, of bacterial and human A-sites are remarkably similar. However, significant differences occur in the helical region just above the A rich bulge (bold in Figure 7). Interestingly, the A-site helices feature short polypurine tracts interrupted by single pyrimidine, U in HRP5 and C in HRP6, which prompted us to explore if the A-site RNA may be recognized via triplex or triplex-invasion mode (Figure 7, Table 3).
In general, PNA and GPNA (L-series) targeting the A-site exhibited modest binding affinity, which was consistent with the impact of a mismatched base triplet observed in our previous study.10 Octamer PNA10 had low sequence selectivity for the bacterial A-site and the binding order indicated potential triplex-invasion (Table 3, entry 1). Surprisingly, guanidine modification resulted in an increased affinity of GPNA10 for human A-site and a dramatic loss in sequence selectivity, which correlated with large increases in binding order (Table 3, entry 2). Shortening the PNA to heptamer (PNA11) and hexamer (PNA12) slightly decreased affinity and increased sequence selectivity. Guanidine modifications in GPNA12 and GPNA13 increased the binding affinity but the sequence selectivity was lost. It is conceivable that the unusually high binding orders for GPNA10 and GPNA13 resulted from non-specific electrostatic association of these GPNA carrying two guanidine modifications with the relatively more flexible (because of the non-canonical base pairs) A-site RNA. Recently, we showed that 2-pyrimidone, as in the novel PNA monomers 5 and 6 (Figure 7) formed a matched triplet with a C-G inversion in the purine rich strand of double helical RNA.12 Replacing the mismatched G in PNA10 with P and Pex (monomers 5 and 6) increased the affinity of PNA13 and PNA14 for the bacterial A-site. The stoichiometry of the complex was close to 1:1 as expected for the triple helix. Most remarkably, the sequence selectivity was excellent – we could not observe any binding to the human A-site (Figures S40 and S42).
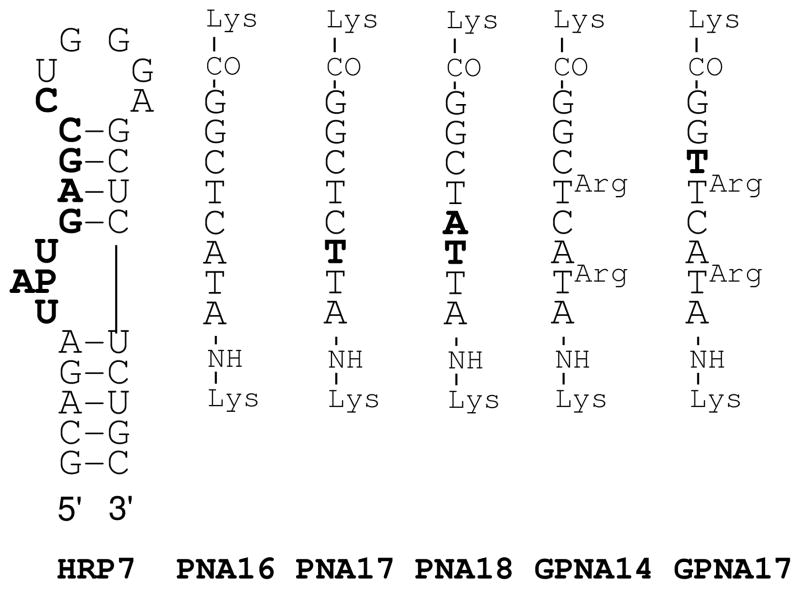
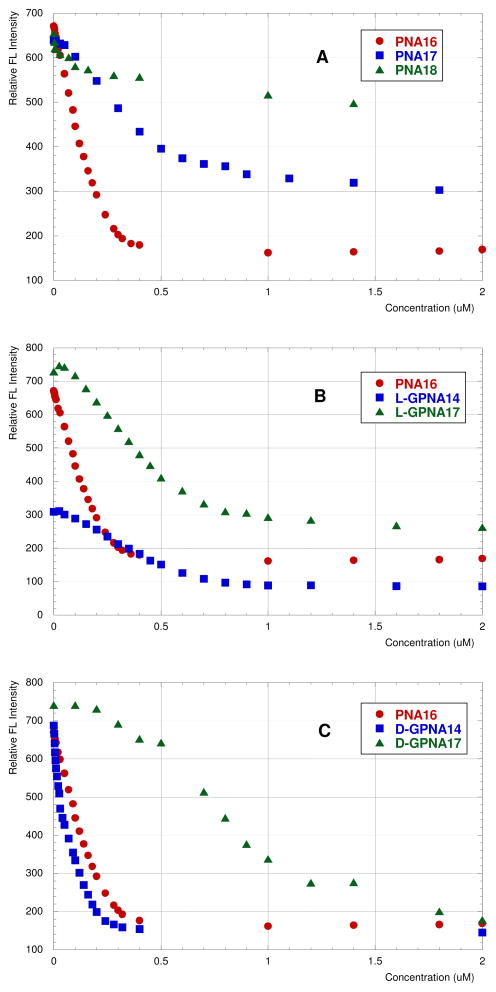
The preference of guanidine-modified PNAs to form strand invasion complexes prompted us to check if GPNAs could bind RNA structures that do not have continuous polypurine tracts. To test this hypothesis we selected a model RNA sequence of the transactivation response element (TAR) of HIV-1 virus (Figure 8). We envisioned that the bulged structure should be thermally less stable than a canonical Watson-Crick helix and may predispose the RNA for strand invasion by PNA. To test this hypothesis we chose PNA16 complementary to the U-rich loop and the stem connecting the U-rich loop and the hairpin loop. This sequence design was similar to that by Pandey24–26 and others,27 except that we decided not to include the G-rich hairpin loop in the recognition site because our focus was on testing duplex invasion as opposed to Watson-Crick binding to the flexible loop. Our initial