Abstract
A fundamental component of brain development is the formation of large-scale networks across the cortex. One such network, the default network, undergoes a protracted development, displaying weak connectivity in childhood that strengthens in adolescence and becomes most robust in adulthood. Little is known about the genetic contributions to default network connectivity in adulthood or during development. Alterations in connectivity between posterior and frontal portions of the default network have been associated with several psychological disorders, including anxiety, autism spectrum disorders, schizophrenia, depression, and attention-deficit/hyperactivity disorder. These disorders have also been linked to variants of the serotonin transporter linked polymorphic region (5-HTTLPR). The LA allele of 5-HTTLPR results in higher serotonin transporter expression than the S allele or the rarer LG allele. 5-HTTLPR may influence default network connectivity, as the superior medial frontal region has been shown to be sensitive to changes in serotonin. Also, serotonin as a growth factor early in development may alter large-scale networks such as the default network. The present study examined the influence of 5-HTTLPR variants on connectivity between the posterior and frontal structures and its development in a cross-sectional study of 39 healthy children and adolescents. We found that children and adolescents homozygous for the S allele (S/S, n = 10) showed weaker connectivity in the superior medial frontal cortex compared to those homozygous for the LA allele (LA/LA, n = 13) or heterozygotes (S/LA, S/LG, n = 16). Moreover, there was an age-by-genotype interaction, such that those with LA/LA genotype had the steepest age-related increase in connectivity between the posterior hub and superior medial frontal cortex, followed by heterozygotes. In contrast, individuals with the S/S genotype had the least age-related increase in connectivity strength. This preliminary report expands our understanding of the genetic influences on the development of large-scale brain connectivity and lays down the foundation for future research and replication of the results with a larger sample.
Keywords: default network, serotonin transporter, genetics, functional connectivity, MRI, self-organizing map
1. Introduction
There is significant interest in examining the functioning and development of large-scale neural networks, such as the default network, as alterations in these systems have been linked to psychopathology (Buckner et al., 2008). Functional connectivity of the default network increases in the absence of a driving task (i.e., during “rest”) and decreases during engagement in a cognitively demanding task (Buckner and Vincent, 2007; Fox et al., 2005; Raichle and Snyder, 2007). Structures involved in the default network include the posterior cingulate, precuneus, retrosplenial, inferior parietal lobule, superior temporal gyrus, parahippocampal gyrus, medial frontal cortex, and superior frontal cortex (Buckner et al., 2008; Fox et al., 2005; Greicius et al., 2003; Shulman et al., 1997). In healthy adults, default network structures are functionally as well as structurally connected (Greicius et al., 2003; Greicius et al., 2009; van den Heuvel et al., 2008). The default network consists of several interconnected subsystems (Buckner et al., 2008; Gusnard and Raichle, 2001), including posterior and frontal subsystems that are generally strongly connected yet distinct from one another (Greicius et al., 2008; Horovitz et al., 2009). Long-range functional connectivity between the posterior hub and anterior regions of the default network, such as the superior medial frontal cortex, is of central interest because connectivity between these two regions is predictive of individual differences in cognitive performance (Hampson et al., 2006), attentional processes (Wang et al., 2006), and psychopathology symptoms (see Buckner et al., 2008 for a review).
Although there is debate as to the default network’s primary function, this network may relate to basic central nervous system functions such as maintaining the balance of excitatory and inhibitory inputs or maintaining and interpreting information from the environment (Raichle and Snyder, 2007). Energy consumption for intrinsic activation of the default network is large compared to typical task related responses (Raichle and Mintun, 2006). Additionally, default network activation persists across differing states of consciousness, including light sleep (Horovitz et al., 2008), deep sleep (albeit split into posterior and anterior subsystems, Horovitz et al., 2009), and under anesthesia (Vincent et al., 2007). The large amount of resources devoted to this system and its ubiquity suggests that the default network is a fundamental brain network. Consequently, identifying factors that may influence variation in the development of this network’s connectivity is of interest.
Imaging genetics is an approach that has proven useful in uncovering influences on important brain networks. In this approach, genetic information is linked to individual variation in brain imaging in order to identify neural networks influenced by genetics and important for normal and abnormal development (Meyer-Lindenberg, 2009). One genetic variant that has been of particular interest in imaging genetics studies is the serotonin transporter-linked polymorphic region (5-HTTLPR) (e.g., Hariri et al., 2002). 5-HTTLPR is a variable number of tandem repeats (VNTR) in the promoter region of the serotonin transporter gene (SLC6A4) that presents as short (S) and long (L) alleles (Lesch et al., 1996). Within the L allele, a single nucleotide polymorphism (A to G substitution, SNP rs25531) generates the LA and LG alleles (Hu et al., 2006). The LA allele results in increased transcriptional efficiency and serotonin transporter expression relative to the S allele and the rarer LG allele (Hu et al., 2006). 5-HTTLPR has been conceptualized as a risk or plasticity genetic variant that contributes to variability in outcomes of psychopathology (Belsky et al., 2009).
There are several reasons to investigate the potential influence of 5-HTTLPR variants on long-range posterior to anterior default network connectivity, particularly in the anterior default network region of the superior medial frontal cortex. First, 5-HTTLPR variants and long-range default network connectivity have been linked to multiple forms of psychopathology. Specifically, 5-HTTLPR variants and altered default network connectivity between the posterior hub and superior medial frontal cortex are both implicated in anxiety (e.g., Liao et al., 2010; Schinka et al., 2004; Sen et al., 2004), depression (e.g., Drevets et al., 2008; Greicius et al., 2007; Zalsman et al., 2006), attention-deficit/hyperactivity disorder (e.g., Castellanos et al., 2008; Manor et al., 2001), autism symptoms (e.g., Brune et al., 2006; Nijmeijer et al., 2010; Wiggins et al., 2011), and psychosis symptoms (e.g., Borroni et al., 2006b; Ezaki et al., 2008; Garrity et al., 2007). Although there is considerable debate, the higher expressing LA allele has generally been implicated as a protective factor compared to the S allele in these and similar studies (particularly in depression, see Levinson, 2006).
Second, among the anterior default network regions that show connectivity with the posterior hub, evidence suggests that the superior medial frontal cortex/Brodmann’s Area (BA) 10, a region implicated in clinical samples (e.g., Castellanos et al., 2008; Garrity et al., 2007; Monk et al., 2009; Weng et al., 2010; Wiggins et al., 2011), may be sensitive to alterations in the serotonin system. For example, following administration of a selective serotonin reuptake inhibitor, healthy adults with the S allele have sharper decreases in cerebral metabolic response in the superior medial frontal cortex compared to those homozygous for the L allele (Smith et al., 2004). Moreover, in the superior medial frontal cortex, adults with the S allele show greater activation levels during a Stroop task as well as larger gray matter volume (Canli et al., 2005). In addition, serotonin transporter binding in the superior medial frontal cortex/BA 10 is altered in individuals with autism (Nakamura et al., 2010) and depression (Mann et al., 2000). Finally, following acute tryptophan depletion (a technique that lowers serotonin levels), resting-state activation in BA 10 decreases (Kunisato et al., 2011). These studies indicate that the superior medial frontal cortex/BA 10 is particularly sensitive to serotonin signaling and potentially to the 5-HTTLPR variant.
Third, serotonin acts as a growth factor during embryogenesis. Serotonin alters the development of fundamental neural networks, including the default network (Sodhi and Sanders-Bush, 2004). The prominence of serotonin in shaping neural networks in early development is consistent with prior work documenting the influence of 5-HTTLPR on the amygdala-ventromedial prefrontal cortex network during social tasks (Hariri et al., 2006) and during rest (Rao et al., 2007). Taken together, these studies suggest that 5-HTTLPR variants may contribute to individual variability in default network connectivity between the posterior hub and superior medial frontal cortex during rest. However, to our knowledge, no study has examined how this genetic variant impacts default network connectivity.
A few studies have investigated the development of the default network. For healthy individuals, long-range functional connectivity in the default network between the posterior hub and the superior medial frontal cortex is weaker during childhood and adolescence compared to adulthood (Fair et al., 2008; Stevens et al., 2009; Wiggins et al., 2011). A study of structural connections between the posterior hub and the superior medial frontal region also revealed weaker connectivity in children versus young adults (Supekar et al., 2010). These studies indicate that connectivity of this network undergoes a protracted developmental time course. However, genetic influences on the development of the default network have not yet been studied.
The present study addresses these two gaps in the literature, 5-HTTLPR’s role in default network connectivity and its development, by directly examining the influence of 5-HTTLPR variants on long-range default network connectivity as well as its developmental time course in a healthy child and adolescent sample. Based on previous research, we hypothesized that youth with the LA/LA genotype have the strongest connectivity between the default network posterior hub and superior medial frontal cortex, whereas youth with the S/S genotype have the weakest connectivity, and other genotypes (S/LA, S/LG, LG/LG, LA/LG) display connectivity strength intermediate between LA/LA and S/S. We also hypothesized that developmental changes in the strength of connectivity between the posterior hub and superior medial frontal cortex differ by genotype group.
2. Materials and Methods
2.1 Participants
Thirty-nine healthy children and adolescents aged 8 to 19 years old were included in this study. These participants are from a larger dataset comprised of 67 individuals. Because genotype frequencies vary by ancestry (e.g., higher S allele frequency in Asian samples compared to Caucasian samples; Ha et al., 2005; Kato et al., 2005; Kim et al., 2007) and can contribute to spurious associations (Pritchard & Rosenberg, 1999), data from 17 non-Caucasian individuals were excluded in this particular study (4 of whom also had excessive movement in the scanner). Limiting the study to participants of Caucasian descent is an approach that has been used with 5-HTTLPR to address population stratification (e.g., Praschak-Rieder et al., 2007; Rao et al., 2007; Zalsman et al., 2006). An additional 11 individuals were excluded from the analyses due to movement greater than 2.5 mm or discomfort in the MRI resulting in an incomplete scan. Participants, ages 8 – 19 years old, were recruited through flyers posted at local community organizations. The University of Michigan Institutional Review Board approved the procedures, and all participants signed consents. All participants above age 18 gave written consent; all minor participants gave assent and their parents gave written consent. To be included, participants must not have had orthodontic braces, any other condition contraindicated for MRI, history of seizures or any neurological disorders. Additionally, participants were screened for psychopathology with parent report (Child Behavior Checklist; Achenbach and Edelbrock, 1981; Social Responsiveness Scale; Constantino et al., 2003; Social Communication Questionnaire; Rutter et al., 2003) as well as self-report (Obsessive Compulsive Inventory - Revised; Foa et al., 2010; Child Depression Inventory; Kovacs, 1992; Multidimensional Anxiety Scale for Children; March et al., 1997; Spence Children’s Anxiety Scales; Spence, 1997) measures. Only children who scored below cutoffs on all of these measures were allowed to participate. Details on these subject characteristics are in Table 1. Twenty-nine participants from Wiggins et al (2011) were included in the present study.
