Abstract
Urokinase-type plasminogen activator (uPA) is expressed by lung epithelial cells and regulates fibrin turnover and epithelial cell viability. PMA, LPS, and TNF-alpha, as well as uPA itself, induce uPA expression in lung epithelial cells. PMA, LPS, and TNF-alpha induce uPA expression through increased synthesis as well as stabilization of uPA mRNA, while uPA increases its own expression solely through uPA mRNA stabilization. The mechanism by which lung epithelial cells regulate uPA expression at the level of mRNA stability is unclear. To elucidate this process, we sought to characterize protein-uPA mRNA interactions that regulate uPA expression. Regulation of uPA at the level of mRNA stability involves the interaction of a ~40 kDa cytoplasmic-nuclear shuttling protein with a 66 nt uPA mRNA 3′UTR sequence. We purified the uPA mRNA 3′UTR binding protein and identified it as ribonucleotide reductase M2 (RRM2). We expressed recombinant RRM2 and confirmed its interaction with a specific 66 nt uPA 3′UTR sequence. Immunoprecipitation of cell lysates with anti-RRM2 antibody and RT-PCR for uPA mRNA confirmed that RRM2 binds to uPA mRNA. Treatment of Beas2B cells with uPA or LPS attenuated RRM2-endogenous uPA mRNA interactions, while overexpression of RRM2 inhibited uPA protein and mRNA expression through destabilization of uPA mRNA. LPS exposure of lung epithelial cells translocates RRM2 from the cytoplasm to the nucleus in a time-dependent manner leading to stabilization of uPA mRNA. This newly recognized pathway could influence uPA expression and a broad range of uPA-dependent functions in lung epithelial cells in the context of lung inflammation and repair.
Keywords: Urokinase, ribonucleotide reductase M2, Urokinase-type plasminogen activator, Acute lung injury
Pulmonary epithelial cells, microvascular endothelial cells and macrophages express urokinasetype plasminogen activator (uPA), a serine protease that catalyzes the conversion of plasminogen to plasmin. By binding to its receptor, uPAR, uPA localizes plasminogen activation at the cell surface. uPA-uPAR complexes serve other functions independent of plasminogen activation, such as cell proliferation, intracellular signaling, differentiation, chemotactic migration and invasion.(1–5) Lung epithelial cells also express PAI-1, which can inhibit uPA activity and promote internalization and recycling of uPA-uPAR-PAI-1 complexes.(6) Previous studies demonstrated rapid increases in circulating and pulmonary concentrations of uPA after endotoxemia or bacterial infection.(7–10) Mice deficient in uPA resist LPS-induced acute lung injury (ALI) and the development of pulmonary edema.(7,11) On the other hand, a defect of uPA-mediated fibrinolysis occurs in Acute Respiratory Distress Syndrome and uPA attenuates the fibrotic response in bleomycin-induced accelerated pulmonary fibrosis. Therefore, uPA appears to play a pivotal role in the pathogenesis of ALI and in pulmonary fibrosis subsequent to ALI.
Recent findings indicated that uPA autoinduction by lung epithelial cells (2,12), endothelial cells and monocytes (13) involve uPA binding to uPAR through its growth factor domain. The potentiation of LPS-induced ALI by uPA (7) and diminished neutrophil recruitment in response to P. aeruginosa pneumonia (14) by uPA- and uPAR-deficient mice underscores the contribution of uPA and uPAR to the development of ALI. Further, increased expression of uPA due to posttranscriptional uPA mRNA stabilization by tumor cells has been implicated in the increased proliferative and invasive potential of cancer cells.(12, 15) We previously reported that uPA expression is upregulated in lung epithelial cells through stabilization of uPA mRNA and that proinflammatory mediators implicated in the pathogenesis of ALI and its repair, stabilizes uPA mRNA.(15) Since elucidation of the underlying mechanism is essential for a better understanding of ALI, we sought to define the regulatory interactions that contribute to the stabilization of uPA mRNA and induce uPA at the posttranscriptional level in lung epithelial cells.
EXPERIMENTAL PROCEDRURES
Materials
Beas2B and small airway epithelial (SAE) cells were purchased from ATCC (Manassas, VA) and Invitrogen (Carlsbad, CA), respectively. Beas2B cell culture (LHC-9) media and SAE cell culture media (SAGM), penicillin, and streptomycin were purchased from Invitrogen. Tissue culture plastics were from Becton Dickinson Labware (Linclon Park, NJ). Tris-base, aprotinin, dithiothreitol (DTT), phenyl-methylsulfonyl fluoride (PMSF), silver nitrate and ammonium persulfate (APS) were from Sigma Chemical Company (St. Louis, MO). Acrylamide, bisacrylamide, and nitrocellulose were from BioRad Laboratories (Richmond, CA). Anti-uPA antibody was purchased from American Diagnostica (Greenwich, CT), anti-RRM2 and anti-β-actin antibodies were obtained from and Santa Cruz Biotechnologies (Santa Cruz, CA). 32P-labeled UTP and dCTP were purchased from DuPont (Wilmington, DE), and X-ray films were purchased from Eastman Kodak (Rochester, NY).
Plasmid construction and in vitro transcription
Human uPA cDNA 3′UTR, and a deletion product containing the previously identified 66 nt uPA mRNA binding protein binding sequence (15) was cloned into pCDNA3.1 vector (Invitrogen) following PCR amplification using full length uPA 3′UTR cDNA as a template. The orientation and sequence of the clones were confirmed by sequencing. The full-length 3′UTR and the deletion product of uPA 3′UTR in pcDNA3.1 vector were linearized with Xba I, purified separately on agarose gels, extracted with phenol-chloroform, and used as a template for in vitro transcription with T7 polymerase. Sense mRNA was transcribed according to the supplier’s (Ambion Inc, Austin TX) protocol, except that 50 μCi (800 Ci/mmol) of [32P] UTP were used to substitute for unlabeled UTP in the reaction mixture. Passage through a NucAway (Ambion) column removed unincorporated radioactivity.
Treatment of lung epithelial cells with LPS and determination of the changes in uPA expression and uPA mRNA binding protein interaction with uPA mRNA 3′UTR sequences
Beas2B cells cultured in 100 mm dishes were treated with LPS (20 μg/ml) for 0–24 h at 37°C. The culture media and the cell lysates were analyzed for changes in uPA and β-actin expression by Western blotting. Total RNA isolated from Beas2B cells treated with LPS for 0–6 h were tested for uPA and β-actin mRNA by RT-PCR using 32P-labeled dCTP in the PCR reaction mixture. The amplified bands were separated on a urea/PAGE using TBE buffer. Afterwards, the gel was dried and autoradiographed. The identity of the amplified PCR product was confirmed by nucleotide sequencing of non-radioactive amplicon. To measure uPA mRNA binding protein activity, the cytosolic and nuclear fractions prepared from the Beas2B cells treated with LPS for 0–24 h were subjected to gel mobility shift assay using the 66 nt uPA mRNA 3′UTR sequence as a probe as we described elsewhere.(15,22) Cytoplasmic and nuclear extracts of LPS exposed Beas2B cells were also immunoblotted for RRM2.
Mouse model of LPS-induced injury
C57B6 mice were kept on a 12:12 h light and dark cycle with free excess to food and water. All experiments were conducted in accordance with institution review board approved protocols. ALI was induced by intratracheal injection of LPS (25 μg/20–25 g mouse) as described previously.(11) Control mice were exposed to saline. BAL fluids and the lung homogenates were prepared 24 h after saline or LPS exposure and analyzed for the changes in uPA expression by Western blotting. Total RNAs isolated from the lung tissues were tested for uPA mRNA expression by RT-PCR as described above. The cytosolic extracts prepared from the lung tissues of mice exposed to PBS or LPS for 24 h were tested for uPA mRNA binding protein activity by gel mobility shift assay using 32P-labled uPA mRNA 3′UTR sequence as probe.
Preparation of lung epithelial cell cytosolic extracts and purification of the uPA mRNA binding protein
Beas2B cells cultured in 150 mm culture dishes containing LHC-9 media were washed with HBSS. The cells were collected and lysed in an extraction buffer (25 mM Tris-HCl, pH 7.9, 0.5 mM EDTA, and 0.1 mM PMSF) with several freeze-thaw cycles. The lysates were then centrifuged at 12,000 × g for 15 minutes at 4°C, and the supernatants were collected. The protein content was measured with a Pierce BCA protein assay kit using various concentrations of serum albumin as standards.
