Abstract
A widely used technique for coordinate-based meta-analysis of neuroimaging data is activation likelihood estimation (ALE), which determines the convergence of foci reported from different experiments. ALE analysis involves modelling these foci as probability distributions whose width is based on empirical estimates of the spatial uncertainty due to the between-subject and between-template variability of neuroimaging data. ALE results are assessed against a null-distribution of random spatial association between experiments, resulting in random-effects inference. In the present revision of this algorithm, we address two remaining drawbacks of the previous algorithm. First, the assessment of spatial association between experiments was based on a highly time-consuming permutation test, which nevertheless entailed the danger of underestimating the right tail of the null-distribution. In this report, we outline how this previous approach may be replaced by a faster and more precise analytical method. Second, the previously applied correction procedure, i.e. controlling the false discovery rate (FDR), is supplemented by new approaches for correcting the family-wise error rate and the cluster-level significance. The different alternatives for drawing inference on meta-analytic results are evaluated on an exemplary dataset on face perception as well as discussed with respect to their methodological limitations and advantages. In summary, we thus replaced the previous permutation algorithm with a faster and more rigorous analytical solution for the null-distribution and comprehensively address the issue of multiple-comparison corrections. The proposed revision of the ALE-algorithm should provide an improved tool for conducting coordinate-based meta-analyses on functional imaging data.
Keywords: fMRI, PET, permutation, inference, cluster-thresholding
Introduction
Over the last decades, neuroimaging research has produced a vast amount of data localising the neural effects of cognitive and sensory processes in the brain of both healthy and diseased populations. In spite of their power to delineate the functional organisation of the human brain, however, neuroimaging also carries several limitations. The most important among these are the rather small sample sizes investigated, the consequently low reliability (Raemaekers et al., 2007) and the inherent subtraction logic which is only sensitive to differences between conditions (Price et al., 2005). Consequently, pooling data from different experiments, which investigate similar questions but employ variations of the experimental design, has become an important task. Such meta-analyses allow the identification of brain regions’ locations that show a consistent response across experiments, collectively involving hundreds of subjects and numerous implementations of a particular paradigm (Laird et al., 2009a; Laird et al., 2009b). Community-wide standards of spatial normalization and the reporting of peak activation locations in stereotaxic coordinates allow researchers to compare results across experiments when the primary data are unavailable or difficult to obtain (Poldrack et al., 2008).
Activation likelihood estimation (ALE; Laird et al., 2005; Turkeltaub et al., 2002) is probably the most common algorithm for coordinate-based meta-analyses (informative review see Wager et al., 2007b). The ALE algorithm is readily available to the neuroimaging community in form of the GingerALE desktop application (http://brainmap.org/ale). This approach treats activation foci reported in neuroimaging studies not as single points but as spatial probability distributions centered at the given coordinates. ALE maps are then obtained by computing the union of activation probabilities for each voxel. As in other algorithms for quantitative meta-analysis, the differentiation between true convergence of foci and random clustering (i.e., noise) is tested by a permutation procedure (Nichols and Hayasaka, 2003). Recently, we have proposed a revised algorithm for ALE analysis (Eickhoff et al., 2009), which models the spatial uncertainty – and thus probability distribution – of each focus using an estimation of the inter-subject and inter-laboratory variability typically observed in neuroimaging experiments, rather than using a pre-specified full-width half maximum (FWHM) for all experiments as originally proposed. In addition, it limits the meta-analysis to an anatomically constrained space specified by a grey matter mask and includes a new method of inference that calculates the above-chance clustering between experiments (i.e., random-effects analysis), rather than between foci (i.e., fixed-effects analysis).
An alternative approach to coordinate-based meta-analysis is kernel density analysis (KDA (Wager and Smith, 2003)). Both algorithms (KDE and ALE) are based on the idea of delineating those locations in the brain where the coordinates reported for a particular paradigm or comparison show an above-chance convergence. However, whereas ALE investigates where the location probabilities reflecting the spatial uncertainty associated with the foci of each experiment overlap in different voxels, KDE tests how many foci are reported close to any individual voxel. Recently, an algorithm for random-effects (RDFX) inference on KDE (termed multi-level kernel density estimation, MKDE) has been proposed (Wager et al., 2007b) which rests on a similar concept as the new random effects approach for ALE meta-analyses (Eickhoff et al., 2009). Both are based on summarising all foci reported for any given study in a single image [the “modelled activation” (MA) map in ALE and “comparison indicator maps” (CIM) in MKDE]. These are then combined across studies, and inference is subsequently sought on those voxels where MA maps (ALE) or CIMs (MKDE) overlap stronger as would be expected if there were a random spatial arrangement, i.e., no correspondence between studies.
The null-distributions for this inference on spatially continuous statistical maps computed by non-linear operations are estimated in both algorithms by using permutation procedures. More precisely, MDKE randomly redistributes the cluster centres throughout the grey matter of the brain, performs the same analysis as computed for the real data and uses the ensuing peak heights to derive FWE corrected voxel-level thresholds. This approach to statistical inference in voxel-wise meta-analysis data has the major advantage that the estimated null-distribution will reflect the spatial continuity of the statistical field of interest without requiring an exact parameterisation of the (non-linear) nature of its properties. That is, algorithms based on random relocation of foci within each experiment, generation of summary images per experiment and quantification of the convergence across these may empirically provide a good estimation on the distribution of statistical features of interest such as cluster size above a given threshold or maximum peak height (Wager et al., 2007b). Here we use this approach to derive a null-distribution of these two measures against which the results of the performed ALE analysis can then be compared for providing FWE or cluster-level corrected statistical inference.
A new approach to coordinate-based meta-analysis has very recently been proposed as signed difference map analysis (SDM; Radua et al., 2010; Radua and Mataix-Cols, 2009). SDM sums the voxel-wise activation probabilities of foci modelled as 3D Gaussian distributions like ALE, instead of counting closely activating experiments like MKDE. As opposed to ALE and MKDE, SDM emphasizes foci that were derived from conservatively corrected analyses. Similar to MKDE, it avoids too high probability values through neighbouring foci in a same experiment by limiting maximum values. This feature has also very recently been introduced to ALE (Turkeltaub et al., 2011) and was incorporated in the present work. Another novel feature of SDM consists in holding positive and negative values in a same map which prevents spurious overlap between those two categories of localization information rarely occurring in ALE. Analogous to MKDE and unrevised ALE implementations, significant convergence is distinguished from noise by computing a whole-brain null-distribution using a permutation procedure. Finally, SDM corrects results by FDR, unlike contemporary variants of ALE and MKDE. Taken together, ALE, MKDE and SDM all represent suitable methods for coordinate-based meta-analysis.
In the present report, we will address two remaining drawbacks of the widely used ALE algorithm. First, the null-distribution for statistical inference, reflecting a random spatial association between experiments is currently based on a permutation procedure. This approach, which has been part of all meta-analysis algorithms proposed up to now, however, has two disadvantages. First, drawing a sufficient estimate of the null-distribution may be rather time-consuming, given that a large number of permutations are required to sufficiently reflect the possible associations between experiments. If the test is underpowered, however, experimental ALE-values may exceed those observed under the null-distribution, indicating an insufficient estimation of its upper tail. Second, statistical inference on the ensuing p- or Z-maps is currently based on either uncorrected thresholds or correction for multiple comparisons using the false discovery rate (FDR) approach (Genovese et al., 2002). While using uncorrected thresholds provides no protection against false positives in a situation of multiple comparisons, FDR is likewise not the optimal approach. It has rather been noted that in cases where the underlying signal is continuous (such as in neuroimaging meta-analyses), controlling the false discovery rate is not equivalent to controlling the false discovery rate of activations (Chumbley and Friston, 2009). FDR corrected inference is therefore not appropriate for inferences on the topological features (regions of activation) of a statistical map as derived from ALE meta-analysis. Finally, in order to avoid spurious clusters consisting of only a few voxels, both of these procedures are commonly combined with an (arbitrary) extent threshold, suppressing clusters that are smaller than, e.g., 50 contiguous supra-threshold voxels. However, this subjective approach neither corresponds to statistical testing nor allows inference on the significance of regional activations. To overcome these limitations and to provide a more valid framework for ALE meta-analyses, we here present an analytical approach for deriving the null-distribution reflecting a random spatial association between experiments and propose algorithms for family-wise error correction and cluster-level inference on ALE data.
Materials and Methods
I. Revised approach for computing the null-distribution
Objective
Activation likelihood estimation (ALE) meta-analysis aims at determining above-chance convergence of activation probabilities between experiments (i.e., not between foci). To this end, ALE seeks to refute the null-hypothesis that the foci of experiments are spread uniformly throughout the brain. More specifically, ALE delineates where in the brain the convergence across all included imaging studies is higher than it would be expected if results were independently distributed (Eickhoff et al., 2009). All foci reported for a given experiment are modelled as Gaussian probability distributions whose width is based on an empirically derived modal of spatial uncertainty associated with neuroimaging foci (Eickhoff et al., 2009). For each voxel within a broadly defined grey matter shell [> 10% probability for grey matter, based on the ICBM tissue probability maps (Evans et al., 1994)] the information provided by the individual foci is then merged by taking the voxel-wise union of their probability values. Hereby, one “modelled activation” (MA) map is computed by merging all the activation foci’s probability distributions reported in a given experiment. The MA maps then contain for each voxel the probability of an activation being located at exactly that position. The MA can hence be conceptualized as a summary of the results reported in that experiment taking into account the spatial uncertainty associated with the reported coordinates. ALE scores are then calculated on a voxel-by-voxel basis by taking the union of these individual MA maps. The possibility of multiple foci from a single experiment jointly influencing the MA value of a single voxel, i.e., within-experiment effects, is controlled as recently proposed (Turkeltaub et al., 2011). Here, voxel-wise MA values are computed by taking the maximum probability associated with any one focus reported by the given experiment. This always corresponds to the probability of the focus with the shortest distance to the voxel in question.