Table 1.
Sample characteristics.
| Caucasian only | S/S | LA/LA | Intermediate Genotypes | F2,36 | p | |||
|---|---|---|---|---|---|---|---|---|
| S/LA | S/LG | LG/LG | LA/LG | |||||
| Number of participants | 10 | 13 | 14 | 2 | 0 | 0 | ||
| Gender (M:F) | 7:3 | 10:3 | 12:4 | |||||
| Handedness* (R:L) | 8:2 | 13:0 | 12:1 | |||||
| Age, mean (SD) | 15.1 (2.29) | 14.4 (3.19) | 14.3 (3.23) | .241 | .787 | |||
| Pubertal Development Scale, mean (SD) | 3.08 (.694) | 2.34 (1.03) | 2.61 (.914) | 1.91 | .163 | |||
| Verbal cognitive functioning, mean (SD) | 117 (12.0) | 118 (13.5) | 115 (12.0) | .152 | .859 | |||
| Nonverbal cognitive functioning, mean (SD) | 104 (9.27) | 105 (9.94) | 104 (12.6) | .015 | .985 | |||
| Spence Children’s Anxiety Scale (SCAS), mean (SD) | 13.1 (5.55) | 15.7 (8.91) | 14.5 (8.78) | .288 | .752 | |||
| Multidimensional Anxiety Scale for Children (MASC), mean (SD) | 34.0 (14.6) | 29.9 (11.0) | 31.5 (19.4) | .174 | .841 | |||
| Obsessive Compulsive Inventory – Revised (OCI-R), mean (SD) | 8.90 (6.49) | 10.1 (8.19) | 10.0 (9.20) | .071 | .932 | |||
| Children’s Depression Inventory (CDI), mean (SD) | 6.60 (4.58) | 5.92 (7.30) | 4.00 (4.68) | .763 | .474 | |||
| Social Communication Questionnaire – Lifetime (SCQ), mean (SD) | 3.40 (2.99) | 1.85 (3.05) | 3.63 (3.61) | 1.17 | .323 | |||
| Social Responsiveness Scale (SRS), mean (SD) | 43.9 (10.1) | 40.4 (5.32) | 43.5 (7.49) | .805 | .455 | |||
| Child Behavior Checklist (CBCL) – Total, mean (SD) | 14.1 (12.7) | 11.3 (7.27) | 13.6 (10.9) | .259 | .773 | |||
| CBCL – Internalizing subscale, mean (SD) | 2.60 (2.41) | 3.56 (3.32) | 4.06 (3.97) | .601 | .554 | |||
| CBCL – Externalizing subscale, mean (SD) | 4.00 (3.68) | 2.08 (2.47) | 3.56 (4.69) | .853 | .434 | |||
| All Ancestries | S/S | LA/LA | Intermediate Genotypes | F2,49 | p | |||
| S/LA | S/LG | LG/LG | LA/LG | |||||
| Number of participants | 17 | 16 | 15 | 3 | 1 | 0 | ||
| Race/Ethnicity | ||||||||
| Asian | 2 | 0 | 0 | 0 | 0 | 0 | ||
| African American | 1 | 2 | 1 | 0 | 1 | 0 | ||
| Hispanic/Latino(a) | 2 | 1 | 0 | 1 | 0 | 0 | ||
| Multi-racial/multi-ethnic | 2 | 0 | 0 | 0 | 0 | 0 | ||
| Caucasian | 10 | 13 | 14 | 2 | 0 | 0 | ||
| Gender (M:F) | 13:4 | 12:4 | 15:4 | |||||
| Handedness* (R:L) | 14:3 | 16:0 | 14:2 | |||||
| Age, mean (SD) | 15.0 (2.36) | 14.5 (3.29) | 14.2 (3.11) | .315 | .731 | |||
| Pubertal Development Scale, mean (SD) | 2.99 (.820) | 2.44 (1.01) | 2.57 (.892) | 1.67 | .198 | |||
| Verbal cognitive functioning, mean (SD) | 112 (13.4) | 115 (13.9) | 114 (12.4) | .166 | .847 | |||
| Nonverbal cognitive functioning, mean (SD) | 105 (10.4) | 103 (12.3) | 103 (13.6) | .236 | .791 | |||
| SCAS, mean (SD) | 15.4 (8.43) | 15.6 (8.5) | 14.6 (8.57) | .067 | .936 | |||
| MASC, mean (SD) | 33.9 (11.7) | 31.3 (10.0) | 31.9 (19.6) | .136 | .873 | |||
| OCI-R, mean (SD) | 10.8 (7.40) | 10.8 (8.06) | 10.1 (9.80) | .037 | .964 | |||
| CDI, mean (SD) | 6.18 (4.25) | 5.15 (5.12) | 4.11 (4.36) | .737 | .484 | |||
| SCQ, mean (SD) | 2.94 (2.77) | 2.00 (2.76) | 3.61 (3.68) | 1.13 | .330 | |||
| SRS, mean (SD) | 43.7 (8.30) | 41.0 (5.56) | 42.6 (6.66) | .605 | .550 | |||
| CBCL – Total, mean (SD) | 13.4 (12.0) | 11.6 (6.85) | 11.4 (9.98) | .220 | .803 | |||
| CBCL – Internalizing subscale, mean (SD) | 2.88 (2.91) | 3.56 (2.87) | 3.28 (3.69) | .189 | .829 | |||
| CBCL – Externalizing subscale, mean (SD) | 3.53 (3.20) | 2.19 (2.48) | 3.06 (4.44) | .617 | .544 | |||
Note: 3 participants were missing handedness data; 4 were missing MASC scores; 2 were missing SCAS; 1 was missing SRS; and 1 was missing the CBCL.
2.2 Genetic Analyses
Participants donated a saliva sample using the Oragene DNA kit (DNA Genotek; Kanata, Canada). S versus L genotype of 5-HTTLPR was determined via PCR following Hu et al’s (2006) procedure with modifications (GC-Rich PCR system, Roche Diagnostics, Indianapolis, IN; forward primer: 5′-GGCGTTGCCGCTCTGAATGC-3′, reverse: 5′-GGGACTGAGCTGGACAACCAC-3′; DNA denatured at 95° for 5 min, followed by two cycles at 95° for 30 sec, 63° for 30 sec, 72° for 1 min; two cycles at 95° for 30 sec, 62° for 30 sec, 72° for 1 min; 36 cycles at 95° for 30 sec, 61° for 30 sec, 72° for 1 min; and final extension at 72° for 10 min). PCR products were then analyzed by agarose gel electrophoresis. DNA was purified from excised gel bands using the QIAquick Gel Extraction kit (Valencia, CA) and then Sanger sequenced to determine the A to G SNP in the L allele (rs25531) and to confirm agarose typing. Previous research has shown that the SNP rs25531 affects the activity level of the 5-HTTLPR variant such that only LA, and not LG, is the truly high expressing allele (Hu et al., 2006). Thus, for subsequent statistical analyses, participants were divided into three groups: homozygous for the L allele with A at rs25531 (denoted LA/LA), homozygous for the S allele (S/S), and heterozygous genotypes (S/LA or S/LG). There were no participants in this cohort with the relatively rare genotypes LA/LG and LG/LG (in Caucasians only; see Table 1 for genotype frequencies for all ancestries). This approach, making the primary comparison homozygotes for the LA allele and the S allele, has been used to illuminate functional brain differences in the past and maximizes the difference in serotonin transporter expression (Roiser et al., 2009). Genotype frequencies were in Hardy-Weinberg equilibrium (χ2 = 1.19, df = 1, p = .276).
2.3 fMRI Data Acquisition
MRI scanning occurred at University of Michigan’s 3 Tesla GE Signa MRI machine. Using a reverse spiral sequence, 300 T2*-weighted blood oxygen level dependent (BOLD) images were collected over 10 minutes for each participant (Glover and Law, 2001; TR=2000 ms, TE=30 ms, flip angle=90°, FOV=22 cm, 64×64 matrix, 40 contiguous axial 3 mm slices). Slices were acquired parallel to the intercommissural (AC-PC) line. A high-resolution 3D T1 axial overlay was acquired for anatomical localization (TR=8.9, TE=1.8, flip angle=15°, FOV=26 cm, slice thickness=1.4 mm, 124 slices; matrix=256 × 160). Additionally, a high-resolution SPGR image acquired in the sagittal plane (flip angle=15°, FOV=26cm, 1.4 mm slice thickness, 110 slices) was used for coregistration of the functional images.
2.4 fMRI Procedures
2.4.1 Participant Instructions
A fixation cross (“plus” symbol) was presented via the projection system to the participant in the MRI scanner while functional data were collected. Participants were instructed to let their minds wander and to not think about anything in particular while they looked at the cross.
2.4.2 Correction for Physiological Noise
Physiological data were collected for subsequent noise correction. An abdominal pressure belt recorded each participant’s respiration, and a pulse oximeter on the participant’s left middle finger recorded oxygenation. The physiological data were synchronized to the fMRI data.
2.5 fMRI Data Analysis
2.5.1 Data Preprocessing
The acquired fMRI data were preprocessed as part of the standard processing stream at the University of Michigan. First, spikes in the raw k-space data lying more than two standard deviations from the mean were replaced with the average of the neighboring time-points. Second, the k-space data were reconstructed to image space with a custom reconstruction program for gridding and inverse 2D Fourier transform. A field map correction was applied to reduce artifacts from susceptibility regions. Third, RETROICOR was utilized to remove noise associated with cardiac and respiratory rhythms (Glover et al., 2000). Fourth, images were corrected for differences in slice timing by phase-shifting and re-sampling the signal (Oppenheim et al., 1999), with the middle slice as the temporal reference point. Finally, the MCFLIRT program in FMRIB Software Library (Jenkinson et al., 2002) corrected for head motion by realigning all images to the tenth functional image. To further address potential differences in head motion among the genotype groups, as similar studies have done (Bunge, et al, 2002; Rubia et al., 1999), an index score was created for each individual by taking the grand mean of head movement measured in each of six rigid body movement modes (3 translation, 3 rotation). This head motion score was compared across genotype groups.
Additional pre-processing of the data was accomplished in-house using the SPM5 Matlab toolbox (Wellcome Department of Cognitive Neurology, London, UK; http://www.fil.ion.ucl.ac.uk). High-resolution T1 anatomical images were co-registered to the functional images. Spatial smoothing was accomplished with an isotropic 8 mm full width at half maximum (FWHM) Gaussian kernel. To exclude higher frequency sources of noise and to isolate the frequency band in which resting state connectivity has previously been observed in fMRI data, the time courses from each voxel were low-pass filtered with a 0.08 Hz cutoff (Biswal et al., 1995; Cordes et al., 2000).