experiments were frustrated by an apparent aggregation of PNAs at high concentrations (ca 100 μM) in the injection syringe of ITC. While PNAs used in the ITC studies above had no more than one purine base (<20%), PNAs complementary to TAR RNA had 50% purines (Figure 8), which may cause some aggregation at high concentrations. In a search for an alternative method, we turned to fluorescence spectroscopy of RNA labeled with the highly fluorescent 2-aminopurine nucleoside.19,28–30 2-Amino-purine fluorescence has been used to characterize binding of small molecules to the rev responsive element RNA of HIV-1,28 aminoglycosides to ribosomal A-site29,30 and, most recently, argininamide, Tat peptide and neomycin to TAR RNA model construct HRP719 (Figure 8, AP = 2-aminopurine).
Figure 8.
Structure of TAR RNA hairpin and the complementary PNAs and GPNAs. The target site on RNA and the mismatched nucleobases are highlighted in bold; AP is 2-aminopurine.
Following the published methodology,19 incremental titration of HRP7 with PNA16 led to decrease of fluorescence intensity (red circles in Figure 9A), as expected for 2-aminopurine moving from a relatively flexible bulge to a more structured PNA-RNA duplex environment. Fitting the data (Figure S44) to a single site, two-state binding model gave Ka ~ 2 ×107 M−1.19 Introduction of a mismatch (highlighted bold in Figure 8) lowered the affinity of PNA17 (blue squares in Figure 9A) to Ka ~ 4 × 106 M−1. Two adjacent mismatches in PNA18 led to a curve (green triangles in Figure 9A) that fit poorly the single site, two-state binding model (Figure S46), however, the relatively small slope clearly indicated a significantly decreased affinity. Binding of L-GPNA14 to HRP7 appeared to be weaker than that of the unmodified PNA16 (Figure 9B). The curve did not fit well the single site, two-state binding model (Figure S47), giving Ka approximately 2 to 5 × 106 M−1, which was comparable to that of PNA17 having a mismatched base pair. Surprisingly, introduction of a mismatch did not significantly change the affinity of L-GPNA17 (Figure 9B, see also Figures S47 and S48). This result might indicate that significant portion of the binding energy came from non-specific electrostatic attraction. In contrast, DGPNA14 had an apparently higher affinity for HRP7 than the unmodified PNA16 (blue squares in Figure 9C). Fitting the data (Figure S49) to a single site, two-state binding model gave Ka ~ 108 M−1.19 Introduction of a mismatch lowered the affinity of D-GPNA17 (green triangles in Figure 9C). The curve did not fit well the single site, two-state binding model (Figure S50), giving Ka approximately 2 × 106 M−1, which was comparable to that of the mismatched PNA17 and L-GPNA17. One potential explanation for the poor fitting of data for mismatched PNA18, L-GPNAs and D-GPNA17 is that more than one equivalent of PNA was binding to HRP7 causing significant deviations from the single site, two-state binding model behavior.