Spatial inference on meta-analysis aims at identifying those voxels where the convergence across experiments (i.e., MA-maps) is higher than expected if the results were independently distributed. Importantly, this independence under the null-distribution only pertains to the relationship between experiments. In contrast, the spatial relationship between the foci reported for any given experiment is considered a given property captured in the MA-map. This distinction entails the difference between fixed-effects (convergence between foci as in earlier meta-analysis algorithms) to random-effects (convergence between experiments) inference. It is important to note that our statistical approach tests random-effects rather than fixed-effects. Only the former allows generalization of the results beyond the analyzed experiments rather than only to experiments considered in the analysis (Penny and Holmes, 2003; Wager et al., 2007b).
Previous algorithm
To enable spatial inference on these ALE scores, random convergence (i.e., noise) needs to be distinguished from locations of true convergence between experiments. Therefore, an empirical null-distribution is computed non-parametrically by a permutation procedure. This step is analogous to other methods for coordinate-based meta-analysis, including multilevel kernel density analysis (MKDA; Wager et al., 2007b) and signed differential mapping (SDM; Radua et al., 2010). In practice, this approach consists of picking a random voxel within the grey-matter mask from the MA map of experiment 1, then picking a (independently sampled) random grey matter voxel from the MA map of experiment 2, experiment 3, etc. until one voxel was selected from each MA map. The union of the respective activation probabilities, which were sampled from random, spatially independent locations, is then computed in the same manner as done for the meta-analysis itself in order to yield an ALE score under the null-hypothesis of spatial independence. This ALE score is recorded and the procedure iterated by selecting a new set of random locations and computing another ALE score under the null-distribution.
Analytical solution - concept
The key idea behind the proposed solution is to abandon the permutation procedure in favour of a non-linear histogram integration, which could be described as a weighted convolution (cf. Hope, 1968). To this end, the computational unit of the revised algorithm are not distinct voxels but distinct MA-values. That is, rather than considering each voxel individually, all voxels showing the same MA-value in a particular experiment are joined into and represented as a single histogram-bin. The entire histogram thus holds the occurrences of all possible MA-values (including those that are zero (voxels not in the vicinity of any reported focus) in form of bins, summarising the MA-map without its spatial information. These histograms are then successively merged throughout the different experiments considered in the meta-analysis to derive the null-distribution of ALE-values under spatial independence.
This approach takes advantage of the fact that the number of unique MA-values in each map is considerably smaller than the number of voxels, i.e., that many voxels show the same MA-value. This property is illustrated by an assessment of the MA-maps resulting from more than 5,500 experiments contained in the BrainMap database (www.brainmap.org; Fox and Lancaster, 2002; Laird et al., 2005). Our assessment of the BrainMap results archive showed that on average 93.6% of all voxels in the MA-maps had a value of zero. That is, across all experiments, only 6.4% of the grey-matter voxels has a non-zero probability of an activation being located at that position. Moreover, this analysis also revealed that the median number of unique values in the MA-maps derived from these 5,500 experiments was only 586. These numbers indicate the substantial advantage in terms of parsimony achieved by pooling MA-values into histogram-bins for further analysis rather than considering each voxel individually. The proposed algorithm thus represents a special case of a permutation test, where each the pool of values that may be drawn from each individual experiment may be represented parsimoniously by the probabilities for the (limited number of) different values. This allows to analytically compute rather than to empirically collect the probabilities of possible outcomes in the permutation test.
Analytical solution - algorithm
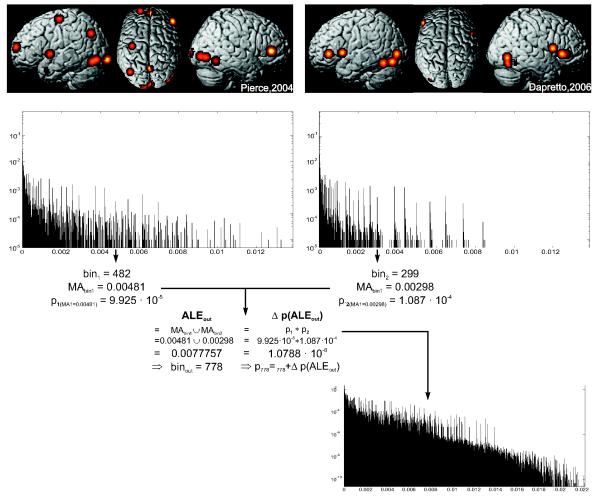
In order to compute the null-distribution of ALE values under spatial independence, each MA-map was first converted into a histogram of observed values (Fig. 1, top). The bin width of these histograms was set to 0.00001 (unit being MA-values, i.e., activation probabilities). Each histogram was then normalised to a sum of one, rendering the histogram-values probabilities of observing the MA-value corresponding to this particular bin in the respective MA-map. The histogram of the null distribution was initialised to correspond to a flat prior with all probabilities being zero. In order to derive final histogram of ALE-values under the null-hypothesis, the histograms corresponding to the MA-maps of the individual experiments were then successively combined. That is, initially, an ALE-histogram was computed by integrating the histograms of the first two experiments (cf. below, Fig. 1). The resulting ALE-histogram is then merged with the normalised histogram representing the MA-values of experiment three. Again, the output histogram is initialised to contain only zeros and the filled as described below. The histogram resulting from the successive integration of the histograms representing the MA-maps of the first three experiments is combined in the same fashion with the one of experiment four and so on. As this integration fulfils associativity like any multiplication, the order of which the MA maps are combined is irrelevant to this calculation. Once all experiments are considered, the final ALE-histogram representing the null-hypothesis for statistical inference is derived.
Figure 1.
Overview on the histogram integration procedure used for computing the null-distribution of ALE scores under the assumption of spatial independence. The top row shows the modelled activation maps of two experiments included in the exemplary face processing dataset. The middle row illustrates the histogram of modelled activation values for these two experiments. The lower panel shows the histogram resulting from the integration of the two histograms displayed in the middle rows. It denotes the probability (y-axis) for observing the different ALE scores (x-axis) when combining voxels from the two modelled activation maps shown above independently of spatial location.
In this context, it is important to note MA- and ALE-values are conceptually identical, as both represent the probability of an activation being present at a given voxel. This equivalence is highlighted by the fact that the probability information of the individual foci reported in a particular experiment is combined into an MA-map in exactly the same fashion (computing the voxel-wise union of probabilities), as MA-maps from different experiments are combined into an ALE-map. The difference in nomenclature thus purely reflects the difference between data pertaining to a single experiment (MA-values) and data computed by the combination of information from different individual experiments (ALE-values).
Analytical solution - implementation
As noted above, all bins of the output-histogram were initialised to a have a probability of zero. The integration algorithm used for combining two MA- or ALE-histograms (here denoted a and b) into a joint (output) histogram c involves cycling through all non-zero bins of both histograms. Each pair of bins is then combined according to the following algorithm. Let bj be the current bin, i.e., MA- or ALE-value, of the first histogram and the corresponding probability. Likewise, bk denotes the current bin, i.e., MA- or ALE-value, of the second histogram and the corresponding probability. The ALE-value l that would be observed in the resulting ALE-map c when voxels drawn from these two bins, bj and bk , are combined is given by the union of these, i.e., l = 1 – [(1 – bj) * (1 – bk)] , while its corresponding bin in the output-histogram is bl (Fig. 1, middle). The probability pl of these two bins being conjointly present in a random association, e.g., when drawing voxels at random from both maps, is given by . This probability pl can be conceptualised as the probability of drawing by chance a voxel from MA- or ALE-map a that has a value of bl, and simultaneously drawing a voxel from MA- or ALE-map b that has a value of bk. As a final step, the probability for the bin bl in the output-histogram is incremented by the observed probability, i.e., (Fig. 1, bottom). This process is continued until all non-zero bins of both input-histograms (representing the result of the previous integration and the next MA-map, respectively) have been combined with each other. The resulting output-histogram now represents the probabilistic distribution of ALE-values resulting from a random combination of the ALE- or MA-maps represented by the two input-histograms, initially derived from two experiments’ sets of activation foci.
II. Revised approach for multiple-comparison corrected inference
Voxel-level inference
In spite of the severe multiple-comparison problem, uncorrected voxel-level inference has long been common in functional neuroimaging (Genovese et al., 2002; Holmes et al., 1996) and is also employed in quantitative meta-analyses since its very beginnings (Laird et al., 2005; Turkeltaub et al., 2002). Inference is performed on the experimental ALE-map computed by taking the voxel-wise, i.e., spatially contingent, union of the MA-maps representing the assessed experiments. Here, the p-value associated with a particular experimental ALE-score is given by the probability of observing this or a more extreme value under the null-hypothesis of spatial independence. In previous implementations based on random sampling techniques, it was provided by the proportion of randomly drawn ALE-scores being at least equal to the experimental ALE-score. In the current algorithm, it is equivalent to the right-sided integral of the null-distribution computed as described above. In other words, computing the p-value of a particular ALE-score involves identifying the corresponding bin in the final histogram reflecting the analytical null-distribution and summing all probability values from this bin to the bin corresponding to the maximum ALE-score observed under the null-distribution (which is equivalent to the union of the highest value observed in each MA-map).