2.5.2 Calculating Connectivity Using a Self-Organizing Map Algorithm
A self-organizing map algorithm was used to identify a data-driven seed for each individual. This seed was cross-correlated with the low-frequency time courses from every other voxel in the individual’s brain (Peltier et al., 2003; Wiggins et al., 2011). Briefly, the self-organizing map algorithm produced a map of exemplar time courses that represented the probability density function of the underlying data while preserving topological properties. In this iterative process, the 10 × 10 exemplar matrix was initialized with random noise. Then, for each iteration, a given voxel’s time course was compared to all the exemplar time courses. The exemplar identified as closest to the data voxel’s time course using a least squares metric, as well as neighboring exemplars, were then updated at each iteration until convergence was achieved. The final exemplar map had a topologically ordered feature map that represented the underlying probability density function of the data with minimal error (Kohonen, 1997). The end product of the self-organizing map algorithm was 16 superclusters representing networks in the brain. A full description of the self-organizing map as implemented for this paper can be found in Wiggins et al. (2011).
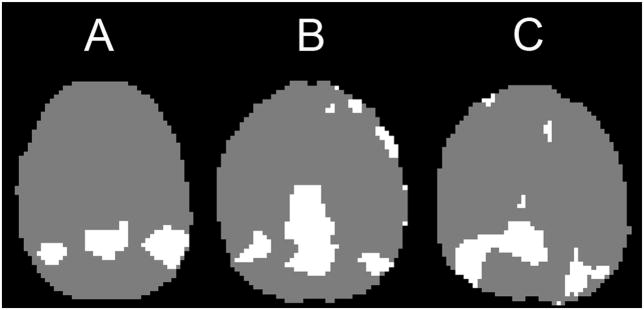
An experienced investigator blind to genotype examined each of the superclusters from the participants. The superclusters were visually compared to a map of the posterior portion of the default network, the posterior cingulate cortex/precuneus and the bilateral inferior parietal lobule, generated by Wake Forest University PickAtlas (Maldjian et al., 2002). The supercluster with structures most similar to the map of the posterior hub of the default network was identified for each participant (Buckner et al., 2008; Horovitz et al., 2009). Three examples from our dataset depicting the range of spatial arrangements of the posterior hubs are shown in Figure 1. All the low-pass filtered BOLD time courses from the voxels in this supercluster were then extracted and averaged to form a reference time course. This reference time course was correlated with all of the low-pass filtered voxels in the brain to form functional connectivity maps for each subject. Lastly, images were normalized to Montreal Neurological Image (MNI) space by estimating the transformation matrix for the SPGR anatomical image to an MNI template image in SPM5, then applying this transformation to the functional images. The Pearson’s r values at each voxel were converted to z values, using Fisher’s r to z transformation. These connectivity images were then used in random effects analyses.
Figure 1. Example posterior hubs from the self-organizing map algorithm.
Superclusters identified as the posterior hubs of the default network by an investigator blind to genotype from three different subjects in this study. This illustrates the range of the spatial arrangement of the posterior hubs across subjects. The majority of individuals had posterior hubs that were easy to recognize (as in A); others had moderately difficult-to-recognize posterior hubs (as in B). A few had difficult-to-recognize posterior hubs (as in C). Note: slices are from approximately the same transverse slice, but brains are not normalized to the same space at this point in the data processing stream.
2.5.3 Second-Level Analyses
Second-level analyses were performed on the images generated by the SOM analysis. We first confirmed that the default network connectivity was present in each of the three genotype groups (LA/LA, S/S, and heterozygous (S/LA and S/LG) genotypes). The default network was defined by the following left and right regions: superior medial frontal cortex, medial frontal gyrus, superior frontal gyrus, retrosplenial cortex, posterior cingulate cortex, precuneus, inferior parietal lobules, temporal lobes, and parahippocampal gyri (Beason-Held et al., 2009; Bluhm et al., 2009; Buckner et al., 2008; Gusnard and Raichle, 2001; Weng et al., 2010). Region of interest (ROI) masks were defined using the Wake Forest University PickAtlas (Maldjian et al., 2002). An ROI analysis was performed for each of the regions and significant clusters in these ROIs for each of the genotype groups are found in Table 2.
Table 2. Default network connectivity for each genotype group.
Functional connectivity in the (A) LA/LA group, (B) heterozygous group, and (C) S/S group between the posterior hub seed identified by the self-organizing map algorithm and other major default network regions. To document all possible connectivity in the default network, the threshold was set at p < 0.05 uncorrected with the number of contiguous voxels set at k ≥ 10. L = left, R = right. A full list of the default network structures used can be found in the Methods.
| (A). LA/LA group
| ||||||
|---|---|---|---|---|---|---|
| Region | Brodmann’s Area | Cluster size | t df = 12 | MNICoordinates | ||
| x | y | z | ||||
| L superior medial frontal cortex | 10 | 2313 | 7.90 | −4 | 72 | 10 |
| R superior medial frontal cortex | 10 | 1696 | 9.71 | 10 | 70 | 14 |
| L medial frontal gyrus | 9 | 1315 | 7.78 | −2 | 50 | 28 |
| 11 | 673 | 6.25 | −6 | 48 | −14 | |
| R medial frontal gyrus | 9 | 1239 | 8.69 | 10 | 54 | 24 |
| 11 | 663 | 6.68 | 2 | 52 | −16 | |
| 6 | 30 | 2.41 | 8 | −18 | 70 | |
| L superior frontal gyrus | 9 | 2788 | 7.56 | −4 | 52 | 28 |
| 11 | 15 | 3.07 | −2 | 56 | −20 | |
| 10 | 11 | 2.53 | −40 | 62 | 0 | |
| R superior frontal gyrus | 10 | 2513 | 9.71 | 10 | 70 | 14 |
| 11 | 23 | 3.98 | 4 | 56 | −20 | |
| 6 | 13 | 2.63 | 10 | −18 | 70 | |
| L retrosplenial cortex | 30 | 29 | 7.56 | −2 | −52 | 20 |
| R retrosplenial cortex | 30 | 36 | 7.10 | 2 | −54 | 20 |
| 30 | 23 | 3.02 | 4 | −60 | 10 | |
| L posterior cingulate | 31 | 585 | 8.60 | −4 | −52 | 24 |
| R posterior cingulate | 23 | 576 | 9.11 | 10 | −50 | 24 |
| L precuneus | 31 | 1656 | 14.43 | −2 | −50 | 36 |
| 19/7 | 129 | 7.83 | −40 | −72 | 42 | |
| R precuneus | 31 | 1776 | 18.43 | 4 | −50 | 34 |
| 39 | 55 | 4.21 | 42 | −68 | 34 | |
| L inferior parietal lobule | 39 | 578 | 11.86 | −44 | −66 | 40 |
| 13 | 60 | 4.26 | −44 | −50 | 24 | |
| R inferior parietal lobule | 40 | 345 | 4.64 | 54 | −62 | 38 |
| 13 | 22 | 3.37 | 44 | −50 | 24 | |
| L parahippocampal gyrus | 35 | 466 | 3.76 | −22 | −28 | −12 |
| R parahippocampal gyrus | 36 | 198 | 3.15 | 34 | −28 | −18 |
| L temporal lobe | 40 | 855 | 7.22 | −58 | −60 | 28 |
| 21 | 3685 | 7.21 | −68 | −26 | −10 | |
| 31 | 30 | 3.98 | −14 | −60 | 18 | |
| R temporal lobe | 21 | 2716 | 8.80 | 60 | −12 | −18 |
| 39 | 1035 | 6.67 | 46 | −60 | 28 | |
| (B). Heterozygous Genotypes (S/LA, S/LG)
| ||||||
|---|---|---|---|---|---|---|
| Region | Brodmann’s Area | Cluster size | t df = 15 | MNI Coordinates | ||
| x | y | z | ||||
| L superior medial frontal cortex | 10 | 2449 | 6.21 | −2 | 60 | 22 |
| R superior medial frontal cortex | 10 | 1844 | 7.71 | 10 | 66 | 16 |
| L medial frontal gyrus | 10 | 1402 | 5.74 | −2 | 60 | 22 |
| 10 | 411 | 4.70 | −10 | 68 | 0 | |
| R medial frontal gyrus | 10 | 1304 | 7.71 | 10 | 66 | 16 |
| 10 | 485 | 4.80 | 2 | 50 | −4 | |
| L superior frontal gyrus | 10 | 2963 | 5.42 | −18 | 62 | 18 |
| R superior frontal gyrus | 10 | 2715 | 6.78 | 12 | 66 | 16 |
| L retrosplenial cortex | 30 | 29 | 4.94 | −2 | −54 | 20 |
| 30 | 12 | 2.61 | −16 | −58 | 10 | |
| R retrosplenial cortex | 30 | 36 | 5.09 | 4 | −54 | 20 |
| L posterior cingulate | 31 | 563 | 5.49 | −8 | −54 | 24 |
| R posterior cingulate | 23 | 522 | 5.65 | 6 | −48 | 24 |
| L precuneus | 31 | 1640 | 6.09 | −2 | −52 | 40 |
| 19 | 189 | 4.27 | −38 | −76 | 38 | |
| R precuneus | 7 | 1803 | 8.25 | 10 | −54 | 44 |
| 39 | 136 | 4.95 | 46 | −76 | 36 | |
| L inferior parietal lobule | 40 | 562 | 4.14 | −46 | −62 | 38 |
| 40 | 111 | 3.79 | −48 | −50 | 24 | |
| R inferior parietal lobule | 13 | 61 | 6.10 | 44 | −50 | 24 |
| 40 | 374 | 4.79 | 56 | −56 | 38 | |
| L parahippocampal gyrus | 35/36 | 263 | 2.91 | −22 | −36 | −8 |
| 30 | 20 | 2.91 | −22 | −42 | 2 | |
| R parahippocampal gyrus | 35 | 113 | 2.53 | 24 | −12 | −26 |
| L temporal lobe | 21 | 310 | 4.75 | −68 | −34 | −8 |
| 39 | 868 | 4.45 | −46 | −56 | 24 | |
| 21 | 643 | 4.08 | −54 | −4 | −20 | |
| 31 | 26 | 3.37 | −14 | −60 | 20 | |
| R temporal lobe | 39 | 1482 | 7.10 | 42 | −52 | 24 |