Figure 9.
Binding of PNA and GPNA to TAR RNA HRP7; change in fluorescence monitored at 365 nm following excitation at 305 nm.
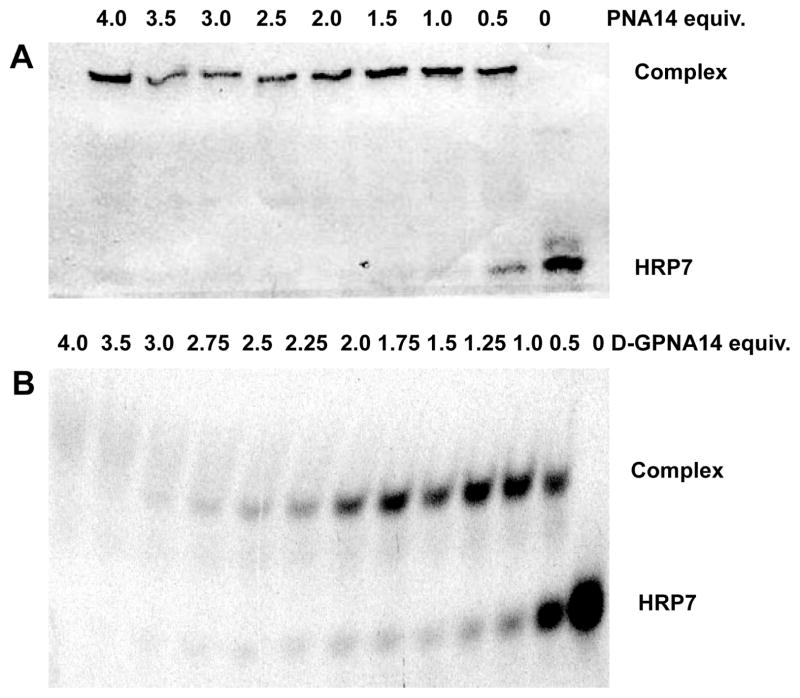
The complex formation between TAR RNA model hairpin and PNA was further confirmed using a gel mobility shift assay (Figure 10). Incremental titration of HRP7 (5′-labeled with fluorescein) with PNA16 (Figure 10A) and D-GPNA14 (Figure 10B) led to disappearance of the RNA band and formation of a slower moving band, which could be assigned to a potential stand invasion complex.27,31 Interestingly and in contrast to PNA16, addition of more than two equivalents of D-GPNA14 led to disappearance of the initial complex band and formation of broad and smeared out bands that were diluted below the detection level. This result suggested that more than one equivalent of guanidine modified PNA may be binding to HRP7 with lower affinity at higher PNA-RNA ratios.
Figure 10.
Binding of PNA16 (A) and D-GPNA14 (B) to 5′-fluorescein labeled TAR RNA HRP7 (5μM) monitored by gel mobility shift assay. Gels run in 89 mM tris-borate buffer, 2 mM EDTA at pH 8.4.