False-discovery rate correction for multiple comparisons
Correction for multiple comparisons using the false discovery rate (FDR) procedure has been used for both fMRI activation data (Genovese et al., 2002) and meta-analyses thereof (Laird et al., 2005). The key idea behind FDR correction is to choose a threshold in such manner that on average no more than a pre-specified proportion of test statistics declared significant can be expected to be false positives. As noted in the introduction, the use of FDR correction has been questioned in the context of (spatially smooth) functional imaging data. Nevertheless, since FDR is widely used in neuroimaging and has been used previously for inference on ALE meta-analyses, its application with the revised version of the algorithm has been included for comparison. Importantly, the statistical (p-value) threshold needed to control the false-discovery rate at a particular level solely depends on the number of parallel tests, i.e., analysed voxels, and the distribution of statistical values observed for these. This implies that FDR correction is readily feasible for meta-analyses performed using the analytical null-distribution detailed above and benefit from the more precise estimation of p-values for higher ALE-scores.
Family-wise error rate correction for multiple comparisons
In the context of neuroimaging data, correcting for the family-wise error rate (FWE) in statistical inference is usually achieved by referring to Gaussian random field theory. These approaches consider a statistical parametric map to be a lattice approximation to an underlying continuous process. Once the smoothness of the underlying field has been estimated, corrected inference becomes possible. Here, a FWE corrected inference at p<0.05 corresponds to choosing a threshold which is exceeded in no more than 5% of random statistical fields of the same size and smoothness as the assessed image. In fMRI and PET analyses, the smoothness of the underlying Gaussian field is conventionally estimated by assessing the residuals of the statistical model under the assumption of normally distributed error. In meta-analyses, however, there is no equivalent to the residuals of a general linear model. Moreover, in spite of the fact that activation foci are modelled by Gaussians, a Gaussian distribution of the statistical field cannot be assumed due to the non-linear operation of computing the ALE-scores. A parametric computation of family-wise error corrected thresholds via Gaussian random field theory for inference on ALE meta-analyses is hence not feasible.
Nevertheless, given that the number of voxels and the entire null-distribution of the statistical field is known, family-wise error corrected thresholds can be computed without reference to the behaviour of random fields. It should be reiterated, that a threshold t0 is considered to correct for multiple comparisons in a set of N (number of voxels) test statistics by controlling the family-wise error rate at αFWE, if under the null-distribution the proportion of random sets containing N test statistics that feature at least one element above αo is less or equal to αFWE. In other words, the threshold αo should be chosen such that the chance of observing a statistic above αo in a set of N realisations of the null-distribution is less than αFWE.
In practice, an upper bound on αo can be derived from the following approach, which is based on the null-distribution histogram c computed as defined above. This approach yields an upper bound rather then the precise value since the calculation below is based on the assumption of independent realisations of the null-distribution across voxels. However, ALE-scores are spatially correlated; the effective number of observations and the corrected threshold should therefore be lower than this upper bound derived from the assumption of independence. For a particular ALE threshold αo, corresponding to the bin bα o, the chance of observing this value or a more extreme one under the null-distribution is given by , i.e., the sum of the probability for this bin and those for all bins corresponding to higher ALE-scores. In turn, the probability of observing at least one ALE-score equal or higher than t0 in a set of N random independent realisations is given by 1 – (1 – Pt0)N. The choice of a family-wise error corrected threshold therefore comes down to identifying the smallest α such that is less or equal to αFWE.
Note that in contrast to random field based approaches, this correction does not consider the signal to be continuous but rather assumes N (number of voxels) independent realisations of the null-distribution. Due to the continuous nature of the data, however, the true number of independent realisations will be substantially lower, reducing the number of multiple comparisons and thus the exponent in the formula stated above. The threshold computed by the approach outlined here can hence be considered the upper bound and hence a conservative estimate to a family-wise error correction of ALE meta-analysis data.
As an alternative to this conservative analytical approach to FWE thresholding, family-wise error corrected thresholds can also be derived from Monte-Carlo analysis as described in detail below in the section “Cluster-level inference – implementation”. The basic idea behind this approach is to simulate random datasets, i.e., “experiments”, with the same characteristics as the real data, compute ALE-scores for these random experiments record the highest ALE-score and iterate the process several times. The FWE corrected threshold for the actual ALE analysis is then given by the ALE-score, which is only exceeded in 5% of the ALE maps based on random data.
Cluster-level inference - concept
The idea behind cluster-level inference on neuroimaging data is to perform topological inference on the statistical maps to be assessed. It addresses a problem that is unique to inference on images such as brain activation maps, in which the underlying signal is continuous, i.e., does not have a compact support. Here inference is strictly only possible on topological features of this image, such as clusters above an ad-hoc threshold. Cluster-level inference does therefore not consider the height of a particular voxel or peak, but rather the spatial extent of the super-threshold clusters treated as single topological entity. In this context, it is important to appreciate that cluster-level inference stand in stark contrast to FDR and voxel-level FDR correction as described above by operating on sets of voxels rather than individual voxels (cf. Chumbley and Friston, 2009).
In fMRI and PET analyses, cluster-level inference is, like FWE correction, conventionally based on the theory of Gaussian random fields. As outlined above, however, the application of corrections derived from random field theory is impeded in the context of ALE meta-analyses for two reasons. First, ALE analyses do not offer the possibility to estimate the smoothness of an underlying random field based on normally distributed residuals and, secondly, a Gaussian distribution of the statistical field cannot be assumed due to the non-linear operation of computing the ALE-scores. Moreover, while FWE correction pertains only to the probability of observing an above-threshold voxel in a random realisation of the statistical field, cluster-level inference necessarily needs to be based on the expected extent of the signal and must therefore consider the non-compact support of the signal, i.e., spatial dependence. In summary, cluster level inference on ALE results can currently neither be based on parametric approaches from random field theory nor on limit-estimates derived under assumptions of spatial independence. Inspired by the recent introduction of cluster-level inference into KDA (Wager et al., 2007b; Wager et al., 2007a), we here propose a Monte-Carlo based approach to cluster-level inference in ALE resembling previous non-parametric approaches to voxel-level inference on ALE data.
Cluster-level inference – implementation
As stated above, the objective of cluster-level inference pertains to a topological feature of the image, more precisely the size of the clusters in the excursion set above a cluster-forming threshold. In theory, this threshold can be arbitrarily chosen, though conventionally, an uncorrected voxel-wise threshold of p<0.001 has been most prevalent in both fMRI and meta-analyses. We will hence use this level as cluster-forming threshold throughout the exemplary analysis while noting that any other uncorrected voxel-wise thresholds would also be perfectly valid. The first step of cluster-level inference is to threshold the statistical image of uncorrected voxel-wise p-values by the cluster-forming threshold. While this procedure is equivalent to conventional uncorrected thresholding, the important subsequent step compares the size of the supra-threshold clusters against a null-distribution of cluster sizes. The p-value associated with each cluster in this procedure is then given by the proportion of clusters arising from random data, which have the same or a larger size as the cluster under investigation. That is, if a cluster is large enough to be only exceeded in size by 1 out of 100 clusters formed by thresholding ALE analyses on random data with the same cluster-forming threshold as used in the true analysis, its p-value will be 0.01. Discarding all clusters that have a p-value of, e.g., less than 0.05, then provides an unbiased estimator for the previously arbitrarily defined extent-threshold.
In order to estimate a null-distribution of cluster sizes given a particular cluster-forming threshold, we propose the following random-simulation algorithm. First, a set of random experiments is simulated using the same characteristics as present in the real data. That is, for every experiment included in the meta-analysis, there is a matching random “experiment” having the same smoothness, i.e., containing the same number of subjects and number of foci. The coordinates of these foci, however, were randomly (and independently across experiments) allocated to any grey matter voxel in MNI space. ALE analysis on this set of random, simulated experiments is then performed in the same fashion as described above for the real data. The statistical map derived from this analysis is thresholded using the same cluster-forming threshold as employed for the actual inference. The size of each cluster above this threshold is recorded, as is the maximum ALE-score observed (for FWE corrected thresholding). Then, a new set of random experiments is generated and the process is iterated several times. In the current analysis, we used 1,000 repetitions, which can be computed in less than one hour. Additionally, we also computed a more extensive null-distribution based on 10,000 repetitions to evaluate the dependence of the derived results on the number of repetitions.
III. Example data
The modified ALE approach is illustrated by a meta-analysis on the brain activity evoked by visually presented faces. Using the BrainMap database (www.brainmap.org), 19 papers reporting 20 individual experiments (305 subjects) and a total of 183 activation foci were obtained (Table 1, cf. Fig. 2). For comparison, meta-analysis on these reported activations was also carried out using the previous version of the random-effects ALE algorithm (Eickhoff et al., 2009) using 106-1012 random samples to establish the null-distribution. For comparison, the results were thresholded at p<0.001 (uncorrected) and at a corrected threshold of p < 0.05 computed using the false discovery rate (FDR) (Genovese et al., 2002; Laird et al., 2005), the family-wise error rate (FWE) and the cluster-level inference described above.