Ammonium sulfate crystals were added to the lysate prepared above to bring it to 40% saturation and the precipitated proteins were discarded after checking the uPA mRNA binding protein activity. Ammonium sulfate crystals were further added to the supernatant to yield a final saturation of 60%. The precipitated proteins were collected, dissolved, and exhaustively dialyzed against the extraction buffer containing 10% glycerol. uPA 3′UTR mRNA binding activity was assessed using aliquot (10 μg) of dialyzed protein. The 40–60% ammonium sulfate fraction was passed through a blue-sepharose column (90 ml bed volume) in the same buffer containing 100 mM NaCl, and the uPA 3′UTR mRNA binding protein was eluted with a linear gradient (200 ml) of 0.1 –1 M NaCl. Positive fractions were pooled and loaded onto a heparin-Affigel column (90 ml bed volume) in the same buffer. After washing the unbound materials, the uPA mRNA binding protein was eluted with a linear gradient (200 ml) of 0–0.5 M NaCl in the extraction buffer. The fractions containing the uPA mRNA binding protein were pooled and loaded onto a DEAE-sephacel column and eluted with a linear NaCl gradient of 0–1 M NaCl in the extraction buffer. Positive fractions were pooled and passed through a phenyl sepharose column and eluted with a reverse gradient buffer. Positive fractions were pooled, dialyzed and concentrated by ultrafiltration, and loaded onto a mono-Q column fitted to a FPLC system. Unbound proteins were removed with 25 mM Tris-HCl (pH, 7.9) buffer, and bound proteins were eluted with a linear gradient (40 ml) of 0–1 M NaCl in 25 mM Tris-HCl (pH 7.9). Fractions containing uPA mRNA binding protein were analyzed by gel mobility shift assays.
Positive fractions were pooled, dialyzed, and subjected to a final round of affinity purification using an RNA affinity column containing biotin-labeled 66 nt uPA mRNA binding protein binding sequences (15) immobilized to streptavidin agarose, and uPA mRNA binding activity was assessed by gel mobility shift assays.(16–19) The uPA mRNA-protein complex was visualized by autoradiography. This band was excised, pooled from several runs, electroeluted and tested for uPA mRNA 3′UTR binding activity by Northwestern assay using 32P-labeled 66 nt uPA mRNA as a probe. SDS-PAGE analyses yielded a protein with an approximate molecular weight of ~40 kDa. The identity of the eluted protein was analyzed by mass spectroscopy. Database analyses revealed the identity of the binding protein. To determine whether the uPA mRNA binding protein specifically binds to uPA mRNA, we expressed it in a prokaryotic system using BL21 cells and purified the GST fusion protein as described previously (E3–E5). Purified recombinant uPA mRNA binding protein was analyzed for uPA mRNA binding using a gel mobility shift assay. To further confirm the specificity of the binding protein interaction with the 66 nt uPA mRNA 3′UTR sequence, we incubated purified recombinant RRM2 with the 32Plabeled 66 nt uPA mRNA binding sequence in the presence of a molar excess of unlabeled 66 nt uPA mRNA binding protein binding sequence or full length uPA or uPAR mRNA 3′UTR sequences and analyzed the interaction by gel mobility shift assay as described previously. (15, 19) The uPA mRNA binding protein cDNA was then subcloned into a eukaryotic expression vector pcDNA 3.1 and transfected to Beas2B cells. Histidine-tagged binding protein, containing a C-terminal V5 epitope, was expressed in Beas2B cells and was affinity purified using a nickel column. (17–18) Expression of the binding protein was confirmed by Western blotting of eluted fractions using anti-V5 and anti-binding protein antibodies. The fractions containing binding proteins were pooled and tested for uPA mRNA 3′UTR binding by a gel mobility shift assay using the 32P-labeled 66 nt sequence as a probe.
Expression of the uPA mRNA binding protein in lung epithelial cells and determination of its role in the regulation of uPA expression
Beas2B cells cultured in LHC-9 media or primary SAE cells maintained in SAGM were transfected with either binding protein cDNA cloned in pcDNA 3.1 or an empty vector using PEI reagent. (20) The cells were lysed and the expression of recombinant protein was confirmed by Western blotting of V5 fusion protein. The conditioned media and cell lysates were tested for uPA expression by Western blotting. Afterwards, the membrane was stripped and sequentially probed with antibody against V5 or β-actin to assess expression of the fusion protein and loading equality. Total RNAs from the Beas2B cells transfected with vector cDNA alone or vector harboring RRM2 cDNA were isolated and analyzed for uPA mRNA by RT-PCR. In order to confirm if inhibition of uPA mRNA by overexpression of RRM2 was due to destabilization of uPA mRNA, Beas2B cells transfected with vector alone or vector harboring RRM2 cDNA were treated with or without LPS for 12 h to induce maximum uPA mRNA. DRB was added to these cells to inhibit ongoing transcription and the total RNA was isolated at different time points. uPA mRNA was analyzed by RT-PCR using 32P-labeled dCTP. Amplified PCR products were separated on a urea/polyacrylamide gel and autoradiographed.
Effect of uPA or LPS on RRM2 interaction with uPA mRNA in lung epithelial cells
Beas2B cells were treated with PBS or uPA for 24 h, or with LPS for 0–24 h, in LHC-9 media. Afterwards, the media were aspirated, cells washed with cold PBS, and cell lysates prepared were cleared with mouse IgG. The precleared lysates were subsequently subjected to immunoprecipation using anti-RRM2 antibody. The immune complexes were washed with the binding buffer, and the RNA isolated and the associated uPA mRNA was amplified by RT-PCR in the presence of 32P-labeled dCTP. The PCR products were separated by urea-PAGE. After electrophoresis, the gel was dried and subjected to autoradiography.
Statistical Analysis
The statistical differences between various experimental conditions were analyzed by Student’s t test.