DISCUSSION
PNAs bearing cationic α- and γ-substituents bind strongly to complementary DNA and RNA and exhibit interesting biological properties.16–18,32–34 Ly and co-workers16–18 have reported that guanidine modification of PNA greatly facilitates crossing of cellular membrane, a highly desirable property for in vivo applications of gene expression control. They found that GPNA induced potent and sequence specific antisense effect and was less toxic to the cells compared to PNA conjugated with polyarginine.32 These favorable properties prompted us to study binding of GPNA to double helical RNA, especially, because we had recently discovered that unmodified PNA binds surprisingly strongly and sequence selectively to double helical RNA.10–12
Ly and co-workers demonstrated that L-GPNA had lower affinity for complementary single stranded DNA16 and RNA17 than unmodified PNA. In contrast, the affinity of D-GPNA was similar or even higher than that of unmodified PNA.16,17 In our hands, both isomers of GPNA sequences optimized for parallel binding to the polypurine tract of double helical RNA had about two orders of magnitude lower binding affinity than the unmodified PNA (c.f., Table 1, entry 6 and 7–12). This result suggested that a PNA backbone derived from α-substituted amino acids instead of glycine might be a poor fit for the PNA-RNA-RNA triple helix. The problem is most likely steric hindrance because the cationic guanidine modification could be expected to enhance the stability of triple helices at the expense of sequence selectivity. For example, conjugation of cationic peptides at PNA termini have been shown to increase the stability of PNA-DNA-DNA triple helices.31 Interestingly, Ly and co-workers18 found that GPNA T10 formed a GPNA-DNA duplex but not a GPNA-DNA-GPNA triplex with the complementary dA10, a result consistent with our findings that the guanidine modification disfavors triple helix formation.
GPNA sequences optimized for anti-parallel binding to the polypurine tract had somewhat higher affinity for double helical RNA than the parallel GPNA sequences (Ka = 106 to 107, depending on sequence and number of modifications). However, the affinity was lower than that of unmodified PNA and decreased with increasing number of guanidine modifications (Table 1, entries 13–15). Multiple modifications derived from L-arginine appeared to be better tolerated leading to higher affinity of LGPNA (Table 1, entry 16) than D-GPNA (entry 15). This was somewhat unexpected because D-GPNA was shown to form more stable duplexes with DNA and RNA than L-GPNA.16,17 However, if the antiparallel GPNA binds RNA by triplex-invasion, both the favored anti-parallel GPNA-RNA duplex and the anti-parallel GPNA-RNA-GPNA triple helix (see Figure 4) contribute to the overall stability of the complex. We recently showed that unmodified PNA formed both parallel and anti-parallel triple helix with RNA (Figure 4A and B), though, the latter was an order of magnitude less stable.10 It is conceivable that the anti-parallel triple helix is disfavored more by the D- than by the L-modification.
For the symmetric sequence CTCCTC, which was optimal for both parallel triple helix and anti-parallel duplex (as required for triplex-invasion) we observed an overall decrease in binding affinity upon guanidine modification (Table 1, entries 1–3). Consistent with more stable duplexes involving D-GPNA 17 and equally destabilized parallel triple helices (Table 1, entries 6–12) D-GPNA1 had slightly higher affinity than L-GPNA1 (c.f., entries 2 and 3). Perhaps the most interesting finding was that guanidine modification shifted the binding mode from 1:1 (as indicated by binding stoichiometry ~1) which we assign to GPNA-RNA-RNA triplex, to a 2:1 complex (binding stoichiometry ~2). We propose that the best explanation for the 2:1 complex is a GPNA-RNA-GPNA triplex-invasion complex (Figure 4C). The relatively higher stability of GPNA-RNA complexes that can form Watson-Crick base pairs (D-GPNA8/HRP3 and D-GPNA9/HRP4) or G-U wobble pair support our hypothesis of triplex-invasion. Thus, guanidine modification of PNA at the α-position appears to enhance the strand invasion of RNA double helix, which is consistent with observations made by others that cationic modifications and α- and γ-substituents predispose PNA for strand invasion of DNA.24,25,35–37 Our experiments with TAR RNA model HRP7 further confirmed that guanidine modification promotes RNA strand invasion. The fact that D-GPNA14 had significantly higher affinity and sequence selectivity than L-GPNA14 was encouraging for future applications and fully consistent with previous findings by Ly and co-workers. 16,17 Overall, the guanidine modification significantly reduced the ability of PNA to form triple helices with complementary double helical RNA. Meanwhile, the affinity of GPNA and unmodified PNA for “mismatched” RNA helices was lowered by about the same extent (Table 2), which resulted in an overall decrease of sequence selectivity for GPNAs.