Table 1.
| Paper | Modality | Exp. | Foci | Subjects | Contrast |
|---|---|---|---|---|---|
| (Benuzzi et al., 2007) | fMRI | 1 | 14 | 24 | neutral faces > parts of neutral faces |
| (Bird et al., 2006) | fMRI | 1 | 5 | 16 | faces > control |
| (Bonner-Jackson et al., 2005) | fMRI | 1 | 5 | 26 | faces > words |
| (Braver et al., 2001) | fMRI | 1 | 4 | 28 | faces > words |
| (Britton et al., 2006) | fMRI | 1 | 6 | 12 | socio-emotional faces > neutral faces |
| (Dapretto et al., 2006) | fMRI | 1 | 14 | 10 | emotional faces > baseline |
| (Dolcos and McCarthy, 2006) | fMRI | 1 | 16 | 15 | faces > scrambled faces |
| (Denslow et al., 2005) | PET | 1 | 10 | 9 | facial identity > spatial position |
| (Hasson et al., 2002) | fMRI | 1 | 4 | 13 | faces > letter strings/buildings |
| (Holt et al., 2006) | fMRI | 1 | 6 | 16 | neutral faces > baseline |
| (Kesler-West et al., 2001) | fMRI | 1 | 17 | 21 | neutral faces > scrambled faces |
| (Kranz and Ishai, 2006) | fMRI | 1 | 21 | 40 | faces > scrambled faces |
| (Kringelbach and Rolls, 2003) | fMRI | 1 | 4 | 9 | emotional faces > neutral faces |
| (Paller et al., 2003) | fMRI | 1 | 1 | 10 | faces > scrambled faces |
| (Pierce et al., 2004) | fMRI | 2 | 25/9 | 9 | familiar faces > baseline familiar faces > strange faces |
| (Platek et al., 2006) | fMRI | 1 | 6 | 12 | faces > scrambled faces |
| (Vuilleumier et al., 2001) | fMRI | 1 | 3 | 12 | faces > houses |
| (Wild et al., 2003) | fMRI | 1 | 10 | 10 | faces > baseline |
| (Williams et al., 2005) | fMRI | 1 | 3 | 13 | faces > houses |
Overview of the individual experiments included in the meta-analysis used to exemplify the revision of the activation likelihood estimation (ALE) algorithm. More than one number is given in the column “Reported foci” if multiple experiments from the same article have been analysed.
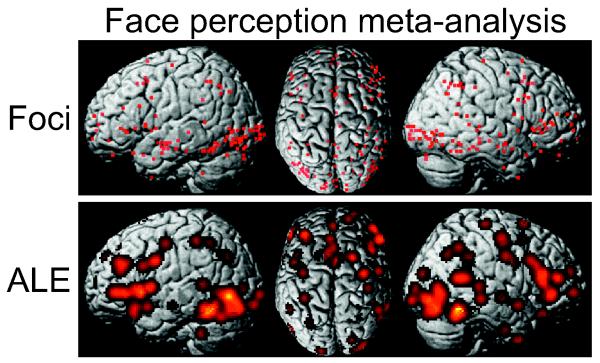
Figure 2.
A real dataset was analysed in order to exemplify the new algorithms. This dataset consisted of 19 papers reporting 20 individual experiments (305 subjects) and a total of 183 activation foci on the brain activity evoked by visually presented faces. The figure shows the distribution of individual foci (upper row) as well as the (un-thresholded) ALE map (lower row) for the exemplary dataset.
Results
I. Revised approach for computing the null-distribution
The analytical null-distributions for inference on both datasets were compared to those derived from the random sampling algorithm described by Eickhoff et al. (2009). For the latter approach we used between 106-1012 random samples. One of the most dramatic differences pertained to the computation time needed to compute the null-distribution. For the face perception dataset, 106 samples were computed in about a minute, 108 samples in about 30 minutes and 1010 samples about 24 hours while 1012 samples took about 3 months to compute on a Intel Core 2 Duo T9300 2.5 GHz computer with 4 GB or RAM. Note that the computation time doesn’t scale linearly due to the smaller relative contribution of reading/writing processes in the higher repetitions. In contrast, the analytical null-distribution was computed in about 10s.
Comparison of randomization-based and analytical null-distributions
A synopsis of the null-distributions (cumulative density functions) for the two analysed datasets yielded by the randomisation approach and the analytical solution, respectively, are displayed in figure 3. It can be noted that the right tail of the randomisation-based null-distributions becomes successively larger as the number of samples increases. This behaviour is associated with lower probabilities for the maximum ALE-scores covered by the null-distribution. Together they reflect the notion that large ALE-scores will only be observed when sampling, by chance, higher and this rarer MA-values in multiple maps. Importantly, notwithstanding the extremely time-consuming computation, even 1012 repetitions of the sampling process fall considerably short of the analytical solution in estimating the p-values of higher ALE-scores.
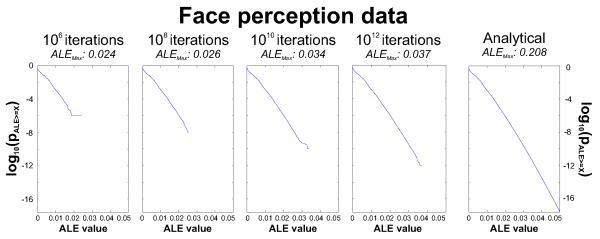
Figure 3.
Quantitative assessment of the differences between computing the null-distribution by the earlier permutation procedure and the proposed analytical solution. Histograms show the null-distribution of ALE scores for the face processing dataset under the assumption of spatial independence between experiments as estimated by the permutation procedure using between 106 to 1012 iterations and computed by the histogram integration (rightmost). It can be noted that as the number of samples increases, the right tail of the randomisation-based null-distributions becomes successively larger, reflecting the notion that large ALE-scores will only be observed when sampling higher and thus rarer MA-values in multiple maps. Importantly, notwithstanding the extremely time-consuming computation, even 1012 repetitions of the sampling process fall considerably short of the analytical solution in estimating the p-values of higher ALE-scores.
This apparently insufficient sampling of the right tail of the null-distribution is reflected by the pronounced difference in the maximum ALE-score covered by the different null-distributions. In the sampling approach, its value is equivalent to the highest ALE-score observed in any of the random drawings. In the analytically computed null-distribution, however, it is equivalent to the union of the highest MA-value in the MAP-map of each individual experiment. For the face perception dataset, the highest ALE-scores observed in the randomisation procedure were 0.024 (106 samples), 0.026 (108 samples), 0.034 (1010 samples) and 0.037 (1012 samples). On the other hand, the highest ALE-score observed in the “real” analysis of the face perception dataset was 0.035. Consequently, a null-distribution based on more than 1010 samples was required to provide an adequate coverage of higher ALE-scores by right tail of the null-distribution. Only such complete coverage, however, can avoid situations where the parametric p-value (fraction of equal or larger random samples) is exactly zero. In contrast to the randomisation procedure, the analytical solution provided a smooth estimation of the null-distribution up to a maximum of 0.208, i.e., well above the highest ALE-score observed experimentally.
Stability of uncorrected thresholds
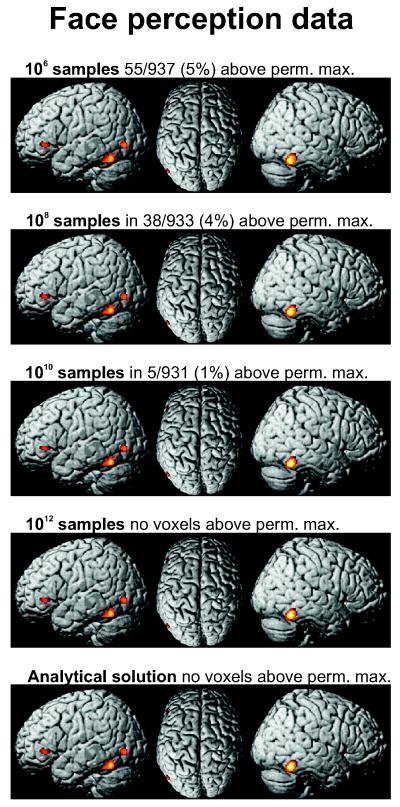
As detailed above, there are considerable differences in the right tails of the null-distributions. In the region of lower ALE-scores, however, all null-distribution show an almost identical shape. A bit surprisingly, this holds true even for that one based on only 106 samples, which corresponds to no more than 5 complete volumes (given that the grey matter mask consists of ~200,000 2×2×2mm3 voxels). This observation is in good agreement with the results of inference on the face perception at a threshold of p < 0.001 (uncorrected). As illustrated (Fig. 4), the supra-threshold clusters are almost completely invariant to the method for computing the null-distribution (sampling vs. analytical) or the number of random ALE-scores sampled. That is, although lower numbers of repetitions generated an incomplete sampling of the right tail of the null-distribution and resulted in a high proportion of voxels exceeding the maximum random sample (i.e., had a p-value of zero), the uncorrected thresholds were almost identical.
Figure 4.
Results of voxel-wise inference on the face processing dataset at p<0.001 uncorrected. The rows correspond to the use of null-distributions derived from different amount of samples of the null-distribution (cf. Fig. 3). For comparison the lowest row shows the result of uncorrected thresholding at p<0.001 using the analytical solution. It can be seen that the results of the uncorrected inference are remarkably stable across the different approaches for deriving the null-distribution. However, as indicated above the individual images, virtually all of the results derived from the random sampling null-distributions show voxels featuring a p-value of 0, corresponding to ALE scores that are higher than any score observed in the sampling procedure.