| 21 | 1312 | 4.81 | 52 | −4 | −18 | |
| (C). S/S group
| ||||||
|---|---|---|---|---|---|---|
| Region | Brodmann’s Area | Cluster size | t df = 9 | MNI Coordinates | ||
| x | y | z | ||||
| L superior medial frontal cortex | 9 | 723 | 3.80 | −2 | 48 | 26 |
| R superior medial frontal cortex | 10 | 1052 | 6.57 | 14 | 56 | 8 |
| L medial frontal gyrus | 10 | 219 | 3.74 | −18 | 68 | −2 |
| 10 | 332 | 3.11 | −2 | 56 | 6 | |
| R medial frontal gyrus | 10 | 1115 | 6.57 | 14 | 56 | 8 |
| 10 | 286 | 5.29 | 12 | 56 | 0 | |
| 6 | 54 | 2.21 | 2 | −28 | 68 | |
| L superior frontal gyrus | 10 | 260 | 4.17 | −20 | 46 | 32 |
| 10 | 416 | 4.16 | −18 | 70 | 0 | |
| R superior frontal gyrus | 9 | 1212 | 5.87 | 12 | 48 | 30 |
| 10 | 111 | 3.30 | 14 | 68 | −6 | |
| L retrosplenial cortex | 30 | 29 | 5.09 | −4 | −50 | 20 |
| 30 | 12 | 3.23 | −16 | −58 | 10 | |
| 30 | 42 | 2.49 | −10 | −46 | 2 | |
| R retrosplenial cortex | 30 | 36 | 4.90 | 6 | −50 | 20 |
| 30 | 14 | 3.48 | 10 | −44 | 2 | |
| 30 | 30 | 3.05 | 4 | −54 | 6 | |
| L posterior cingulate | 30 | 656 | 7.24 | −12 | −58 | 14 |
| R posterior cingulate | 23 | 764 | 6.75 | 8 | −46 | 24 |
| L precuneus | 7/31 | 1885 | 8.26 | −14 | −48 | 40 |
| 39 | 89 | 4.68 | −40 | −68 | 36 | |
| R precuneus | 31/39 | 2542 | 12.35 | 18 | −48 | 34 |
| L inferior parietal lobule | 39 | 679 | 5.46 | −42 | −66 | 38 |
| 39/40 | 94 | 3.90 | −42 | −48 | 28 | |
| R inferior parietal lobule | 40 | 399 | 4.05 | 60 | −54 | 42 |
| 40 | 10 | 2.41 | 48 | −50 | 24 | |
| 40 | 10 | 2.37 | 68 | −46 | 26 | |
| L parahippocampal gyrus | 28 | 354 | 4.45 | −20 | −26 | −14 |
| R parahippocampal gyrus | 30 | 53 | 3.82 | 8 | −42 | 2 |
| 28 | 58 | 3.64 | 28 | −26 | −14 | |
| 28 | 46 | 3.29 | 24 | −12 | −24 | |
| L temporal lobe | 31 | 1136 | 6.22 | −14 | −60 | 16 |
| 21 | 27 | 3.34 | −58 | −10 | −14 | |
| 21 | 48 | 2.64 | −58 | −26 | −16 | |
| 38 | 25 | 2.29 | −42 | 14 | −18 | |
| R temporal lobe | 40 | 1161 | 7.29 | 64 | −58 | 28 |
| 21 | 1366 | 7.17 | 66 | −22 | −14 | |
| 38 | 366 | 4.29 | 34 | 8 | −26 | |
| 31 | 121 | 2.96 | 20 | −66 | 24 | |
To address our first hypothesis, a voxel-wise group-level ANOVA was estimated in SPM5 to examine connectivity within the default network by genotype group: S/S, LA/LA, and heterozygous genotypes. ROI analyses were then performed to examine long-range functional connectivity between the posterior hubs of the default network and the intersection of the left and right superior medial frontal mask, from the AAL (Anatomical Automatic Labeling) digital atlas (Tzourio-Mazoyer, et al., 2002) as implemented in Wake Forest University PickAtlas (Maldjian et al., 2002), with Brodmann’s Area 10. The superior medial frontal cortical area overlapping BA 10 represents the default network area implicated in clinical samples (e.g., Castellanos et al., 2008; Garrity et al., 2007; Monk et al., 2009; Weng et al., 2010; Wiggins et al., 2011) and has been shown to be sensitive to changes in serotonin levels (Canli et al., 2005; Kunisato et al., 2011; Mann et al., 2000; Nakamura et al., 2010; Smith et al., 2004). The purpose of the ROI analyses was to restrict the search area for significant voxels to the superior medial frontal/BA 10. First, significance values from the F-test of differences in connectivity among the three genotype groups were small volume-corrected for multiple comparisons within each ROI (left and right superior medial frontal/BA 10) using family wise error (FWE) correction (Worsley et al., 1996). Post-hoc contrasts of the connectivity maps among the three genotype groups used the same superior medial frontal cortex/BA 10 mask for the ROI analyses. Those family wise error-corrected p values from the post-hoc contrasts were then subjected to an additional correction (Bonferroni) based on comparisons among the three genotype groups (family wise error corrected α = .05/3 = .0167).
To address our second hypothesis, an age by genotype interaction model was estimated using multiple regression in SPM5. The three levels of genotype were dummy coded. The dummy-coded genotype variables and age were entered into the model, as well as the two dummy-coded genotype by age interaction variables. An F test of the change in model fit after including the two dummy coded interaction variables indicated the overall interaction between genotype and age (Allison, 1977; Irwin and McClellan, 2001). To further examine developmental change, the same interaction model was estimated for puberty as well. ROI analyses were performed to examine differences between genotype groups in age-related connectivity changes between the posterior hub and superior medial frontal cortex. The same superior medial frontal cortex/BA 10 mask used in the first hypothesis was also used in ROI analyses for this hypothesis.
3. Results
All participants scored below clinical cutoffs on the measures of depression, anxiety, and autism symptoms as well as other internalizing and externalizing behaviors; moreover, the three genotype groups did not differ significantly on any of the measures (Child Behavior Checklist, Achenbach and Edelbrock, 1981; Social Responsiveness Scale, Constantino et al., 2003; Social Communication Questionnaire, Rutter et al., 2003; Obsessive Compulsive Inventory – Revised, Foa et al., 2010; Child Depression Inventory, Kovacs, 1992; Multidimensional Anxiety Scale for Children, March et al., 1997; Spence Children’s Anxiety Scales, Spence, 1997; Table 1). Genotype groups also did not significantly differ in average head motion (F2,36 = 1.02, p = .373). All three genotype groups showed default network connectivity patterns (Table 2).
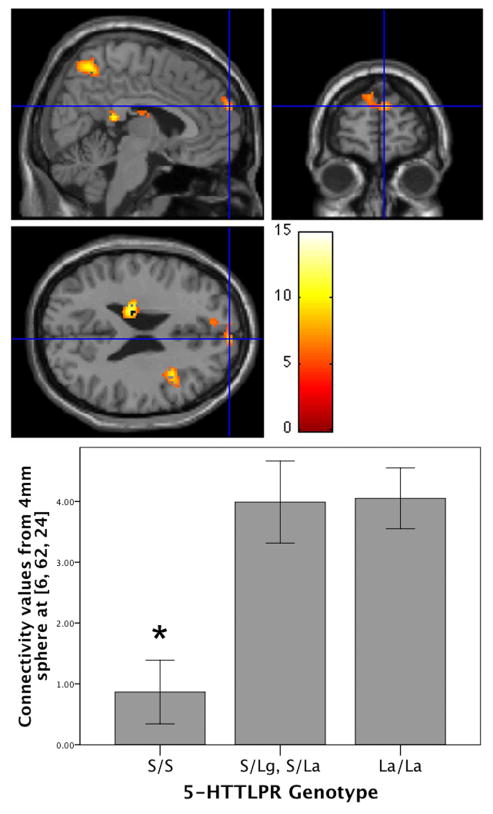
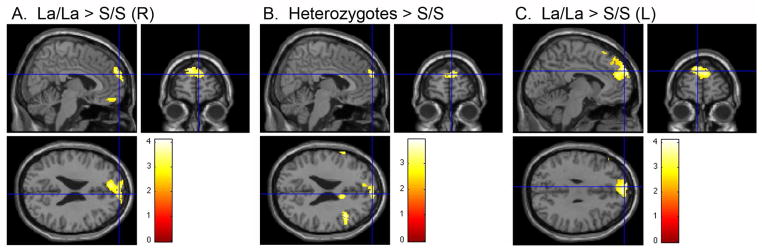
Our first hypothesis was confirmed: a one-way ANOVA revealed that the genotype groups differed in their connectivity strength in the right superior medial frontal cortex (xyz = 6, 62, 24, F2,36 = 8.84, p = .028 corrected for multiple comparisons within the right superior medial frontal cortex/BA 10; Figure 2). Bonferroni-corrected post-hoc comparisons of the connectivity maps of the three genotype groups indicated that the S/S group had significantly weaker connectivity in the right superior medial frontal cortex than both the LA/LA group (xyz = 8, 62, 26, t36 = 3.70, p = .012; Figure 3) and the heterozygous group (S/LA, S/LG; xyz = 6, 62, 24, t36 = 3.75, p = .011, both corrected for multiple comparisons within the right superior medial frontal cortex/BA 10; Figure 3). The LA/LA versus heterozygotes comparison was not significant (xyz = 16, 70, 12, t36 = 2.11, p = .233, corrected for multiple comparisons within the right superior medial frontal cortex/BA 10).
Figure 2. Posterior-anterior connectivity by genotype.
Connectivity between the posterior default network hub and superior medial frontal cortex varies by genotype (xyz = 6, 62, 24, F2,36 = 8.84, p = .028 corrected for multiple comparisons within the right superior medial frontal cortex/BA 10 mask, k = 98). Unless otherwise specified, all brain images in this article are presented with orthogonal views in the sagittal (top left), coronal (top right), and transverse (bottom left) planes; threshold set to p < 01, k ≥ 100 for illustration purposes. Color bar represents F values. Below, in the bar graph, functional connectivity values from a 4 mm sphere around the peak voxel in the cross-hairs (xyz = 6, 62, 24) were extracted and averaged. Significantly different groups in the post-hoc Bonferroni-corrected contrasts are indicated with an asterisk (*). Error bars indicate standard error of the mean.
Figure 3. Post-hoc contrasts show weaker connectivity in S/S group.
Connectivity between the posterior default network hub and the superior medial frontal cortex is significantly weaker in the S/S group compared to: (A) the LA/LA group (xyz = 8, 62, 26, t36 = 3.44, p = .012, corrected for multiple comparisons in the right superior medial frontal cortex/BA 10, k = 159), (B) the heterozygous group (xyz = 6, 62, 24, t36 = 3.75, p = .011, corrected for multiple comparisons in the right superior medial frontal cortex/BA 10, k = 122), and (C) the LA/LA group in the left superior medial frontal cortex (xyz = −6, 60, 30, t36 = 3.78, p = .012, corrected for multiple comparisons within the left superior medial frontal cortex/BA 10, k = 159). Color bar indicates t values.