Binding of PNA to A-site RNA had not been studied prior to our recent work.12 Our results show that unmodified PNA was able to bind the polypurine tract of bacterial A-site RNA in preference to human A-site RNA. The relatively low affinity and modest sequence selectivity of binding was most likely due to inability of Hoogsteen triplets to recognize the pyrimidine interruption in the polypurine tract of HRP6. The results with PNA11 and PNA12 are encouraging for triplex recognition of A-site RNA, providing that a modified heterocycle could be designed that would recognize the pyrimidine interruption in the polypurine tract and restore binding affinity and sequence selectivity.38–40 Consistent with this notion, incorporation of a modified heterocyles (P and Pex, Figure 7), recently developed by us to recognize cytosine in G-C inversion,12 significantly increased the sequence selectivity while maintaining excellent affinity in the triple helical binding mode (Table 3). Brief review of secondary structure databases of non-coding RNAs reveals that it is relatively common to find short homopurine tracts of eight and more contiguous purines, sometimes interrupted by one or two pyrimidines, in bacterial ribosomal RNAs (http://www.rna.ccbb.utexas.edu/) and micro RNAs (http://www.mirbase.org/). Our preliminary results with PNA13 and PNA14 suggest the possibility of designing relatively small PNA analogues to recognize such binding sites. It is conceivable that further development of chemical modifications may allow general recognition of isolated pyrimidines in the context of homopurine triple helix at physiological pH, which may open a novel way to recognize and interfere with function of non-coding RNAs.
Because of the need for cytosine (pKa~ 4.5) protonation to form the Hoogsteen C*G-C triplets, the experiments on triple helical recognition of RNA were performed at pH 5.5. Consistent with this requirement, we did not observe binding of GPNAs to RNA hairpins at physiologically relevant pH. This problem is beyond the scope of the present study and may be addressed in future work by designing PNAs containing more basic cytoisine analogues and/or alternative cationic modifications.31 While the α-guanidine-modified PNAs did not improve triple helix formation with double helical RNA, related cationic modifications, such as γ-guanidine34 and γ-lysine33 modified PNA still are interesting alternatives, which may allow effective triple helical recognition and enhanced cellular uptake to be realized in a modified PNA analogue.
Promising result was obtained using D-GPNA14 in a strand invasion mode with TAR RNA hairpin. Binding of PNA to TAR RNA has previously been demonstrated by gel mobility shift analysis and by blocking the Tat-mediated transactivation in cell culture.26,27 The data are consistent with formation of 1:1 PNA-RNA strand invasion duplex.26,27 Similarly, PNA14 and especially D-GPNA14 showed excellent affinity and sequence selectivity for TAR RNA. The gel mobility shift assay and good fit of the fluorescence data to a single site, two-state binding model suggested formation of a 1:1 PNA-RNA complex, most likely a strand invasion duplex, as previously reported by others.27 However, gel mobility experiments also indicated that guanidine modifications may cause non-selective binding of additional PNA molecules to RNA hairpin at higher concentrations. Consistent with the literature data,16,17 guanidine modification in L-GPNA14 decreased the binding affinity and sequence selectivity. Overall, D-GPNA may be promising compounds to explore for strand invasion recognition of biologically relevant RNAs featuring hairpin structures that are thermally weaker due to non-canonical base pairs, bulges and internal loops. Finally, it should be noted that the present study was done on relatively short (hexamer to octamer) PNAs. It is conceivable that longer PNAs will be required to recognize the target RNAs sequences selectively in the presence of other DNA and RNA species in cells. Such studies will be future priority after strong binding at physiologically relevant conditions is achieved by additional chemical modifications.