Finally, the comparison between the results yielded by inference on ALE-analyses using the previous and the revised version of the algorithm at the same threshold (p<0.001) also provides a valuable cross-validation of both approaches. In spite of the considerable conceptual differences between them, randomisation-based and analytical inference at a conventional uncorrected threshold produce nearly identical results. This stability indicates a good robustness of inference on ALE data, and furthermore provides added validity to the analytical solution to the computation of the null-distribution.
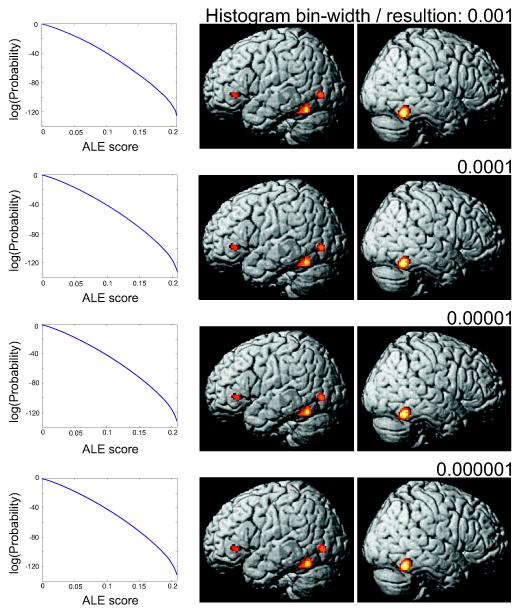
Effect of histogram bin-size
In the above description of the new algorithm for deriving the null-distribution, we proposed a bin-size for the histograms of 0.00001 (units: MA- or ALE-values). In order to assess the dependence of the results on the bin-size, i.e., resolution, used when computing the histograms of the individual MA-maps and, eventually, the null-distribution on the ALE-scores, we repeated the analyses with several different bin-sizes ranging from 0.001 to 0.000001. It can be observed (Fig. 5), that the choice of the bin-width did not have any noticeable effect on either the resulting histogram or the results of the statistical inference. Likewise, the increase in computation time caused by a finer bin-size of the histograms was only minimal, as even at the highest resolution the full null-distribution was computed in about a minute. The proposed algorithm may therefore be considered very robust across a wide range of bin-widths. We nevertheless chose to keep the resolution at 0.00001, as there is no evident advantage of wider bins but the (theoretical, though never observed) potential for additive rounding errors in very large meta-analyses involving hundreds of experiments.
Figure 5.
In order to assess the dependence of the results on the bin-size, i.e., resolution, used when computing the histograms of the individual MA-maps and, eventually, the null-distribution on the ALE-scores, we repeated the analyses with several different bin-sizes ranging from 0.001 to 0.000001. As shown here for the face processing dataset, it can be observed that the choice of the bin-width during histogram integration did not have any noticeable effect on either the resulting histogram or the results of the statistical inference.
II. Revised approach for multiple-comparison corrected inference
FWE corrected thresholding
When performing inference on a continuous statistical map a threshold αo is considered to correct for multiple comparisons at a voxel-level FWE of αFWE if under the null-distribution the proportion of random analyses that feature at least one element above αo is less or equal to αFWE. In the context of ALE analyses, this means that αo should be chosen such that in a complete dataset obtained under the null-distribution, the probability of observing a single ALE score above αo is less than αFWE. Here, we proposed two approaches to derive these voxel-level FWE corrected thresholds, either analytically by reference to the computed null-distribution or by Monte-Carlo analysis, i.e., permutation testing. It has to be noted, that the former approach is based on the assumption of independence between voxels and should hence provide a conservative upper bound on the corrected threshold αo.
For the face perception dataset this upper bound as computed from the analytical null-distribution corresponded to an ALE-threshold of 0.0216 to control the FWE rate at p<0.05. The FWE corrected thresholds derived from a Monte-Carlo analysis were based on recording the maximum ALE-score for each of ALE-maps reflecting a random relocation of activation foci within each experiment (cf. Fig. 6) and correspond to the ALE-score that was exceeded in only a fraction of all realisations corresponding to αFWE. As expected from the theoretical considerations, the FWE thresholds obtained from this randomisation-approach were lower than the bounds given by the analytical solution. The ALE-threshold needed to control the voxel-level FWE at p<0.05 in the face dataset was 0.0196 when based on 1,000 repetitions while 10,000 repetitions yielded a threshold of 0.0198. The randomisation-based FWE thresholds seem to be highly stable even after only 1,000 repetitions of random relocation, which can be computed in about 2-5 minutes (depending on the number of experiments in the analysis) by the approach outlined above.
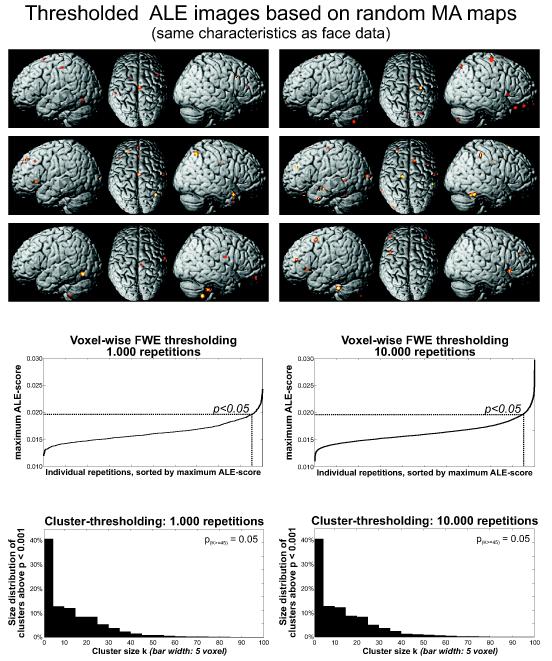
Figure 6.
Illustration of the approach for computing cluster-level and voxel-wise FWE thresholds based on randomization. The top row illustrates 6 ALE maps based on independent random relocation of cluster foci for each experiment of the face processing dataset (keeping the number of foci and FWHM identical to the real data) after applying an uncorrected threshold of p<0.001.
The middle row illustrates the maximum ALE scores observed in the noise datasets obtained from 1,000 (left) or 10,000 (right) iterations of the random relocation procedure. The ALE-threshold needed to control the voxel-level FWE at p<0.05 in the face dataset was almost identical between both cases (1,000 repetitions: 0.0196, 10,000 repetitions 0.0198).
The bottom row illustrates the distribution of cluster sizes in the excursion set (above p<0.001 uncorrected) following 1,000 (left) or 10,000 (right) iterations of the random relocation procedure. In both cases the cluster-level threshold needed to correct at p<0.05 corresponded to a cluster extent of at least 45 voxels.
Cluster-level thresholding by randomization
Due to the unavailability of random field models for the topology of ALE maps, cluster level thresholds were derived from the same permutation-approach as used for the randomisation-based voxel-level FWE thresholding. As noted above, cluster-level thresholding is equivalent to first applying a (uncorrected) cluster-forming threshold to the ALE-analysis. Subsequently, it is assessed how likely clusters of the obtained size may have arisen by chance, i.e., when applying the same cluster-forming threshold to random data. The cluster-level corrected threshold corresponding to p<0.05 is equivalent to the cluster size, which is reached or surpassed by only 5% of the clusters observed when applying the cluster-forming threshold to the ALE-maps, reflecting a random relocation of activation foci within each experiment. For the face dataset, 1,000 repetitions of this randomisation approach yielded a cluster-level threshold of 45 voxels when the cluster-forming (uncorrected) threshold was p<0.001 (Fig. 6). Exactly the same cluster-level threshold of 45 voxels was also found when the null-distribution of cluster-sizes was based on 10,000 ALE-analyses of randomly relocated foci with the same properties as the actual data. Like the voxel-level FWE thresholds, also the cluster-level thresholds seem to be reliably estimated after 1,000 repetitions of the random relocation.
Comparison of thresholding approaches
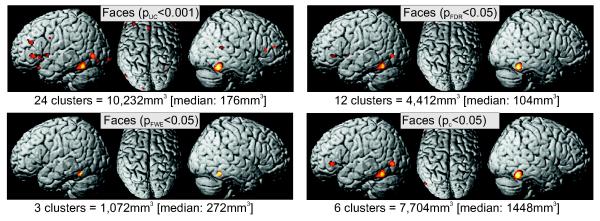
In order to compare the results yielded by the different methods for dealing with the problem of multiple comparisons when performing inference on ALE maps, we applied each of them to the dataset on face processing. In particular, we thresholded the ALE maps derived from these meta-analyses at i) p<0.001 (uncorrected); ii) p<0.05 (FDR corrected); iii) p<0.05 (voxel-level FWE corrected); iv) p<0.05 (cluster-level inference using p<0.001 at voxel-level as cluster-forming threshold).
As illustrated (Fig. 7), the uncorrected voxel-level inference yielded the most extensive activation, with regard to activated volume as well as to the number of clusters, in the two performed meta-analyses. In particular, in both datasets, the number of clusters is about three times that obtained from any other approach. In contrast, FDR and especially FWE thresholding resulted in the most conservative delineation of activation, yielding both fewer and smaller significant clusters. Finally, cluster-level thresholding takes an intermediate position. On the one hand, the number of significant clusters in the face processing dataset is smaller as compared to the uncorrected results. On the other hand, the total size of the ensuing activations is close to that yielded by uncorrected thresholding and substantially exceeds the very restricted results obtained from FDR or FWE thresholding. This is also reflected in the median size of the individual clusters, which are considerably larger when using cluster-level thresholding as compared to the very small foci yielded by the FDR and FWE approaches.