RESULTS
LPS induces uPA expression and inhibits the uPA mRNABp interaction with uPA mRNA in lung epithelial cells
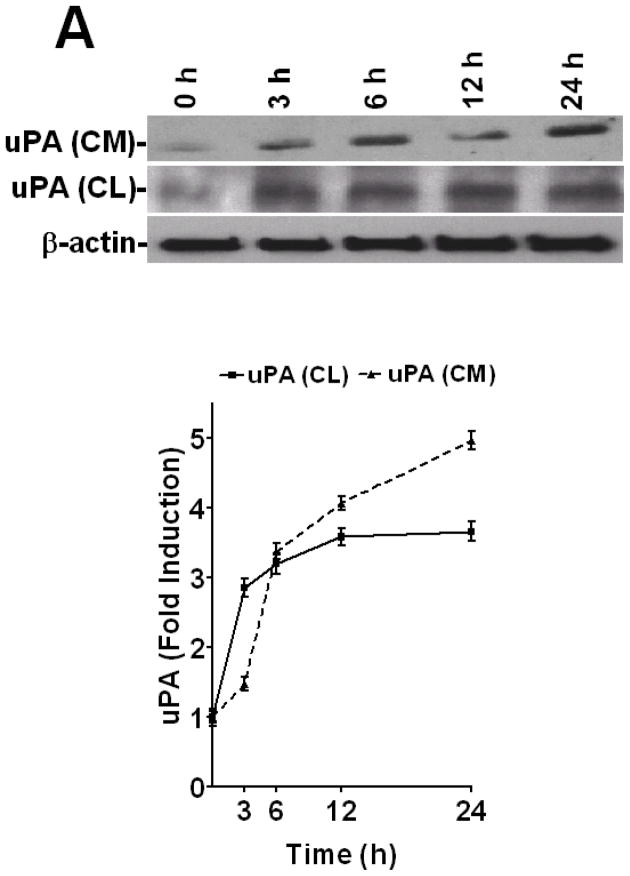
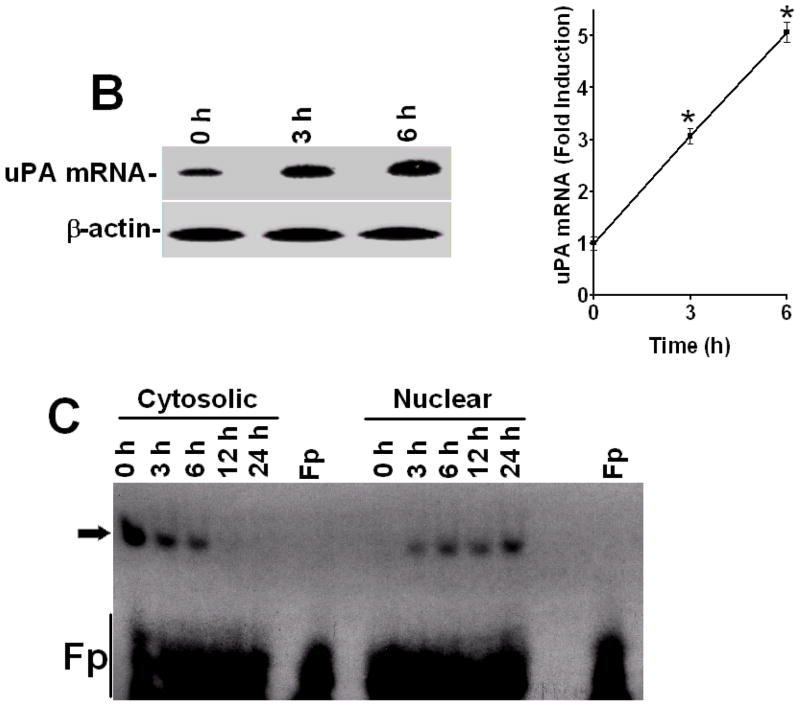
To determine how LPS alters uPA expression in human lung epithelial cells, we treated Beas2B cells with LPS for 0–24 h and tested both conditioned media and cell lysates for uPA antigen level by Western blotting. The results showed that LPS induced uPA expression in a time-dependent manner with maximum effect between 3–24 h after LPS exposure (Fig 1A). LPS treatment likewise induced uPA mRNA within 3–6 h (Fig 1B). We previously reported that treatment of Beas2B and primary SAE cells with LPS, uPA or TNF-α, induced uPA protein and mRNA expression in a similar manner.(21) We further reported that uPA induction by TNF-α involved, at least in part, stabilization of uPA mRNA (15,21), whereas uPA induced its own expression solely through uPA mRNA stabilization at the posttranscriptional level.(2) Additionally, we found that posttranscriptional control of uPA expression involves specific binding of a ~40 kDa uPA mRNABp to a 66 nt uPA mRNA 3′UTR destabilization determinant.(15,22) We hypothesized that altered interaction of the uPA mRNABp with the uPA mRNA 3′UTR in the cytoplasm may contribute to the induction of uPA expression by LPS since the latter induces both TNF-α and uPA expression during sepsis.(23–25) To test this postulate, we prepared cytosolic and nuclear extracts from Beas2B cells exposed to LPS for 0–24 h, and analyzed them for uPA mRNABp binding activity. Cytosolic fractions of LPS treated Beas2B cells showed decreased uPA mRNABp interaction with the 66 nt uPA mRNA 3′UTR binding sequence in a temporal fashion (Fig 1C). In contrast, uPA mRNA binding activity in the nuclear fractions gradually increased and reached a plateau between 6–24 h after LPS exposure, indicating nuclear translocation of uPA mRNABp. To determine if LPS induces uPA expression in vivo, we analyzed the BAL fluids and lung homogenates of mice exposed to intratracheal LPS for uPA protein and mRNA expression. Our results clearly showed the induction of both uPA protein and uPA mRNA after LPS exposure (Fig 2). Further analysis of cytoplasmic preparations of LPS-exposed mouse lung tissues for uPA mRNABp and uPA mRNA interaction by gel mobility shift assay revealed significant inhibition of the uPA mRNABp interaction with the 66 nt uPA 3′UTR mRNA sequence after LPS exposure (Fig 2C).
Figure 1.
LPS induces time dependent uPA expression in lung epithelial cells. Beas2B cells cultured in 100 mm dishes were treated with LPS (20 μg/ml) for 0–24 h. A. The conditioned media (CM) and cytoplasmic extracts cell lysates (CL) of Beas2B cells were analyzed for uPA expression by Western blotting. The lower panel shows corresponding β-actin loading controls. The line graph shows fold induction relative to time 0 calculated from the densities of the individual bands from the three Western blots. In the case of CL, fold induction was calculated after normalization of the densities of each uPA bands with the corresponding densities of β-actin from three independent experiments. B. Total RNA isolated from Beas2B cells treated with LPS for 0, 3 and 6 h were analyzed for uPA and β-actin mRNA by RT-PCR using 32P-labeled dCTP. 32P-labeled PCR products were separated on a urea/PAGE, dried and autoradiographed. The fold induction relative to time 0 was calculated from the densities of uPA mRNA bands after normalization with that of β-actin mRNA is depicted in line graph. The * indicates that the mean values from three independent experiments are statistically significant (p<0.0001) compared to time 0. C. Cytosolic and nuclear extracts prepared from the Beas2B cells exposed to LPS for 0–24 h as described in Fig 1A were subjected to uPA mRNA 3′UTR binding by gel mobility shift assay using 32P-labeled 66 nt 3′UTR sequence as a probe. The reaction mixtures were incubated with RNaseT1 at 37° C for 30 minute followed by heparin treatment at room temperature for 10 min. RNaseT1-resistant uPA mRNABp and uPA mRNA complexes were separated by PAGE using TBE buffer, dried and autoradiographed. Fp = reaction mixture in cytoplasmic or nuclear extraction buffers without Beas2B cell proteins. Arrow indicates the uPA mRNA-uPA mRNABp complex.
Figure 2.
LPS induces expression of uPA in mouse lungs. C57BL-6 mice were treated with PBS or 25 μg of LPS intratracheally and sacrificed after 24 h. A. Brochoalveolar lavage fluid obtained at the time of sacrifice (b) and lung homogenates (h) were analyzed for uPA protein by Western blotting. Membrane containing proteins from the lung homogenates were stripped and tested for β-actin proteins to assess loading equality. Fold induction in uPA levels of LPS versus PBS treated control mice (presented as bar graph) were calculated based on the densities of bands integrated from four independent experiments. In the case of lung homogenates, densities of the uPA bands were normalized to the densities of the corresponding β-actin. The mean values are statistically (**p<0.001, *p<0.01) significant compared to those of PBS controls. B. Total RNA was isolated from lung tissues of mice exposed to PBS or LPS and analyzed for uPA mRNA expression as described in Fig 2B. Fold induction of the uPA/β-actin mRNA ratio presented as bar graphs are from four independent experiments. The * symbol indicates that the mean values are statistically (p<0.001) significant compared to PBS controls. C. Cytosolic extracts prepared from lung tissues of mice exposed to PBS or LPS, as described in Fig 2A, were subjected to uPA mRNA 3′UTR binding by gel mobility shift assays using 32P-labeled 66 nt uPA 3′UTR sequence as a probe. RNAseT1-resistant uPA mRNABp-uPA mRNA complexes were separated by PAGE and autoradiography. Fp = free probe. Arrow indicates uPA mRNA-uPA mRNABp complex.