Supplementary Material
Acknowledgments
We thank Prof. Susannah Gal (Biological Sciences, Binghamton University) for help with gel mobility shift assay.
Funding: We thank NIH (R01 GM071461), Binghamton University and Clifford D. Clark Graduate Fellowship (to O. M.) for financial support of this research.
Footnotes
Supporting Information Available. Details of PNA synthesis, ITC experiments and data, CD spectra, fitting of fluorescence data and copies of the NMR spectra. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Thomas JR, Hergenrother PJ. Targeting RNA with Small Molecules. Chem Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 2.Sucheck SJ, Wong CH. RNA as a target for small molecules. Curr Opin Chem Biol. 2000;4:678–686. doi: 10.1016/s1367-5931(00)00142-3. [DOI] [PubMed] [Google Scholar]
- 3.Chow CS, Bogdan FM. A Structural Basis for RNA-Ligand Interactions. Chem Rev. 1997;97:1489–1513. doi: 10.1021/cr960415w. [DOI] [PubMed] [Google Scholar]
- 4.Fox KR, Brown T. An extra dimension in nucleic acid sequence recognition. Q Rev Biophys. 2005;38:311–320. doi: 10.1017/S0033583506004197. [DOI] [PubMed] [Google Scholar]
- 5.Roberts RW, Crothers DM. Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition. Science. 1992;258:1463–1466. doi: 10.1126/science.1279808. [DOI] [PubMed] [Google Scholar]
- 6.Han H, Dervan PB. Sequence-specific recognition of double helical RNA and RNA-DNA by triple helix formation. Proc Natl Acad Sci U S A. 1993;90:3806–3810. doi: 10.1073/pnas.90.9.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Escude C, Francois JC, Sun JS, Ott G, Sprinzl M, Garestier T, Helene C. Stability of triple helixes containing RNA and DNA strands: experimental and molecular modeling studies. Nucleic Acids Res. 1993;21:5547–5553. doi: 10.1093/nar/21.24.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Semerad CL, Maher LJ., III Exclusion of RNA strands from a purine motif triple helix. Nucleic Acids Res. 1994;22:5321–5325. doi: 10.1093/nar/22.24.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 10.Li M, Zengeya T, Rozners E. Short Peptide Nucleic Acids Bind Strongly to Homopurine Tract of Double Helical RNA at pH 5.5. J Am Chem Soc. 2010;132:8676–8681. doi: 10.1021/ja101384k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zengeya T, Li M, Rozners E. PNA containing isocytidine nucleobase: Synthesis and recognition of double helical RNA. Bioorg Med Chem Lett. 2011;21:2121–2124. doi: 10.1016/j.bmcl.2011.01.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta P, Zengeya T, Rozners E. Triple helical recognition of pyrimidine inversions in polypurine tracts of RNA by nucleobase-modified PNA. Chem Commun. 2011;47:11125–11127. doi: 10.1039/c1cc14706d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nielsen PE. Sequence-selective targeting of duplex DNA by peptide nucleic acids. Curr Opin Mol Ther. 2010;12:184–191. [PubMed] [Google Scholar]
- 14.Shiraishi T, Nielsen PE. Enhanced delivery of cell-penetrating peptide-peptide nucleic acid conjugates by endosomal disruption. Nature Protocols. 2006;1:633–636. doi: 10.1038/nprot.2006.92. [DOI] [PubMed] [Google Scholar]
- 15.Nielsen PE. Addressing the challenges of cellular delivery and bioavailability of peptide nucleic acids (PNA) Q Rev Biotech. 2005;38:345–350. doi: 10.1017/S0033583506004148. [DOI] [PubMed] [Google Scholar]