Figure 7.
This figure illustrates the results of thresholding the face processing meta-analysis using the four different approaches for dealing with multiple comparisons when performing inference on ALE maps. Within the display for each of the two datasets, the applied methods are (in clockwise order starting at the top left): i) p<0.001 (uncorrected); ii) p<0.05 (FDR corrected); iii) p<0.05 (voxel-level FWE corrected using randomisation analysis); iv) p<0.05 (cluster-level corrected inference using p<0.001 uncorrected at voxel-level as the cluster-forming threshold).
Evidently, the number of true activations is unknown in the face processing dataset. There was, however, a good correspondence of FWE, FDR and cluster-level thresholding and a much higher number of activation clusters obtained by the uncorrected inference. These observations therefore also points to a low specificity of uncorrected inference on ALE data. Between FWE, FDR and cluster-level thresholding, all approaches revealed correspondence in the bilateral posterior fusiform gyrus and the right amygdala. Using FDR and cluster-level thresholding, additional foci of convergence became significant in the amygdala, MT/V5 and inferior frontal gyrus (just anterior to BA 45) on the left side. Thresholding for cluster-level significance revealed additional activation in the right anterior fusiform gyrus.
Discussion
Here we outlined a revision of the activation likelihood estimation (ALE) algorithm for coordinate-based meta-analyses of neuroimaging experiments that address two potential shortcomings of the current implementation of this approach. These pertain to how the null-distribution reflecting the expected ALE values under the assumption of spatial independence is computed and to the methods for correcting the statistical inference for multiple comparisons. In summary, we demonstrated in an analytical fashion that histogram integration allows a faster and more complete estimation of the null-distribution than achievable with permutation testing, and that cluster-level correction for multiple comparisons provides higher sensitivity than FDR or FWE thresholding while still providing stringent protection against false positives.
Revised approach for computing the null-distribution
Classically, all approaches for coordinate-based meta-analysis have based the statistical inference on randomisation procedures. For example, the original ALE algorithm derived a null-distribution of ALE scores through random relocation of all foci analysed in the current study throughout the brain (Turkeltaub et al., 2002). In addition to ALE analyses, meta-analyses using kernel density analysis (KDA; Wager and Smith, 2003) or random-effects multilevel KDA (Wager et al., 2007b) are also based the statistical inference of the convergence of reported foci on empirically estimated null-distributions. In the current MKDA algorithm, this permutation procedure involves a random relocation of cluster foci throughout the grey matter and estimating the convergence across those simulated studies as a reference for thresholding the real data. Finally, the random-effects approach to ALE meta-analysis (Eickhoff et al., 2009) also applies a permutation procedure for statistical inference. In contrast to the above-mentioned approaches, however, random-effects inference in ALE was not based on random relocation of foci but rather random sampling of the modelled activation map for each study. That is, this algorithm employed a null-distribution of spatial independence across studies rather than a null-distribution of random cluster location. Following the assumption of spatial independence is the notion that the permutation procedure does not have to accommodate spatial correlations. Rather, the null-distribution should reflect for each ALE score the probability of obtaining this value through a random combination of the modelled activation maps. As outlined here and validated against different numbers of iterations in the permutation procedure, the ensuing null-distribution can be computed analytically. This is achieved by successive integration of the conceptually equivalent “modelled activation” or ALE histograms and integration of the probabilities across the possible combinations in order to derive a complete null-distribution of ALE scores under spatial independence. Comparative analysis demonstrated that this approach yields results that are highly comparable to those derived from the permutation approach. However, the computational time taken by the analytical solution is more bearable than in previously suggested random drawing procedures. Moreover, the new approach also allows the computation of the entire right tail of the distribution up to the probability for the ALE score that would result from taking the union of the maximum MA value for each experiment. The revised algorithm hence excludes the occurrence of situations where experimental values exceed those covered by the null-distribution and would hence have to be assigned a p-value of exactly zero.
Correcting for multiple comparisons
When performing coordinate-based meta-analysis convergence of activation foci reported in the literature is assessed individually at each voxel of the reference space. That is, an ALE score is computed for each voxel and then compared against a null-distribution of ALE-scores that would be expected given the currently analysed set of experiments but a random spatial association between these. At a resolution of 2 × 2 × 2 mm3 the applied grey matter mask results in approximately 200,000 individual tests that are performed in parallel. This situation of massive univariate inference poses a considerable multiple comparison problem, similar to the situation faced in general linear model analysis of neuroimaging data (Kiebel and Holmes, 2003).
The statistical inference must account for the presence of parallel tests, i.e., a correction for multiple comparisons has to be performed. However, ALE analyses possess several properties, which potentially invalidate the application of established multiple-comparison corrections. On the one hand, ALE data does not have compact support but rather consists of spatially correlated observations. This invalidates the assumptions necessary for FDR procedures, which were designed for families of discrete, i.e., independent, tests (Benjamini and Hochberg, 1995) and hence are not applicable for continuous signals. As recently outlined by Chumbley et al. (2009), approaches that are aimed at controlling the voxel-wise false discovery rate should consequently be inadequate in controlling the false positives among topological features, i.e., clusters. That is, although FDR corrections seem to yield reasonable results and have enjoyed considerable popularity in both neuroimaging (Genovese et al., 2002) and meta-analyses (Laird et al., 2005), their interpretation in a sense of “clusters of observed activations” leads to a problematic underestimation of the false discovery rate (Chumbley and Friston, 2009).
Cluster-level thresholding by randomization
The problems associated with applying approaches intended for discrete data on neuroimaging results has prompted the development of methods based on random field theory that allow topological inference on the statistical maps (Worsley et al., 1996). The key idea of these approaches is to consider the data a lattice approximation to an underlying continuous process and peruse topological inference based on random field properties. That is, one is not making an inference about a voxel, but a topological feature attributed to a region or cluster, i.e., a connected excursion set above a certain a-priori (cluster-forming) threshold (Worsley et al., 1996; Worsley, 2003). Using these methods, cluster-level correction becomes feasible, which allow controlling the family-wise error rate at cluster-level. Here inference is based on first setting a cluster-forming threshold and then computing for each cluster the probability for finding a set of connected voxels in the excursion set of the same size as the cluster given the underlying random statistical field. In other words, cluster-level thresholding aims at excluding those regions, which are small enough to be found above threshold by chance if no true signal was present in the data (Chumbley and Friston, 2009).
Importantly, in random field theory, the extent-threshold needed to correct the inference at cluster-level only depends on the type of the statistical field (F/T/Chi2), the size of the search volume and the smoothness of the field. In the assessment of fMRI and PET data, the latter is estimated from the spatial derivative of the residual field, i.e., by the smoothness of the noise term in the general linear model (Worsley et al., 1996; Worsley, 2003). In contrast to fMRI and PET experiments, however, ALE analyses do not yield a parametric residual field from which the smoothness of the underlying random field can be computed. Moreover, given the non-linear nature of ALE, classical concepts from random field theory should not hold in the case of inference on ALE analyses as the distribution of ALE scores does not follow classical formulations for random fields based on F- / T- or Chi2-statistics.
Given these limitations prohibiting the application of random field theory, we here propose to derive empirical thresholds for cluster-level correction based on a randomisation procedure. The main advantage of this approach is its potential to provide a reliable estimation of the null-distribution of topological features of the excursion set without necessitating assumptions on the nature of the statistical field or its analytical description. The datasets derived from the random relocation of coordinates are based on the same number of individual foci as well as the same size of the FWHM as the original data and are processed by the same algorithm for the computation of ALE maps and uncorrected thresholding. This approach should reflect the topology of the statistical field in the absence of true convergence, allowing the estimation of null-distributions for cluster-sizes in the excursion set as well maximum ALE scores, which can be applied for multiple-comparison corrected thresholding of the real data.
In this context, it is interesting to note that while the current revision replaces the previously applied permutation procedure for the estimation of voxel-level significance, it introduces a randomisation approach for correcting the inference for multiple comparisons. While this may sound illogical at first, these two changes are closely dependent on each other. By deriving the null-distribution of ALE-scores (and hence uncorrected thresholds) analytically, the computation of thresholded ALE maps from a set of (real or randomly relocated) foci becomes expedient enough to allow for the simulation of noise datasets within a reasonable time. That is, cluster-level and voxel-wise FWE thresholding of ALE datasets depend on a randomisation procedure which only becomes feasible through the replacement of permutation based approaches for deriving uncorrected voxel-wise p-values by a considerably faster analytical solution.
Apart from the integration of fMRI and PET data, ALE (Nickl-Jockschat et al., 2011a, b; Schroeter et al., 2007) and SDM (Radua et al., 2010; Radua and Mataix-Cols, 2009) have also been repeatedly used to summarize findings from voxel-based morphometry (VBM) studies. Given that VBM studies report grey matter differences in form of peak coordinates, cluster-level correction may analogously be applied on ALE of VBM data. It is, however, a topic of debate whether cluster-based inference is at all conceptually appropriate for VBM data (Ashburner and Friston, 2000).