In the left superior medial frontal cortex, whereas the main effect of genotype on connectivity was a trend (xyz = −6, 60, 30, F2,36 = 7.14, p = .087 corrected for multiple comparisons within the left superior medial frontal cortex/BA 10 mask), Bonferroni-corrected post-hoc comparisons of the groups revealed that the S/S group had significantly weaker connectivity than the LA/LA group (xyz = −6, 60, 30, t36 = 3.78, p = .012, corrected for multiple comparisons within the left superior medial frontal cortex/BA 10; Figure 3). Neither the post-hoc contrast of heterozygotes versus the S/S group (xyz = −2, 62, 24, t36 = 3.00, p = .071) nor the LA/LA group versus heterozygotes reached significance (xyz = −6, 62, 32, t36 = 2.14, p = .320, both corrected for multiple comparisons within the left superior medial frontal cortex/BA 10).
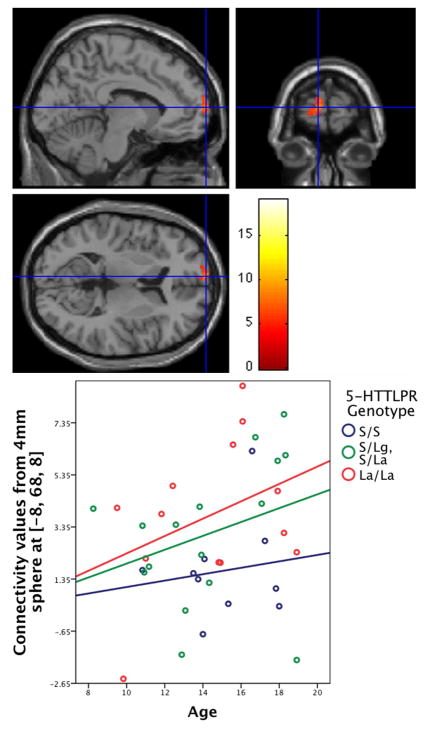
Our second hypothesis was also confirmed. A significant interaction between genotype and age was detected in the superior medial frontal cortex of the default network. Specifically, connectivity between the posterior hub and the left superior medial frontal cortex strengthened with age across individuals the most in the LA/LA group, but less so for heterozygous genotypes (S/LA, S/LG), and the least for the S/S group (xyz = −8, 68, 8, F2,33 = 8.71, p = .039, corrected for multiple comparisons within the left superior medial frontal cortex/BA 10; Figure 4). The interaction effect was not significant in the right superior medial frontal cortex (xyz = 12, 70, 2, F2,33 = 6.21, p = .128, corrected for multiple comparisons within the right superior medial frontal cortex/BA 10). To corroborate our findings of an age-by-genotype interaction, we used an additional measure of development, pubertal state, to investigate a puberty-by-genotype interaction. Similar to our findings with age, a puberty-by-genotype interaction pattern was found in the superior medial frontal cortex, albeit non-significant (xyz = 6, 58, 24, F2,33 = 5.97, p = .146, corrected for multiple comparisons within the right superior medial frontal cortex/BA 10).
Figure 4. Age-by-genotype interaction predicts connectivity.
Connectivity between the posterior hub and the left superior medial frontal cortex strengthens with age the most in the LA/LA group, less so for heterozygous genotypes (S/LA, S/LG), and the least for the S/S group (xyz = −8, 68, 8, F2,33 = 8.71, p = .039, corrected for multiple comparisons within the left superior medial frontal cortex/BA 10). For the scatter plot, functional connectivity values from a 4 mm sphere around the peak voxel were extracted and averaged. The best-fit lines for the three genotype groups were overlaid on the scatter plot to illustrate the interaction.
3.1 Additional Analyses
Since including individuals of multiple ancestries introduces heterogeneity into the genetic analyses, this study was restricted to individuals of Caucasian descent. Nonetheless, we conducted additional analyses to determine how including individuals of all ancestries affected our main findings. We repeated all of the previous analyses with an additional 13 non-Caucasian individuals included, for a total of 52 individuals (see Table 1 for subject characteristics). Because of the increased difficulty of detecting genotype effects with the added noise, we utilized a significance threshold of p < .05 without family-wise error correction, but with a Bonferroni correction for post-hoc contrasts of α = .05/3 = .0167.
With participants of all ancestries included, our first hypothesis, weaker connectivity in the S/S group, was confirmed. With all individuals, we still found that the genotype groups differed in their connectivity strength in the right superior medial frontal cortex (xyz = 18, 70, 14, F2,49 = 5.71, p = .006). Bonferroni-corrected post-hoc comparisons among the three genotype groups still indicated that the S/S group had significantly weaker connectivity in the right superior medial frontal cortex than both the LA/LA group (xyz = 18, 70, 14, t49 = 3.37, p = .0007) and the intermediate genotypes group (S/LA, S/LG, LG/LG; xyz = 6, 62, 26, t49 = 2.49, p = .008). As in the analysis with only Caucasian participants, the LA/LA versus intermediate genotypes comparison was not significant after Bonferroni correction (xyz = 18, 70, 12, t49 = 1.97, p = .027).
In Caucasian participants only, we found a trend for a the main effect of genotype on connectivity in the left superior medial frontal cortex; in participants of all ancestries, the main effect of genotype on connectivity in the left superior medial frontal cortex was significant at the uncorrected threshold (xyz = −8, 64, 32, F2,49 = 3.56, p = .035). Bonferroni-corrected post-hoc comparisons of the genotype groups including all ancestries corroborated the original findings with Caucasians only: the S/S group had weaker connectivity than the LA/LA group (xyz = −8, 64, 32, t49 = 2.62, p = .006). As in the original finding, neither the post-hoc contrast of intermediate genotypes versus the S/S group (xyz = −2, 62, 24, t49 = 2.10, p = .020) nor the LA/LA group versus intermediate genotypes reached significance after Bonferroni correction (xyz = −8, 64, 32, t49 = 1.79, p = .039).
Our second hypothesis, positing differential developmental changes in connectivity strength by genotype, was also confirmed with participants of all ancestries. There was still an interaction between genotype and age in left BA 10 of the default network (xyz = −36, 62, 16, F2,46 = 8.12, p = .0009) with all ancestries included. As with the original finding, the interaction effect was not significant in the right superior medial frontal cortex (xyz = 8, 62, 22, F2,46 = 1.85, p = .156). Corroborating the findings with age, a puberty-by-genotype interaction pattern was found in the superior medial frontal cortex, in the left hemisphere, with all ancestries included (xyz = −10, 62, 30, F2,46 = 5.97, p = .030). To summarize, when including individuals of all ancestries, the pattern of findings was largely similar to the original results with just Caucasian participants, albeit at a more lenient significance threshold.
4. Discussion
To our knowledge, this is the first study to examine the influence of 5-HTTLPR on resting-state default network connectivity. To summarize, children and adolescents with the S/S genotype have the weakest connectivity between the posterior hub and superior medial frontal cortex in the default network compared to other 5-HTTLPR genotypes. Additionally, examining connectivity in children and adolescents revealed that group differences among genotypes are driven by the older participants: although all three genotype groups show similar levels of connectivity at a young age, the genotype groups show markedly different connectivity levels in older participants. Specifically, across individuals, connectivity strengthens most with age in the LA/LA group, followed by heterozygotes. The S/S group has the least increase in connectivity strength with age.
Most studies on 5-HTTLPR and human brain function have focused on emotion and have utilized tasks with emotional stimuli (e.g., faces modeling happiness, sadness, etc.; for reviews, see Dannlowski et al., 2010; Hariri and Holmes, 2006). Although much less well-researched, cognition, particularly executive function, has also been shown to be influenced by 5-HTTLPR variants in a few studies. Specifically, healthy S or LG carriers have decreased cognitive flexibility and altered conflict- and error-related anterior cingulate activation (Holmes et al., 2010) as well as alterations in attention allocation (Enge et al., 2011; Roiser et al., 2006). The present study provides additional evidence that 5-HTTLPR-related differences in brain function generalize beyond specific emotional tasks to non-emotionally charged activities as well, such as during rest. Our findings suggest that alterations in brain activation patterns related to 5-HTTLPR may be ubiquitous and not manifest only in specific situations, such as the presentation of emotional stimuli.
To ensure that genotype is not acting as a proxy for psychopathology in the present study, we confirmed that all participants had levels of symptoms and problem behaviors that did not meet threshold for a clinical diagnosis. Nevertheless, we examined whether genotype groups differed in subclinical levels of symptoms or other behaviors. The three genotype groups did not differ significantly on any of the measures of depression, anxiety, and autism symptoms as well as other internalizing and externalizing behaviors. Specifically, we did not find significant differences among genotype groups on the following measures: Child Behavior Checklist (Achenbach and Edelbrock, 1981); Social Responsiveness Scale (Constantino et al., 2003); Social Communication Questionnaire (Rutter et al., 2003); Obsessive Compulsive Inventory – Revised (Foa et al., 2010); Child Depression Inventory (Kovacs, 1992); Multidimensional Anxiety Scale for Children (March et al., 1997); Spence Children’s Anxiety Scales (Spence, 1997; Table 1). There are multiple possibilities as to why we do not see symptom differences among the genotype groups. These possibilities include reduced variability of symptoms in our sample, which was screened for psychological disorders. However, the small sample size prevents us from identifying the specific reasons. The focus of the present study was on building a normative foundation with a healthy sample; it will be important for future research to examine the impact of 5-HTTLPR on default network connectivity in more heterogeneous samples, with greater variability in symptoms and problem behaviors.
The present findings suggest that it may be fruitful for future research to examine the effects of gene and environmental interactions on brain development. Our finding that the neural effects of genotype become more pronounced over the course of development is consistent with the concept that the brain is the ongoing product of gene and environmental interactions in development (Monk, 2008). Thus, exposure to the environment, with both beneficial and detrimental effects, compounds over time and may lead to greater differences in brain function with age. Moreover, brain differences among the genotype groups are most marked during adolescence, a crucial transition period associated with multiple opportunities and challenges that could interact with genetic differences. This transition includes physical changes due to puberty, as well as increased autonomy to carve out one’s own environment and identity (Grotevant, 1992; McLellan and Pugh, 1999; Petersen, 1988; Steinberg and Silverberg, 1986). In addition, there is an increase in risk-taking and antisocial behaviors (Rutter et al., 2006a; Rutter et al., 2006b), as well as heightened incidence of some forms of psychopathology, such as depression and social anxiety (Cohen et al., 1993; Costello et al., 2003; Costello et al., 2011). Genes interacting with environmental factors at sensitive periods in development (Uher and McGuffin, 2008), such as adolescence, may alter the trajectories of key brain networks and impact behavioral outcomes.