- 16.Zhou P, Dragulescu-Andrasi A, Bhattacharya B, O’Keefe H, Vatta P, Hyldig-Nielsen JJ, Ly DH. Synthesis of cell-permeable peptide nucleic acids and characterization of their hybridization and uptake properties. Bioorg Med Chem Lett. 2006;16:4931–4935. doi: 10.1016/j.bmcl.2006.06.052. [DOI] [PubMed] [Google Scholar]
- 17.Dragulescu-Andrasi A, Zhou P, He G, Ly DH. Cell-permeable GPNA with appropriate backbone stereochemistry and spacing binds sequence-specifically to RNA. Chem Commun. 2005:244–246. doi: 10.1039/b412522c. [DOI] [PubMed] [Google Scholar]
- 18.Zhou P, Wang M, Du L, Fisher GW, Waggoner A, Ly DH. Novel Binding and Efficient Cellular Uptake of Guanidine-Based Peptide Nucleic Acids (GPNA) J Am Chem Soc. 2003;125:6878–6879. doi: 10.1021/ja029665m. [DOI] [PubMed] [Google Scholar]
- 19.Bradrick TD, Marino JP. Ligand-induced changes in 2-aminopurine fluorescence as a probe for small molecule binding to HIV-1 TAR RNA. RNA. 2004;10:1459–1468. doi: 10.1261/rna.7620304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wojciechowski F, Hudson RHE. A Convenient Route to N-[2-(Fmoc)aminoethyl]glycine Esters and PNA Oligomerization Using a Bis-N-Boc Nucleobase Protecting Group Strategy. J Org Chem. 2008;73:3807–3816. doi: 10.1021/jo800195j. [DOI] [PubMed] [Google Scholar]
- 21.Kleiner RE, Brudno Y, Birnbaum ME, Liu DR. DNA-Templated Polymerization of Side-Chain-Functionalized Peptide Nucleic Acid Aldehydes. J Am Chem Soc. 2008;130:4646–4659. doi: 10.1021/ja0753997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wittung P, Nielsen P, Norden B. Direct Observation of Strand Invasion by Peptide Nucleic Acid (PNA) into Double-Stranded DNA. J Am Chem Soc. 1996;118:7049–7054. [Google Scholar]
- 23.Wittung P, Nielsen P, Norden B. Extended DNA-recognition repertoire of peptide nucleic acid (PNA): PNA-dsDNA triplex formed with cytosine-rich homopyrimidine PNA. Biochemistry. 1997;36:7973–7979. doi: 10.1021/bi963136b. [DOI] [PubMed] [Google Scholar]
- 24.Kaushik N, Basu A, Palumbo P, Myers RL, Pandey VN. Anti-TAR polyamide nucleotide analog conjugated with a membrane-permeating peptide inhibits human immunodeficiency virus type 1 production. J Virol. 2002;76:3881–3891. doi: 10.1128/JVI.76.8.3881-3891.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tripathi S, Chaubey B, Ganguly S, Harris D, Casale RA, Pandey VN. Anti-HIV-1 activity of anti-TAR polyamide nucleic acid conjugated with various membrane transducing peptides. Nucleic Acids Res. 2005;33:4345–4356. doi: 10.1093/nar/gki743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayhood T, Kaushik N, Pandey PK, Kashanchi F, Deng L, Pandey VN. Inhibition of Tat-Mediated Transactivation of HIV-1 LTR Transcription by Polyamide Nucleic Acid Targeted to TAR Hairpin Element. Biochemistry. 2000;39:11532–11539. doi: 10.1021/bi000708q. [DOI] [PubMed] [Google Scholar]
- 27.Belousoff MJ, Gasser G, Graham B, Tor Y, Spiccia L. Binding of HIV-1 TAR mRNA to a peptide nucleic acid oligomer and its conjugates with metal-ion-binding multidentate ligands. J Biol Inorg Chem. 2009;14:287–300. doi: 10.1007/s00775-008-0448-6. [DOI] [PubMed] [Google Scholar]