Cluster-level inference in coordinate-based meta-analyses
Some potentially important conceptual caveats of cluster-level inference on any coordinate-based meta-analysis, including ALE, should not go unnoted. First, above-threshold cluster size increases when more studies report foci near each other, yet it decreases when the correspondence between those foci improves as their Gaussians will overlap more tightly. Counter-intuitively, a better convergence (closer proximity) of foci from different experiments may thus leads to a reduction in cluster-size. Moreover, the width of the Gaussians, modeling the uncertainty of each focus, is inversely related to (the square root of) the sample size of the original experiment. Consequently, convergence between experiments with fewer subjects may lead to more extensive, and hence significant, clusters than the same convergence between an equivalent number of experiment with large sample sizes. Finally, even though the modification presented by Turkeltaub et al. (2011) corrects for the effects of within-experiment clustering on the MA values of each voxel, the extent of high values in the ensuing MA, and hence ensuing ALE maps, may still be influenced by the amount of closely co-localized, i.e., clustered, foci in a particular experiment. Consequently, cluster extent thresholding may seem to reintroduce the recently addressed effects of within-experiment clustering of foci.
Taken together, these reflections might converge to the notion that cluster-extent thresholding may allow voxels with relatively low probabilities of representing true convergence between experiments to become significant if they are distributed enough by virtue of less tight correspondence, smaller sample sizes or within-experiment clustering of foci. However, the relevance of such clusters in which most, if not all, voxels feature only moderately high ALE values and hence significance, evidently has to be questioned.
Indeed, it should be noted that most of these theoretical concerns may not be practically relevant in standard ALE analyses, especially when performed with sufficiently high cluster-forming thresholds. First, although in the case of close proximity between foci from different experiments the overall extent of the cluster will be lower than in the case of more disperse foci, the former scenario will in turn yield a larger area of high ALE values given the better overlap of higher probability values close to the centers of the respective Gaussians. If the cluster-forming threshold is sufficiently high, closer proximity between foci from different experiments should thus yield larger not smaller above-threshold clusters. Second, while experiments featuring a lower number of subjects and hence potentially larger clusters, it should be noted that the ALE values throughout these clusters will be lower given the lower probability values due to wider Gaussians. Overlap between experiments featuring low numbers of subjects will thus only become extendedly above-threshold if either there is a convergence across a higher number of experiments (which should be biologically relevant) or a low cluster-forming threshold has been used (which should increase the likelihood of observing larger spurious clusters). Third, clustering of foci within a particular experiment may indeed increase the size of above-threshold clusters if other experiments also show activation within the same general region. On the other hand, however, a high number of foci and hence higher values in the MA map will also affect the null-distribution for inference on the ensuing ALE map and generally reduce significance of the respective ALE values.
If not used with extremely liberal cluster-forming thresholds, extent-thresholding may therefore represent a rational and unbiased way of setting a cluster threshold after an appropriate voxel-level threshold is has been applied. Moreover, cluster-level thresholding seems to provide a better balance between sensitivity and specificity than the highly conservative voxel-level FWE correction, as illustrated by the presented exemplary analysis. In summary, cluster-level inference may thus represent a compromise between uncorrected thresholding with additional arbitrary extent-filters and voxel-level corrected inference. In light of the above considerations, however, an exhaustive assessment of the behavior of cluster-level corrected thresholds under different levels of correspondence (proximity) between peaks of different experiments, different amount of within-experiment clustering of peaks, different sample sizes and different cluster-forming thresholds is highly warranted, yet far beyond the scope of the present paper.
Conclusions
The present revision of the activation likelihood estimation (ALE) algorithm was aimed at improving two aspects of this method. First, we showed how an analytical solution based on histogram permutation might provide a faster and more precise approach to computing the null-distribution of ALE scores under the assumption of spatial independence. Secondly, we outlined a framework for correcting for multiple comparison correction in the inference on ALE data, which accommodates the spatially contiguous nature of the underlying signal. As this framework has to deal with non-linear data, it is necessarily dependent of a permutation test. The application of such a permutation could only be facilitated by the fast analytical solution for computing the distribution of ALE-values for all permutations. We conclude that cluster-level thresholding is the most appropriate replacement for thresholding approaches based on uncorrected inference or FDR correction. In light of these advances, the revised ALE algorithm will provide an improved tool for conducting coordinate-based meta-analyses on functional imaging data, which in turn should influence the growing importance of summarising the multitude of results obtained by neuroimaging research.
Highlights.
The permutation procedure of ALE is replaced by faster and more accurate approach
Family-wise error correction and cluster-level inference are introduced into ALE
The current and revised implementation of ALE yield comparable results
Acknowledgements
We acknowledge funding by the Human Brain Project (R01-MH074457-01A1; PTF, ARL, SBE), the DFG (IRTG 1328; SBE, DB) and the Helmholtz Initiative on Systems-Biology “The Human Brain Model” (SBE).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Amunts K, Weiss PH, Mohlberg H, Pieperhoff P, Eickhoff S, Gurd JM, Marshall JC, Shah NJ, Fink GR, Zilles K. Analysis of neural mechanisms underlying verbal fluency in cytoarchitectonically defined stereotaxic space--the roles of Brodmann areas 44 and 45. Neuroimage. 2004;22:42–56. doi: 10.1016/j.neuroimage.2003.12.031. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Voxel-based morphometry - The methods. Neuroimage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate-a practical and powerful approach to multiple testing. J.R.Stat.Soc., B.Methodol. 1995;57:289–300. [Google Scholar]
- Benuzzi F, Pugnaghi M, Meletti S, Lui F, Serafini M, Baraldi P, Nichelli P. Processing the socially relevant parts of faces. Brain Res.Bull. 2007;74:344–356. doi: 10.1016/j.brainresbull.2007.07.010. %19. [DOI] [PubMed] [Google Scholar]
- Bird G, Catmur C, Silani G, Frith C, Frith U. Attention does not modulate neural responses to social stimuli in autism spectrum disorders. Neuroimage. 2006;31:1614–1624. doi: 10.1016/j.neuroimage.2006.02.037. [DOI] [PubMed] [Google Scholar]
- Bonner-Jackson A, Haut K, Csernansky JG, Barch DM. The influence of encoding strategy on episodic memory and cortical activity in schizophrenia. Biol.Psychiatry. 2005;58:47–55. doi: 10.1016/j.biopsych.2005.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braver TS, Barch DM, Kelley WM, Buckner RL, Cohen NJ, Miezin FM, Snyder AZ, Ollinger JM, Akbudak E, Conturo TE, Petersen SE. Direct comparison of prefrontal cortex regions engaged by working and long-term memory tasks. Neuroimage. 2001;14:48–59. doi: 10.1006/nimg.2001.0791. [DOI] [PubMed] [Google Scholar]
- Britton JC, Taylor SF, Sudheimer KD, Liberzon I. Facial expressions and complex IAPS pictures: common and differential networks. Neuroimage. 2006;31:906–919. doi: 10.1016/j.neuroimage.2005.12.050. [DOI] [PubMed] [Google Scholar]
- Caspers S, Eickhoff SB, Geyer S, Scheperjans F, Mohlberg H, Zilles K, Amunts K. The human inferior parietal lobule in stereotaxic space. Brain Struct.Funct. 2008;212:481–495. doi: 10.1007/s00429-008-0195-z. [DOI] [PubMed] [Google Scholar]
- Chumbley JR, Friston KJ. False discovery rate revisited: FDR and topological inference using Gaussian random fields. Neuroimage. 2009;44:62–70. doi: 10.1016/j.neuroimage.2008.05.021. [DOI] [PubMed] [Google Scholar]
- Dapretto M, Davies MS, Pfeifer JH, Scott AA, Sigman M, Bookheimer SY, Iacoboni M. Understanding emotions in others: mirror neuron dysfunction in children with autism spectrum disorders. Nat.Neurosci. 2006;9:28–30. doi: 10.1038/nn1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denslow S, Lomarev M, George MS, Bohning DE. Cortical and subcortical brain effects of transcranial magnetic stimulation (TMS)-induced movement: an interleaved TMS/functional magnetic resonance imaging study. Biol.Psychiatry. 2005;57:752–760. doi: 10.1016/j.biopsych.2004.12.017. [DOI] [PubMed] [Google Scholar]