Purification of the uPA mRNABp and determination of its ability to bind to the 66 nt uPA mRNA 3′UTR sequence
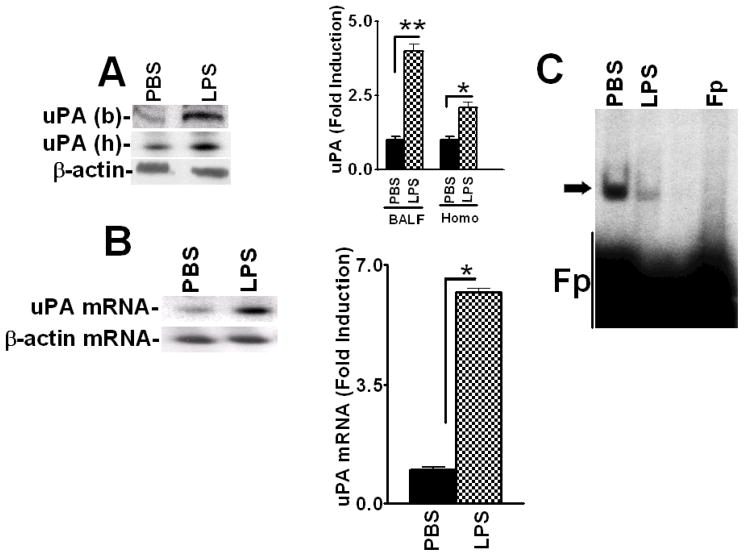
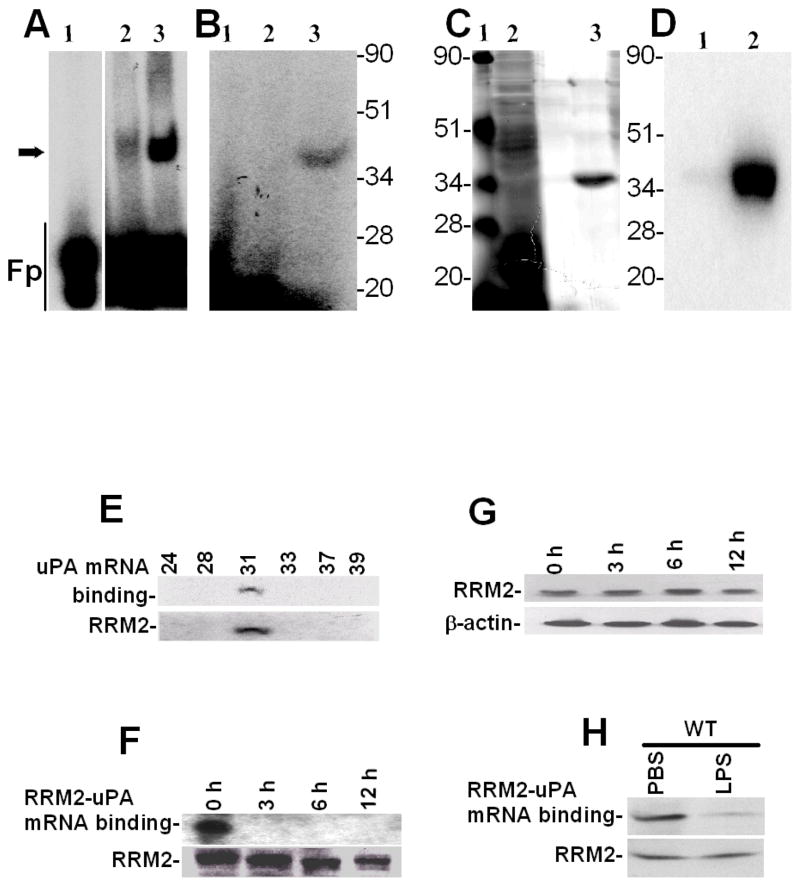
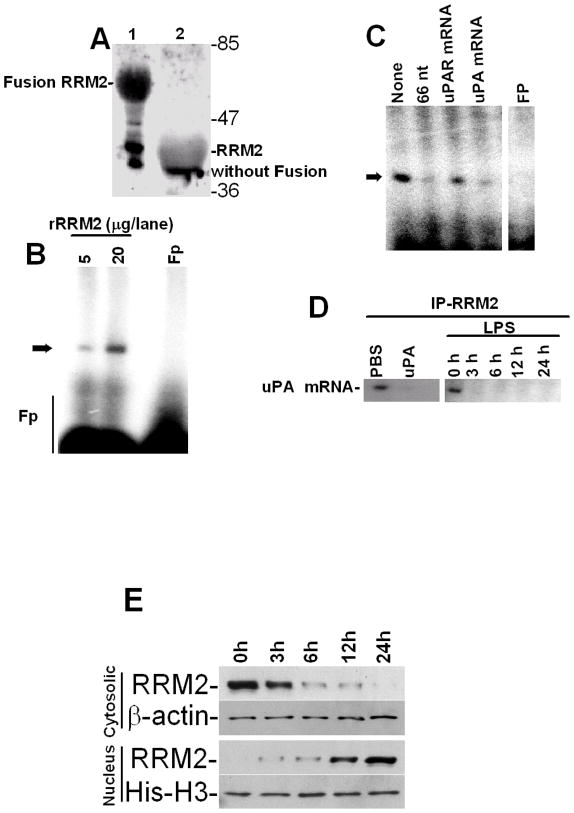
To further characterize the mechanism by which uPA expression is controlled at the posttranscriptional level in lung epithelial cells, we purified the uPA mRNABp from Beas2B cell lysates using conventional as well as uPA mRNA affinity column chromatography. Purification yielded a preparation enriched with a ~40 kDa protein having uPA mRNA binding activity, as confirmed by gel mobility shift (Fig 3A) and UV cross-linking (Fig 3B) assays, SDS-PAGE and silver staining (Fig 3C) or Northwestern assay (Fig 3D). Mass spectrophotometric analysis and database search indicated a high homology (69%) of this polypeptide to RRM2. To further evaluate the identity of the purified uPA mRNABp as RRM2, we subjected various protein fractions eluted from the Mono-Q column to Northwestern blotting using 32P-labeled 66 nt uPA mRNA binding sequence as a probe, and found that one specific fraction showed uPA mRNA binding activity at the approximate molecular weight position of ~40 kDa. To confirm that the enriched fraction co-migrates with RRM2, we stripped the same membrane and subjected it to Western blotting using anti-RRM2 antibody as a probe. As shown in Fig 3E, RRM2 antigens were indeed detected in the same fraction that exhibited uPA 3′UTR mRNA binding activity.
Figure 3.
Purification of the uPA mRNABp from lung epithelial cells. Cytosolic extracts from Beas2B cells were subjected to sequential ammonium sulfate fractionation. Protein fractions containing uPA mRNA binding activity were purified by passage through heparin sepharose, phenyl sepharose, Mono-Q and uPA mRNA affinity columns. Positive fractions were then subjected to gel mobility shift assay using 32P-labeled 66 nt uPA mRNA 3′UTR sequence as a probe. The RNA-protein complex was visualized by autoradiography, after which the bands were excised from the gel, pooled, electroeluted, and analyzed for uPA mRNA binding activity. A. Gel mobility shift assay of the uPA mRNA probe alone (lane 1), semipurified fractions from the mono-Q column (lane 2) or the uPA mRNABp fraction electroeluted from the gel (lane 3). B. Corresponding fractions from the mono-Q (lane 2) or uPA mRNABp fraction electroeluted from the gel (lane 3) were subjected to uPA mRNA 3′UTR binding using a 32P-labeled 66 nt uPA mRNA probe. Lane 1, uPA mRNA probe alone. These reaction mixtures were subjected to UV cross-linking after RNaseT1 and heparin digestion. Immobilized uPA mRNA-uPA mRNABp complexes were resolved on SDS-PAGE and autoradiographed. C. SDS-PAGE and silver staining of protein fractions from a Mono-Q column (lane 2), electroelution (lane 3) and Molecular weight standards (Lane 1). D. Protein fractions (lanes 2 and 3) from SDS-PAGE as described in Fig 3C were transferred to nitrocellulose and subjected to Northwestern assay using 32P-labeled uPA mRNA transcripts. E. Identification of RRM2 as an uPA mRNA 3′UTR binding protein. Protein fractions collected from the mono-Q column were separated on a SDS-PAGE and subjected to uPA mRNA 3′UTR binding by Northwestern assay using the 32P-labeled 66 nt uPA 3′UTR sequence as a probe. uPA mRNA 3′UTR binding activity in the fractions was confirmed by autoradiography. The same membrane was stripped and immunoblotted for RRM2 using anti-RRM2 antibody. F. Beas2B cells cultured in 100 mm dishes were treated with LPS (20 μg/ml) for 0–12 h as described in Fig 1A. The cells were detached and the cytoplasmic extracts were immunoprecipitated using anti-RRM2 antibody, separated on a SDS-PAGE and transferred to nitrocellulose membrane. The membrane was subjected to uPA mRNA 3′UTR binding by Northwestern assay using the 32P-labeled 66 nt 3′UTR sequence as a probe. The same membrane was stripped and developed by Western blotting using anti-RRM2 antibody. G. Beas2B cell lysates prepared after LPS treatment for 0–12 h as described in Fig 1A were separated on SDS-PAGE and subjected to Western blotting using anti-RRM2 antibody. The same membrane was stripped and developed with anti-β-actin antibody to assess equal loading. H. Cytoplasmic fractions of the lung homogenates of mice exposed PBS and LPS as described in Fig 2A were immunoprecipitated with anti-RRM2 antibody. The immune-complexes were separated on a SDS-PAGE, transferred to nitrocellulose membrane and subjected to Northwestern assay using 32P-labeled 66 nt uPA mRNA 3′UTR probe. The same membrane was stripped and Western blotted for RRM2 using anti-RRM2 antibody.