- 28.Lacourciere KA, Stivers JT, Marino JP. Mechanism of Neomycin and Rev Peptide Binding to the Rev Responsive Element of HIV-1 As Determined by Fluorescence and NMR Spectroscopy. Biochemistry. 2000;39:5630–5641. doi: 10.1021/bi992932p. [DOI] [PubMed] [Google Scholar]
- 29.Kaul M, Barbieri CM, Pilch DS. Fluorescence-Based Approach for Detecting and Characterizing Antibiotic-Induced Conformational Changes in Ribosomal RNA: Comparing Aminoglycoside Binding to Prokaryotic and Eukaryotic Ribosomal RNA Sequences. J Am Chem Soc. 2004;126:3447–3453. doi: 10.1021/ja030568i. [DOI] [PubMed] [Google Scholar]
- 30.Blount KF, Zhao F, Hermann T, Tor Y. Conformational Constraint as a Means for Understanding RNA-Aminoglycoside Specificity. J Am Chem Soc. 2005;127:9818–9829. doi: 10.1021/ja050918w. [DOI] [PubMed] [Google Scholar]
- 31.Hansen ME, Bentin T, Nielsen PE. High-affinity triplex targeting of double stranded DNA using chemically modified peptide nucleic acid oligomers. Nucleic Acids Res. 2009;37:4498–4507. doi: 10.1093/nar/gkp437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dragulescu-Andrasi A, Rapireddy S, He G, Bhattacharya B, Hyldig-Nielsen JJ, Zon G, Ly DH. Cell-Permeable Peptide Nucleic Acid Designed to Bind to the 5′-Untranslated Region of E-cadherin Transcript Induces Potent and Sequence-Specific Antisense Effects. J Am Chem Soc. 2006;128:16104–16112. doi: 10.1021/ja063383v. [DOI] [PubMed] [Google Scholar]
- 33.Englund EA, Appella DH. γ-substituted peptide nucleic acids constructed from L-lysine are a versatile scaffold for multifunctional display. Angew Chem, Int Ed. 2007;46:1414–1418. doi: 10.1002/anie.200603483. [DOI] [PubMed] [Google Scholar]
- 34.Sahu B, Chenna V, Lathrop KL, Thomas SM, Zon G, Livak KJ, Ly DH. Synthesis of Conformationally Preorganized and Cell-Permeable Guanidine-Based γ-Peptide Nucleic Acids (γ-GPNAs) J Org Chem. 2009;74:1509–1516. doi: 10.1021/jo802211n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishizuka T, Tedeschi T, Corradini R, Komiyama M, Sforza S, Marchelli R. SSB-Assisted Duplex Invasion of Preorganized PNA into Double-Stranded DNA. Chem Bio Chem. 2009;10:2607–2612. doi: 10.1002/cbic.200900381. [DOI] [PubMed] [Google Scholar]
- 36.He G, Rapireddy S, Bahal R, Sahu B, Ly DH. Strand Invasion of Extended, Mixed-Sequence B-DNA by γ-PNAs. J Am Chem Soc. 2009;131:12088–12090. doi: 10.1021/ja900228j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaihatsu K, Braasch DA, Cansizoglu A, Corey DR. Enhanced strand invasion by peptide nucleic acid-peptide conjugates. Biochemistry. 2002;41:11118–11125. doi: 10.1021/bi0263659. [DOI] [PubMed] [Google Scholar]
- 38.Rusling DA, Broughton-Head VJ, Brown T, Fox KR. Towards the targeted modulation of gene expression by modified triplex-forming oligonucleotides. Curr Chem Biol. 2008;2:1–10. [Google Scholar]
- 39.Rusling DA, Powers VEC, Ranasinghe RT, Wang Y, Osborne SD, Brown T, Fox KR. Four base recognition by triplex-forming oligonucleotides at physiological pH. Nucleic Acids Res. 2005;33:3025–3032. doi: 10.1093/nar/gki625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buchini S, Leumann CJ. Recent improvements in antigene technology. Curr Opin Chem Biol. 2003;7:717–726. doi: 10.1016/j.cbpa.2003.10.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.