Our study has some limitations. First, we used a cross-sectional design to examine developmental changes in the default network between genotype groups. It is possible that cohort differences across the 8- to 19-year-old age span may be driving our effects. A longitudinal study involving repeated scans on individuals through this age range is necessary to confirm our findings.
Second, sources of potential artifacts in functional neuroimaging data include excessive head movement and physiological rhythms. All youth included in this study exhibited less than 2.5 mm of movement in any direction, and genotype groups did not differ in their average head motion. Subsequent pre-processing steps included realignment of the functional images to reduce remaining head motion artifact. To address noise due to physiological rhythms, cardiac and respiratory data were collected on each participant. A correction based on these data was applied to the images. Noise due to artifacts is inherent in fMRI studies, but our efforts to reduce their impact make it unlikely that head motion and physiological rhythms are driving our findings.
Third, the primary analyses included data only from children and adolescents of Caucasian descent in order to reduce the likelihood of spurious relationships due to population stratification and to reduce the amount of heterogeneity introduced by multiple ancestries. This approach has been used in other studies of 5-HTTLPR (e.g., Praschak-Rieder et al., 2007; Rao et al., 2007; Zalsman et al., 2006). Nonetheless, we conducted additional analyses including the non-Caucasian individuals to examine how they affected our findings (“3.1 Additional Analyses”). We found that although including multiple ancestries added noise to the data, the patterns found with Caucasian participants only are still evident. To draw conclusions about the effects of 5-HTTLPR on default network connectivity in other ancestry groups, however, future studies focusing on each ethnic group may be necessary.
Fourth, our findings should be considered preliminary, as we have a small sample size (N=39 overall, with 10 participants in the S/S group, 13 in the LA/LA group, and 16 in the heterozygous genotypes group). This distribution is to be expected, as our sample is within Hardy-Weinberg equilibrium (χ2 = 1.19, df = 1, p = .276). However, future studies are needed to confirm the results from the present study with a larger sample size.
In conclusion, we demonstrate here, for the first time, the influence of 5-HTTLPR variants on long-range default network connectivity and on the developmental time course of connectivity in a healthy child and adolescent sample. We found that youth with the S/S genotype have the weakest connectivity between the posterior hub and superior medial frontal cortex in the default network compared to youth with the LA/LA genotype and heterozygous genotypes (S/LA, S/LG). We also found that brain differences among the genotype groups become more pronounced with increasing age: among the youngest participants, the groups are similar in connectivity levels, but in older adolescents, connectivity is markedly different among genotype groups. Specifically, connectivity between the posterior hub and superior medial frontal region in the LA/LA group strengthens the most with age across individuals, followed by heterozygotes. Connectivity in the S/S group strengthens the least with age. Our findings have implications for understanding how genetic influences on large-scale brain networks may compound over time and result in marked differences in a healthy developing cohort of individuals.
Acknowledgments
This research was supported in part by an Autism Speaks Predoctoral Fellowship (4773) to J.L.W.; an Autism Speaks grant (2573) to C.S.M.; National Institutes of Health grants to D.M.M. (R01 NS54784, R01 DC009410), S.J.P. (MH079871), and J.K.B. (K12 HD028820); a Michigan Institute for Clinical and Health Research pilot award to D.M.M. (U024600); and an Elizabeth E. Kennedy (Children’s Research) Fund Award from the Department of Pediatrics, University of Michigan, to J.K.B. We thank Dr. Douglas Noll for methodological advice and the staff of the University of Michigan Functional MRI Center and DNA Sequencing Core for technical support. Lastly, we thank the families who participated.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achenbach TM, Edelbrock CS. Behavioral problems and competencies reported by parents of normal and disturbed children aged four through sixteen. Monogr Soc Res Child Dev. 1981;46:1–82. [PubMed] [Google Scholar]
- Allison P. Testing for interaction in multiple regression. The American Journal of Sociology. 1977;83:144–153. [Google Scholar]
- Beason-Held LL, Kraut MA, Resnick SM. Stability Of Default-Mode Network Activity In The Aging Brain. Brain Imaging Behav. 2009;3:123–131. doi: 10.1007/s11682-008-9054-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky J, Jonassaint C, Pluess M, Stanton M, Brummett B, Williams R. Vulnerability genes or plasticity genes? Mol Psychiatry. 2009;14:746–754. doi: 10.1038/mp.2009.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky J, Pluess M. Beyond diathesis stress: differential susceptibility to environmental influences. Psychol Bull. 2009;135:885–908. doi: 10.1037/a0017376. [DOI] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Bluhm RL, Miller J, Lanius RA, Osuch EA, Boksman K, Neufeld RW, Theberge J, Schaefer B, Williamson P. Spontaneous low-frequency fluctuations in the BOLD signal in schizophrenic patients: anomalies in the default network. Schizophr Bull. 2007;33:1004–1012. doi: 10.1093/schbul/sbm052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluhm RL, Williamson PC, Osuch EA, Frewen PA, Stevens TK, Boksman K, Neufeld RW, Theberge J, Lanius RA. Alterations in default network connectivity in posttraumatic stress disorder related to early-life trauma. J Psychiatry Neurosci. 2009;34:187–194. [PMC free article] [PubMed] [Google Scholar]
- Borroni B, Grassi M, Agosti C, Archetti S, Costanzi C, Cornali C, Caltagirone C, Caimi L, Di Luca M, Padovani A. Cumulative effect of COMT and 5-HTTLPR polymorphisms and their interaction with disease severity and comorbidities on the risk of psychosis in Alzheimer disease. Am J Geriatr Psychiatry. 2006a;14:343–351. doi: 10.1097/01.JGP.0000192491.50802.c3. [DOI] [PubMed] [Google Scholar]
- Borroni B, Grassi M, Agosti C, Costanzi C, Archetti S, Franzoni S, Caltagirone C, Di Luca M, Caimi L, Padovani A. Genetic correlates of behavioral endophenotypes in Alzheimer disease: role of COMT, 5-HTTLPR and APOE polymorphisms. Neurobiol Aging. 2006b;27:1595–1603. doi: 10.1016/j.neurobiolaging.2005.09.029. [DOI] [PubMed] [Google Scholar]
- Brune CW, Kim SJ, Salt J, Leventhal BL, Lord C, Cook EH., Jr 5-HTTLPR Genotype-Specific Phenotype in Children and Adolescents With Autism. Am J Psychiatry. 2006;163:2148–2156. doi: 10.1176/ajp.2006.163.12.2148. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Andrews-Hanna JR, Schacter DL. The brain’s default network: anatomy, function, and relevance to disease. Ann N Y Acad Sci. 2008;1124:1–38. doi: 10.1196/annals.1440.011. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Vincent JL. Unrest at rest: default activity and spontaneous network correlations. Neuroimage. 2007;37:1091–1096. doi: 10.1016/j.neuroimage.2007.01.010. discussion 1097–1099. [DOI] [PubMed] [Google Scholar]
- Bunge SA, Dudukovic NM, Thomason ME, Vaidya CJ, Gabrieli JD. Immature frontal lobe contributions to cognitive control in children: evidence from fMRI. Neuron. 2002;33:301–311. doi: 10.1016/s0896-6273(01)00583-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canli T, Omura K, Haas BW, Fallgatter A, Constable RT, Lesch KP. Beyond affect: a role for genetic variation of the serotonin transporter in neural activation during a cognitive attention task. Proc Natl Acad Sci U S A. 2005;102:12224–12229. doi: 10.1073/pnas.0503880102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi A, Hariri AR, Holmes A, Uher R, Moffitt TE. Genetic sensitivity to the environment: the case of the serotonin transporter gene and its implications for studying complex diseases and traits. Am J Psychiatry. 2010;167:509–527. doi: 10.1176/appi.ajp.2010.09101452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, McClay J, Mill J, Martin J, Braithwaite A, Poulton R. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- Castellanos FX, Margulies DS, Kelly C, Uddin LQ, Ghaffari M, Kirsch A, Shaw D, Shehzad Z, Di Martino A, Biswal B, Sonuga-Barke EJ, Rotrosen J, Adler LA, Milham MP. Cingulate-precuneus interactions: a new locus of dysfunction in adult attention-deficit/hyperactivity disorder. Biol Psychiatry. 2008;63:332–337. doi: 10.1016/j.biopsych.2007.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantino JN, Davis SA, Todd RD, Schindler MK, Gross MM, Brophy SL, Metzger LM, Shoushtari CS, Splinter R, Reich W. Validation of a brief quantitative measure of autistic traits: comparison of the social responsiveness scale with the autism diagnostic interview-revised. J Autism Dev Disord. 2003;33:427–433. doi: 10.1023/a:1025014929212. [DOI] [PubMed] [Google Scholar]
- Cordes D, Haughton VM, Arfanakis K, Wendt GJ, Turski PA, Moritz CH, Quigley MA, Meyerand ME. Mapping functionally related regions of brain with functional connectivity MR imaging. AJNR Am J Neuroradiol. 2000;21:1636–1644. [PMC free article] [PubMed] [Google Scholar]
- Dannlowski U, Konrad C, Arolt V, Suslow T. Neurogenetics of emotional processes. Neuroimaging findings as endophenotypes for depression. Nervenarzt. 2010;81:24–31. doi: 10.1007/s00115-009-2828-8. [DOI] [PubMed] [Google Scholar]
- Dolzan V, Serretti A, Mandelli L, Koprivsek J, Kastelic M, Plesnicar BK. Acute antipyschotic efficacy and side effects in schizophrenia: association with serotonin transporter promoter genotypes. Prog Neuropsychopharmacol Biol Psychiatry. 2008;32:1562–1566. doi: 10.1016/j.pnpbp.2008.05.022. [DOI] [PubMed] [Google Scholar]