We then immunoprecipitated RRM2 proteins from the cytoplasmic fractions of Beas2B cells treated with LPS for 0–12 h. Northwestern blotting of immunoprecipitated RRM2 using 32Plabeled 66 nt uPA mRNA 3′UTR binding sequence as a probe demonstrated that LPS treatment inhibits RRM2 binding with the uPA 3′UTR mRNA sequence in Beas2B cells in a timedependent manner (Fig 3F). However, when LPS treated Beas2B cell lysates were tested for RRM2 expression, no significant inhibition of RRM2 levels by LPS was observed (Fig 3G). We next isolated RRM2 proteins using cytoplasmic extracts prepared from the lungs of mice exposed to PBS or LPS, and tested them for uPA 3′UTR mRNA binding activity by Northwestern assay (Fig 3H). Consistent with the responses found in Beas2B cells, RRM2 proteins isolated from mice exposed to intratracheal LPS showed reduced uPA mRNA binding activity compared to control mice exposed to PBS.
Expression of rRRM2 and verification of its uPA mRNA 3′UTR binding activity
To independently determine that RRM2 binds to the uPA mRNA 3′UTR, we next expressed RRM2 in E. coli and affinity-purified rRRM2 using a GST Sepharose column (Fig 4A). Purified rRRM2 protein was tested for uPA mRNA 3′UTR binding activity. Gel mobility shift assay demonstrated specific binding of rRRM2 with a 66 nt 3′UTR determinant of uPA mRNA (Fig 4B). We expressed RRM2 in Beas2B cells and likewise confirmed its ability to specifically interact with the 66 nt uPA mRNA 3′UTR sequence. To confirm the specificity of the interaction of RRM2 with the uPA mRNA, we incubated rRRM2 protein with 32P-labeled 66 nt uPA mRNA 3′UTR sequence in the presence of a 200-fold molar excess of unlabeled 66 nt uPA 3′UTR binding sequence, full length uPAR or uPA 3′UTR sequence. The results showed that rRRM2 binds the uPA mRNA 3′UTR through the 66 nt sequence, that the interaction was specific and that it could be inhibited by self competition (Fig 4C).
Figure 4.
Expression of recombinant RRM2 (rRRM2) and determination of its uPA mRNA 3′UTR binding activity. A. Expression and purification of rRRM2 in a prokaryotic system. rRRM2 expressed in E. Coli was affinity purified using a glutathione-sepharose column. Purified rRRM2 with GST-fusion (lane 1) or without GST-fusion (lane 2) were separated on a SDS-PAGE and stained with Coomassie blue. B. Purified rRRM2 protein (5 and 20 μg) without GST fusion was subjected to uPA mRNA 3′UTR binding by gel mobility shift assay using 32P-labeled 66 nt uPA mRNA 3′UTR as a probe. Fp = 32P-labeled probe alone. C. Cold competition experiments. Purified rRRM2 were incubated with or without a 200-fold molar excess of unlabeled 66 nt uPA RRM2 binding sequence or full length uPAR or uPA 3′UTR sequences before incubation with 32P-labeled 66 nt uPA 3′UTR sequences. The reaction mixtures were subjected to RNAseT1 and heparin digestion and the uPA mRNA-RRM2 complexes were separated by PAGE and autoradiographed. Fp = reaction mixture containing all the reagents except rRRM2. Arrow indicates uPA mRNA-rRRM2 complex. D. Beas2B cells were treated with PBS or uPA for 24 h, or LPS for 0–24 h and the cell lysates were immunoprecipitated with anti-RRM2 antibody. uPA mRNA associated with RRM2 immune-complex was amplified by RT-PCR in the presence of 32P-dCTP using uPA specific primers. The 32P-labeled PCR products were separated on a urea/polyacrylamide gel, dried and autoradiographed. E. The cytoplasmic and nuclear extracts prepared from Beas2B cells treated with LPS for 0–24 h as described in Fig 1C were subjected to Western blotting using anti-RRM2 antibody. The nitrocellulose membranes containing proteins from cytoplasmic and nuclear extracts were stripped and reprobed with anti-β-actin and anti-histone H3 antibodies respectively to assess similar loading. Experiments were repeated twice with identical results.
We then sought to confirm whether RRM2 binds to endogenous uPA mRNA in Beas2B cells and if the increased uPA expression due to posttranscriptional uPA mRNA stabilization involves altered RRM2-uPA mRNA interaction. RRM2 protein was immunoprecipitated from the cytoplasmic extracts of Beas2B cells treated with PBS, uPA for 24 h, or LPS for 0–24 h using non-specific mouse IgG or an anti-RRM2 monoclonal antibody. Total RNAs isolated from the mouse IgG and anti-RRM2-immune complexes were analyzed for associated uPA mRNA by RT-PCR. As shown in Fig 4D, uPA mRNA was found to co-precipitate with the RRM2-anti- RRM2-immune complex in PBS treated Beas2B cells. Interestingly, treatment of cells with either uPA or LPS suppressed uPA mRNA co-precipitation with the RRM2 protein. No uPA mRNA was detected when immunoprecipitation reactions were performed with non-specific mouse IgG, indicating that RRM2 specifically interacts with endogenous uPA mRNA. Since LPS inhibits RRM2 interaction with uPA mRNA without inhibiting RRM2 expression, we next analyzed the cytoplasmic and nuclear extracts of Beas2B cell exposed to LPS for varying time for RRM2 by Western blotting. Our results showed translocation of RRM2 from the cytoplasm to the nucleus in a temporal fashion after LPS treatment. These results also suggest that increased uPA expression by lung epithelial cells following exogenous uPA or LPS treatment may be associated with reduced RRM2 interaction with cytoplasmic uPA mRNA due to relocation of RRM2 to the nucleus.
Effect of RRM2 overexpression on uPA protein and mRNA expression, and uPA mRNA stabilization in lung epithelial cells
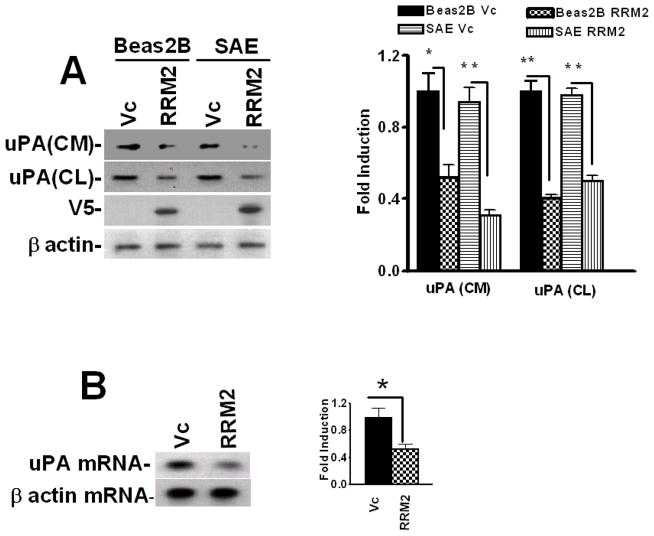
To determine whether RRM2 inhibits uPA expression in lung epithelial cells, we overexpressed RRM2 in Beas2B cells and tested for changes in the expression of uPA protein. We found that the expression of RRM2 in Beas2B cells led to the attenuation of uPA expression, compared to control Beas2B cells transfected with empty vector DNA (Fig 5A). RRM2 overexpression similarly inhibited uPA expression in primary SAE cells. We next tested if RRM2 overexpression by lung epithelial cells inhibits uPA mRNA. As shown in Fig 5B, transfection of Beas2B cells with RRM2 cDNA suppressed uPA mRNA expression, indicating that the effect was controlled at the mRNA level.
Figure 5.