- Dolcos F, McCarthy G. Brain systems mediating cognitive interference by emotional distraction. J.Neurosci. 2006;26:2072–2079. doi: 10.1523/JNEUROSCI.5042-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff SB, Laird AR, Grefkes C, Wang LE, Zilles K, Fox PT. Coordinate-Based Activation Likelihood Estimation Meta-Analysis of Neuroimaging Data: A Random-Effects Approach Based on Empirical Estimates of Spatial Uncertainty. Hum Brain Mapp. 2009;30:2907–2926. doi: 10.1002/hbm.20718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans AC, Kamber M, Collins DL, MacDonald D. An MRI based probabilistic atlas of neuroanatomy. In: Shorvon S, Fish D, Andermann F, Bydder GM, editors. Magnetic Resonance Scanning and Epilepsy. 1994. pp. 263–274. [Google Scholar]
- Genovese CR, Lazar NA, Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage. 2002;15:870–878. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- Hasson U, Levy I, Behrmann M, Hendler T, Malach R. Eccentricity bias as an organizing principle for human high-order object areas. Neuron. 2002;34:479–490. doi: 10.1016/s0896-6273(02)00662-1. [DOI] [PubMed] [Google Scholar]
- Holmes AP, Blair RC, Watson JD, Ford I. Nonparametric analysis of statistic images from functional mapping experiments. J.Cereb.Blood Flow Metab. 1996;16:7–22. doi: 10.1097/00004647-199601000-00002. [DOI] [PubMed] [Google Scholar]
- Holt DJ, Kunkel L, Weiss AP, Goff DC, Wright CI, Shin LM, Rauch SL, Hootnick J, Heckers S. Increased medial temporal lobe activation during the passive viewing of emotional and neutral facial expressions in schizophrenia. Schizophr.Res. 2006;82:153–162. doi: 10.1016/j.schres.2005.09.021. [DOI] [PubMed] [Google Scholar]
- Hope ACA. A Simplified Monte Carlo Significance Test Procedure. J R Stat Soc Series B Stat Methodol. 1968;30:582–598. [Google Scholar]
- Kesler-West ML, Andersen AH, Smith CD, Avison MJ, Davis CE, Kryscio RJ, Blonder LX. Neural substrates of facial emotion processing using fMRI. Brain Res.Cogn Brain Res. 2001;11:213–226. doi: 10.1016/s0926-6410(00)00073-2. [DOI] [PubMed] [Google Scholar]
- Kiebel S, Holmes AP. The General Linear Model. In: Frackowiak RS, Friston KJ, Frith CD, Dolan RJ, Price CJ, Ashburner J, Penny WD, Zeki S, editors. Human Brain Function. 2 ed. Academic Press; 2003. pp. 725–760. [Google Scholar]
- Kranz F, Ishai A. Face perception is modulated by sexual preference. Curr.Biol. 2006;16:63–68. doi: 10.1016/j.cub.2005.10.070. [DOI] [PubMed] [Google Scholar]
- Kringelbach ML, Rolls ET. Neural correlates of rapid reversal learning in a simple model of human social interaction. Neuroimage. 2003;20:1371–1383. doi: 10.1016/S1053-8119(03)00393-8. [DOI] [PubMed] [Google Scholar]
- Laird AR, Eickhoff SB, Kurth F, Fox PM, Uecker AM, Turner JA, Robinson JL, Lancaster JL, Fox PT. ALE Meta-Analysis Workflows Via the Brainmap Database: Progress Towards A Probabilistic Functional Brain Atlas. Front Neuroinformatics. 2009a;3:23. doi: 10.3389/neuro.11.023.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird AR, Eickhoff SB, Li K, Robin DA, Glahn DC, Fox PT. Investigating the Functional Heterogeneity of the Default Mode Network Using Coordinate-Based Meta-Analytic Modeling. J Neurosci. 2009b;29:14496–14505. doi: 10.1523/JNEUROSCI.4004-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird AR, Fox PM, Price CJ, Glahn DC, Uecker AM, Lancaster JL, Turkeltaub PE, Kochunov P, Fox PT. ALE Meta-Analysis: Controlling the False Discovery Rate and Performing Statistical Contrasts. Hum Brain Mapp. 2005;25:155–164. doi: 10.1002/hbm.20136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malikovic A, Amunts K, Schleicher A, Mohlberg H, Eickhoff SB, Wilms M, Palomero-Gallagher N, Armstrong E, Zilles K. Cytoarchitectonic analysis of the human extrastriate cortex in the region of V5/MT+: A probabilistic, stereotaxic map of area hOc5. Cereb Cortex. 2007;17:562–574. doi: 10.1093/cercor/bhj181. [DOI] [PubMed] [Google Scholar]
- Nichols T, Hayasaka S. Controlling the familywise error rate in functional neuroimaging: a comparative review. Stat.Methods Med.Res. 2003;12:419–446. doi: 10.1191/0962280203sm341ra. [DOI] [PubMed] [Google Scholar]
- Paller KA, Ranganath C, Gonsalves B, LaBar KS, Parrish TB, Gitelman DR, Mesulam MM, Reber PJ. Neural correlates of person recognition. Learn.Mem. 2003;10:253–260. doi: 10.1101/lm.57403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny WD, Holmes AP. Random effects analysis. In: Frackowiak RS, Friston KJ, Frith CD, Dolan RJ, Price CJ, Ashburner J, Penny WD, Zeki S, editors. Human Brain Function. 2 ed. Academic Press; 2003. pp. 843–850. [Google Scholar]
- Pierce K, Haist F, Sedaghat F, Courchesne E. The brain response to personally familiar faces in autism: findings of fusiform activity and beyond. Brain. 2004;127:2703–2716. doi: 10.1093/brain/awh289. [DOI] [PubMed] [Google Scholar]
- Platek SM, Loughead JW, Gur RC, Busch S, Ruparel K, Phend N, Panyavin IS, Langleben DD. Neural substrates for functionally discriminating self-face from personally familiar faces. Hum Brain Mapp. 2006;27:91–98. doi: 10.1002/hbm.20168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack RA, Fletcher PC, Henson RN, Worsley KJ, Brett M, Nichols TE. Guidelines for reporting an fMRI study. Neuroimage. 2008;40:409–414. doi: 10.1016/j.neuroimage.2007.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CJ, Devlin JT, Moore CJ, Morton C, Laird AR. Meta-analyses of object naming: effect of baseline. Hum Brain Mapp. 2005;25:70–82. doi: 10.1002/hbm.20132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radua J, Mataix-Cols D. Voxel-wise meta-analysis of grey matter changes in obsessive-compulsive disorder. Br.J.Psychiatry. 2009;195:393–402. doi: 10.1192/bjp.bp.108.055046. [DOI] [PubMed] [Google Scholar]
- Radua J, van den Heuvel OA, Surguladze S, Mataix-Cols D. Meta-analytical comparison of voxel-based morphometry studies in obsessive-compulsive disorder vs other anxiety disorders. Arch.Gen.Psychiatry. 2010;67:701–711. doi: 10.1001/archgenpsychiatry.2010.70. [DOI] [PubMed] [Google Scholar]
- Raemaekers M, Vink M, Zandbelt B, van Wezel RJ, Kahn RS, Ramsey NF. Test-retest reliability of fMRI activation during prosaccades and antisaccades. Neuroimage. 2007 doi: 10.1016/j.neuroimage.2007.03.061. [DOI] [PubMed] [Google Scholar]
- Schroeter ML, Raczka K, Neumann J, Yves v.C. Towards a nosology for frontotemporal lobar degenerations-a meta-analysis involving 267 subjects. Neuroimage. 2007;36:497–510. doi: 10.1016/j.neuroimage.2007.03.024. [DOI] [PubMed] [Google Scholar]
- Turkeltaub PE, Eden GF, Jones KM, Zeffiro TA. Meta-analysis of the functional neuroanatomy of single-word reading: method and validation. Neuroimage. 2002;16:765–780. doi: 10.1006/nimg.2002.1131. [DOI] [PubMed] [Google Scholar]
- Turkeltaub PE, Eickhoff SB, Laird AR, Fox M, Wiener M, Fox P. Minimizing within-experiment and within-group effects in activation likelihood estimation meta-analyses. Human Brain Mapp. 2011 doi: 10.1002/hbm.21186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuilleumier P, Armony JL, Driver J, Dolan RJ. Effects of attention and emotion on face processing in the human brain: an event-related fMRI study. Neuron. 2001;30:829–841. doi: 10.1016/s0896-6273(01)00328-2. [DOI] [PubMed] [Google Scholar]
- Wager TD, Barrett LF, Bliss-Moreau E. The neuroimaging of emotion. In: Lewis M, editor. Handbook of Emotion. 2007a. [Google Scholar]
- Wager TD, Lindquist M, Kaplan L. Meta-analysis of functional neuroimaging data: current and future directions. Soc.Cogn Affect.Neurosci. 2007b;2:150–158. doi: 10.1093/scan/nsm015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wager TD, Smith EE. Neuroimaging studies of working memory: a meta-analysis. Cogn Affect.Behav.Neurosci. 2003;3:255–274. doi: 10.3758/cabn.3.4.255. [DOI] [PubMed] [Google Scholar]
- Wild B, Erb M, Eyb M, Bartels M, Grodd W. Why are smiles contagious? An fMRI study of the interaction between perception of facial affect and facial movements. Psychiatry Res. 2003;123:17–36. doi: 10.1016/s0925-4927(03)00006-4. [DOI] [PubMed] [Google Scholar]
- Williams MA, McGlone F, Abbott DF, Mattingley JB. Differential amygdala responses to happy and fearful facial expressions depend on selective attention. Neuroimage. 2005;24:417–425. doi: 10.1016/j.neuroimage.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Wilms M, Eickhoff SB, Specht K, Amunts K, Shah NJ, Malikovic A, Fink GR. Human V5/MT+: comparison of functional and cytoarchitectonic data. Anat.Embryol.(Berl) 2005;210:485–495. doi: 10.1007/s00429-005-0064-y. [DOI] [PubMed] [Google Scholar]
- Worsley KJ. Developments in Random Field Theory. In: Frackowiak RS, Friston KJ, Frith CD, Dolan RJ, Price CJ, Ashburner J, Penny WD, Zeki S, editors. Human Brain Function. 2 ed. Academic Press; 2003. pp. 881–886. [Google Scholar]
- Worsley KJ, Marrett S, Neelin P, Vandal AC, Friston KJ, Evans AC. A unified statistical approach for determining significant signals in images of cerebral activation. Human Brain Mapp. 1996;4:58–74. doi: 10.1002/(SICI)1097-0193(1996)4:1<58::AID-HBM4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]