- Drevets WC, Price JL, Furey ML. Brain structural and functional abnormalities in mood disorders: implications for neurocircuitry models of depression. Brain Struct Funct. 2008;213:93–118. doi: 10.1007/s00429-008-0189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enge S, Fleischhauer M, Lesch KP, Strobel A. On the role of serotonin and effort in voluntary attention: evidence of genetic variation in N1 modulation. Behav Brain Res. 2011;216:122–128. doi: 10.1016/j.bbr.2010.07.021. [DOI] [PubMed] [Google Scholar]
- Ezaki N, Nakamura K, Sekine Y, Thanseem I, Anitha A, Iwata Y, Kawai M, Takebayashi K, Suzuki K, Takei N, Iyo M, Inada T, Iwata N, Harano M, Komiyama T, Yamada M, Sora I, Ujike H, Mori N. Short allele of 5-HTTLPR as a risk factor for the development of psychosis in Japanese methamphetamine abusers. Ann N Y Acad Sci. 2008;1139:49–56. doi: 10.1196/annals.1432.011. [DOI] [PubMed] [Google Scholar]
- Fair DA, Cohen AL, Dosenbach NU, Church JA, Miezin FM, Barch DM, Raichle ME, Petersen SE, Schlaggar BL. The maturing architecture of the brain’s default network. Proc Natl Acad Sci U S A. 2008;105:4028–4032. doi: 10.1073/pnas.0800376105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foa EB, Coles M, Huppert JD, Pasupuleti RV, Franklin ME, March J. Development and validation of a child version of the obsessive compulsive inventory. Behav Ther. 2010;41:121–132. doi: 10.1016/j.beth.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci U S A. 2005;102:9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity AG, Pearlson GD, McKiernan K, Lloyd D, Kiehl KA, Calhoun VD. Aberrant “default mode” functional connectivity in schizophrenia. Am J Psychiatry. 2007;164:450–457. doi: 10.1176/ajp.2007.164.3.450. [DOI] [PubMed] [Google Scholar]
- Glover GH, Law CS. Spiral-in/out BOLD fMRI for increased SNR and reduced susceptibility artifacts. Magn Reson Med. 2001;46:515–522. doi: 10.1002/mrm.1222. [DOI] [PubMed] [Google Scholar]
- Glover GH, Li TQ, Ress D. Image-based method for retrospective correction of physiological motion effects in fMRI: RETROICOR. Magn Reson Med. 2000;44:162–167. doi: 10.1002/1522-2594(200007)44:1<162::aid-mrm23>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Flores BH, Menon V, Glover GH, Solvason HB, Kenna H, Reiss AL, Schatzberg AF. Resting-state functional connectivity in major depression: abnormally increased contributions from subgenual cingulate cortex and thalamus. Biol Psychiatry. 2007;62:429–437. doi: 10.1016/j.biopsych.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Kiviniemi V, Tervonen O, Vainionpaa V, Alahuhta S, Reiss AL, Menon V. Persistent default-mode network connectivity during light sedation. Hum Brain Mapp. 2008;29:839–847. doi: 10.1002/hbm.20537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Krasnow B, Reiss AL, Menon V. Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proc Natl Acad Sci U S A. 2003;100:253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Supekar K, Menon V, Dougherty RF. Resting-state functional connectivity reflects structural connectivity in the default mode network. Cereb Cortex. 2009;19:72–78. doi: 10.1093/cercor/bhn059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotevant HD. Assigned and chosen identity components: A process perspective on their integration. In: Adams GR, Gullotta TP, Montemayor R, editors. Adolescent Identity Formation (Advances in Adolescent Development) Sage Publications, Inc; Thousand Oaks, CA: 1992. pp. 73–90. [Google Scholar]
- Gusnard DA, Raichle ME. Searching for a baseline: functional imaging and the resting human brain. Nat Rev Neurosci. 2001;2:685–694. doi: 10.1038/35094500. [DOI] [PubMed] [Google Scholar]
- Ha TM, Cho DM, Park SW, Joo MJ, Lee BJ, Kong BG, Kim JM, Yoon JS, Kim YH. Evaluating associations between 5-HTTLPR polymorphism and Alzheimer’s disease for Korean patients. Dement Geriatr Cogn Disord. 2005;20:31–34. doi: 10.1159/000085071. [DOI] [PubMed] [Google Scholar]
- Hampson M, Driesen NR, Skudlarski P, Gore JC, Constable RT. Brain connectivity related to working memory performance. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2006;26:13338–13343. doi: 10.1523/JNEUROSCI.3408-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri AR, Drabant EM, Weinberger DR. Imaging genetics: perspectives from studies of genetically driven variation in serotonin function and corticolimbic affective processing. Biol Psychiatry. 2006;59:888–897. doi: 10.1016/j.biopsych.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Hariri AR, Holmes A. Genetics of emotional regulation: the role of the serotonin transporter in neural function. Trends Cogn Sci. 2006;10:182–191. doi: 10.1016/j.tics.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Hariri AR, Mattay VS, Tessitore A, Kolachana B, Fera F, Goldman D, Egan MF, Weinberger DR. Serotonin transporter genetic variation and the response of the human amygdala. Science. 2002;297:400–403. doi: 10.1126/science.1071829. [DOI] [PubMed] [Google Scholar]
- Holmes AJ, Bogdan R, Pizzagalli DA. Serotonin transporter genotype and action monitoring dysfunction: a possible substrate underlying increased vulnerability to depression. Neuropsychopharmacology. 2010;35:1186–1197. doi: 10.1038/npp.2009.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horovitz SG, Braun AR, Carr WS, Picchioni D, Balkin TJ, Fukunaga M, Duyn JH. Decoupling of the brain’s default mode network during deep sleep. Proc Natl Acad Sci U S A. 2009;106:11376–11381. doi: 10.1073/pnas.0901435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horovitz SG, Fukunaga M, de Zwart JA, van Gelderen P, Fulton SC, Balkin TJ, Duyn JH. Low frequency BOLD fluctuations during resting wakefulness and light sleep: a simultaneous EEG-fMRI study. Hum Brain Mapp. 2008;29:671–682. doi: 10.1002/hbm.20428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu XZ, Lipsky RH, Zhu G, Akhtar LA, Taubman J, Greenberg BD, Xu K, Arnold PD, Richter MA, Kennedy JL, Murphy DL, Goldman D. Serotonin transporter promoter gain-of-function genotypes are linked to obsessive-compulsive disorder. Am J Hum Genet. 2006;78:815–826. doi: 10.1086/503850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin J, McClellan G. Misleading heuristics and moderated multiple regression models. Journal of Marketing Research. 2001;38:100–109. [Google Scholar]
- Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Karg K, Burmeister M, Shedden K, Sen S. The Serotonin Transporter Promoter Variant (5-HTTLPR), Stress, and Depression Meta-analysis Revisited: Evidence of Genetic Moderation. Arch Gen Psychiatry. 2011 doi: 10.1001/archgenpsychiatry.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Ikenaga Y, Wakeno M, Okugawa G, Nobuhara K, Fukuda T, Fukuda K, Azuma J, Kinoshita T. Controlled clinical comparison of paroxetine and fluvoxamine considering the serotonin transporter promoter polymorphism. Int Clin Psychopharmacol. 2005;20:151–156. doi: 10.1097/00004850-200505000-00005. [DOI] [PubMed] [Google Scholar]
- Kennedy DP, Courchesne E. The intrinsic functional organization of the brain is altered in autism. Neuroimage. 2008;39:1877–1885. doi: 10.1016/j.neuroimage.2007.10.052. [DOI] [PubMed] [Google Scholar]
- Kim JM, Stewart R, Kim SW, Yang SJ, Shin IS, Kim YH, Yoon JS. Interactions between life stressors and susceptibility genes (5-HTTLPR and BDNF) on depression in Korean elders. Biol Psychiatry. 2007;62:423–428. doi: 10.1016/j.biopsych.2006.11.020. [DOI] [PubMed] [Google Scholar]
- Kohonen T. Self-organizing maps. 2. Springer; Heidelberg: 1997. [Google Scholar]
- Kovacs M. Children’s Depression Inventory (CDI) Manual. Multi-Health Systems; North Tonawanda, NY: 1992. [Google Scholar]
- Kunisato Y, Okamoto Y, Okada G, Aoyama S, Demoto Y, Munakata A, Nomura M, Onoda K, Yamawaki S. Modulation of default-mode network activity by acute tryptophan depletion is associated with mood change: a resting state functional magnetic resonance imaging study. Neurosci Res. 2011;69:129–134. doi: 10.1016/j.neures.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Lesch KP, Bengel D, Heils A, Sabol SZ, Greenberg BD, Petri S, Benjamin J, Muller CR, Hamer DH, Murphy DL. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science. 1996;274:1527–1531. doi: 10.1126/science.274.5292.1527. [DOI] [PubMed] [Google Scholar]
- Levinson DF. The genetics of depression: a review. Biol Psychiatry. 2006;60:84–92. doi: 10.1016/j.biopsych.2005.08.024. [DOI] [PubMed] [Google Scholar]
- Liang M, Zhou Y, Jiang T, Liu Z, Tian L, Liu H, Hao Y. Widespread functional disconnectivity in schizophrenia with resting-state functional magnetic resonance imaging. Neuroreport. 2006;17:209–213. doi: 10.1097/01.wnr.0000198434.06518.b8. [DOI] [PubMed] [Google Scholar]
- Liao W, Chen H, Feng Y, Mantini D, Gentili C, Pan Z, Ding J, Duan X, Qiu C, Lui S, Gong Q, Zhang W. Selective aberrant functional connectivity of resting state networks in social anxiety disorder. Neuroimage. 2010;52:1549–1558. doi: 10.1016/j.neuroimage.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Maldjian J, Laurienti P, Burdette J, Kraft R. An Automated Method for Neuroanatomic and Cytoarchitectonic Atlas-based Interrogation of fMRI Data Sets. Neuroimage. 2002;19:1233–1239. doi: 10.1016/s1053-8119(03)00169-1. [DOI] [PubMed] [Google Scholar]
- Mann JJ, Huang YY, Underwood MD, Kassir SA, Oppenheim S, Kelly TM, Dwork AJ, Arango V. A serotonin transporter gene promoter polymorphism (5-HTTLPR) and prefrontal cortical binding in major depression and suicide. Arch Gen Psychiatry. 2000;57:729–738. doi: 10.1001/archpsyc.57.8.729. [DOI] [PubMed] [Google Scholar]
- Manor I, Eisenberg J, Tyano S, Sever Y, Cohen H, Ebstein RP, Kotler M. Familybased association study of the serotonin transporter promoter region polymorphism (5-HTTLPR) in attention deficit hyperactivity disorder. Am J Med Genet. 2001;105:91–95. [PubMed] [Google Scholar]