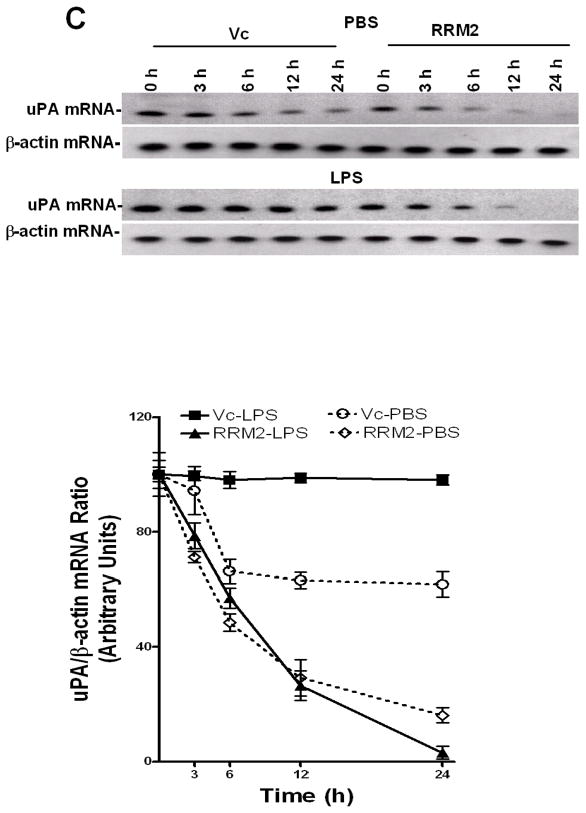
Inhibition of uPA expression by RRM2 in lung epithelial cells. Beas2B and primary SAE cells were transfected with vector pcDNA3.1 alone (Vc) or RRM2 cDNA in pcDNA3.1. A. The conditioned media (CM) and cell lysates (CL) were collected and analyzed for uPA expression by Western blotting. The membrane containing proteins from the CL were stripped and immunoblotted using anti-V5 and anti-β-actin antibody to assess expression of the fusion protein and equal loading, respectively. Experiments were repeated at least three times. The bar graph represents fold changes in uPA protein level in CM and CL. In the case of CL, uPA fold induction was calculated after normalization with the densities of β-actin proteins. The changes in the expression levels between cells transfected with vector DNA and RRM2 cDNA are statistically significant (**p<0.001 and *p<0.01). B. Total RNA isolated from the Beas2B cells treated with vector DNA or cDNA coding for RRM2 were analyzed for uPA mRNA by RT-PCR using 32P-labeled dCTP in the reaction mixture. Amplified PCR products were separated on a urea/polyacrylamide gel using TBE buffer, dried and autoradiographed. Fold induction of mRNA ratio (uPA/β-actin density) presented as bar graph from three experiments. The * symbol indicates that the mean values are statistically (p<0.001) significant when compared between Vc and RRM2. C. Beas2B cells expressing pcDNA3.1 or RRM2 cDNA were treated with PBS or LPS for 12 h to induce maximum uPA mRNA expression. These cells were then exposed to DRB (20 μg/ml) to inhibit ongoing transcription. Total RNA was isolated from these cells 0–24 h after treatment with DRB and was analyzed for uPA mRNA by RT-PCR in the presence of 32P-dCTP. The PCR products were separated on a urea/polyacrylamide gel and autoradiographed. The line graph represents the percentage of mRNA decay relative to Time 0 (designated 100%), calculated from the mean values obtained by integrating the densities of individual bands from three independent experiments.
We previously reported that uPA mRNABp (15,22) regulates uPA mRNA expression at the posttranscriptional level which is now identified as RRM2. We therefore sought to clarify whether inhibition of uPA mRNA and protein expression following RRM2 overexpression in lung epithelial cells involves destabilization of uPA mRNA at the posttranscriptional level. Beas2B cells, with or without LPS treatment, were treated with DRB to inhibit uPA mRNA synthesis, and the posttranscriptional uPA mRNA decay was analyzed. Our results showed that overexpression of RRM2 destabilized basal uPA mRNA compared to the lung epithelial cells stably transfected with empty vector DNA (Fig 5C). RRM2 overexpression also prevented LPS-induced stabilization of uPA mRNA in Beas2B cells compared to control cells transfected with vector DNA only.
Role of p53 in RRM2-mediated control of uPA expression
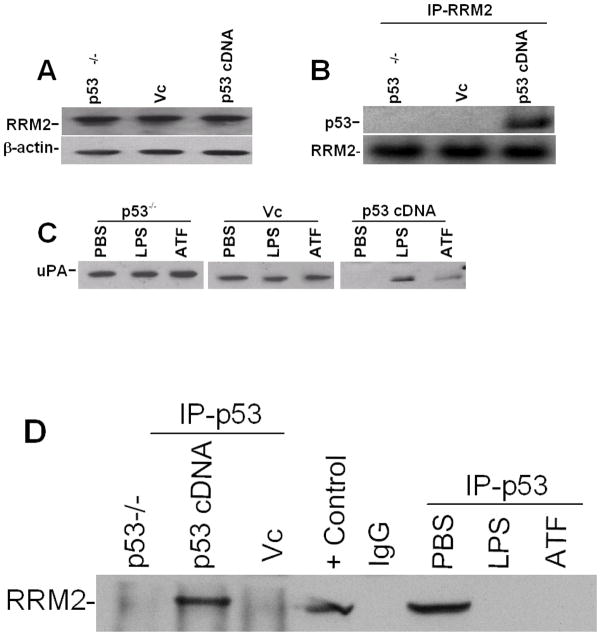
We recently reported that p53 inhibits uPA protein and mRNA expression in lung epithelial cells. (26) The process involves binding of p53 with a 35 nt 3′UTR sequences of uPA mRNA and destabilization of the uPA mRNA transcript. Our earlier findings (15) indicate that RRM2 binding with a 66 nt 3′UTR determinant resides upstream of the 35 nt p53 binding 3′UTR sequence. In addition, earlier reports (27–28) demonstrated that p53 controls RRM2 through a protein-protein interaction. We therefore hypothesized that the inhibitory effect of RRM2 on uPA expression at the posttranscriptional level is controlled by p53. To test this possibility, we initially analyzed basal RRM2 expression in p53-deficient (H1299) cells transfected with pcDNA3.1 harboring p53 cDNA to determine if reintroduction of p53 in p53-deficient cells affects RRM2 expression. These responses were compared with naïve p53-deficient cells and H1299 cells transfected with pcDNA3.1 only. As shown in Fig 6A, transfection of p53-deficient cells with p53 cDNA failed to induce RRM2 expression. We then immunoprecipitated RRM2 and analyzed for associated p53 by Western blotting, confirming the interaction of p53 with RRM2 protein in p53-deficient cells transfected with p53 cDNA (Fig 6B). To clarify if LPS treatment alters p53-induced inhibition of uPA expression, we exposed naïve p53-deficient cells or p53-deficient cells transfected with vector DNA or p53 cDNA to PBS, LPS or the amino-terminal fragment (ATF) of uPA for 24 h. The conditioned media were analyzed for uPA by Western blotting. Consistent with our earlier report, (26) reintroduction of p53 in p53-deficient cells inhibited uPA expression. However, treatment with either LPS or ATF, which mimics full length uPA in terms of uPA expression, partially reversed the inhibitory effects of p53 (Fig 6C). We next immunoprecipitated p53 from the cell lysates of naïve p53-deficient H1299 cells, or the same cells expressing vector DNA (pcDNA 3.1) alone or p53 cDNA in pcDNA 3.1 and tested for RRM2 by Western blotting. We also exposed p53 cDNA transfected H1299 cells to PBS, LPS, or ATF of uPA and cytosolic extracts were immunoprecipitated for p53 and immunoblotted for RRM2 to assess changes in p53-RRM2 interaction. We found p53 and RRM2 interactions in the cytosolic fractions of H1299 cells transfected with p53 cDNA. However, treatment of these with either LPS or ATF abolished p53 and RRM2 interaction (Fig 6D). These results demonstrate that the changes in the interaction between p53 and RRM2 following LPS injury are associated with the induction of uPA expression at the posttranscriptional level in lung epithelial cells.
Figure 6.
Role of p53 in RRM2 expression and RRM2-uPA 3′UTR mRNA binding. A. cytosolic extracts of naïve p53-deficient (H1299) cells or H1299 cells transfected with vector pcDNA3.1 alone (Vc) or p53 cDNA in pcDNA3.1 were analyzed for RRM2 expression by Western blotting using anti-RRM2 antibody. The same membrane was stripped and immunoblotted for β-actin to assess equal loading. B. Cytoplasmic extracts of H1299 cells or H1299 cells transfected with vector pcDNA 3.1 (Vc) or p53 cDNA in pcDNA3.1 as described in Fig 6A were immunoprecipitated with anti-RRM2 antibody and immunoblotted for p53 (upper panel). The same membrane was stripped and developed for RRM2 (lower panel) by Western blotting. C. Naïve H1299 cells transfected with or without pcDNA3.1 alone (Vc) or p53 cDNA in pcDNA3.1 were treated with PBS or LPS (20 μg/ml) or ATF of uPA (1 μg/ml) for 24 h at 37°C. The conditioned media were analyzed for uPA expression by Western blotting using anti-uPA antibody. D. Naïve H1299 cells or H1299 cells transfected with p53 cDNA in pcDNA 3.1 or pcDNA 3.1 alone (Vc) were immunoprecipitated with anti-p53 antibody and immunoblotted for RRM2. H1299 cells transfected with p53 cDNA in pcDNA3.1 were treated with PBS, ATF or LPS as described in Fig 6C and the cytosolic extracts were immunoprecipitated with p53 and developed for RRM2 by Western blotting. H1299 cell lysate was used as a positive control and immunoprecipitation without H1299 cell lysates (mouse IgG) was as a negative control for Western blotting.
DISCUSSION
Proinflammatory cytokines such as TNF-α induce uPA expression in multiple cell types, including lung epithelial cells. uPA likewise induces its own expression in lung epithelial cells (2), endothelial and myeloid cells. (13) LPS induces both TNF-α and uPA expression in multiple cell types including lung epithelial cells.(15,23–25) Stabilization of uPA mRNA at the posttranscriptional level, at least in part in the case of TNF-α (15, 21) and exclusively in the case of uPA (2, 26), contributes to increased uPA mRNA and protein expression. Posttranscriptional regulation of uPA involves the interaction of a ~40 kDa cytoplasmic-nuclear shuttling protein with a 66 nt destabilization determinant present in the uPA mRNA 3′UTR. (15) Here, we extended our earlier study, purified the uPA mRNABp from the lysates of lung epithelial cells, identified it as RRM2 and characterized the role of the RRM2-uPA mRNA 3′UTR in the regulation of uPA mRNA stability and uPA expression by lung epithelial cells.
Expression of RRM2 inhibited uPA protein expression by lung epithelial cells and this response involves the suppression of uPA mRNA expression. LPS and uPA inhibited RRM2 binding to uPA mRNA 3′UTR, suggesting that RRM2 attenuated uPA expression. This is consistent with our earlier observation that TNF-α-mediated induction of uPA in lung epithelial cells is associated with parallel inhibition of the cytoplasmic uPA mRNABp interaction with the 66 nt uPA mRNA 3′UTR sequence due to translocation of uPA mRNABp to the nucleus.(15) The uPA mRNA destabilizing effect of the RRM2-uPA mRNA interaction was confirmed by the time dependent translocation of uPA mRNA binding activity from cytoplasm to the nucleus with accumulation of RRM2 proteins in the latter compartment after LPS treatment. This is further supported by the destabilization of uPA mRNA in resting as well as LPS-treated lung epithelial cells that overexpressed RRM2.
Ribonucleotide reductase is an important enzyme involved in the synthesis of DNA and responsible for the reduction of ribonucleotides to their corresponding deoxyribonucleotides. Three subunits of ribonucleotide reductase, RRM1, RRM2 and p53R2, provide a balanced supply of nucleotide precursors for DNA synthesis. RRM2 interacts with RRM1 to form a heterotertramer complex which is catalytically active. We recently reported that tumor suppressor protein, p53 inhibits uPA expression through destabilization of uPA mRNA.(26) The process involves sequence specific interaction of p53 with a 35 nucleotide destabilization determinant present in the uPA mRNA 3′UTR. Previous studies (27–28) suggested that p53 and RRM2 directly interact to control ribonucleotide reductase activity during DNA damage. Interestingly, the RRM2 binding sequence on uPA mRNA 3′UTR resides adjacent but upstream to the p53 binding sequence, supporting their potential for coordinate interaction. Increased translocation of RRM2 from the cytoplasm to the nucleus after UV irradiation (29–30) is consistent with our earlier report that uPA mRNABp shuttles to the nucleus following stimulation of lung epithelial cells with TNF-α, leading to stabilization of uPA mRNA and induction of uPA expression. RRM2 and its mode of regulation of uPA expression through cytoplasmic-nuclear shuttling are illustrated in a schematic diagram (Fig 7).
We reported earlier that uPA induces its own expression as well as that of uPAR through obliteration of p53. We therefore inferred that p53 must have contributed to RRM2-mediated inhibition of uPA expression by lung epithelial cells. Cells lacking p53 express a significant amount of uPA and reintroduction of p53 inhibits uPA expression through destabilization of uPA mRNA.(26) p53 and RRM2 independently interact with unique 3′UTR sequences that contain information for mRNA degradation (12,26). p53 interacts with RRM2 without significant changes in RRM2 expression. However, treatment of the cells with LPS inhibits cytosolic RRM2 and p53 interaction due to translocation of RRM2 to the nucleus. These changes are associated with a parallel induction of uPA expression through mRNA stabilization. Based on our present findings and prior reports (15,21,26) it is unlikely that p53 directly interferes with RRM2 binding to uPA mRNA since they independently bind to two destabilization determinants present in the 3′UTR (15,26). However, the possibility of p53 being involved in the translocation of RRM2 from the cytoplasm to the nucleus after LPS treatment is not yet ruled out and could be a subject of future study. This observation demonstrates that RRM2 maintains a complex network of cellular functions depending upon the external stimuli, metabolic state of the cell and the localization of RRM2.
This newly recognized paradigm is to our knowledge the first description of RRM2 regulating uPA expression in any cell type. If operative in vivo, this pathway could contribute to the regulation of uPA expression under normal conditions and in pathophysiologic states including various forms of ALI and its most severe form; acute respiratory distress syndrome.
Acknowledgments
This work was supported in part by grants from Flight Attendant Medical Research Institute Clinical Innovator Award (FAMRI-ID-082380) and NHLBI R21-HL093547.
The abbreviations used are
- UTR
untranslated region
- uPA
urokinase-type plasminogen activator
- LPS
lipopolysacharide
- TNF-α
tumor necrosis factor-alpha
- mRNA
messenger RNA
- RRM2
ribonucleotide reductase M2
- PAI-1
plasminogen activator inhibitor-1
- uPAR
uPA receptor
- ALI
acute lung injury
- RT-PCR
reverse transcription-polymerase reaction
Footnotes
Author disclosure: S.S. has received from the sponsored grants from FAMRI and National Institutes of Health for more than $100,001.