- March JS, Parker JD, Sullivan K, Stallings P, Conners CK. The Multidimensional Anxiety Scale for Children (MASC): factor structure, reliability, and validity. Journal of the American Academy of Child and Adolescent Psychiatry. 1997;36:554–565. doi: 10.1097/00004583-199704000-00019. [DOI] [PubMed] [Google Scholar]
- McLellan JA, Pugh MJV. The role of peer groups in adolescent social identity: exploring the importance of stability and change. Jossey-Bass Publishers; San Francisco: 1999. [Google Scholar]
- Meyer-Lindenberg A. Neural connectivity as an intermediate phenotype: brain networks under genetic control. Hum Brain Mapp. 2009;30:1938–1946. doi: 10.1002/hbm.20639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk CS, Peltier SJ, Wiggins JL, Weng SJ, Carrasco M, Risi S, Lord C. Abnormalities of intrinsic functional connectivity in autism spectrum disorders. Neuroimage. 2009;47:764–772. doi: 10.1016/j.neuroimage.2009.04.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Sekine Y, Ouchi Y, Tsujii M, Yoshikawa E, Futatsubashi M, Tsuchiya KJ, Sugihara G, Iwata Y, Suzuki K, Matsuzaki H, Suda S, Sugiyama T, Takei N, Mori N. Brain serotonin and dopamine transporter bindings in adults with high functioning autism. Arch Gen Psychiatry. 2010;67:59–68. doi: 10.1001/archgenpsychiatry.2009.137. [DOI] [PubMed] [Google Scholar]
- Nijmeijer JS, Hartman CA, Rommelse NN, Altink ME, Buschgens CJ, Fliers EA, Franke B, Minderaa RB, Ormel J, Sergeant JA, Verhulst FC, Buitelaar JK, Hoekstra PJ. Perinatal risk factors interacting with catechol O-methyltransferase and the serotonin transporter gene predict ASD symptoms in children with ADHD. J Child Psychol Psychiatry. 2010;51:1242–1250. doi: 10.1111/j.1469-7610.2010.02277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheim A, Schafer R, Buck J. Discrete-time signal processing. 2. Prentice Hall; Upper Saddle River, NJ: 1999. [Google Scholar]
- Peltier SJ, Polk TA, Noll DC. Detecting low-frequency functional connectivity in fMRI using a self-organizing map (SOM) algorithm. Hum Brain Mapp. 2003;20:220–226. doi: 10.1002/hbm.10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen AC. Adolescent development. Annu Rev Psychol. 1988;39:583–607. doi: 10.1146/annurev.ps.39.020188.003055. [DOI] [PubMed] [Google Scholar]
- Praschak-Rieder N, Kennedy J, Wilson AA, Hussey D, Boovariwala A, Willeit M, Ginovart N, Tharmalingam S, Masellis M, Houle S, Meyer JH. Novel 5-HTTLPR allele associates with higher serotonin transporter binding in putamen: a [(11)C] DASB positron emission tomography study. Biol Psychiatry. 2007;62:327–331. doi: 10.1016/j.biopsych.2006.09.022. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Rosenberg NA. Use of unlinked genetic markers to detect population stratification in association studies. American journal of human genetics. 1999;65:220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME, Mintun MA. Brain work and brain imaging. Annu Rev Neurosci. 2006;29:449–476. doi: 10.1146/annurev.neuro.29.051605.112819. [DOI] [PubMed] [Google Scholar]
- Raichle ME, Snyder AZ. A default mode of brain function: a brief history of an evolving idea. Neuroimage. 2007;37:1083–1090. doi: 10.1016/j.neuroimage.2007.02.041. discussion 1097–1089. [DOI] [PubMed] [Google Scholar]
- Rao H, Gillihan SJ, Wang J, Korczykowski M, Sankoorikal GM, Kaercher KA, Brodkin ES, Detre JA, Farah MJ. Genetic variation in serotonin transporter alters resting brain function in healthy individuals. Biol Psychiatry. 2007;62:600–606. doi: 10.1016/j.biopsych.2006.11.028. [DOI] [PubMed] [Google Scholar]
- Roiser JP, de Martino B, Tan GC, Kumaran D, Seymour B, Wood NW, Dolan RJ. A genetically mediated bias in decision making driven by failure of amygdala control. J Neurosci. 2009;29:5985–5991. doi: 10.1523/JNEUROSCI.0407-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roiser JP, Rogers RD, Cook LJ, Sahakian BJ. The effect of polymorphism at the serotonin transporter gene on decision-making, memory and executive function in ecstasy users and controls. Psychopharmacology (Berl) 2006;188:213–227. doi: 10.1007/s00213-006-0495-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubia K, Overmeyer S, Taylor E, Brammer M, Williams SC, Simmons A, Bullmore ET. Hypofrontality in attention deficit hyperactivity disorder during higher-order motor control: a study with functional MRI. The American journal of psychiatry. 1999;156:891–896. doi: 10.1176/ajp.156.6.891. [DOI] [PubMed] [Google Scholar]
- Rutter M, Bailey A, Berument S, Le Couteur A, Lord C, Pickles A. Social Communication Questionnaire (SCQ); Los Angeles, Calif: Western Psychological Services; 2003. [Google Scholar]
- Rutter M, Kim-Cohen J, Maughan B. Continuities and discontinuities in psychopathology between childhood and adult life. J Child Psychol Psychiatry. 2006a;47:276–295. doi: 10.1111/j.1469-7610.2006.01614.x. [DOI] [PubMed] [Google Scholar]
- Rutter M, Moffitt TE, Caspi A. Gene-environment interplay and psychopathology: multiple varieties but real effects. J Child Psychol Psychiatry. 2006b;47:226–261. doi: 10.1111/j.1469-7610.2005.01557.x. [DOI] [PubMed] [Google Scholar]
- Rutter M, Sroufe LA. Developmental psychopathology: concepts and challenges. Dev Psychopathol. 2000;12:265–296. doi: 10.1017/s0954579400003023. [DOI] [PubMed] [Google Scholar]
- Schinka JA, Busch RM, Robichaux-Keene N. A meta-analysis of the association between the serotonin transporter gene polymorphism (5-HTTLPR) and trait anxiety. Mol Psychiatry. 2004;9:197–202. doi: 10.1038/sj.mp.4001405. [DOI] [PubMed] [Google Scholar]
- Sen S, Burmeister M, Ghosh D. Meta-analysis of the association between a serotonin transporter promoter polymorphism (5-HTTLPR) and anxiety-related personality traits. Am J Med Genet B Neuropsychiatr Genet. 2004;127B:85–89. doi: 10.1002/ajmg.b.20158. [DOI] [PubMed] [Google Scholar]
- Shulman GL, Fiez JA, Corbetta M, Buckner RL, Miezin FM, Raichle ME, Petersen SE. Common blood flow changes across visual tasks: II. Decreases in cerebral cortex. Journal of Cognitive Neuroscience. 1997;9:648–663. doi: 10.1162/jocn.1997.9.5.648. [DOI] [PubMed] [Google Scholar]
- Smith GS, Lotrich FE, Malhotra AK, Lee AT, Ma Y, Kramer E, Gregersen PK, Eidelberg D, Pollock BG. Effects of serotonin transporter promoter polymorphisms on serotonin function. Neuropsychopharmacology. 2004;29:2226–2234. doi: 10.1038/sj.npp.1300552. [DOI] [PubMed] [Google Scholar]
- Sodhi MS, Sanders-Bush E. Serotonin and brain development. Int Rev Neurobiol. 2004;59:111–174. doi: 10.1016/S0074-7742(04)59006-2. [DOI] [PubMed] [Google Scholar]
- Spence SH. Structure of anxiety symptoms among children: a confirmatory factoranalytic study. J Abnorm Psychol. 1997;106:280–297. doi: 10.1037//0021-843x.106.2.280. [DOI] [PubMed] [Google Scholar]
- Steinberg L, Silverberg SB. The vicissitudes of autonomy in early adolescence. Child development. 1986;57:841–851. doi: 10.1111/j.1467-8624.1986.tb00250.x. [DOI] [PubMed] [Google Scholar]
- Stevens MC, Pearlson GD, Calhoun VD. Changes in the interaction of resting-state neural networks from adolescence to adulthood. Hum Brain Mapp. 2009;30:2356–2366. doi: 10.1002/hbm.20673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supekar K, Uddin LQ, Prater K, Amin H, Greicius MD, Menon V. Development of functional and structural connectivity within the default mode network in young children. Neuroimage. 2010;52:290–301. doi: 10.1016/j.neuroimage.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Uher R, McGuffin P. The moderation by the serotonin transporter gene of environmental adversity in the aetiology of mental illness: review and methodological analysis. Molecular psychiatry. 2008;13:131–146. doi: 10.1038/sj.mp.4002067. [DOI] [PubMed] [Google Scholar]
- van den Heuvel M, Mandl R, Luigjes J, Hulshoff Pol H. Microstructural organization of the cingulum tract and the level of default mode functional connectivity. J Neurosci. 2008;28:10844–10851. doi: 10.1523/JNEUROSCI.2964-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JL, Patel GH, Fox MD, Snyder AZ, Baker JT, Van Essen DC, Zempel JM, Snyder LH, Corbetta M, Raichle ME. Intrinsic functional architecture in the anaesthetized monkey brain. Nature. 2007;447:83–86. doi: 10.1038/nature05758. [DOI] [PubMed] [Google Scholar]
- Wang L, Zang Y, He Y, Liang M, Zhang X, Tian L, Wu T, Jiang T, Li K. Changes in hippocampal connectivity in the early stages of Alzheimer’s disease: evidence from resting state fMRI. Neuroimage. 2006;31:496–504. doi: 10.1016/j.neuroimage.2005.12.033. [DOI] [PubMed] [Google Scholar]
- Weng SJ, Carrasco M, Swartz JR, Wiggins JL, Kurapati N, Liberzon I, Risi S, Lord C, Monk CS. Neural activation to emotional faces in adolescents with autism spectrum disorders. J Child Psychol Psychiatry. 2011;52:296–305. doi: 10.1111/j.1469-7610.2010.02317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng SJ, Wiggins JL, Peltier SJ, Carrasco M, Risi S, Lord C, Monk CS. Alterations of resting state functional connectivity in the default network in adolescents with autism spectrum disorders. Brain Res. 2010;1313:202–214. doi: 10.1016/j.brainres.2009.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggins JL, Peltier SJ, Ashinoff S, Weng SJ, Carrasco M, Welsh RC, Lord C, Monk CS. Using a self-organizing map algorithm to detect age-related changes in functional connectivity during rest in autism spectrum disorders. Brain Res. 2011;1380:187–197. doi: 10.1016/j.brainres.2010.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsley KJ, Marrett S, Neelin P, Vandal AC, Friston KJ, Evans AC. A unified statistical approach for determining significant signals in images of cerebral activation. Hum Brain Mapp. 1996;4:58–73. doi: 10.1002/(SICI)1097-0193(1996)4:1<58::AID-HBM4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Zalsman G, Huang YY, Oquendo MA, Burke AK, Hu XZ, Brent DA, Ellis SP, Goldman D, Mann JJ. Association of a triallelic serotonin transporter gene promoter region (5-HTTLPR) polymorphism with stressful life events and severity of depression. Am J Psychiatry. 2006;163:1588–1593. doi: 10.1176/ajp.2006.163.9.1588. [DOI] [PubMed] [Google Scholar]