References
- 1.Dumler I, Petri T, Schleuning WD. Interaction of urokinase-type plasminogenactivator (u-PA) with its cellular receptor (u-PAR) induces phosphorylation on tyrosine of a 38 kda protein. FEBS Lett. 1993;322:37–40. doi: 10.1016/0014-5793(93)81106-a. [DOI] [PubMed] [Google Scholar]
- 2.Shetty S, Pendurthi UR, Halady PK, Azghani AO, Idell S. Urokinase induces its own expression in Beas2b lung epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2002;283:L319–L328. doi: 10.1152/ajplung.00395.2001. [DOI] [PubMed] [Google Scholar]
- 3.Koopman JL, Slomp J, de Bart AC, Quax PH, Verheijen JH. Effects of urokinase on melanoma cells are independent of high affinity binding to the urokinase receptor. J Biol Chem. 1998;273:33267–33272. doi: 10.1074/jbc.273.50.33267. [DOI] [PubMed] [Google Scholar]
- 4.Koshelnick Y, Ehart M, Hufnagl P, Heinrich PC, Binder BR. Urokinase receptor is associated with the components of the JAK1/STAT1 signaling pathway and leads to activation of this pathway upon receptor clustering in the human kidney epithelial tumor cell line tcl-598. J Biol Chem. 1997;272:28563–28567. doi: 10.1074/jbc.272.45.28563. [DOI] [PubMed] [Google Scholar]
- 5.Bhat GJ, Gunaje JJ, Idell S. Urokinase-type plasminogen activator induces tyrosine phosphorylation of a 78-kDa protein in H-157 cells. Am J Physiol. 1999;277:L301–L309. doi: 10.1152/ajplung.1999.277.2.L301. [DOI] [PubMed] [Google Scholar]
- 6.Shetty S, Bdeir K, Cines DB, Idell S. Induction of plasminogen activator inhibitor-1 by urokinase in lung epithelial cells. J Biol Chem. 2003;278:18124–18131. doi: 10.1074/jbc.M207445200. [DOI] [PubMed] [Google Scholar]
- 7.Abraham E, Gyetko MR, Kuhn K, Arcaroli J, Strassheim D, Park JS, Shetty S, Idell S. Urokinase-type plasminogen activator potentiates lipopolysaccharide-induced neutrophil activation. J Immunol. 2003;170:5644–5651. doi: 10.4049/jimmunol.170.11.5644. [DOI] [PubMed] [Google Scholar]
- 8.Abraham E, Matthay MA, Dinarello CA, Vincent JL, Cohen J, Opal SM, Glauser M, Parsons P, Fisher CJ, Jr, Repine JE. Consensus conference definitions for sepsis, septic shock, acute lung injury, and acute respiratory distress syndrome: time for a reevaluation. Crit Care Med. 2000;28:232–235. doi: 10.1097/00003246-200001000-00039. [DOI] [PubMed] [Google Scholar]
- 9.Welty-Wolf KE, Carraway MS, Ortel TL, Ghio AJ, Idell S, Egan J, Zhu X, Jiao JA, Wong HC, Piantadosi CA. Blockade of tissue factor-factor X binding attenuates sepsis-induced respiratory and renal failure. Am J Physiol Lung Cell Mol Physiol. 2006;290:L21–L31. doi: 10.1152/ajplung.00155.2005. [DOI] [PubMed] [Google Scholar]
- 10.Robbie LA, Dummer S, Booth NA, Adey GD, Bennett B. Plasminogen activator inhibitor 2 and urokinase-type plasminogen activator in plasma and leucocytes in patients with severe sepsis. Br J Haematol. 2000;109:342–348. doi: 10.1046/j.1365-2141.2000.02057.x. [DOI] [PubMed] [Google Scholar]
- 11.Bhandary YP, Velusamy T, Shetty P, Shetty RS, Idell S, Cines DB, Jain D, Bdeir K, Abraham E, Tsuruta Y, Shetty S. Post-transcriptional regulation of urokinase-type plasminogen activator receptor expression in lipopolysaccharide-induced acute lung injury. Am J Respir Crit Care Med. 2009;179:288–298. doi: 10.1164/rccm.200712-1787OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shetty S, Idell S. Urokinase induces expression of its own receptor in Beas2b lung epithelial cells. J Biol Chem. 2001;276:24549–24556. doi: 10.1074/jbc.M101605200. [DOI] [PubMed] [Google Scholar]
- 13.Li C, Zhang J, Jiang Y, Gurewich V, Chen Y, Liu JN. Urokinase-type plasminogen activator up-regulates its own expression by endothelial cells and monocytes via the u-PAR pathway. Thromb Res. 2001;103:221–232. doi: 10.1016/s0049-3848(01)00322-x. [DOI] [PubMed] [Google Scholar]
- 14.Gyetko MR, Aizenberg D, Mayo-Bond L. Urokinase-deficient and urokinase receptor-deficient mice have impaired neutrophil antimicrobial activation in vitro. J Leukoc Biol. 2004;76:648–656. doi: 10.1189/jlb.0104023. [DOI] [PubMed] [Google Scholar]
- 15.Shetty S, Idell S. Identification of a novel urokinase mRNA-binding protein in human lung epithelial cells in vitro. J Biol Chem. 2000;275:13771–13779. doi: 10.1074/jbc.275.18.13771. [DOI] [PubMed] [Google Scholar]
- 16.Shetty S, Velusamy T, Shetty RS, Marudamuthu AS, Shetty SK, Florova G, Tucker T, Koenig K, Shetty P, Bhandary YP, Idell S. Post-transcriptional regulation of plasminogen activator inhibitor type-1 expression in human pleural mesothelial cells. Am J Respir Cell Mol Biol. 2010;43:358–367. doi: 10.1165/rcmb.2009-0046OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shetty S, Ganachari M, Liu MC, Azghani A, Muniyappa H, Idell S. Regulation of urokinase receptor expression by phosphoglycerate kinase is independent of its catalytic activity. Am J Physiol Lung Cell Mol Physiol. 2005;289:L591–L598. doi: 10.1152/ajplung.00319.2004. [DOI] [PubMed] [Google Scholar]
- 18.Shetty S, Muniyappa H, Halady PK, Idell S. Regulation of urokinase receptor expression by phosphoglycerate kinase. Am J Respir Cell Mol Biol. 2004;31:100–106. doi: 10.1165/rcmb.2003-0104OC. [DOI] [PubMed] [Google Scholar]
- 19.Shetty S, Kumar A, Idell S. Posttranscriptional regulation of urokinase receptor mRNA: identification of a novel urokinase receptor mRNA binding protein in human mesothelioma cells. Mol Cell Biol. 1997;17:1075–1083. doi: 10.1128/mcb.17.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erbacher P, Remy JS, Behr JP. Gene transfer with synthetic virus-like particles via the integrin-mediated endocytosis pathway. Gene Ther. 1999;6:138–145. doi: 10.1038/sj.gt.3300783. [DOI] [PubMed] [Google Scholar]
- 21.Shetty S. Cytoplasmic-nuclear shuttling of the urokinase mRNA binding protein regulates message stability. Mol Cell Biochem. 2002;237:55–67. doi: 10.1023/a:1016558200199. [DOI] [PubMed] [Google Scholar]
- 22.Shetty S. Protein synthesis and urokinase mRNA metabolism. Mol Cell Biochem. 2005;271:13–22. doi: 10.1007/s11010-005-3453-x. [DOI] [PubMed] [Google Scholar]
- 23.Philippe J, Offner F, Declerck PJ, Leroux-Roels G, Vogelaers D, Baele G, Collen D. Fibrinolysis and coagulation in patients with infectious disease and sepsis. Thromb Haemost. 1991;65:291–295. [PubMed] [Google Scholar]
- 24.Wygrecka M, Markart P, Ruppert C, Kuchenbuch T, Fink L, Bohle RM, Grimminger F, Seeger W, Gunther A. Compartment- and cell-specific expression of coagulation and fibrinolysis factors in the murine lung undergoing inhalational versus intravenous endotoxin application. Thromb Haemost. 2004;92:529–540. doi: 10.1160/TH04-02-0126. [DOI] [PubMed] [Google Scholar]
- 25.Kwak SH, Mitra S, Bdeir K, Strassheim D, Park JS, Kim JY, Idell S, Cines D, Abraham E. Abraham E. The kringle domain of urokinase-type plasminogen activator potentiates LPS-induced neutrophil activation through interaction with {alpha}v{beta}3 integrins. J Leukoc Biol. 2005;78:937–945. doi: 10.1189/jlb.0305158. [DOI] [PubMed] [Google Scholar]
- 26.Shetty P, Velusamy T, Bhandary YP, Shetty RS, Liu MC, Shetty S. Urokinase expression by tumor suppressor protein p53: a novel role in mRNA turnover. Am J Respir Cell Mol Biol. 2008;39:364–372. doi: 10.1165/rcmb.2007-0406OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, Takei Y, Nakamura Y. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 28.Zhang YW, Jones TL, Martin SE, Caplen NJ, Pommier Y. Implication of checkpoint kinase-dependent up-regulation of ribonucleotide reductase R2 in DNA damage response. J Biol Chem. 2009;284:18085–18095. doi: 10.1074/jbc.M109.003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou B, Liu X, Mo X, Xue L, Darwish D, Qiu W, Shih J, Hwu EB, Luh F, Yen Y. The human ribonucleotide reductase subunit hRRM2 complements p53R2 in response to UV-induced DNA repair in cells with mutant p53. Cancer Res. 2003;63:6583–6594. [PubMed] [Google Scholar]
- 30.Xue L, Zhou B, Liu X, Qiu W, Jin Z, Yen Y. Wild-type P53 regulates human ribonucleotide reductase by protein-protein interaction with P53R2 as well as hRRM2 subunits. Cancer Res. 2003;63:980–986. [PubMed] [Google Scholar]